Abstract
Rap1b has been implicated in the transduction of the cAMP mitogenic response. Agonists that increase intracellular cAMP rapidly activate (i.e. GTP binding) and phosphorylate Rap1b on Ser179 at its C terminus. cAMP-dependent protein kinase (PKA)-mediated phosphorylation of Rap1b is required for cAMP-dependent mitogenesis, tumorigenesis, and inhibition of AKT activity. However, the role of phosphorylation still remains unknown. In this study, we utilized amide hydrogen/deuterium exchange mass spectroscopy (DXMS) to assess potential conformational changes and/or mobility induced by phosphorylation. We report here DXMS data comparing exchange rates for PKA-phosphorylated (Rap1-P) and S179D phosphomimetic (Rap1-D) Rap1b proteins. Rap1-P and Rap1-D behaved exactly the same, revealing an increased exchange rate in discrete regions along the protein; these regions include a domain around the phosphorylation site and unexpectedly the two switch loops. Thus, local effects induced by Ser179 phosphorylation communicate allosterically with distal domains involved in effector interaction. These results provide a mechanistic explanation for the differential effects of Rap1 phosphorylation by PKA on effector protein interaction.
Rap1b, a member of the Ras superfamily of small G proteins, is a GTPase that acts as a molecular on/off switch for the transduction of several external stimuli by alternating from an inactive GDP-bound to an active GTP-bound state (1, 2). Rap1 activation is mediated by several second messengers, growth factors, cytokines, and cell adhesion molecules. The steady-state level of Rap1-GTP is tightly regulated by a family of guanine nucleotide exchange factors that catalyze the otherwise slow dissociation of GDP (i.e. activation) and GTPase-activating proteins, which stimulate the rather slow intrinsic GTPase catalytic activity (i.e. inactivation) (3). GTP binding is coupled to conformational changes in two well defined regions, the switch I (residues 30–40) and switch II (residues 60–76) domains, responsible for high affinity interaction with effector molecules (4, 5) and thus downstream signal transduction.
cAMP is one among several pathways leading to Rap1 activation (6). cAMP exerts both mitogenic and anti-mitogenic responses in different cell types, and Rap1 activation is required downstream of cAMP in both scenarios (7, 8). Elevation of intracellular cAMP levels activates cAMP-dependent protein kinase (PKA)4 and Epac (exchange protein activated by cAMP), a Rap guanine nucleotide exchange factor (9). Expression of Rap1b in cells where cAMP is mitogenic is associated with an increase in cAMP-mediated G1/S phase entry (7, 10), and both biochemical events, Rap activation and phosphorylation at Ser179, are synergistically required for this action (11).
PKA substrates able to modulate Rap1 activity (i.e. Src/C3G recruitment and GTPase-activating protein) were recently reported (12, 13). However, the role of PKA-dependent Rap1 phosphorylation at Ser179 is still unknown. Rap1 phosphorylation does not affect its overall intracellular localization, its basal GTP/GDP exchange reaction, its intrinsic rate of GTP hydrolysis, or its ability to be stimulated by a cytosolic Rap GTPase-activating protein (10); however, several reports suggest that Rap1 phosphorylation is able to modulate its association with some binding partners, namely cytochrome b558 (14) and Raf1 (15). The mechanism by which a modification of Ser179 at the C-terminal end of the molecule affects the regions involved with effector interaction at its N terminus is for the moment unclear.
In this study, we report a global assessment of the effects of Ser179 phosphorylation on conformational change/mobility analyzed by hydrogen/deuterium exchange mass spectrometry (DXMS). The results are consistent with an allosteric effect of the C terminus (containing Ser179) to the switch loops/effector domain.
EXPERIMENTAL PROCEDURES
Expression Plasmids
Rap1b-G12V, Rap1b-G12V/S179D, and Rap1b-G12V/S179A were subcloned as NdeI-XhoI fragments into pET28c (Novagen). All Rap1 constructs contained a C181G mutation. For cotransformation, the catalytic subunit of PKA (cPKA) was subcloned into pACYCDuet-1 (Novagen) as a NcoI-SalI fragment.
