Abstract
In this study, we investigated the mechanism by which the CUX1 transcription factor can stimulate cell migration and invasion. The full-length p200 CUX1 had a weaker effect than the proteolytically processed p110 isoform; moreover, treatments that affect processing similarly impacted cell migration. We conclude that the stimulatory effect of p200 CUX1 is mediated in part, if not entirely, through the generation of p110 CUX1. We established a list of putative transcriptional targets with functions related to cell motility, and we then identified those targets whose expression was directly regulated by CUX1 in a cell line whose migratory potential was strongly stimulated by CUX1. We identified 18 genes whose expression was directly modulated by p110 CUX1, and its binding to all target promoters was validated in independent chromatin immunoprecipitation assays. These genes code for regulators of Rho-GTPases, cell-cell and cell-matrix adhesion proteins, cytoskeleton-associated proteins, and markers of epithelial-to-mesenchymal transition. Interestingly, p110 CUX1 activated the expression of genes that promote cell motility and at the same time repressed genes that inhibit this process. Therefore, the role of p110 CUX1 in cell motility involves its functions in both activation and repression of transcription. This was best exemplified in the regulation of the E-cadherin gene. Indeed, we uncovered a regulatory cascade whereby p110 CUX1 binds to the snail and slug gene promoters, activates their expression, and then cooperates with these transcription factors in the repression of the E-cadherin gene, thereby causing disorganization of cell-cell junctions.
The molecular mechanisms by which transformed cells become migratory and invasive during tumor progression are beginning to be unraveled (reviewed in Ref. 1). Some events are reminiscent of an important developmental process termed epithelial-to-mesenchymal transition (EMT)3 (reviewed in Refs. 2, 3). During EMT, tumor cells redistribute or down-regulate their epithelium-specific proteins such as adherent and tight-junction proteins, including E-cadherin and occludin, and start to express mesenchymal proteins, such as vimentin and N-cadherin. As a result, cell-cell contacts are disrupted causing a loss of apico-basal polarity, and cells acquire mesenchymal and migratory properties necessary for invasion. Transcriptional repression has emerged as a fundamental mechanism for silencing of E-cadherin and occludin, and several transcriptional repressors have been identified (reviewed in Ref. 4). Snail and Slug, which belong to the Snail superfamily of zinc finger transcriptional repressors, are the most characterized E-cadherin repressors (5–9). The zinc fingers present at the carboxyl terminus of the proteins function as the sequence-specific DNA-binding domains that recognize consensus E2 box-type elements. Their repressor capacity is mediated by the SNAG domain present at the amino-terminal part of the proteins (reviewed in Ref. 10).
A requirement for the CUX1 homeodomain protein in cell motility was originally revealed from a high throughput RNA interference screen (11). CDP/Cux/Cut (CCAAT-displacement protein/cut homeobox) proteins are a family of transcription factors present in all metazoans and involved in the control of proliferation and differentiation (reviewed in Refs. 12, 13). The mammalian gene and protein are now called Cux1 and CUX1, respectively, in accordance with new nomenclature rules. In a panel of human cancer cell lines, siRNA-mediated knockdown of CUX1 expression caused a decrease in cell motility and formation of lung metastasis as measured in a wound healing assay, an invasion assay through Matrigel, and in a pulmonary colonization assay after caudal vein injection (11). In addition, CUX1 silencing was later found to delay cell spreading, but not the adhesive properties of cells, upon seeding on a fibronectin-coated substrate (14).
CUX1 is primarily regulated by post-translational modifications, including phosphorylation by protein kinase C (15), CKII (16), protein kinase A (17), and cyclin A/Cdk1 (18), dephosphorylation by Cdc25A (19), acetylation by PCAF (20), and proteolytic processing by cathepsin L (21–23) and a caspase (24). Proteolytic processing by cathepsin L generates the p110 isoform (21, 25) and, in some types of cells, the p90 isoform (26). In contrast to the full-length p200 protein, which functions exclusively as a repressor via its CCAAT displacement activity (27), the amino-terminally processed isoforms of CUX1 function as transcriptional repressors on some promoters and as transcriptional activators on other promoters (24, 28–30). The available evidence suggests that p110 CUX1 associates with distinct partners on specific promoters, and that the type of protein complex that forms determines whether the promoter is activated or repressed (31–35).
In this study, we investigated the mechanism by which CUX1 can stimulate cell migration and invasion. We first determined that the effect of p200 CUX1 on cell migration was weaker than that of p110 CUX1 and might be mediated in part through the generation of its processed isoform. We performed genome-wide location analysis to establish a list of putative transcriptional targets of CUX1. We identified those targets that may be involved in cell motility. Independent ChIP and reporter assays confirmed the binding of CUX1 to the promoter of these genes and the resultant regulatory effect. Our findings indicate that the role of p110 CUX1 in cell motility involves its dual function as a transcriptional repressor or activator of specific targets. For example, p110 CUX1 activated transcription from the vimentin and N-cadherin gene promoters but repressed transcription from the occludin and E-cadherin gene promoters. Further analysis revealed that CUX1 activates transcription of snail and slug genes and then cooperates with these transcription factors in the repression of the E-cadherin gene. We show that the end point of this regulation at the cellular level is the disorganization of cell-cell junctions.
EXPERIMENTAL PROCEDURES
Cell Culture
Hs578T, MGT/p110.1, and Madin-Darby canine kidney (MDCK) cells were cultured in Dulbecco's modified minimum essential medium (DMEM) supplemented with penicillin/streptomycin, glutamine, and 10% fetal bovine serum (FBS) (Invitrogen). The Hs578T human breast tumor cell line has been described previously (22, 36). The MGT/p110.1 cell line was established in our laboratory from a mammary gland tumor that developed in a transgenic mouse expressing p110 CUX1 under the control of regulatory sequences from the mouse mammary tumor virus. NMuMG and NMuMG-NYPD cells were cultured in DMEM supplemented with 10% FBS and 10 μg/ml insulin (37). Mouse embryo fibroblasts were prepared as described previously (38).
