Abstract
CCN3 (NOV), a putative ligand for integrin receptors, is tightly associated with the extracellular matrix and mediates diverse cellular functions, including cell adhesion and proliferation. CCN3 has been shown to negatively regulate growth although it promotes migration in a cell type-specific manner. In this study, overexpression of CCN3 reduces growth and increases intercellular adhesion of breast cancer cells. Interestingly, CCN3 overexpression also led to the formation of multiple pseudopodia that are enriched in actin, CCN3, and vinculin. Breast cancer cells preincubated with exogenous CCN3 protein also induced the same phenotype, indicating that secreted CCN3 is sufficient to induce changes in cell morphology. Surprisingly, extracellular CCN3 is internalized to the early endosomes but not to the membrane protrusions, suggesting pseudopodia-enriched CCN3 may derive from a different source. The presence of an intracellular variant of CCN3 will be consistent with our finding that the cytoplasmic tail of the gap junction protein connexin43 (Cx43) associates with CCN3. Cx43 is a channel protein permitting intercellular communication to occur. However, neither the channel properties nor the protein levels of Cx43 are affected by the CCN3 protein. In contrast, CCN3 proteins are down-regulated in the absence of Cx43. Finally, we showed that overexpression of CCN3 increases the activity of the small GTPase Rac1, thereby revealing a pathway that links Cx43 directly to actin reorganization.
The CCN (CYR61/Connective Tissue Growth Factor/Nephroblastoma Overexpressed) family of multimodular proteins mediates diverse cellular functions, including cell adhesion, migration, and proliferation (1–3). Overexpression of CCN3, one of the founding members of the family, inhibits proliferation in most types of tumors such as glioblastoma and Ewing sarcoma (4, 5). Similarly, down-regulation of CCN3 has been suggested to promote melanoma progression (6). On the other hand, CCN3 can also promote migration in sarcoma and glioblastoma (4, 7), although a separate study shows that it decreases the invasion of melanoma (6). Therefore, in contrast to its role in growth suppression, the role of CCN3 signaling in cell motility is less clear.
Most evidence suggests CCN3 mediates its effects by binding to the integrin proteins, such as the αVβ3 receptors (8, 9), and that CCN3 alters cell adhesion in an integrin-dependent fashion (4, 10). In melanocytes, the discoidin domain receptor 1 mediates CCN3-dependent adhesion (11). CCN3 has also been observed to associate with Notch1 (12), fibulin 1C (13), S100A4 (14), and the gap junction protein Cx433 (15, 16), suggesting that CCN3 may also modulate cell growth via non-integrin signaling pathways.
Gap junction proteins are best known for forming channels between cells, contributing to intercellular communication by allowing the exchange of small ions and molecules (17, 18). Consequently, attenuated intercellular communication has been implicated in promoting carcinogenesis (19, 20). Recent evidence has indicated that connexins can mediate channel-independent growth control through interaction of their C-terminal cytoplasmic tail with various intracellular signaling molecules (21–23). In addition, many Cx43-interacting proteins, including ZO-1 (zonula occludens-1) (24), Drebrin (25), and N-cadherin (26) associate with F-actin, thus placing Cx43 in close proximity to the actin cytoskeleton.
In this study, we show for the first time that CCN3 reorganizes the actin cytoskeleton of the breast cancer cells MDA-MB-231 with the formation of multiple cell protrusions, possibly by activating the small GTPase Rac1. Our results also suggest an alternative route by which Cx43 may be functionally linked to actin cytoskeletal signaling via CCN3.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfections
(i) Plasmid DNA transfection of MDA-MB-231 cells (American Type Culture Collection) expressing pcDNA (Invitrogen), SP-NH25, NH25, NH35, NH45, and NH24 (27) was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol, and the resultant stable lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum. (ii) Retroviral infection of Hs578T cells (American Type Culture Collection) with Cx43shRNA was carried out with retroviral particles of Cx43shRNA-1 (a gift from Dr. Dale Laird, University of Western Ontario, London, Canada) and scrambled shRNA in the pH1.1QCXIH retroviral vector (28) as described previously (29). Both parental and transfected cell lines were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Sigma) at 37 °C in the presence of 5% CO2. (iii) Transient transfection of siRNAs against CCN3 was carried out by transfecting Hs578T cells with siRNAs (Dharmacon) to a final concentration of 20 nm with DharmaFECT 1 (Dharmacon) following the manufacturer's protocol. The best CCN3 knockdown was obtained with siRNA-3 of sequence 5′-CACACCAACTGTCCTAAGATT-3′ (siGENOME siRNA, Dharmacon) targeted against the C-terminal cysteine knot (CT) region of human CCN3 (GenBankTM accession number NM_002514). (iv) Exogenous recombinant human CCN3 (R & D Systems) was used to coat the culture dish or glass coverslips at 10 μg/ml as suggested by the manufacturer. In some instances, CCN3 proteins were fluorescently labeled with Alexa Fluor 568 protein labeling kit (Invitrogen), according to manufacturer's instruction, and added to the breast cells at a final concentration of 3 μg/ml.
Generation of Multicellular Tumor Spheroids
Multicellular tumor spheroids were generated based on the method described by Nielsen and co-workers (30) with some modifications. Briefly, MDA-MB-231 cells were plated at the density of 10,000 cells in 100 μl of medium (Dulbecco's modified Eagle's medium/Ham's F-12 in the presence of 1 mm sodium pyruvate and N2 supplement) per well in an ultra-low attachment 96-well dish (Corning Glass) for 7 days. Spheroids were then visualized with a Zeiss Axioskop microscope.
Protein Isolation and Western Analysis
For detection of CCN and Cx43 proteins present in the cytoplasm, breast cells were lysed in RIPA buffer containing 0.1% SDS, 1% IGEPAL, 0.5% Sarkosyl, 50 mm Tris-HCl (pH 8.0), 150 mm NaCl supplemented with MiniComplete protease inhibitors (Roche Applied Science) and phosphatase inhibitors (Sigma). Precipitation of proteins from conditioned medium was carried out as described previously (16). Briefly, conditioned media were collected following culturing of breast cells in serum-free media for 24 h. Methanol precipitation was performed on 200 μl of each sample. Proteins were then air-dried and resuspended in RIPA buffer. Equal loading of proteins from conditioned medium was ensured by normalizing the protein samples to the number of cells in the culture dish determined by a hemocytometer. Protein samples were then separated on a 12% polyacrylamide gel and Western-transferred onto polyvinylidene difluoride membrane (Amersham Biosciences). Rabbit anti-CCN1 (Santa Cruz Biotechnology), rabbit anti-CCN3 (16) or goat anti-CCN3 (R & D Systems), rabbit anti-Cx43 (Sigma), mouse anti-GAPDH (Hytest, Turku, Finland), and mouse anti-GFP (StressGen) were used as the primary antibodies, followed by either anti-rabbit horseradish peroxidase, anti-mouse horseradish peroxidase, or anti-goat horseradish peroxidase (Cedarlane, Hornby, Canada) as secondary antibodies. Protein bands were detected with SuperSignal chemiluminescent substrate (Pierce). Quantification of protein bands was carried out by using ImageJ software (31) available at rsb.info.nih.gov.