Protein Expression and Purification
Escherichia coli BL21(DE3) cells were chemically transformed with pET28c-Rap1b vectors. To achieve the phosphorylation of Rap1b-G12V (Rap1-P), cells were cotransformed with pET28c-Rap1b-G12V and pACYCDuet-1-cPKA and cultured in LB plates containing kanamycin and chloramphenicol. A single colony was inoculated into LB medium and grown overnight at 37 °C. Bacterial cultures were diluted and grown up to A600 ∼ 0.6. Protein expression was induced by the addition of 1 mm isopropyl β-d-thiogalactopyranoside for 1 h at 37 °C. Cells were harvested by centrifugation at 5000 × g for 10 min and resuspended in lysis buffer containing 50 mm Tris-HCl (pH 7), 50 mm NaCl, 10 mm MgCl2, 1 mm benzamidine HCl, protease inhibitor mixture (Set I, Calbiochem), and phosphatase inhibitors (50 mm sodium fluoride, 2 mm sodium orthovanadate, 1 mm benzamidine, and 1 mm β-glycerophosphate). Cells were lysed by the addition of lysozyme (1 mg/ml), β-mercaptoethanol (5 mm), Sarkosyl (1%), and Triton X-100 (2%) for 30 min at room temperature and sonicated (four cycles of 20 s). Soluble fraction was recovered after centrifugation at 9000 rpm for 20 min at 4 °C. Lysates were incubated with nickel-nitrilotriacetic acid (Qiagen) according to the manufacturer's instructions. Unbound protein was washed with 50 mm Tris-HCl, 150 mm NaCl, 10 mm MgCl2, 50 mm imidazole, and 5 mm β-mercaptoethanol, and proteins were eluted with 3 ml of buffer containing 50 mm Tris-HCl (pH 7), 150 mm NaCl, 10 mm MgCl2, 300 mm imidazole, and 5 mm β-mercaptoethanol. Upon a size exclusion step in an Econo-Pac P6 cartridge (Bio-Rad), purified proteins were concentrated and stored in 25% glycerol at −80 °C.
Nucleotide Analysis
Nucleotide-bound Rap1 was determined by fast protein liquid chromatography using a Mono Q 5/50 GL column (GE Healthcare) in a GE Healthcare ÄKTA purifier system. The column was equilibrated with 20 mm Tris-HCl (pH 7.5), and the nucleotide was eluted using a 0–250 mm NaCl gradient at a flow rate of 1 ml/min. GDP and GTP retention times were 14.50 and 19.60 min, respectively. In a typical experiment, an aliquot of a sample containing the purified protein was boiled for 3 min at 95 °C and centrifuged at high speed for 10 min to remove denatured protein. An aliquot of the supernatant was injected into the column. The column was calibrated with solutions containing increasing concentrations of pure guanine nucleotides.
Circular Dichroism
CD spectra for purified Rap1b proteins were recorded in phosphate-buffered saline (pH 7.4) using an Aviv 202 series CD spectrophotometer. Spectra were recorded at 25 °C with a thermostatically controlled cell holder in a fused quartz cell with a path length of 0.1 cm and scanned at 1-nm intervals in the far-UV region (195–280 nm). Each spectrum is the average of two independent experiments (Rap1 at ∼10 μm), each consisting of three scans averaged and base line-corrected by blank buffer subtraction. Spectra are expressed in units of mean molar ellipticity per residue, [θ]. Melting curves (Tm) were generated by measuring [θ]220 as a function of temperature (Rap1 at ∼25 μm). Because of protein denaturation at high temperatures, no attempts were made to extract thermodynamic information.
Rap Activation Assay (RalGDS Ras-binding Domain Pulldown)
Approximately 400 μg of lysates from PCCL3 cells (or 1 μg of purified Rap1b, Rap1b-P, or Rap1b-D protein) was mixed with 10 μg of GST-RalGDS Ras-binding domain (RBD) precoupled to glutathione-Sepharose beads (Amersham Biosciences) in 500 μl of lysis buffer and incubated at 4 °C for 60 min with agitation. Beads were washed four times with lysis buffer. After the final wash, Laemmli sample buffer was added to the samples. Proteins were then fractionated on a 12% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane for Western blot analysis.
DXMS
Optimization of the Fragmentation Conditions
To optimize the conditions for efficient peptide coverage, a guanidine hydrochloride curve was performed. 15 μl of stock solution of Rap1b mutant (5 mg/ml in 10 mm Tris-HCl (pH 7.0) and 50 mm NaCl) was diluted with 45 μl of water, quenched with 90 μl of 0.8% (v/v) formic acid containing various concentrations of guanidine hydrochloride (0.8, 1.6, 3.2, and 6.4 m) at 0 °C, and stored frozen at −80 °C. Samples were thawed on ice and immediately passed over a protease column (66-μl bed volume) filled with porcine pepsin (Sigma; immobilized on Poros 20 AL medium at 30 mg/ml following the manufacturer's instructions) at a flow rate of 100 μl/min with 0.05% trifluoroacetic acid. The duration of digestion was 40 s. The proteolytic products were directly collected on a C18 column (Vydac catalog no. 218MS5105) and then eluted with a linear gradient of 8–48% solvent B for 30 min (solvent A is 0.05% trifluoroacetic acid in water, and solvent B is 80% (v/v) acetonitrile and 0.01% trifluoroacetic acid). The eluate was transferred to a Finnigan LCQ Classic mass spectrometer (with ion trap) for mass spectrometric analyses, with the electrospray ionization sprayer voltage set at 5 kV, the capillary temperature at 200 °C, and data acquisition in either MS1 profile mode or data-dependent tandem mass spectrometric mode. The SEQUEST software program (Thermo Finnigan) was used to identify the sequence of the pepsin-generated peptide ions. This set of peptides was then further examined by specialized software, DXMS Explorer (Sierra Analytics Inc., Modesto, CA), to determine whether the measured isotopic envelope of peptides was qualified for accurate measurement of the geometric centroid of isotopic envelopes on deuterated samples. The peptide coverage maps for various concentrations of guanidine hydrochloride were compared, and the condition with the best coverage map was used for the deuterium exchange experiments.