Retroviral Infections
Retroviruses were produced by transfecting 293 VSV cells with different isoforms of CUX1 (Myc tagged at the amino terminus and hemagglutinin (HA) tagged at the carboxyl terminus) inserted in the pLXSN plasmid (Clontech). The supernatant was applied on NIH 3T3, NMuMG, or NMuMG-NYPD cells at an equivalent titer, along with 8 μg/ml Polybrene (Roche Applied Science), and the plates were centrifuged at 300 × g for 1 h. After 48 h, infected cells were selected for 5 days in G418, and at least 500 resistant clones were pooled together for each population. Cell lines were infected with empty vector or vectors expressing either p200 CUX1 (amino acids 1–1505) or p110 CUX1 (amino acids 747–1505). For conditional knockdown of CUX1, we took advantage of Addgene plasmid 11643. NMuMG-NYPD cells were infected with pLVCT shCUX1-(5326–5348)-tTRKRAB lentivirus at a multiplicity of infection of 10 as described previously (39). 48 h after infection, cells were split and cultured with or without doxycycline at a final concentration of 2.5 μg/ml. Cells were used for experiments after 5 days of treatment.
Migration and Invasion Assays
In vitro migration assays were performed in modified Boyden chambers (8-μm pores, BD Falcon). Briefly, 80,000 cells in 500 μl of serum-free DMEM were added to the upper part of the inserts. The lower chambers were filled with DMEM supplemented with 10% FBS. The chambers were incubated at 37 °C in 5% CO2 atmosphere for 16 h. Cells were then fixed using 10% neutral buffered formalin for 20 min at room temperature, washed three times with phosphate-buffered saline (PBS), and then stained with 0.1% crystal violet in 20% methanol for 20 min. The cells that did not migrate through the filter were removed from the inside of the inserts using cotton swab. Migrating cells were examined under a microscope and quantified by evaluating the number of pixels using Scion software. Invasion assays were performed similarly using BD Matrigel invasion chambers (8-μm pores BD Falcon). Each experiment was done in triplicate, and the graphs represent an average of the 3 wells. All the experiments were repeated at least three times, and similar results were obtained. For all experiments * means 0.05 > p value > 0.01, ** means 0.01 > p value > 0.001, and *** means p value < 0.001.
Wound Healing Assay
Confluent cell cultures were grown on 60-mm culture plates for 48 h. Wounds were made with the tip of a micropipette. Cells were maintained in DMEM supplemented with 10% FBS. To analyze cell motility, phase contrast microscopy was done, and images were collected every 6 h.
Cell Spreading and Adhesion Assay
96-Well plates were coated overnight at room temperature with Matrigel (1:50 in PBS) or with collagen (40 μg/ml in DMEM). Wells were rinsed and blocked for 1 h with 1% BSA. Logarithmic phase cells were harvested with trypsin and plated at 60,000 cells per well. After 45, 60, 75, and 90 min of incubation at 37 °C, wells were rinsed to remove nonadherent cells. Adhered cells were fixed in 10% formalin for 5 min and stained with 0.1% crystal violet for 5 min. Cells were then rinsed and dried. Adherent cells were examined under a microscope and quantified by evaluating the number of pixels using Scion software. For actin cytoskeleton staining, cells were fixed for 10 min in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS for 10 min, blocked in 2% BSA for 30 min, and incubated for 1 h with Alexa 488-coupled phalloidin. Nuclei were counterstained with DAPI. Images were acquired using a Zeiss confocal microscope.
RNA and Real Time PCR
RNA was extracted using TRIzol reagent (Invitrogen), and cDNA was prepared using Superscript II RNase H-reverse transcriptase kit (Invitrogen) following the manufacturer's instructions. Real time PCR was performed on a LightCycler instrument using the FastStart DNA Master SYBR Green kit (Roche Applied Science) and specific primer pairs for each gene (see sequences in supplemental Table 3).
ChIP
2 × 108 Hs578T cells were used for ChIP analysis. Immunoprecipitation of endogenous CUX1 was done using anti-CUX1 antibodies 861 and 1300. Nuclei were purified as described previously (40), then lysed in RIPA-M buffer (10 mm Tris-HCl, pH 8, 1 mm EDTA, 0.5 mm EGTA, 150 mm NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 1 mm phenylmethylsulfonyl fluoride, protease inhibitors), and sonicated on ice to obtain 250–800-bp-long DNA fragments. Un-enriched input chromatin was put aside as a control. After preclearing for 1 h and incubation with antibodies overnight, immunocomplexes were washed three times each in wash buffer I (20 mm Tris-HCl, pH 8, 2 mm EDTA, 2 mm EGTA, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.2% SDS), wash buffer II (20 mm Tris-HCl, pH 9, 2 mm EDTA, 2 mm EGTA, 500 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS), wash buffer III (50 mm Tris-HCl, pH 7.5, 2 mm EDTA, 1 mm EGTA, 0.5 m LiCl, 1% Nonidet P-40, 0.7% deoxycholate), and then washed once in Tris-EDTA. Cross-linked DNA was eluted with 1% SDS, 10 mm Tris-HCl, pH 8, 10 mm EDTA at 65 °C for 30 min. PCR was performed using specific primers pairs specific for each promoter (see sequences in supplemental Table 4).
Scanning Chromatin Affinity Purification (ChAP) or ChIP
ChAP was preformed using Hs578T cells stably expressing p110-Tag2 as described previously (28), and ChIP was performed as described above. Primer pairs specific for different regions of the snail (−2.2 to −2.0 kb, −1.9 to −1.6 kb, −1.4 to −1.1 kb, and −0.9 to −0.6 kb) and Slug (−3.2 to −3.0 kb, −2.7 to −2.4 kb, −2.0 to −1.7 kb, −1.6 to −1.4 kb, and −0.8 to −0.5 kb) promoters were designed (see sequences in supplemental Table 5). Enrichment was calculated using the G6PDH locus as a reference and is shown relative to the DNA obtained by purification on Sepharose beads without IgG.
Genome-wide Location Analysis
ChIP and ChAP were performed as described above. Probe generation, microarray design and hybridization, and data analysis were performed as described previously (28).