Immunocytochemistry
Cells grown on coverslips were fixed in cold 80% methanol at −20 °C for 10 min or 4% paraformaldehyde at room temperature for 10 min depending on the primary antibodies used. Coverslips were blocked in 2% bovine serum albumin in phosphate-buffered saline for 30 min. Primary antibodies used include mouse anti-αV integrin (BD Transduction Laboratories), mouse anti-vinculin (Sigma), mouse anti-EEA1 (BD Transduction Laboratories), and goat anti-CCN3 (R & D Systems). Generally, primary antibodies were incubated with the cells overnight at 4 °C, followed by sequential incubation with secondary antibodies anti-rabbit Alexa 568 and/or anti-mouse Alexa 488 (Invitrogen) for 1 h at room temperature. Actin was visualized by staining with Alexa 568-conjugated phalloidin (Invitrogen). Coverslips were mounted onto glass slides using ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen) and viewed with a Zeiss Axioskop epifluorescence microscope.
Cell Growth Analysis
MDA-MB-231 breast cancer cells were seeded at a density of 20,000 cells per well of a 12-well plate in triplicate. Cells were counted daily using a Coulter Counter R Z1 series particle counter (Coutler Electronics, Ontario, Canada). Mean values were assessed for significance using analysis of variance, followed by Tukey-Kramer multiple comparison test to determine significant difference (p < 0.01).
Wound Healing Assay
Migration rate of MDA-MB-231 cells was determined by scraping a confluent monolayer of cells (1 × 106 cells/35-mm dish) with a pipette tip, and the closure of the wound because of migrating cells was monitored after 8 h in culture and quantified using Zeiss Axiovision 4.6 software.
Preloading/Dye Coupling
Intercellular coupling between cells was evaluated according to the established preloading method described previously (32). Briefly, donor cells were bathed in 5 μm calcein-AM (Invitrogen) and 10 μm 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (Sigma) in an isotonic (0.3 m) glucose solution for 20 min, rinsed several times with isotonic glucose, trypsinized, seeded onto confluent unlabeled recipient sister cultures at a ratio of 1:500, and maintained for 3.5 h in the incubator. Cells were examined by epifluorescence microscopy, and gap junctional coupling was assessed and quantified by the number of cells that received passage of calcein from donor cells. All donor cells that were coupled to at least one other cell were included in the analysis.
GST Protein Expression and Purification
The cDNAs encoding various deletions within amino acids 236–382 of Cx43 were cloned into a pGEX-2TK vector to generate GST-Cx43Ctail recombinant fusion proteins. GST fusion proteins were produced in Escherichia coli BL21 and solubilized with lysis buffer containing 1% Triton X-100, 50 mm Tris (pH 8.0), and 150 mm NaCl in the presence of the protease inhibitor 1 mm phenylmethylsulfonyl fluoride. The fusion protein was purified with glutathione-Sepharose beads (Amersham Biosciences) incubated with bacterial cell lysates for 2.5 h at 4 °C. The beads were subsequently washed in lysis buffer, and the GST tag of these fusion proteins was removed by thrombin protease, and the cleaved proteins were used as competitive antigens in co-immunoprecipitations.
Co-immunoprecipitation Competition Assay
Hs578T breast cells were lysed in RIPA buffer containing 0.1% SDS, 1% IGEPAL, 0.5% Sarkosyl, 50 mm Tris-HCl (pH 8.0), 150 mm NaCl supplemented with MiniComplete protease inhibitors (Roche Applied Science) and phosphatase inhibitors (Sigma). Insoluble material was removed by centrifugation at 10,000 × g for 10 min, and the resultant supernatant was mixed with 1 μg of various C-terminal Cx43 deleted proteins as competitive antigens of endogenous Cx43. Following preclearing with 10 μl of preimmune serum and 10 μl of protein G Plus-agarose (Santa Cruz Biotechnology) at 4 °C for 30 min, cell lysates were incubated with 10 μl of rabbit anti-CCN3 (16) for 1 h and 20 μl of protein G-Plus beads for 2 h at 4 °C with gentle agitation. Beads were washed three times in RIPA buffer and boiled in SDS sample buffer to release bound proteins. Co-immunoprecipitated endogenous Cx43, if any, was detected by Western analysis as described above.
FRET Analysis
The use of pRaichu-1026X (Rac1), pRaichu-1069X (Cdc42), and pRaichu-1298X (RhoA) biosensors to detect activated small GTPases was originally reported by Matsuda and co-workers (33, 34). The presence of activated GTP-bound GTPases will bring YFP and cyan fluorescent protein in close proximity allowing FRET to occur. pRaichu plasmids were introduced into MDA-MB-231 breast cancer cells by standard Lipofectamine 2000 (Invitrogen) transfection as mentioned above for 48 h. FRET images were collected using an Olympus FV1000 laser-scanning confocal microscope. The occurrence of FRET was determined by the acceptor photobleaching method where FRET efficiency (E%) = 1 − (quenched donor/unquenched donor). Briefly, the image of the donor cyan fluorescent protein was captured before and after photobleaching the acceptor YFP at 515 nm. FRET efficiency was calculated using an algorithm developed by Periasamy and co-workers (35) as a plug-in for the ImageJ software (31). Only data points showing more than 80% bleaching of the acceptor YFP were included in the final analysis.