Deuterium Exchange Experiments
Deuterated samples were prepared at 0 °C by diluting 15 μl of Rap1b mutant stock solution with 45 μl of deuterated buffer (8.3 mm Tris-HCl (pH 7.2), 5 mm MgCl2, and 50 mm NaCl), followed by on-exchange incubation for varying times (10, 30, 100, 300, 1000, and 3000 s) prior to quenching in 90 μl of 0.8% formic acid and 3.2 m guanidine hydrochloride at 0 °C and then stored frozen at −80 °C. These deuterated samples were processed as described above, along with control samples of non-deuterated and fully deuterated Rap1b proteins (incubated in 0.5% formic acid in 100% D2O for 12 h at room temperature). The centroids of isotopic envelopes of non-deuterated, partially deuterated, and fully deuterated peptides were measured using DXMS Explorer and then converted to deuteration level with corrections for back-exchange. The deuteron recovery of fully deuterated sample was, on average, 80%.
Modeling the C Terminus of Rap1b
Loop predictions were performed using PLOP (protein local optimization program) (16). The algorithm uses a hierarchical method involving loop structure buildup, clustering, side chain optimization, and energy minimization of the selected loops, including side chains. Energy evaluations were performed using an atom-based molecular mechanics force field (OPLS) (17), with a surface generalized Born solvation model (18, 19). A three-step hierarchical protocol was utilized here; the first 6 amino acids (residues 167–172) were predicted first, yielding 30 possible structures that were used next to predict structures of residues 170–176. The 10 lowest energy structures from each of the 30 calculations were used as starting coordinates for the final step of predicting the structure of residues 176–181. In all three steps, side chains within 3 Å of the predicted loop were optimized.
RESULTS AND DISCUSSION
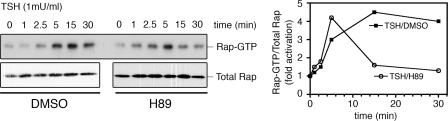
We have recently reported that, in the PCCL3 thyroid cell line, both PKA and Epac are synergistically involved in thyroid-stimulating hormone (TSH) action (11). Closer examination of the kinetic profile of Rap1 activation (Fig. 1) confirmed that TSH rapidly activates Rap1, remaining in its active state for up to 30 min; however, pharmacological inhibition of PKA by H89 renders it only transient, indicating a potential role for PKA in the sustained phase of TSH-mediated Rap1 activation, a condition not assessed in our original studies.
FIGURE 1.
Kinetic profile of TSH-mediated Rap activation. PCCL3 thyroid cells were starved for 16 h in basal Coon's medium and 0.2% bovine serum albumin, followed by agonist stimulation as indicated. For PKA inhibition, cells were preincubated for 20 min with H89 (10 μm) or dimethyl sulfoxide (DMSO) as a control. TSH (1 milliunits/ml) was added, and at different times, cells were washed and lysed. An aliquot was taken for total Rap (left, lower panels), and lysate was subjected to RalGDS RBD pulldown assay (left, upper panel). The ratio of Rap-GTP to total Rap is presented (right), normalized to basal conditions.