Immunoblotting
Nuclear extracts were prepared according to the procedure of Lee et al. (41), except that nuclei were obtained by subjecting cells to three freeze/thaw cycles in buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 1 mm dithiothreitol) along with a protease inhibitor tablet (Roche Applied Science). Total protein extracts were prepared using RIPA buffer (150 mm NaCl, 50 mm Tris-HCl, pH 8, 1% Nonidet P-40, 0.01% SDS, 0.5% deoxycholate) along with a protease inhibitor tablet. SDS-PAGE was performed, and after electrophoretic transfer to polyvinylidene difluoride, membranes were washed in Tris-buffered saline, 0.1% Tween 20 (TBS 0.1% T) and blocked in TBS 0.1% T containing 5% milk and 0.5% BSA. Membranes were probed with antibodies directed against the following: HA (1:2000, MMS-101R; Covance); CUX1 (1:3000, anti-1300; 1:2000, anti-861) (25); E-cadherin (1:1000; BD Transduction Laboratories); vimentin (1:500; Epitomics); N-cadherin (1:1000; BD Transduction Laboratories); actin (1:1000, I-19; Santa Cruz Biotechnology); occludin (1:1000; Zymed Laboratories Inc.); Snail (1:1000; Santa Cruz Biotechnology); γ-tubulin (1:10,000; Sigma). Primary antibodies were incubated in TBS 0.1% T, and detection was done using a horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibody in TBS 0.1% T. Immunoreactive proteins were visualized by chemiluminescence with an ECL Western blotting detection kit (Amersham Biosciences).
Luciferase Assay
PCR amplification was performed to obtain fragments of genomic DNA from vimentin, N-cadherin, E-cadherin, and occludin promoters containing the sequence present in the ChIP microarray, plus at least 250 bp on each side. The obtained fragments were cloned into the luciferase reporter vector, pGL3 (Promega). Luciferase assays were performed as described previously (29).
Immunofluorescence
48 h after seeding on coverslips, MDCK cells were fixed with 2% paraformaldehyde for 20 min at room temperature and permeabilized for 10 min with 0.5% Triton X-100. After three rinses in 100 mm glycine, cells were incubated for 30 min in blocking buffer (2% BSA, 0.2% Triton X-100, 0.05% Tween 20). Anti-E-cadherin (1:200) or anti-occludin (1:100) antibodies were added together with anti-HA antibody (1:1000) for 1 h at room temperature in the blocking buffer. Cells were than rinsed three times in immunofluorescent buffer (0.1% BSA, 0.2% Triton X-100, 0.05% Tween 20 in PBS) and incubated with anti-mouse Alexa 594 (1:1000) and anti-rabbit Alexa 488 (1:1000) secondary antibodies in blocking buffer for 45 min. Nuclei were counterstained with 0.5 ng/ml DAPI for 5 min at room temperature, and coverslips were mounted on slides using Immu-MountTM (Thermo Scientific). Cells were visualized using a Zeiss LSM 510 confocal microscope with a ×100 objective.
RESULTS
p200 CUX1 Mediates Its Effects on Migration in Part through the Generation of p110 CUX1
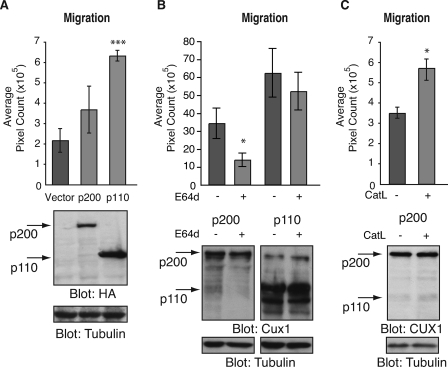
In preliminary experiments, we established that the stimulatory effect of CUX1 on cell migration and invasion was greater in epithelial than in fibroblastic cell lines (supplemental Fig. 1, A–D). We noted that the defect in migration of mouse embryo fibroblasts derived from the Cux1Z/Z knock-out was rescued partially by p200 CUX1 but completely by p110 CUX1 (supplemental Fig. 1, E and F). The greatest effect was observed in NMuMG-NYPD cells, which are normal mouse mammary cells that express a mutated version of the Neu receptor tyrosine kinase in which many of the tyrosine autophosphorylation sites were replaced with alanine (42). The ability of p200 and p110 CUX1 to stimulate migration was then compared in the NMuMG-NYPD epithelial cells. Again, the stimulatory effect of p200 CUX1 was less than that of p110 CUX1 (Fig. 1A). Because the p110 isoform is generated by proteolytic processing of p200 CUX1, it is not clear whether p200 CUX1 has a weak activity on its own or merely provides a source of p110 CUX1. To investigate this question, we measured migration of cells expressing p200 CUX1 following treatments that would either decrease or increase the production of p110 CUX1. Incubation of cells with the cell-permeable cathepsin L inhibitor, E64D, reduced both the steady-state level of p110 CUX1 and the migratory properties of cells, whereas the same treatment did not significantly reduce the migration of cells expressing a recombinant p110 CUX1 protein (Fig. 1B). In contrast, ectopic expression of cathepsin L stimulated both the production of p110 CUX1 and the migratory properties of cells (Fig. 1C). Although we cannot entirely exclude an activity of p200 CUX1 on its own, these results strongly suggest that p200 CUX1 mediates its effects on migration, at least in part, through the generation of p110 CUX1. This finding together with the stronger effect of p110 CUX1 led us to focus on this CUX1 isoform in subsequent experiments.
FIGURE 1.
p200 CUX1 mediates its effects on migration, at least in part, through the generation of p110 CUX1. Populations of NMuMG-NYPD mouse mammary epithelial cells stably carrying a retroviral vector, either empty or expressing p110-HA or p200-HA CUX1, were established. Following the indicated treatment, CUX1 protein expression was analyzed by Western blotting using an anti-CUX1 or anti-HA antibody as indicated, and cell motility was measured in a two-chamber migration assay. The migration assays were all performed in triplicate and repeated at three independent times. A, no treatment. B, cells were incubated for 48 h in the presence of the cell-permeable cysteine protease inhibitor, E64d, or the carrier. C, cells were infected with a retroviral vector, either empty or expressing cathepsin L, and were analyzed 48 h later.