RESULTS
Overexpression of CCN3 Reduces Growth and Increases Intercellular Adhesion of Breast Cancer Cells
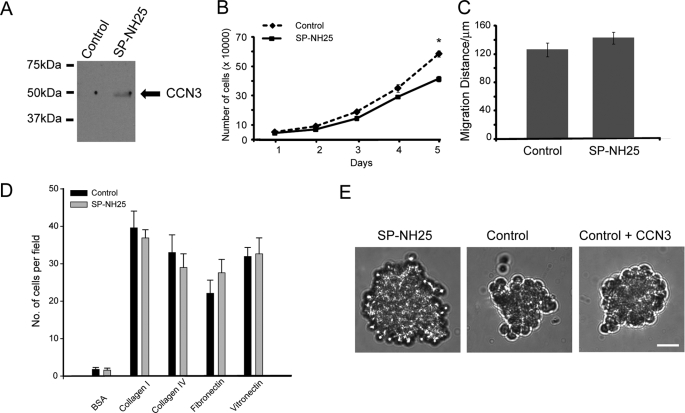
We manipulated the protein levels of CCN3 in MDA-MB-231 (MDA-231) cells that expressed negligible levels of CCN3 (Fig. 1A and supplemental Fig. 1A). Forced expression of full-length CCN3 (SP-NH25) in MDA-231 cells increased the secretion of CCN3 (Fig. 1A). As expected, expression of CCN3 reduced the growth rate of the breast cells (Fig. 1B), which is consistent with it being a known negative growth regulator. In contrast, the motility of SP-NH25 cells was not significantly altered (Fig. 1C). CCN3 has been shown to modify the adhesive affinities of Ewing sarcoma (4) and melanoma cells (10, 11) to various extracellular substrates. We therefore investigated whether CCN3 also affects the adhesiveness of MDA-231 cells. However, we did not find significant changes in the adhesive properties of SP-NH25 cells to the extracellular matrix components in a two-dimensional monolayer culture (Fig. 1D). In contrast, these cells behaved very differently from the control cells in a three-dimensional culture system (Fig. 1E). SP-NH25 cells formed large multicellular tumor spheroids, whereas control cells formed small loose cell aggregates (Fig. 1E). As SP-NH25 cells had reduced growth rate (Fig. 1B), the difference in their sizes is not likely to be due to an increase in cell number. Instead, overexpression of CCN3 presumably enhances the intercellular adhesion because a compact spheroid morphology is often due to an increased number of cell-cell contacts (36, 37). However, addition of recombinant CCN3 minimally affected the ability of control MDA-231 cells to form three-dimensional spheroids (Fig. 1E), indicating that exogenous CCN3 is not sufficient for facilitating adhesion.
FIGURE 1.
Overexpression of CCN3 reduces growth and increases intercellular adhesion of breast cancer cells. A, Western analysis of the conditioned medium of MDA-MB-231 cells transfected with control vector pcDNA or full-length human CCN3 (SP-NH25). B, growth curve of CCN3-overexpressing MDA-MB-231 (SP-NH25) cells compared with control cells (* = p < 0.001, one-way analysis of variance). C, migration distance of SP-NH25 cells compared with control cells as determined by wound healing assay. D, adhesive properties of MDA-MB-231 cells transfected with control vector pcDNA or full-length human CCN3 (SP-NH25) on various substrates. BSA, bovine serum albumin. E, representative picture of a typical multicellular tumor spheroid of SP-NH25 cells after 7 days in culture. Control cells formed smaller loose cell aggregates. Addition of CCN3 minimally affected spheroid formation. In all cases, 10,000 cells were plated per well of an ultra-low attachment 96-well dish. Scale bar, 50 μm.
CCN3 Induces Reorganization of Actin Filaments and Focal Adhesions
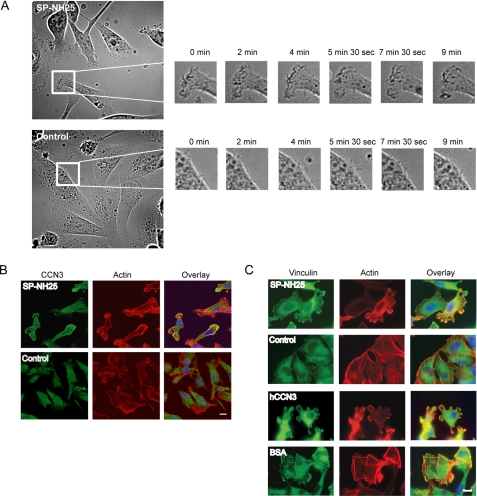
To extend our previous observation, we examined the morphology of control and SP-NH25 cells in a two-dimensional monolayer culture. SP-NH25 cells possessed multiple membrane protrusions not observed in control cells (Fig. 2A). Time-lapse live imaging of these breast cancer cells showed rapid cell membrane dynamics in SP-NH25 cells not apparent in control cells within the same time period (Fig. 2A; see also supplemental movies CCN3 and Control). The alteration in cell shape because of forced expression of CCN3 in MDA-231 cells suggests a rearrangement of cytoskeleton by CCN3. Reorganization of actin in CCN3-expressing MDA-231 cells was readily apparent when these cells were stained with phalloidin to visualize the actin cytoskeleton (Fig. 2, B and C). Control cells typically possessed actin stress fibers that are aligned to the long axis of the cell body (Fig. 2, B and C), in contrast CCN3 overexpressors (SP-NH25) possessed multiple cell protrusions that are actin-rich but devoid of stress fibers. Interestingly, CCN3 proteins were also localized to the leading edges of multiple actin-rich protrusions (Fig. 2B). In addition, staining the cells with anti-vinculin, a component of the mature focal adhesions, showed that overexpression of CCN3 induced a redistribution of vinculin from focal adhesions scattered throughout the entire cell to the lamellipodia protrusions (Fig. 2C). Interestingly, a similar alteration in actin staining was also observed in parental MDA-231 cells in the presence of exogenous recombinant CCN3 (Fig. 2C), suggesting that extracellular CCN3 is sufficient to induce cell protrusions and acts in an autocrine fashion.
FIGURE 2.
CCN3 induces reorganization of actin filaments and focal adhesions. A, time-lapse live imaging of CCN3 (SP-NH25)-expressing and control MDA-231 cells. Images within the white boxes are magnified showing regions with active ruffling only in SP-NH25 cells (also see supplemental movie CCN3). B, CCN3 protein was detected in actin-rich protrusions (as visualized by Alexa 568-conjugated phalloidin) of MDA-231 cells overexpressing CCN3 (SP-NH25). Scale bar, 10 μm. C, overexpression of CCN3 (SP-NH25) led to dissolution of stress fibers and vinculin-containing focal adhesion. MDA-231 cells plated on recombinant human hCCN3-coated, but not bovine serum albumin (BSA)-coated, coverslips showed a similar phenotype as the CCN3 overexpressors. Scale bar, 10 μm.