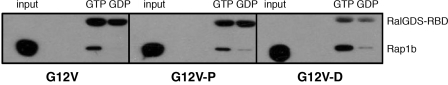
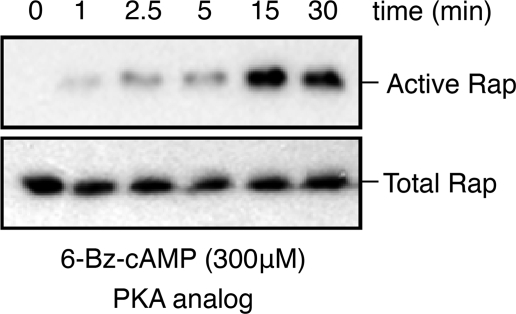
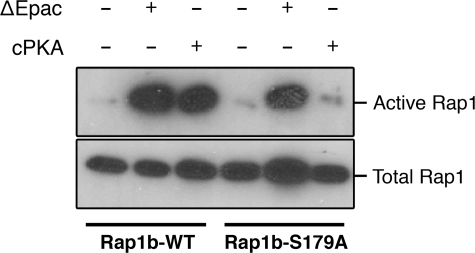
We investigated a potential effect of Rap1b phosphorylation on effector interaction using the RalGDS RBD binding assay, utilizing recombinant purified proteins expressed in E. coli. Rap1b-G12V, phosphomimetic G12V/S179D (G12V-D), and in vitro PKA-phosphorylated G12V (G12V-P) were loaded with GDP or GTP, and their activation state was monitored by pulldown assays with the RalGDS RBD (Fig. 2). The results confirmed that GTP-bound Rap1 proteins bind with similar affinity to the RalGDS RBD, regardless of their Ser179 phosphorylation status. Guanine nucleotide disassociation inhibitor assays (20) using mant-GTP-loaded proteins confirmed no differences in affinity (Kd ∼ 90 nm) (data not shown). However, unexpectedly, contrary to G12V, where only GTP-bound protein interacted with the RBD, G12V-D and G12V-P showed some interaction when loaded with GDP. These results indicate the possibility of conformational exchange between phospho-Ser179 and the switch loops/effector domain. A limitation of the studies presented above is that they involved recombinant proteins without the known post-translational modifications that occur in mammalian cells. To assess the potential effects of Ser179 phosphorylation on Rap activation in vivo, we investigated whether conditions able to activate PKA, but not Epac, result in increased affinity for the RalGDS RBD. First, we addressed pharmacologically whether a cAMP analog able to activate only PKA (and not Epac) is able to activate Rap1 in thyroid PCCL3 cells; as shown in Fig. 3, the PKA-specific analog N6-benzoyl-cAMP activated Rap1 with kinetics consistent with the sustained phase described above. We then turned to a genetic approach, exploiting the observation that cAMP is not able to activate Rap1 in human embryonic kidney cells unless Epac is expressed (11). This model provides us the opportunity to express constitutively active forms of Epac and PKA and to assess their ability to activate Rap1 in the absence of cAMP. Upon cotransfection with hemagglutinin (HA)-Rap constructs (wild-type or S179A) and either constitutively active Epac (ΔEpac) or cPKA, HA-Rap activity was assessed by RalGDS RBD pulldown assays and HA blots. Although ΔEpac was able to activate both wild-type Rap1b and Rap1b-S179A, activation by cPKA was not observed in Rap1-S179A (Fig. 4). This result indicates that PKA is able to activate post-translationally modified Rap1 in vivo in a manner strictly dependent on its phosphorylation status. The combined results suggested the hypothesis that Ser179 phosphorylation at the C terminus might communicate allosterically with the effector domain of Rap1b, thus providing an explanation for the differential effects of Rap1 phosphorylation by PKA on its association with binding partners. To directly assess the potential effects of Ser179 phosphorylation on the switch loops/effector domain, we decided to monitor phospho-Ser179-dependent conformational changes using amide DXMS.
FIGURE 2.
Effect of phosphorylation of Rap1b on its interaction with RalGDS. Purified proteins were loaded in vitro with GTP or GDP, and its interaction with RalGDS was analyzed by an RBD pulldown assay utilizing GST-RalGDS RBD precoupled to glutathione-Sepharose beads. An aliquot of the nucleotide-loaded proteins was run in parallel lanes (input) before the pulldown affinity step (RalGDS). Western blots revealed with anti-GST (RalGDS RBD) and anti-Rap (Rap1b) antibodies are shown.
FIGURE 3.
PKA activates Rap1 in thyroid cells. PCCL3 thyroid cells were starved for 16 h in basal Coon's medium and 0.2% bovine serum albumin, followed by stimulation with the PKA-specific cAMP analog N6-benzoyl-cAMP (300 μm). Lysates were prepared and processed for RalGDS RBD pulldown assays as described in the legend to Fig. 1.
FIGURE 4.
Phospho-Ser179 is required for PKA (but not Epac)-mediated Rap1 activation. Human embryonic kidney cells were cotransfected with HA-Rap1 (wild-type (WT)) or HA-Rap1-S179A and constitutively active Epac (ΔEpac) or cPKA as indicated. 24 h later, lysates were prepared and processed for RalGDS RBD pulldown assays as described in the legend to Fig. 1.