p110 CUX1 Stimulates Cell Spreading and Cell Adhesion
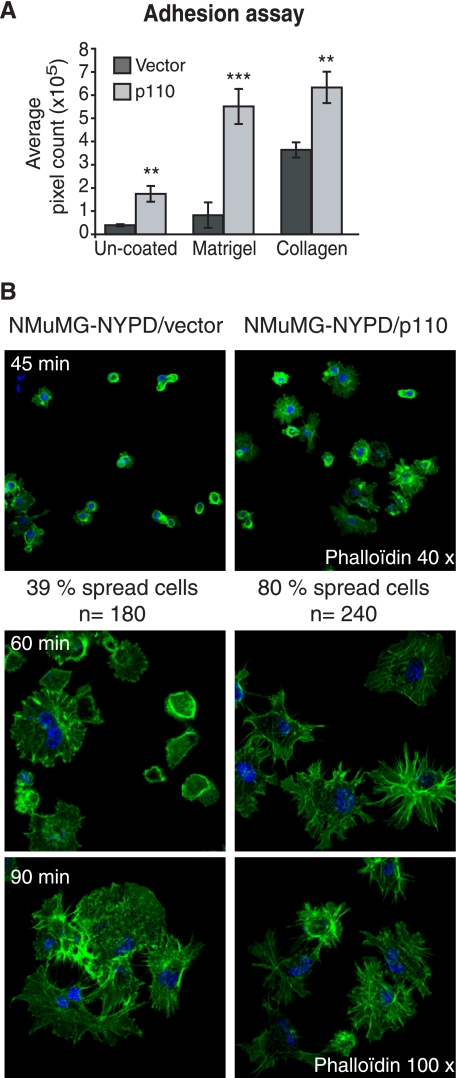
In a previous study, cell adhesion was not significantly altered following the knockdown of CUX1 expression with siRNA, although a spreading defect was observed (14). We therefore tested whether overexpression of p110 CUX1 would affect the ability of NMuMG-NYPD cells to adhere to plastic that was either un-coated or coated with Matrigel or collagen. As compared with cells carrying the empty vector, cells expressing p110 CUX1 exhibited an increase in adhesion, from 1.5-fold on un-coated plastic up to 6-fold on Matrigel after 45 min (Fig. 2A).
FIGURE 2.
p110 CUX1 stimulates cell spreading and cell adhesion. A, adhesion ability of NMuMG-NYPD/vector and p110 CUX1-expressing cells was analyzed using plates that were either uncoated or coated with Matrigel or collagen. After 45 min, adherent cells were fixed in 10% formalin and stained with 0.1% crystal violet, and pixel counts were measured using Scion software. B, spreading of NMuMG-NYPD/vector or p110-expressing cells was analyzed at 45, 60, and 90 min after plating on collagen-coated slides. At the indicated time, cells were fixed, stained with phalloidin, and visualized by fluorescence microscopy. Nuclei were counterstained with DAPI. The proportion of cells that were spread was counted at 45 min.
When cells adhere to a substrate in culture, they reorganize their actin filaments following an ordered sequence of events. When cells first attach, they begin to spread, organizing actin in peripheral filopodia and lamellipodia. Actin stress fibers and mature focal adhesion appear only later (43). To verify the effect of CUX1 on cell spreading, we analyzed the actin cytoskeleton at various time points using phalloidin staining followed by fluorescence microscopy (Fig. 2B). Results are shown after plating on collagen, but similar results were observed on Matrigel (data not shown). A large proportion of control cells that were attached after 45 min remained round, and only 39% of them presented membrane extensions (Fig. 2B, top left panel). In contrast, 80% of the attached p110 CUX1-expressing cells had spread and presented many extensions (Fig. 2B, top right panel). At 60 min, the actin cytoskeleton was still not organized in the vector cells, whereas long extensions were clearly visible in p110 CUX1 cells (Fig. 2B, 60 min panel). We noted, however, that the differences diminished at later time points (Fig. 2B, 90 min panel). Altogether, these results showed that cell spreading is accelerated in cells that overexpress p110 CUX1.
Identification of CUX1 Targets Implicated in Cell Migration, Invasion, and Adhesion
We used the following strategy to identify transcriptional targets of CUX1 that mediate its effects on cell migration, invasion, and adhesion. First, we established an extensive list of transcriptional targets by performing genome-wide location analysis in several cell lines (supplemental Table 1); second, we verified which of the relevant targets exhibited changes in expression in NMuMG-NYPD cells stably expressing p110 CUX1 (Table 1), and third, we performed independent ChIP experiments to validate the binding of CUX1 to the promoters of these genes (Fig. 3).
TABLE 1.
Expression of target genes in cells stably expressing p110 CUX1
RNA was extracted from populations of NMuMG-NYPD cells stably carrying a p110 CUX1 or an empty retroviral vector. Quantitative real time reverse transcription-PCR analysis was performed using primer pairs specific for each target. Values were normalized using G6PDH and represent an average of three independent measurements. The 3rd column indicates the positive or negative fold difference in p110 CUX1-expressing cells as compared with vector cells, and the right column indicates the corresponding p values.
| Function | Gene symbol | p110 versus vector |
|
|---|---|---|---|
| -Fold | p value | ||
| GTPase-related proteins | ARHGDIB | 3.4 | 4.9E-03 |
| RAB36 | 1.5 | 3.4E-02 | |
| RAIN | 2.5 | 6.2E-03 | |
| RHOBTB3 | 3.0 | 6.0E-03 | |
| RICS/GC-GAP | −72.9 | 1.1E-07 | |
| Cell adhesion proteins | ITGB2 | 3.4 | 3.6E-02 |
| PNN | −1.7 | 1.5E-02 | |
| PCDH10 | −1.7 | 1.5E-02 | |
| EMT proteins | CDH2/NCAD | 2.1 | 3.7E-03 |
| ECAD | −2.3 | 2.2E-03 | |
| OCLN | −2.5 | 5.0E-03 | |
| VIM | 2.4 | 4.4E-02 | |
| Cytoskeleton proteins | EPLIN | −2.5 | 3.5E-02 |
| FLNA | 1.8 | 5.1E-02 | |
| MARCKS | 15.9 | 5.8E-02 | |
| PTK2/FAK | 1.5 | 9.4E-03 | |
| Others | THBS1/TSP1 | 28.1 | 3,9E-02 |
| WISP-2 | 391.4 | 2.9E-02 | |
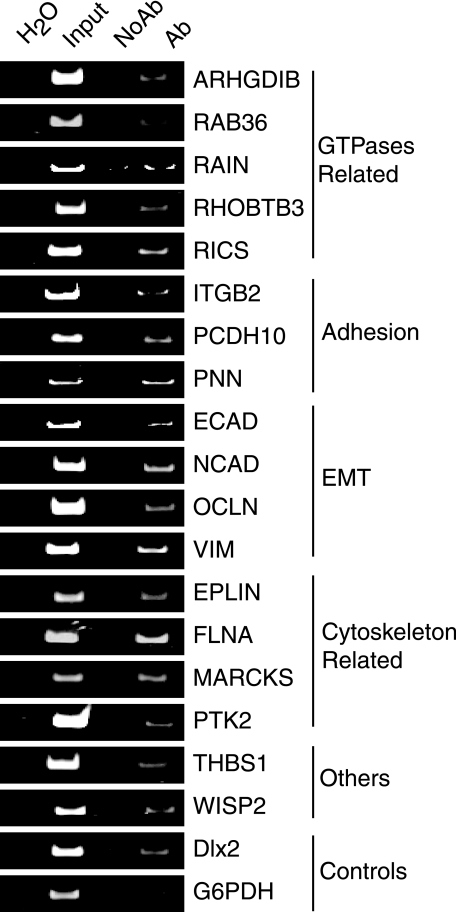
FIGURE 3.