Extracellular CCN3 Is Internalized to Early Endosomes but Not to the Membrane Protrusions
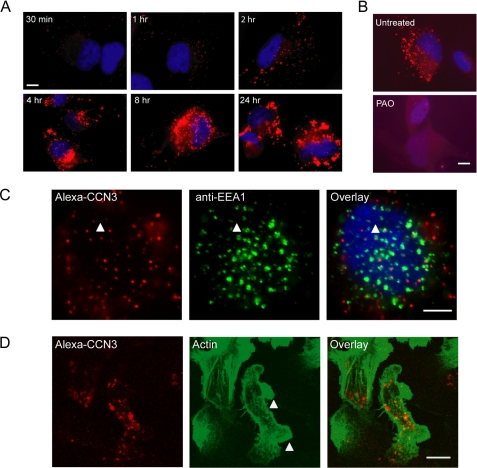
Our results so far suggested that the secreted isoform of CCN3 is responsible for regulating actin cytoskeletal reorganization. Extracellular CCN2/CTGF has been shown to be internalized and routed to an intracellular compartment, namely the nucleus (38). To determine whether extracellular CCN3 can be similarly routed to the cellular protrusions (Fig. 2B), we introduced Alexa 568-labeled recombinant CCN3 protein into the culture medium and monitored its uptake by the breast cells. We first used Hs578T cells expressing endogenous CCN3 (supplemental Fig. 1A) to optimize our experimental conditions. Punctate staining of Alexa 568-labeled CCN3 was apparent as early as 30 min, and the signal intensity continued to increase for as long as 24 h after addition of labeled CCN3 (Fig. 3A). Addition of an unrelated Alexa 568-labeled protein showed no punctate staining in the cells (data not shown), indicating that the protein uptake is a specific event. In addition, the punctate CCN3 staining was abolished with the addition of phenylarsine oxide, an endocytosis inhibitor (Fig. 3B). Indeed, a high percentage of internalized CCN3 appeared to co-stain with EEA1, an early endosome marker (39) (Fig. 3C). Thus extracellular CCN3 was actively internalized by these cells into early endosomes. We subsequently added Alexa 568-labeled recombinant CCN3 to MDA-231 breast cancer cells and similarly observed punctate staining of CCN3 inside the cells (Fig. 3D). In addition, we also observed a change in cell morphology with the introduction of exogenous CCN3 in the medium, consistent with our previous observation with unlabeled CCN3 (Fig. 2C). However, we did not observe the accumulation of endocytosed CCN3 in the actin-rich protrusions within the time frame that morphological changes had occurred (Fig. 3D), suggesting a separate pool of cytoplasmic CCN3 proteins was compartmentalized to the cellular protrusions observed in SP-NH25 cells (Fig. 2B).
FIGURE 3.
Extracellular CCN3 is internalized to the early endosomes but not to the membrane protrusions. A, images of Hs578T breast cancer cells fixed at different time intervals following addition of Alexa 568-labeled CCN3 into the culture medium. B, addition of phenylarsine oxide (PAO), an endocytosis inhibitor, abolished the internalization of CCN3. C, co-localization of CCN3 punctate staining with EEA1, an early endosome marker (white arrowhead). Scale bar, 5 μm. D, internalized CCN3 in MDA-231 cells did not localize to the actin-rich protrusions (white arrowheads). Scale bar, 10 μm.
Gap Junction Protein Cx43 Regulates CCN3 Expression
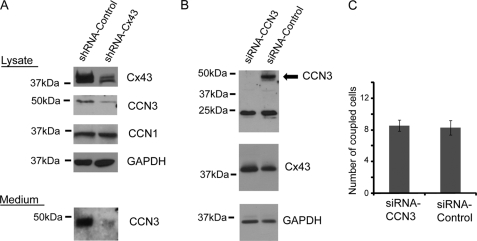
We next explored one possible pathway by which CCN3 can be regulated. We first observed an up-regulation of CCN3 in glioma cells overexpressing the gap junction protein Cx43 (16, 40, 41). To determine whether Cx43 similarly regulates CCN3 levels in breast cancer cells, we reduced endogenous Cx43 in Hs578T cells by generating a stable line with a Cx43shRNA plasmid that had been successfully used to knock down Cx43 in the same cell type (28). Cx43shRNA-expressing cells had a significant reduction of Cx43 compared with parental cells (Fig. 4A). We observed a corresponding reduction of secreted (complete knockdown) and intracellular CCN3 (75% knockdown) only in cells stably transduced with shRNA-Cx43 (Fig. 4A). As it often appears that members of the CCN family interact with each other and potentially modulate cell growth in a concerted manner (40, 42), we also checked for alternation of CCN1 expression but observed no correlation of CCN1 with Cx43 protein levels in breast cancer cells (Fig. 4A). Therefore, Cx43 specifically regulates the expression of CCN3. In contrast, knockdown of CCN3 with an siRNA against the CT region of the protein did not affect Cx43 expression levels (Fig. 4B). Accordingly, the coupling ability of Hs578T as measured by fluorescent dye transfer was also not significantly different between CCN3 knockdown and control cells (Fig. 4C).
FIGURE 4.
Gap junction protein Cx43 regulates CCN3 expression. A, Western analysis showing the reduction of Cx43 protein (shRNA-Cx43) in Hs578T breast cancer cells decreased CCN3 but not CCN1 protein expression. GAPDH was used as a loading control. B, Western analysis of the cell lysate and conditioned medium of Hs578T cells using anti-CCN3 and anti-Cx43 antibodies showing the level of Cx43 was not altered when CCN3 protein was eliminated with siRNA. siRNA against CCN3 had no effect on the smaller 25-kDa band that was also recognized by the CCN3 antibody in the cell lysate. Anti-GAPDH antibody was used as a loading control. C, quantification on the number of coupled cells per donor cells in Hs578T breast cancer cells is described under “Experimental Procedures.”
CCN3 Binds to the Cytoplasmic Tail of Gap Junction Protein Cx43 in Breast Cancer Cells
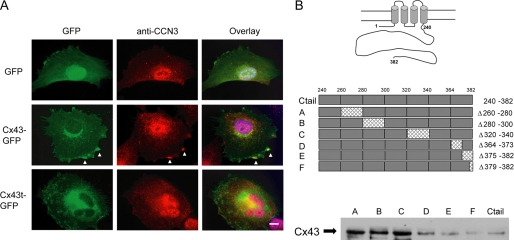
The detection of CCN3 in the lamellipodia protrusions in SP-NH25 cells and the absence of exogenously added CCN3 in the same regions appear to suggest the presence of an intracellular CCN3 variant. In addition, we have shown previously that CCN3 associates with the cytoplasmic tail of the gap junction protein Cx43 in C6 glioma cells (16). Similarly, overexpression of full-length Cx43 in breast cancer cells resulted in the localization of CCN3 in punctate staining with GFP-tagged Cx43 along the cell edges (Fig. 5A). Expression of truncated Cx43 (Cx43t) exhibited no obvious co-staining of the GFP-tagged truncated Cx43 and CCN3, indicating that the cytoplasmic tail of Cx43 is necessary for the interaction (Fig. 5A). We further narrowed down the interaction sites between Cx43 and CCN3 by performing a co-immunoprecipitation (co-IP) assay with Hs578T cell lysate using anti-CCN3 antibody to precipitate endogenous Cx43 in the presence of competing antigens. These competing antigens were a series of Cx43 C-terminal deletion constructs each containing specific amino acid deletions (i.e. amino acids 260–280, 280–300, 320–340, 364–373, 375–382, and 379–382) expressed as GST recombinant proteins, as shown schematically in Fig. 5B. The presence of endogenous Cx43 in the immunoprecipitates implies that the exogenous recombinant proteins containing the deletions were unable to bind CCN3 and therefore could not compete with the binding of endogenous Cx43 to CCN3. The wild type C-tail of Cx43 (amino acids 246–382; Ctail) was used as the positive control showing the near complete removal of endogenous Cx43 in the anti-CCN3 co-IP (Fig. 5B). Similarly, the presence of mutants D, E, and F in the breast lysates resulted in the absence of Cx43 in the co-IP, indicating the D, E, and F mutants, like the Cx43 Ctail, still retain the CCN3-interacting sites. In contrast, endogenous Cx43 was co-immunoprecipitated with CCN3 in the presence of mutants A, B, and C, indicating that these three mutants lost their abilities to bind CCN3, suggesting residues 260–340 of Cx43 contain the CCN3-binding site(s).