DXMS is a very powerful approach to attain a global assessment of potential conformational changes induced by binding and/or post-translational modifications (21). DXMS monitors exchange in backbone amide hydrogens; those that are involved in hydrogen bonds or buried inside the protein are protected, manifesting a slow exchange rate, whereas those that are solvent-exposed and present in flexible mobile loops usually show a high exchange rate. Thus, DXMS exchange rate determination can be used as a conformationally sensitive assay to monitor the effects of phosphorylation. The main goal of the next set of experiments was to exploit this technique in the context of PKA-mediated Rap1b phosphorylation. For this purpose, constitutively active Rap1b (Rap1b-G12V) constructs with wild-type Ser179, phosphomimetic S179D (Rap1-D), or PKA-phosphorylated Ser179 (Rap1-P) were purified from E. coli as His-tagged proteins. For the latter, both substrate (His-Rap1b-G12V) and kinase (untagged cPKA) were coexpressed in independent compatible vectors before affinity purification. Rap phosphorylation was confirmed by Western blotting with anti-Rap1b phospho-Ser179 antiserum recently developed in-house (supplemental Fig. S1). Because of the high level of constitutive cPKA expression attained, isopropyl β-d-thiogalactopyranoside induction conditions had to be properly optimized to avoid nonspecific phosphorylation events. The identity of phospho-Ser179 under the conditions chosen was fully characterized by mass spectroscopy (supplemental Fig. S2). Purified Rap1-P was digested with clostripain, a proteinase that cleaves proteins on the carboxyl peptide bond of arginine, releasing the C-terminal KKSSGQLL peptide. The expected mass of the non-phosphorylated protonated peptide was 860.52. The observed peptide mass of 940.52 was therefore an indication of a phosphorylation event (addition of 80 Da) on either Ser179 or Ser180. Further tandem mass spectrometric analysis of this ion confirmed that Rap1-P was phosphorylated at Ser179 (supplemental Fig. S2), confirming previous results obtained in vivo (22).
Proper folding of the purified proteins was assessed by CD spectroscopy, nucleotide content, and effector interaction before proceeding to the DXMS studies. Similar far-UV CD spectra and melting curves were observed for all the purified proteins, indicating that phosphorylation does not have a major impact on secondary structure (supplemental Fig. S3). Accordingly, their nucleotide-bound content (as assessed by fast protein liquid chromatography) indicates no differences among the purified proteins; under the current conditions of purification, ∼70–75% of the protein was loaded with GTP (supplemental Fig. S4). Rap1 proteins bind in a GTP-dependent manner to effectors; as reported previously (23), Rap1-GTP interacted with higher affinity with RalGDS than with Raf1 RBDs (data not shown). Moreover, Rap1 phosphorylation affected Raf1 interaction (data not shown), whereas its affinity for RalGDS was not modified (Fig. 2). Thus, these studies indicate that all the purified proteins are properly folded and show their expected functional properties.
Next, the proteins were subjected to DXMS analysis. Identification of regions with differential rate of deuterium exchange among the different proteins by mass spectrometry depends strictly on our ability to fragment and generate a good sequence coverage map. Optimal proteolysis conditions were determined by digesting Rap1b proteins in the presence of increasing concentrations of the denaturant guanidine hydrochloride (0.8, 1.6, 3.2, and 6.4 m) over a pepsin column. The proteolytic products were collected by a C18 column, and the resulting peptides were subjected to mass spectrometric analyses. The 3.2 m guanidine hydrochloride condition gave us the best coverage map, with no significant gaps; 43 peptides were analyzed with a coverage of >98% of Rap1 sequence (supplemental Fig. S5). This condition was thus used in the next set of deuterium exchange experiments.
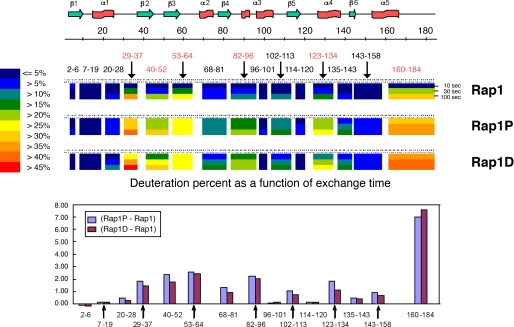
Deuterium exchange was monitored as a function of time, and the reaction was rapidly quenched in acid conditions before proteolysis and analysis. The kinetic profile of the peptides analyzed is shown in supplemental Fig. S6. Similar behavior was observed for Rap1-P and Rap1-D, validating the use of the latter as a phosphomimetic construct. In all regions where differences were observed, exchange rates increased upon phosphorylation. Peptides showing the highest difference in exchange rates between the phosphorylated and non-phosphorylated states encompass discrete regions of the protein. C-terminal fragment 160–184, containing the phosphorylation site (including α5 and the hypervariable region), slightly inaccessible in the non-phosphorylated form, showed an increased exchange rate in the phosphorylated form. Interestingly, the flexible loops corresponding to the switch I (positions 32–40, α1-β2 loop) and switch II (positions 60–76, β3-α2 loop and α2) domains presented an increased deuterium exchange upon phosphorylation, as evidenced by peptides 29–37 and 53–64. Similarly, four other regions represented by peptides 40–52 (β2 and β2-β3 loop), 83–96 (β4-α3 loop and α3), 102–113 (α3-β5 loop), and 123–134 (β5-α4 loop and α4) manifested changes in conformation/mobility upon phosphorylation.
The exchange difference plot (Fig. 5, lower panel) showed differences in the deuterium accumulation between Rap1-P/Rap1-D and Rap1 (only selected peptides upon 10-s exchange are shown). The profiles for both Rap1-D and Rap1-P are identical, and overall, a higher exchange is observed in the phosphorylated or phosphomimetic state compared with the non-phosphorylated state. The main differences were mapped onto the GppNHp-bound Rap1a crystal structure (24) and are presented in Fig. 6.