Validation of p110 CUX1 targets. Chromatin from invasive human ductal carcinoma-derived cell line Hs578T was submitted to immunoprecipitation using anti-CUX1 antibodies (Ab) or IgG as a control, and analyzed by PCR using primers specific for the promoter of each potential CUX1 target gene. G6PDH and Dlx2 were used as negative and positive controls, respectively. Input DNA (0.02%) was used as control. Note that the region to be amplified was chosen to be approximately in the middle of the sequence spotted on the location array.
In total, 22 independent location analyses experiments were performed (supplemental Table 1). In some cases, the chromatin was purified by immunoprecipitation using CUX1 antibodies, as detailed previously (28, 29, 33). In other cases, a doubly tagged recombinant CUX1 protein corresponding to a specific isoform was expressed at physiological levels, and chromatin was purified by tandem affinity purification using a strategy previously described and validated (28). Based on validation experiments, we considered as putative targets those genes with a p value smaller than 0.005 because our estimated false-positive rate was less than 3% using this threshold (28). Classification into functional categories using programs from DAVID (see “Experimental Procedures”) enabled us to make a list of 66 putative targets with functions related to cell migration, invasion, or adhesion processes (supplemental Table 2).
Using quantitative real time RT-PCR analysis, we compared the expression of these genes in NMuMG-NYPD cells carrying an empty vector or stably expressing p110 CUX1. We found 18 genes whose expression was either higher or lower in cells expressing p110 CUX1 (Table 1). These genes code for different categories of proteins with a function related to migration and invasion. These include regulators of Rho-GTPases implicated in cytoskeleton remodeling required for cell motility (44, 45), cell-cell and cell-matrix adhesion proteins such as integrins, focal adhesion kinase, E-cadherin, and occludin. Others are markers of epithelial-to-mesenchymal transition such as vimentin and N-cadherin or cytoskeleton-associated proteins like filamin and EPLIN. Interestingly, expression of genes promoting cell migration and invasion such as focal adhesion kinase, N-cadherin, and vimentin was elevated in p110-expressing cells when compared with control cells (Table 1), whereas genes inhibiting cell migration and invasion such as E-cadherin, occludin, RICS, and EPLIN were down-regulated in p110-expressing cells (Table 1). These results suggested that a role for p110 CUX1 in cell motility involves its dual function as a transcriptional repressor or activator of specific targets.
Validation of CUX1 Targets
Because CUX1 is elevated in human cancers (reviewed in Ref. 13), we verified whether CUX1 can directly interact with the promoters of these genes in human cancer cells. Chromatin immunoprecipitation (ChIP) was performed using CUX1-specific antibodies and chromatin isolated from cells of the Hs578T breast tumor cell line, which was previously shown to express all CUX1 isoforms (22, 24, 46). As a control, the chromatin was immunoprecipitated in parallel using IgG alone. Standard PCR analysis was performed using primer pairs derived from each of the gene promoters. Because our promoter array contained promoter sequences ranging approximately from 800 bp upstream to 200 bp downstream of the transcription start sites, primer pairs were chosen to amplify a region located in the middle of these sequences (see sequences in supplemental Table 4). G6PDH and DLX2 served as negative and positive controls, respectively (28). We observed enrichment of all 18 genes listed in Table 1 and conclude that these genes are direct targets of CUX1 (Fig. 3).
Protein Expression of p110 CUX1 Targets
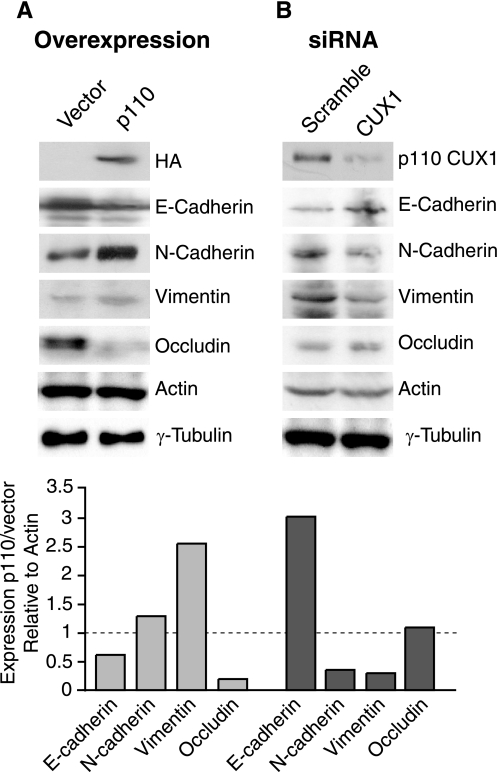
We next performed immunoblotting analysis to verify whether the changes in transcription imparted by p110 CUX1 in NMuMG-NYPD cells were also reflected at the protein level. In cells stably expressing p110 CUX1, we observed a decrease in the steady-state level of E-cadherin and occludin, and an increase in N-cadherin and vimentin (Fig. 4A). Remarkably, siRNA-mediated knockdown of p110 CUX1 had the opposite effect, an increase in the steady-state level of E-cadherin and occludin and a decrease in N-cadherin and vimentin (Fig. 4B).