FIGURE 5.
CCN3 binds to the cytoplasmic tail of gap junction protein Cx43. A, immunofluorescence image showing the co-staining of endogenous CCN3 with Cx43 only in Hs578T cells overexpressing full-length Cx43 (white arrowheads) and not in C-tail truncated Cx43 (Cx43) or GFP-expressing cells. Blue, 4′,6-diamidino-2-phenylindole staining. Scale bar, 10 μm. B, Cx43 deletion mutants used as competing antigens in a co-IP assay using anti-CCN3 antibody. The presence of endogenous Cx43 in A–C showed that these proteins cannot replace endogenous Cx43 in the co-immunoprecipitation and thereby most likely have lost the CCN3-interacting sites.
Intracellular CCN3 Is Functionally Active
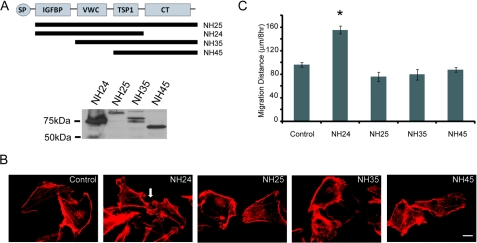
CCN3 proteins are composed of five modules commonly found in the extracellular matrix proteins (43). The N-terminal signal peptide is followed by a putative insulin growth factor binding domain, a von Willebrand type C module, a thrombospondin 1 domain, and a cysteine-rich knot (CT) at the C-terminal tail (Fig. 6A). CCN3 mutants containing various deletions (NH25, NH24, NH35, and NH45) were expressed in MDA-231 cells (Fig. 6A). Interestingly, the full-length NH25 lacking the secretory signal did not induce membrane protrusions reminiscent of those induced by the secretory domain-containing full-length CCN3 (SP-NH25) (Fig. 6B). Instead, it is the truncated NH24 protein lacking the CT domain that exhibits actin-rich protrusions (Fig. 6B). NH24 also had significantly higher motility compared with mutants with an intact CT domain, including NH25, NH35, and NH45 (Fig. 6C). Our results show that intracellular CCN3 has the capacity to alter cellular characteristics. In addition, the enhancement of motility in NH24 is in agreement with our previous result showing that the CT region is the “inhibitory” domain of CCN3 (27, 44).
FIGURE 6.
Intracellular CCN3 lacking the CT domain promotes migration. A, top panel, schematic diagram showing the CCN3 deletion mutants. Bottom panel, Western analysis with anti-GFP antibody showing the expression of recombinant GFP-CCN3 fusion proteins expressed in MDA-231 cells. B, actin cytoskeleton of CCN3 mutant-expressing cells as visualized by Alexa 568-conjugated phalloidin. White arrow indicates the membrane protrusions present in NH24-expressing cells. Scale bar, 10 μm. C, migration distance of CCN3 mutants-expressing cells compared with control as determined by wound healing assays (* = p < 0.001, one-way analysis of variance).
Overexpression of CCN3 Increases the Activity of Small GTPase Rac1
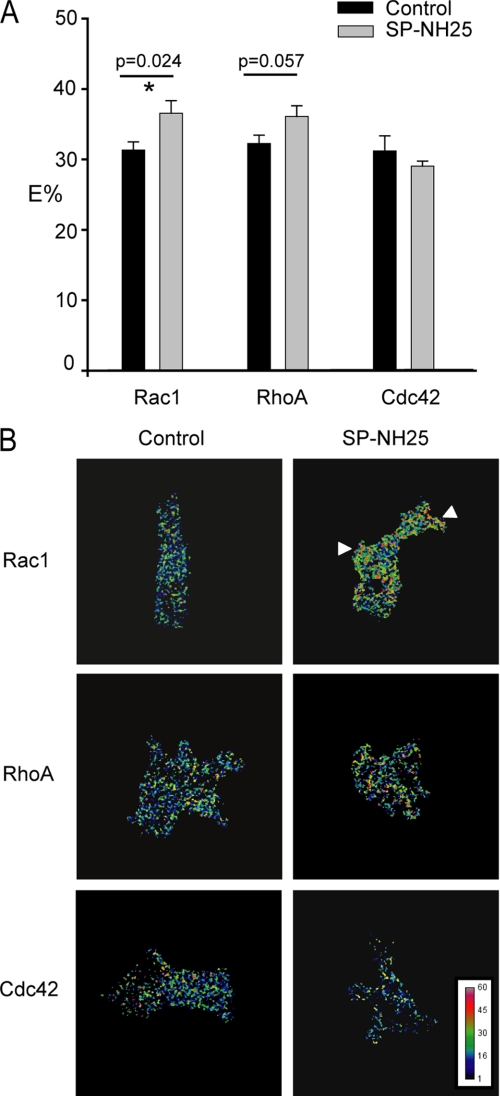
To investigate the possible signaling mechanisms downstream of CCN3, we looked into the Rho GTPase family of proteins, which mediate diverse cellular functions but primarily act on the actin cytoskeleton (45–47). The founding members RhoA, Rac1 and Cdc42 induce the formation of stress fibers, lamellipodia, and filopodia in fibroblasts/epithelial cells, respectively. In addition, numerous reports indicate that the epithelial cell-cell adhesion is facilitated or preceded by Rac-dependent lamellipodia formation (48–50). We therefore carried out Förster resonance energy transfer (FRET) analysis using three well characterized Rho GTPase biosensors, pRaichu-Rac1, pRaichu-Cdc42, and pRaichu-RhoA (33, 34), to determine the level of activated small GTPases in these breast cells. We observed a significant increase in the level of activated Rac1 in SP-NH25 cells (Fig. 7, A and B), with localized high Rac activity in the membrane protrusions. Interestingly, there was also a slight increase in RhoA activity in SP-NH25 cells although the increase was not statistically significant. In contrast, Cdc42 activity was not affected by the expression of CCN3. Therefore, our results suggest that CCN3 works upstream of Rac1 to regulate the actin cytoskeleton.
FIGURE 7.