FIGURE 5.
Summary of hydrogen/deuterium exchange rates induced by phosphorylation. Deuteration levels of representative peptides at various exchange time points (10, 30, and 100 s) are shown as a pseudo color scale (middle panel) along the secondary structure of full-length Rap1b (upper panel). The difference in the number of deuterons at 10 s between Rap1b-G12V/S179D (Rap1-D) or PKA-phosphorylated Rap1b-G12V (Rap1-P) and Rap1b-G12V is shown (lower panel). Similar results were obtained for three independent protein purification/DXMS assays.
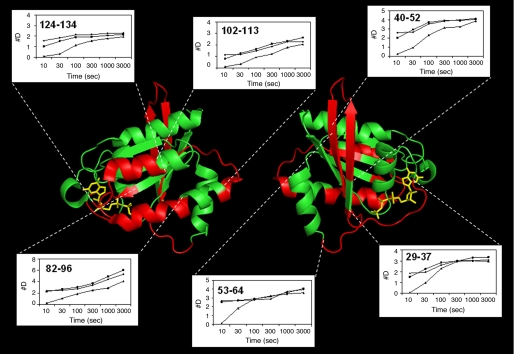
FIGURE 6.
Mapping the regions of high exchange rates induced by phosphorylation. Kinetic profiles (10–3000s) of peptides with the highest exchange rate differences are shown and mapped onto the known x-ray structure of the Rap1a-(1–166)·GppNHp complex (protein backbone in green, corresponding peptides in red, and nucleotide in yellow). The Left and right images represent a 180° rotation for aid in visualization. The figure was generated using PyMOL (38).
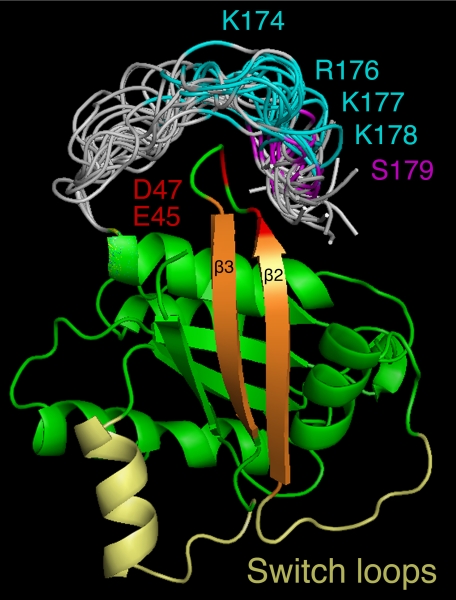
Unfortunately, all available Rap1 structures are based on truncated proteins lacking the C-terminal fragment (Rap1-(1–166)) and thus the phosphorylation site (Ser179). This prevents us from comparing the changes observed by deuterium exchange with the known structure of the low energy conformers. However, reasonable C-terminal structures were modeled using loop prediction algorithms. On the basis of recent computer simulations of membrane-bound K-Ras that showed that the C terminus (very similar in sequence to the C-terminal loop of Rap1b) converges to a conformation in which the midsection of the loop is close to the β2-β3 turn,5 we focused on the possibility that the Rap1b C-terminal tail might interact with the same region. Shown in Fig. 7 is a collection of low energy structures where positive charges in the C terminus were restrained at 5–8 Å of the negatively charged residues Glu45 and Asp47 in the β2-β3 turn. According to this model, the polybasic domain at the flexible C terminus dynamically interacts with the negative environment of the β2-β3 turn (i.e. Glu45 and Asp47). Phosphorylation of Ser179 next to the polybasic domain would incorporate a repulsive negative charge, increasing β2-β3 turn flexibility that could be transduced to the switch I and II domains via β2 and β3, respectively. This model is consistent with our experimental results; apart from the C-terminal peptide (containing the polybasic region and phosphorylation site), peptides 29–37 (switch I), 53–64 (switch II), and 40–52 (containing the negatively charged β2-β3 turn) represent the regions with the largest changes in hydrogen/deuterium exchange rates upon phosphorylation. It is noteworthy to mention that, consistent with this model, molecular dynamics simulations on Ras proteins suggested the presence of an isoform-specific network of amino acids communicating the C terminus with the β2-β3 turn, allosterically affecting the nucleotide-binding and effector domains (25), and accordingly, removal of the corresponding negatively charged residues in H-Ras (i.e. Asp47 and Glu49) controls Ras orientation at the membrane and signaling (26). Current mutagenesis studies on Rap1b involving the elimination of the negative charges in Glu45/Asp47 is ongoing to test some of these predictions. Utilizing the approach described in Fig. 4, we predict mutations could be identified affecting the ability of cPKA, but not ΔEpac, to activate Rap1.
FIGURE 7.