FIGURE 4.
Protein expression of CUX1 target genes. A, total protein extract was prepared from logarithmic growing populations of NMuMG-NYPD cells stably carrying a p110 CUX1 or an empty retroviral vector. Equal amount of proteins was loaded on SDS-polyacrylamide gel for Western blot analysis using specific antibodies raised against different CUX1 target genes. HA antibody was used to verify the expression of p110 CUX1, and actin and γ-tubulin serve as loading controls. B, primary cell line established from a p110 transgenic mouse mammary gland tumor was transfected with scrambled or anti-CUX1 siRNA. 5 days after transfection, total protein extract was prepared. Equal amount of proteins was loaded on SDS-polyacrylamide gel for Western blot analysis using specific antibodies raised against different CUX1 target genes. Anti-CUX1 antibody was used to verify p110 CUX1 expression, and β-actin and γ-tubulin serve as a loading controls. In the histogram, bands from A (light gray) and B (dark gray) were quantified using the Scion software, and expression of targets in p110-expressing cells compared with control cells was calculated relative to actin.
Immediate Promoter Region Is Sufficient for Regulation by CUX1
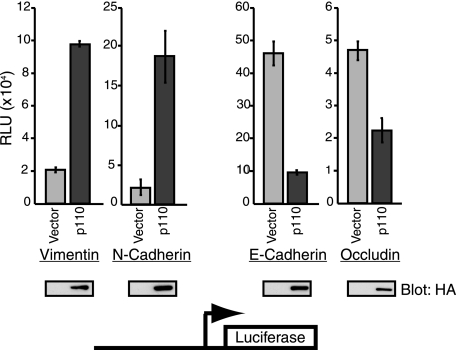
To confirm the role of p110 CUX1 as a repressor or activator of specific genes, and to verify whether the immediate promoter region of these genes was sufficient to enable regulation by CUX1, we performed reporter assays using luciferase reporter plasmids for four target genes as follows: those coding for vimentin, N-cadherin, E-cadherin, and occludin. Each reporter plasmid included the promoter sequences that were present on the promoter microarray plus ∼250 bp on either side. In the presence of p110 CUX1, the vimentin and N-cadherin reporters were activated ∼5- and 8-fold, whereas the E-cadherin and occludin reporters were repressed ∼5- and 2-fold, respectively (Fig. 5). We conclude that the immediate promoters of these genes contain all the regulatory sequences required for their regulation by CUX1. Moreover, these results demonstrated that p110 CUX1 can either activate or repress genes depending on promoter context.
FIGURE 5.
Luciferase reporter assays. The promoter regions of vimentin, N-cadherin, E-cadherin, and occludin were cloned into a luciferase reporter plasmid. Hs578T cells were transfected with each reporter plasmid together with a vector expressing p110 CUX1 or with an empty vector. The experiments were done in triplicate and performed independently at least three times. RLU, relative luciferase unit.
p110 CUX1 Activates Snail and Slug Expression and Cooperates in the Repression of E-cadherin
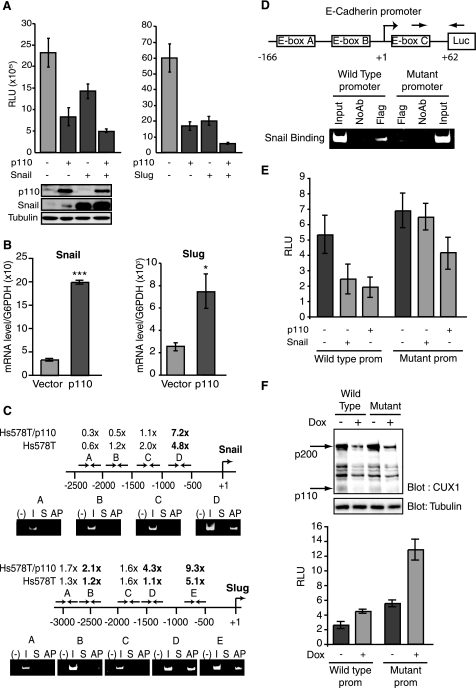
As Snail and Slug are two of the best characterized transcriptional repressors involved in EMT (reviewed in Ref. 4), we considered the possibility that p110 CUX1 might cooperate with one or the other of these factors in transcriptional repression. In cotransfection assays using the E-cadherin reporter plasmid together with suboptimal amounts of effector plasmids, we observed a cooperation between p110 CUX1 and Snail and between p110 CUX1 and Slug (Fig. 6A). In each case, reporter gene expression was lower in the presence of two transcription factors than either alone.
FIGURE 6.
p110 CUX activates Snail and Slug expression and cooperates with them in the repression of E-cadherin. A, luciferase reporter assays. NMuMG-NYPD cells were cotransfected with a reporter plasmid containing nucleotides −166 to + 62 of the E-cadherin promoter and a vector expressing p110 CUX1, either alone or together with Snail or Slug. As a control, cells were transfected with vector alone. Values are means of three measurements, and error bars represent standard deviation. B, snail and slug mRNA expression was measured by quantitative real time PCR in NMuMGNYPD/vector and NMuMG-NYPD/p110 CUX1 cells. The values are the mean of three measurements, and the error bars represent standard deviation. *, p < 0.05; ***, p < 0.001. C, scanning ChAP and ChIP within the snail and slug gene promoters. Chromatin from Hs578T/p110CUX1-Tag2 and from Hs578T cells was submitted to affinity purification or immunoprecipitation and analyzed by quantitative real time PCR using primer pairs specific for different regions of the promoters. Templates for the PCRs were 0.1% total input DNA (I), nonspecific DNA from Sepharose beads alone (S), and ChAP- (AP) or ChIP-purified DNA. The positions of amplified fragments are indicated over the maps, and primer sequences are given under “Experimental Procedures.” The respective fold enrichment of the different DNA fragments are indicated relative to the DNA obtained by purification on Sepharose beads without IgG (S). Enrichment was calculated using the G6PDH locus as a reference. D, replacement mutations were made to change the first and sixth nucleotide of each of the three E boxes (CANNTG) within a reporter plasmid containing nucleotides −166 to + 62 of the E-cadherin promoter as follows: box A, CAGGTG to AAGGTA; box B, CACCTG to AACCTA; and box C, CACCTG to AACCTA. NMuMG-NYPD cells were transfected with either the wild type or mutant E-cadherin reporter plasmid together with a vector expressing a FLAG-tagged Snail protein. The chromatin was immunoprecipitated with an anti-FLAG antibody and qRT-PCR was performed with the indicated primers to compare the recruitment of Snail to the wild type and mutated reporter. E, luciferase reporter assays. NMuMG-NYPD cells were cotransfected either the wild type or mutated (−166/+62) E-cadherin reporter plasmid together with expression vectors expressing either nothing (vector), p110 CUX1 or FLAG-tagged Snail. F, NMuMG-NYPD cells were infected with a Tet-On lentiviral vector expressing a CUX1-specific shRNA in the presence of doxycycline. Cell were either treated with doxycycline or left untreated, and CUX1 protein levels were assessed by Western blotting after 5 days. In parallel, doxycycline-treated or untreated cells were transfected with the wild type or mutated (−166/+62) E-cadherin reporter plasmid, and a luciferase assay was performed. RLU, relative luciferase unit.