Overexpression of CCN3 increases the activity of small GTPase Rac1. Control or CCN3-expressing cells (SP-NH25) were transfected with either pRaichu-1026X (Rac1), pRaichu-1069X (Cdc42), or pRaichu-1298X (RhoA) biosensors. FRET was determined by acceptor photobleaching. A, quantification of the FRET efficiencies (E%) from three independent experiments. FRET efficiencies were determined on a pixel-by-pixel basis as described under “Experimental Procedures.” Only pixels showing more than 80% bleaching of the acceptor YFP were included in the final analysis. Results are expressed as means ± S.E. (* = statistically significant with Student's t test). B, representative images illustrating FRET efficiencies (displayed using a color scale). Arrowheads indicate the protrusive regions of SP-NH25 cells with high Rac activities.
DISCUSSION
The multimodular CCN family of proteins perform diverse functions that are often cell type-specific (1–3). Expression of CCN3, however, has been mostly associated with a reduction in cell growth (4, 5) and an increase in adhesion to various extracellular matrix substrates in an integrin-dependent manner (4, 10). Our study demonstrated that CCN3 similarly reduces cell proliferation and increases adhesiveness of MDA- 231 breast cancer cells.
There are several possibilities by which CCN3 can exert its cytoskeletal effects. First, CCN3 might act through the integrin signaling pathways. CCN3 has been shown to be a putative ligand for αV integrin receptor (8), which was also expressed in the MDA-231 cells used in this study (51) (supplemental Fig. 1B). However, both endocytosis of CCN3 and the resultant change in cell morphology are not inhibited by anti-integrin αV antibodies (data not shown). We were also not able to detect αV staining in the CCN3-containing early endosomes within the time frame under our experimental conditions (data not shown), indicating αV integrin may not be the “receptor” that assists in CCN3 internalization in MDA-231 breast cells. In addition, the same neutralizing αVβ5 (P1F6) and αVβ3 (23C6) antibodies previously demonstrated to successfully block CCN3-dependent increase in adhesiveness in melanoma cells (10) had no effect on the intercellular adhesion of CCN3-expressing MDA-231 breast cells (data not shown). The observation that the addition of CCN3 directly to the medium only has a minimal effect on the adhesiveness of control cells also argues against the involvement of ligand-receptor binding in promoting intercellular adhesion. This is in agreement with an earlier report stating that melanoma cells pre-incubated with recombinant CCN3 did not show an increase in adhesion, which is in contrast to the increase in adhesion manifested by CCN3-expressing cells (10), indicating that the CCN3 must be made by the cells to affect adhesion. Likewise, the C-terminal region of CCN3 alone is reported to be sufficient to support adhesion in myoblasts (52). Second, CCN3 might affect adhesion by activation of the Rho GTPase pathways. Rac-induced spreading has been shown to promote the formation of adhesive contacts between cells (48–50). Interestingly, Rac-induced spreading is accompanied by integrin clustering (50), which is also observed in CCN3 overexpressors (supplemental Fig. 1B). Third, CCN3 might influence adhesion through the association with non-integrin proteins such as Notch1 (12) and Cx43 (15, 16). Cx43 is the only protein that has been shown to both regulate and associate with CCN3. Reduced Cx43 levels cause a substantial decrease in the levels of transcription factors in Cx43 null heart tissue (53), which indicates the capacity of a gap junction protein to regulate cell growth by altering gene expression. Although Cx43 is more well known as a channel protein allowing exchange of small molecules between cells, recent evidence has emerged that Cx43 can have a structural role by promoting cell adhesion directly via its extracellular domains (54, 55).
In addition, the cytoplasmic tail of Cx43 interacts with signaling molecules such as ZO-1 (24), N-cadherin (26), p120catenin (26), microtubules (56), and Drebrin, an actin-binding protein (25), providing a context for Cx43 to be involved in signaling events leading to cytoskeletal rearrangement. The interaction of CCN3 with the cytoplasmic tail of Cx43 indicates either there is an intracellular variant of CCN3 or that the secreted CCN3 is somehow recycled back inside the cell to be available for interaction with Cx43. Our study tentatively points toward the existence of an intracellular variant. This is because recombinant CCN3 endocytosed by the breast cells did not localize to the membrane protrusions, whereas CCN3 was detected at the protrusions in CCN3 overexpressors, suggesting that CCN3 detected at the membrane protrusions may derive from an alternative intracellular route.
Intracellular CCN3 variants have been previously detected in the cytoplasm and in the nucleus of some cancer cell lines (3, 40, 57, 58). A smaller protein has been detected using a C-terminal specific but not an N-terminal specific antibody, and it is widely believed that the presence of intracellular CCN3 is either because of a proteolytic cleavage event or derived from an alternately spliced mRNA (3, 27, 58, 59). Although we did detect a smaller protein at 25 kDa in the breast cell lysate but not in the culture medium using a C-terminal CCN3-specific antibody (Fig. 4B), its resistance to siRNA-CCN3 treatment led us to believe that the smaller 25-kDa form may be a closely related CCN3-like protein translated from a distinct transcript. However, we could not rule out that truncated CCN3 may have a relatively long half-life and so remained detectable even in the absence of the full-length CCN3.
Taken together, CCN3 appears to exert its cellular effects in MDA-231 breast cancer cells in multiple ways. Secreted CCN3 reorganizes the actin cytoskeleton via a yet-to-be identified cell surface receptor, resulting in the disappearance of stress fibers and mature focal adhesions containing vinculin, and the formation of cortical actin-rich pseudopodia. The relocalization of vinculin and integrins probably facilitated membrane protrusions and cell spreading (60) by serving as an anchor at the leading edge of CCN3-expressing cells. The interaction of CCN3 with Cx43 is another potential route to alter cytoskeletal organization. Interestingly, Rac1 activity is necessary for proper Cx43 localization (61). It is therefore tempting to speculate that increased Rac activity because of CCN3 expression helps to localize Cx43 to proper subcellular compartment and acts in synergy with the gap junction protein to modify cell adhesion.
Actin cytoskeleton architecture is often dependent on the proliferative or metastatic properties of the cancer cells. Metastatic MDA-231 cells form active stress fibers, and quiescent cells contain cortical actin staining (62). Thus, the reduced proliferation and increased cortical actin staining in CCN3-expressing MDA-231 cells is reminiscent of nonmetastatic breast cancer cells, confirming the well established anti-oncogenic property of CCN3. Our study showed for the first time that CCN3 directly regulates the actin cytoskeleton, possibly via the activation of small Rho GTPase Rac1. CCN3 also appears to slightly increase the activity of RhoA. It is interesting to note that activated RhoA has been recently shown to be enriched in the pseudopodia of tumor cells (63). The next step is to determine how CCN3 exerts its effect on the activation states of the small Rho GTPases. Our finding also reveals a possible pathway by which a gap junction protein can affect actin reorganization. Although the growth suppression effect of overexpressed Cx43 is well documented in breast cells (28, 64–66), we have also observed a change in cell morphology in gliomas expressing high levels of Cx43 (40, 67). Recent evidence in human gliomas indicates that the cytoplasmic tail is sufficient to induce Cx43-dependent actin rearrangement.4 The ability of Cx43 to place itself as a nexus of interacting proteins (21) and the association of CCN3 and Cx43 reveal an unexplored pathway by which a gap junction protein can facilitate actin cytoskeletal reorganization.