Proposed model for phosphorylation-induced allostery. Modeled C-terminal conformations (gray) as an extension of the known Rap1a structure (green) highlight the polybasic region (cyan) and the position of the Ser179 phosphorylation site (magenta). Putative electrostatic interactions between the polybasic region and the acidic residues Glu45 and Asp47 (red) and the role of the β2-β3 strands (orange) communicating to the switch loops (yellow) are shown. The figure was generated using PyMOL (38).
A coupling of the nucleotide-binding domain with the C terminus membrane-anchoring region was originally proposed in Ras proteins to explain the GTP-dependent lateral segregation on the plasma membrane (27). Ras proteins are not homogeneously distributed along the membrane compartment; a fixed fraction of Ras-GTP at the plasma membrane (∼40%) is organized in nanoclusters, immobile complex structures composed of approximately seven Ras proteins, whereas the remaining ∼60% represents a mobile fraction of monomeric Ras (28). The assembly of Ras-GTP nanoclusters is required for effector recruitment and efficient downstream signaling (29–31). Interestingly, phosphorylation of K-Ras at Ser181 (pK-Ras), next to its C-terminal polybasic domain, has a negative effect on plasma membrane interaction and cluster formation and thus indirectly on effector recruitment. Unexpectedly, however, pK-Ras showed an increased effect on output signaling (30). Moreover, protein kinase C-mediated phosphorylation of K-Ras at Ser181 translocated pK-Ras from the plasma membrane to endomembranes including mitochondria, where interaction with Bcl-xL triggered an apoptotic response (32). Thus, consistent with our results on Rap1b, K-Ras might also be subjected to a similar allosteric effect, associated with a phospho-dependent switch in effector interaction.
For many years, the GTPase cycle represented a simple two-state conformational switch. It is becoming clear, however, that even in the native states (both GDP- and GTP-bound), a multiple conformational ensemble with distinct rates of interchange on specific time scales has to be considered (25, 33–37); effector binding then “selects” one of the pre-existing conformers, thus displacing the equilibrium. Our results suggest the intriguing possibility that phosphorylation, either directly by “fixing” a conformational state or indirectly by altering the interstate exchange rates, might act as an allosteric discriminator of conformational/dynamic states, i.e. a phosphorylation-dependent switch acting in parallel to the canonical nucleotide-dependent switch. Further structural, dynamic, and computational studies are required to understand the trajectory of the allosteric pathway governing the effects of phosphorylation and the detailed amino acid network involved.
Supplementary Material
This work was supported by National Institutes of Health Grants R01 CA071649 and DK063069 (to D. L. A.); Innovative Technologies for the Molecular Analysis of Cancer (IMAT) Program Grants CA099835 and CA118595 (to V. L. W.); National Institutes of Health Grants AI076961, AI081982, AI2008031, AI072106, AI068730, GM037684, GM020501, and GM066170 (to V. L. W.); and Discovery Grant UC10591 from the University of California IUCRP Program, BiogenIDEC corporate sponsor (to V. L. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
A. A. Gorfe, B. J. Grant, and J. A. McCammon, unpublished data.
- PKA
- cAMP-dependent protein kinase
- DXMS
- hydrogen/deuterium exchange mass spectroscopy
- cPKA
- catalytic subunit of PKA
- RBD
- Ras-binding domain
- GST
- glutathione S-transferase
- TSH
- thyroid-stimulating hormone
- HA
- hemagglutinin
- GppNHp
- guanosine 5′-(β,γ-imido)triphosphate.