We next verified whether expression of Snail and/or Slug was altered in cells overexpressing p110 CUX1. Using quantitative real time PCR, we observed a 6- and a 3-fold increase in snail and slug expression, respectively, in p110 CUX1-expressing cells as compared with cells carrying an empty vector (Fig. 6B). These findings raised the possibility that p110 CUX1 might directly regulate the snail and slug genes. These gene promoters had not been identified as putative targets in our genome-wide location arrays, which contained promoter regions from approximately −800 to +200 bp relative to the transcription start site. Upon examination of the slides, however, we realized that these genes were among the ∼20% of sequences for which the DNA spot was either nonexistent or defective. We therefore performed scanning ChAP and ChIP analyses using multiple primer pairs derived from the snail and slug gene promoter sequences (see sequences in supplemental Table 5). The analysis was performed both in Hs578T cells and in Hs578T cells stably expressing a recombinant p110 CUX1 protein. The results indicated that p110 CUX1 is recruited to a specific region of each of these promoters (Fig. 6C). Note that the fold enrichment was increased in cells overexpressing p110 CUX1, and in the case of the Slug promoter, the region bound by p110 CUX1 was also enlarged. This observation suggests cooperativity in the recruitment of CUX1 to an adjacent region of this promoter.
The above results suggest that p110 CUX1 may regulate E-cadherin expression by two mechanisms as follows: through the activation of E-cadherin repressors, Snail and Slug, and by direct repression of the E-cadherin promoter. The following strategy was used to verify whether p110 CUX1 can have a direct effect on the E-cadherin promoter. Scanning ChIP analysis indicated that p110 CUX1 binds to the immediate promoter region of the E-cadherin gene (data not shown). We therefore used a shorter version of the E-cadherin reporter plasmid, containing nucleotides −166 to +62, and including only three E boxes, the consensus binding site for Snail. Replacement mutations were introduced to change the first and sixth nucleotide of each of these three E boxes (Fig. 6D). Following cotransfection of either the wild type or the mutated E-cadherin reporter plasmid together with a vector expressing Snail with a FLAG tag, the chromatin was immunoprecipitated with the α-FLAG antibody, and PCR amplification was performed using primers specific for the E-cadherin reporter plasmid. The results show that the replacement mutations caused a decrease in the recruitment of Snail to the E-cadherin promoter (Fig. 6D). In agreement with this finding, Snail repressed the wild type but not the mutated E-cadherin promoter (Fig. 6E). In contrast to the lack of effect of Snail, overexpression of p110 CUX1 was still associated with the repression of the mutated E-cadherin promoter (Fig. 6E). Moreover, shRNA-mediated knockdown of CUX1 led to an increase in expression of both the wild type and mutated E-cadherin reporter (Fig. 6F). We conclude that p110 CUX1 can directly repress the E-cadherin promoter. Altogether these results indicate that p110 CUX1 binds to the snail and slug gene promoters, activates their expression, and then cooperates with these transcription factors in the repression of the E-cadherin gene.
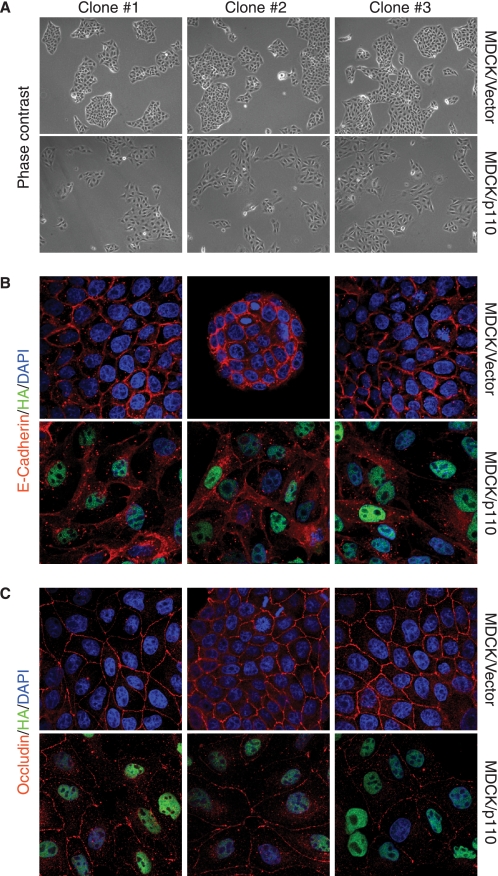
Elevated Expression of p110 CUX1 Causes Loss of E-cadherin and Occludin from Cell-Cell Junctions
To evaluate the consequences of E-cadherin and occludin gene repression in cells expressing elevated levels of p110 CUX1, we took advantage of the MDCK epithelial kidney cells whose ability to form well organized adherens and tight junctions has been extensively characterized (6, 47, 48). Clones of MDCK cells stably expressing p110 CUX1 or carrying an empty retroviral vector were analyzed by phase contrast microscopy and indirect immunofluorescence for components of each of these junctions, E-cadherin for adherens junctions and occludin for tight junctions. When compared with control cells, p110 CUX1-expressing cells displayed an elongated shape and formed looser colonies (Fig. 7A). In control MDCK cells, staining for E-cadherin and occludin revealed a well organized structure at cell-cell junctions (Fig. 7, B and C). In contrast, in p110 expressing CUX1 cells E-cadherin was reduced and was no longer localized at cell-cell contacts. Similarly, staining of occludin was reduced at cell-cell contacts (Fig. 7, B and C). These results support that the transcriptional repression of the E-cadherin and occludin genes by p110 CUX1 can significantly alter the ability of epithelial cells to form organized cell-cell junctions.