Supplementary Material
This work was supported by Operating Grant MOP 81202 (to C. C. N. and W. C. S.) from the Canadian Institutes of Health Research and European PROTHETS (Prognosis and Therapeutic Targets of Ewing Family of Tumors) FP6 Contract 503036 and the French Ministry of Education (to B. P.). This work was also supported in part by National Institutes of Health Grant GM55632 (to P. D. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and two movies.
S. Crespin, J. Bechberger, M. Mesnil, C. C. Naus, and W.-C. Sin, manuscript in preparation.
- Cx43
- connexin43
- siRNA
- small interfering RNA
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- YFP
- yellow fluorescent protein
- co-IP
- co-immunoprecipitation
- FRET
- Forster resonance energy transfer
- shRNA
- short hairpin RNA
- CT
- C-terminal cysteine knot.
REFERENCES
- 1.Leask A., Abraham D. J. (2006) J. Cell Sci. 119, 4803–4810 [DOI] [PubMed] [Google Scholar]
- 2.Brigstock D. R. (2003) J. Endocrinol. 178, 169–175 [DOI] [PubMed] [Google Scholar]
- 3.Perbal B. (2004) Lancet 363, 62–64 [DOI] [PubMed] [Google Scholar]
- 4.Benini S., Perbal B., Zambelli D., Colombo M. P., Manara M. C., Serra M., Parenza M., Martinez V., Picci P., Scotlandi K. (2005) Oncogene 24, 4349–4361 [DOI] [PubMed] [Google Scholar]
- 5.Gupta N., Wang H., McLeod T. L., Naus C. C., Kyurkchiev S., Advani S., Yu J., Perbal B., Weichselbaum R. R. (2001) Mol. Pathol. 54, 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukunaga-Kalabis M., Martinez G., Telson S. M., Liu Z. J., Balint K., Juhasz I., Elder D. E., Perbal B., Herlyn M. (2008) Oncogene 27, 2552–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laurent M., Martinerie C., Thibout H., Hoffman M. P., Verrecchia F., Le Bouc Y., Mauviel A., Kleinman H. K. (2003) FASEB J. 17, 1919–1921 [DOI] [PubMed] [Google Scholar]
- 8.Lin C. G., Leu S. J., Chen N., Tebeau C. M., Lin S. X., Yeung C. Y., Lau L. F. (2003) J. Biol. Chem. 278, 24200–24208 [DOI] [PubMed] [Google Scholar]
- 9.Lin C. G., Chen C. C., Leu S. J., Grzeszkiewicz T. M., Lau L. F. (2005) J. Biol. Chem. 280, 8229–8237 [DOI] [PubMed] [Google Scholar]
- 10.Vallacchi V., Daniotti M., Ratti F., Di Stasi D., Deho P., De Filippo A., Tragni G., Balsari A., Carbone A., Rivoltini L., Parmiani G., Lazar N., Perbal B., Rodolfo M. (2008) Cancer Res. 68, 715–723 [DOI] [PubMed] [Google Scholar]
- 11.Fukunaga-Kalabis M., Martinez G., Liu Z. J., Kalabis J., Mrass P., Weninger W., Firth S. M., Planque N., Perbal B., Herlyn M. (2006) J. Cell Biol. 175, 563–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakamoto K., Yamaguchi S., Ando R., Miyawaki A., Kabasawa Y., Takagi M., Li C. L., Perbal B., Katsube K. (2002) J. Biol. Chem. 277, 29399–29405 [DOI] [PubMed] [Google Scholar]
- 13.Perbal B., Martinerie C., Sainson R., Werner M., He B., Roizman B. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C. L., Martinez V., He B., Lombet A., Perbal B. (2002) Mol. Pathol. 55, 250–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gellhaus A., Dong X., Propson S., Maass K., Klein-Hitpass L., Kibschull M., Traub O., Willecke K., Perbal B., Lye S. J., Winterhager E. (2004) J. Biol. Chem. 279, 36931–36942 [DOI] [PubMed] [Google Scholar]
- 16.Fu C. T., Bechberger J. F., Ozog M. A., Perbal B., Naus C. C. (2004) J. Biol. Chem. 279, 36943–36950 [DOI] [PubMed] [Google Scholar]
- 17.Scemes E., Suadicani S. O., Dahl G., Spray D. C. (2007) Neuron Glia Biol. 3, 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simon A. M., Goodenough D. A. (1998) Trends Cell Biol. 8, 477–483 [DOI] [PubMed] [Google Scholar]
- 19.Mesnil M., Crespin S., Avanzo J. L., Zaidan-Dagli M. L. (2005) Biochim. Biophys. Acta 1719, 125–145 [DOI] [PubMed] [Google Scholar]
- 20.Naus C. C. (2002) Can. J. Physiol. Pharmacol. 80, 136–141 [DOI] [PubMed] [Google Scholar]
- 21.Hervé J. C., Bourmeyster N., Sarrouilhe D. (2004) Biochim. Biophys. Acta 1662, 22–41 [DOI] [PubMed] [Google Scholar]
- 22.Stout C., Goodenough D. A., Paul D. L. (2004) Curr. Opin. Cell Biol. 16, 507–512 [DOI] [PubMed] [Google Scholar]
- 23.Giepmans B. N. (2004) Cardiovasc. Res. 62, 233–245 [DOI] [PubMed] [Google Scholar]
- 24.Giepmans B. N., Moolenaar W. H. (1998) Curr. Biol. 8, 931–934 [DOI] [PubMed] [Google Scholar]
- 25.Butkevich E., Hülsmann S., Wenzel D., Shirao T., Duden R., Majoul I. (2004) Curr. Biol. 14, 650–658 [DOI] [PubMed] [Google Scholar]
- 26.Xu X., Li W. E., Huang G. Y., Meyer R., Chen T., Luo Y., Thomas M. P., Radice G. L., Lo C. W. (2001) Cell Commun. Adhes. 8, 321–324 [DOI] [PubMed] [Google Scholar]
- 27.Planque N., Long Li C., Saule S., Bleau A. M., Perbal B. (2006) J. Cell Biochem. 99, 105–116 [DOI] [PubMed] [Google Scholar]
- 28.Shao Q., Wang H., McLachlan E., Veitch G. I., Laird D. W. (2005) Cancer Res. 65, 2705–2711 [DOI] [PubMed] [Google Scholar]
- 29.Bates D. C., Sin W. C., Aftab Q., Naus C. C. (2007) Glia 55, 1554–1564 [DOI] [PubMed] [Google Scholar]
- 30.Kelm J. M., Timmins N. E., Brown C. J., Fussenegger M., Nielsen L. K. (2003) Biotechnol. Bioeng. 83, 173–180 [DOI] [PubMed] [Google Scholar]