REFERENCES
- 1.Caron E. (2003) J. Cell Sci. 116, 435–440 [DOI] [PubMed] [Google Scholar]
- 2.Hattori M., Minato N. (2003) J. Biochem. 134, 479–484 [DOI] [PubMed] [Google Scholar]
- 3.Bos J. L., Rehmann H., Wittinghofer A. (2007) Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 4.Herrmann C. (2003) Curr. Opin. Struct. Biol. 13, 122–129 [DOI] [PubMed] [Google Scholar]
- 5.Stieglitz B., Bee C., Schwarz D., Yildiz O., Moshnikova A., Khokhlatchev A., Herrmann C. (2008) EMBO J. 27, 1995–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altschuler D. L., Peterson S. N., Ostrowski M. C., Lapetina E. G. (1995) J. Biol. Chem. 270, 10373–10376 [DOI] [PubMed] [Google Scholar]
- 7.Altschuler D. L., Ribeiro-Neto F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7475–7479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitt J. M., Stork P. J. (2001) Mol. Cell. Biol. 21, 3671–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng X., Ji Z., Tsalkova T., Mei F. (2008) Acta Biochim. Biophys. Sin. 40, 651–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribeiro-Neto F., Urbani J., Lemee N., Lou L., Altschuler D. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochbaum D., Hong K., Barila G., Ribeiro-Neto F., Altschuler D. L. (2008) J. Biol. Chem. 283, 4464–4468 [DOI] [PubMed] [Google Scholar]
- 12.McAvoy T., Zhou M. M., Greengard P., Nairn A. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3531–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitt J. M., Stork P. J. (2002) Mol. Cell 9, 85–94 [DOI] [PubMed] [Google Scholar]
- 14.Bokoch G. M., Quilliam L. A., Bohl B. P., Jesaitis A. J., Quinn M. T. (1991) Science 254, 1794–1796 [DOI] [PubMed] [Google Scholar]
- 15.Hu C. D., Kariya K., Okada T., Qi X., Song C., Kataoka T. (1999) J. Biol. Chem. 274, 48–51 [DOI] [PubMed] [Google Scholar]
- 16.Jacobson M. P., Pincus D. L., Rapp C. S., Day T. J., Honig B., Shaw D. E., Friesner R. A. (2004) Proteins 55, 351–367 [DOI] [PubMed] [Google Scholar]
- 17.Kaminski G., Friesner R. A., Tirado-Rives J., Jorgensen W. (2001) J. Phys. Chem. B 105, 6474–6487 [Google Scholar]
- 18.Gallicchio E., Zhang L. Y., Levy R. M. (2002) J. Comput. Chem. 23, 517–529 [DOI] [PubMed] [Google Scholar]
- 19.Ghosh A., Rapp C. S., Friesner R. A. (2002) J. Phys. Chem. B 102, 10983–10990 [Google Scholar]
- 20.Herrmann C., Martin G. A., Wittinghofer A. (1995) J. Biol. Chem. 270, 2901–2905 [DOI] [PubMed] [Google Scholar]
- 21.Tsutsui Y., Wintrode P. L. (2007) Curr. Med. Chem. 14, 2344–2358 [DOI] [PubMed] [Google Scholar]
- 22.Altschuler D., Lapetina E. G. (1993) J. Biol. Chem. 268, 7527–7531 [PubMed] [Google Scholar]
- 23.Herrmann C., Horn G., Spaargaren M., Wittinghofer A. (1996) J. Biol. Chem. 271, 6794–6800 [DOI] [PubMed] [Google Scholar]
- 24.Nassar N., Horn G., Herrmann C., Scherer A., McCormick F., Wittinghofer A. (1995) Nature 375, 554–560 [DOI] [PubMed] [Google Scholar]
- 25.Gorfe A. A., Grant B. J., McCammon J. A. (2008) Structure 16, 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abankwa D., Hanzal-Bayer M., Ariotti N., Plowman S. J., Gorfe A. A., Parton R. G., McCammon J. A., Hancock J. F. (2008) EMBO J. 27, 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prior I. A., Harding A., Yan J., Sluimer J., Parton R. G., Hancock J. F. (2001) Nat. Cell Biol. 3, 368–375 [DOI] [PubMed] [Google Scholar]
- 28.Harding A., Hancock J. F. (2008) Cell Cycle 7, 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murakoshi H., Iino R., Kobayashi T., Fujiwara T., Ohshima C., Yoshimura A., Kusumi A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7317–7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plowman S. J., Ariotti N., Goodall A., Parton R. G., Hancock J. F. (2008) Mol. Cell. Biol. 28, 4377–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian T., Harding A., Inder K., Plowman S., Parton R. G., Hancock J. F. (2007) Nat. Cell Biol. 9, 905–914 [DOI] [PubMed] [Google Scholar]
- 32.Bivona T. G., Quatela S. E., Bodemann B. O., Ahearn I. M., Soskis M. J., Mor A., Miura J., Wiener H. H., Wright L., Saba S. G., Yim D., Fein A., Pérez de Castro I., Li C., Thompson C. B., Cox A. D., Philips M. R. (2006) Mol. Cell 21, 481–493 [DOI] [PubMed] [Google Scholar]
- 33.Arai Y., Iwane A. H., Wazawa T., Yokota H., Ishii Y., Kataoka T., Yanagida T. (2006) Biochem. Biophys. Res. Commun. 343, 809–815 [DOI] [PubMed] [Google Scholar]
- 34.Ito Y., Yamasaki K., Iwahara J., Terada T., Kamiya A., Shirouzu M., Muto Y., Kawai G., Yokoyama S., Laue E. D., Wälchli M., Shibata T., Nishimura S., Miyazawa T. (1997) Biochemistry 36, 9109–9119 [DOI] [PubMed] [Google Scholar]
- 35.O'Connor C., Kovrigin E. L. (2008) Biochemistry 47, 10244–10246 [DOI] [PubMed] [Google Scholar]
- 36.Spoerner M., Herrmann C., Vetter I. R., Kalbitzer H. R., Wittinghofer A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4944–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spoerner M., Nuehs A., Herrmann C., Steiner G., Kalbitzer H. R. (2007) FEBS J. 274, 1419–1433 [DOI] [PubMed] [Google Scholar]
- 38.DeLano W. L. (2002) PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.