- 31.Girish V., Vijayalakshmi A. (2004) Ind. J. Cancer 41, 47. [PubMed] [Google Scholar]
- 32.Goldberg G. S., Bechberger J. F., Naus C. C. (1995) BioTechniques 18, 490–497 [PubMed] [Google Scholar]
- 33.Yoshizaki H., Ohba Y., Kurokawa K., Itoh R. E., Nakamura T., Mochizuki N., Nagashima K., Matsuda M. (2003) J. Cell Biol. 162, 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itoh R. E., Kurokawa K., Ohba Y., Yoshizaki H., Mochizuki N., Matsuda M. (2002) Mol. Cell. Biol. 22, 6582–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y., Elangovan M., Periasamy A. (2005) in Molecular Imaging FRET Microscopy and Spectroscopy (Periasamy A., Day R. N. eds) pp. 126–145, Oxford University Press, New York [Google Scholar]
- 36.Powers M. J., Griffith L. G. (1998) Microsc. Res. Tech. 43, 379–384 [DOI] [PubMed] [Google Scholar]
- 37.Hedlund T. E., Duke R. C., Miller G. J. (1999) Prostate 41, 154–165 [DOI] [PubMed] [Google Scholar]
- 38.Wahab N. A., Brinkman H., Mason R. M. (2001) Biochem. J. 359, 89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mu F. T., Callaghan J. M., Steele-Mortimer O., Stenmark H., Parton R. G., Campbell P. L., McCluskey J., Yeo J. P., Tock E. P., Toh B. H. (1995) J. Biol. Chem. 270, 13503–13511 [DOI] [PubMed] [Google Scholar]
- 40.Sin W. C., Bechberger J. F., Rushlow W. J., Naus C. C. (2008) J. Cell. Biochem. 103, 1772–1782 [DOI] [PubMed] [Google Scholar]
- 41.McLeod T. L., Bechberger J. F., Naus C. C. (2001) Cell Commun. Adhes. 8, 441–445 [DOI] [PubMed] [Google Scholar]
- 42.Bleau A. M., Planque N., Perbal B. (2005) Front. Biosci. 10, 998–1009 [DOI] [PubMed] [Google Scholar]
- 43.Holbourn K. P., Acharya K. R., Perbal B. (2008) Trends Biochem. Sci. 33, 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bleau A. M., Planque N., Lazar N., Zambelli D., Ori A., Quan T., Fisher G., Scotlandi K., Perbal B. (2007) J. Cell. Biochem. 101, 1475–1491 [DOI] [PubMed] [Google Scholar]
- 45.Jaffe A. B., Hall A. (2005) Annu. Rev. Cell Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 46.Burridge K., Wennerberg K. (2004) Cell 116, 167–179 [DOI] [PubMed] [Google Scholar]
- 47.Van Aelst L., D'Souza-Schorey C. (1997) Genes Dev. 11, 2295–2322 [DOI] [PubMed] [Google Scholar]
- 48.Ehrlich J. S., Hansen M. D., Nelson W. J. (2002) Dev. Cell 3, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasioukhin V., Bauer C., Yin M., Fuchs E. (2000) Cell 100, 209–219 [DOI] [PubMed] [Google Scholar]
- 50.D'Souza-Schorey C., Boettner B., Van Aelst L. (1998) Mol. Cell. Biol. 18, 3936–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong N. C., Mueller B. M., Barbas C. F., Ruminski P., Quaranta V., Lin E. C., Smith J. W. (1998) Clin. Exp. Metastasis 16, 50–61 [DOI] [PubMed] [Google Scholar]
- 52.Lafont J., Thibout H., Dubois C., Laurent M., Martinerie C. (2005) Cell Commun. Adhes. 12, 41–57 [DOI] [PubMed] [Google Scholar]
- 53.Iacobas D. A., Iacobas S., Li W. E., Zoidl G., Dermietzel R., Spray D. C. (2005) Physiol. Genomics 20, 211–223 [DOI] [PubMed] [Google Scholar]
- 54.Cotrina M. L., Lin J. H., Nedergaard M. (2008) Glia 56, 1791–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elias L. A., Wang D. D., Kriegstein A. R. (2007) Nature 448, 901–907 [DOI] [PubMed] [Google Scholar]
- 56.Giepmans B. N., Verlaan I., Hengeveld T., Janssen H., Calafat J., Falk M. M., Moolenaar W. H. (2001) Curr. Biol. 11, 1364–1368 [DOI] [PubMed] [Google Scholar]
- 57.Perbal B. (2006) Cell Commun. Signal. 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Planque N., Perbal B. (2003) Cancer Cell Int. 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lazar N., Manara C., Navarro S., Bleau A. M., Llombart-Bosch A., Scotlandi K., Planque N., Perbal B. (2007) J. Cell Commun. Signal. 1, 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeMali K. A., Barlow C. A., Burridge K. (2002) J. Cell Biol. 159, 881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matsuda T., Fujio Y., Nariai T., Ito T., Yamane M., Takatani T., Takahashi K., Azuma J. (2006) J. Mol. Cell. Cardiol. 40, 495–502 [DOI] [PubMed] [Google Scholar]
- 62.Barkan D., Kleinman H., Simmons J. L., Asmussen H., Kamaraju A. K., Hoenorhoff M. J., Liu Z. Y., Costes S. V., Cho E. H., Lockett S., Khanna C., Chambers A. F., Green J. E. (2008) Cancer Res. 68, 6241–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stuart H. C., Jia Z., Messenberg A., Joshi B., Underhill T. M., Moukhles H., Nabi I. R. (2008) J. Biol. Chem. 283, 34785–34795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McLachlan E., Shao Q., Wang H. L., Langlois S., Laird D. W. (2006) Cancer Res. 66, 9886–9894 [DOI] [PubMed] [Google Scholar]
- 65.Qin H., Shao Q., Curtis H., Galipeau J., Belliveau D. J., Wang T., Alaoui-Jamali M. A., Laird D. W. (2002) J. Biol. Chem. 277, 29132–29138 [DOI] [PubMed] [Google Scholar]
- 66.Hirschi K. K., Xu C. E., Tsukamoto T., Sager R. (1996) Cell Growth & Differ. 7, 861–870 [PubMed] [Google Scholar]
- 67.Naus C. C., Zhu D., Todd S. D., Kidder G. M. (1992) Cell. Mol. Neurobiol. 12, 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.