FIGURE 7.
Elevated expression of p110 CUX1 causes loss of E-cadherin and occludin from cell-cell junctions. Clones of MDCK epithelial cells stably expressing p110 CUX1 or carrying the empty retroviral vector were seeded on coverslips and analyzed by phase-contrast microscopy and indirect immunofluorescence. A, phase-contrast microscopy: note the larger size and elongated shape of p110 CUX1 cells and the loose contacts they form between each other, as compared with vector cells. B, cells were stained for E-cadherin (red) and HA (green), and nuclei were stained with DAPI. C, cells were stained for occludin (red) and HA (green), and nuclei were stained with DAPI. B and C, original magnification was ×100.
DISCUSSION
In this study, we presented results suggesting that the effect of p200 CUX1 on cell motility and EMT was mediated in part, if not entirely, through the generation of p110 CUX1 (Fig. 1 and supplemental Fig. 1F). This notion is further supported by considerations regarding the regulatory properties of these two CUX1 isoforms. Among the 18 target genes investigated in this study, 6 were down-regulated and 12 were up-regulated in cells overexpressing p110 CUX1 (Table 1). We can envisage that the p200 isoform would be able to repress those targets that were down-regulated; however, activation by p200 CUX1 has never been observed (reviewed in Refs. 12, 13). In contrast, the p110 CUX1 isoform was previously shown to repress or activate distinct genes, at least in the context of cell cycle progression (18, 25, 28, 29, 38). Here we demonstrated that, depending on promoter context, p110 CUX1 can also function as a repressor or activator of genes that play a role in cell migration and EMT (Fig. 5).
Overexpression of p110 CUX1 led to an increase in cell adhesion and an acceleration in cell spreading (Fig. 2). These results are in apparent contradiction with the observations made in a previous study where cell adhesion was not affected following the inhibition of CUX1 expression with siRNA (14). The discrepancy between these results could be attributed to the different substrates used in the two studies or more likely to the cells chosen for the experiments, as the PANC1 pancreatic cells adhered very rapidly and were almost completely spread after as little as 16 min, making it more difficult to see a difference in adhesion ability (14).
From previous studies little is known about the mechanism(s) by which CUX1 could stimulate the migratory properties of cells. Expression profiling following the knockdown of CUX1 allowed the identification of genes whose normal level of expression requires the presence of CUX1 (11). This approach did not distinguish between direct and indirect effects and could not reveal which genes are affected in cancer cells that overexpress CUX1. In a subsequent study, Ripka et al. (49) used reporter assays to demonstrate that Wnt5A is a transcriptional target of CUX1. Finally, another study identified a link between CUX1 and Src signaling (14). Cells in which CUX1 expression was stably knocked down displayed an increase in proteasome-mediated degradation of Src that was associated with a reduction in Src-regulated downstream signaling proteins such as RhoA, Rac1, Cdc42, and Rock. Because expression of the carboxyl-terminal Src kinase Csk paralleled that of CUX1, it was proposed that the effect of CUX1 on the stability of Src, and thus on cell motility, could be mediated through the transcriptional regulation of the Csk gene, although whether Csk is a direct transcriptional target of CUX1 remains to be verified.
We considered it unlikely that a transcription factor, in particular a homeodomain-containing protein, would impact on such a complex process as cell motility just by regulating a single or even a handful of genes. Although the function of transcription factors has for a long period of time been investigated through the analysis of a few candidate targets, the development of the ChIP assay together with the availability of genomic microarrays has enabled the unbiased identification of transcriptional targets. From the analysis of ChIP-chip assays came the dual realization primarily that individual transcription factors bind to a large repertoire of genes and secondarily that the regulation of a single gene must be accomplished through the integration of multiple inputs that converge to regulatory sequences in a combinatorial fashion. Our current understanding of transcriptional regulation is forcibly limited; however, a few paradigms have already emerged, for example the existence of regulatory cascades whereby a transcription factor induces the expression of other transcription factors that will cooperate with the first one in subsequent waves of regulation.
In epithelial cells, E-cadherin is a key component of the establishment and maintenance of cell polarity. The loss of E-cadherin-mediated cell-cell adhesion is currently thought to be a prerequisite for tumor cell invasion and metastasis formation (reviewed in Ref. 50). Regulation of E-cadherin expression during malignant progression has been extensively studied, and transcriptional repression has emerged as a fundamental mechanism for its silencing. In a number of human cancer types, the loss of E-cadherin function during tumor progression is accompanied by the de novo expression of mesenchymal N-cadherin, constituting a “cadherin switch” (reviewed in Ref. 51). Our results identified p110 CUX1 as a transcription factor that cannot only directly interact with both the E-cadherin and N-cadherin gene promoters, but moreover can repress the E-cadherin gene while activating the N-cadherin gene. Moreover, we obtained evidence that p110 CUX1 can bind to the snail and slug gene promoters and activate their expression (Fig. 6, B and C). These data are of great interest as, to date, little is known about the transcriptional regulation of these two major players of EMT. NF-κB and β-catenin have been proposed as transcriptional activators of snail or slug, but the evidence was limited only to reporter assays (52–55). Interestingly, our results have uncovered a transcriptional regulatory cascade whereby p110 CUX1 activates the expression of the Snail and Slug transcription factors which, in turn, cooperate with CUX1 in the regulation of downstream targets involved in cell migration as well as EMT, thus highlighting CUX1 as an important regulator of genes associated with tumor progression and metastases.
Supplementary Material
Acknowledgments
We thank Dr. Amparo Cano for the kind gift of Snail and Slug expression vectors and Dr. Peter Siegel for the kind gift of NMuMG and NMuMG-NYPD cell lines. We thank Lam Leduy and Ginette Bérubé for their technical assistance and Dong Mei Zuo for help with confocal microscopy.
This work was supported in part by Team Grant CTP-79857 from the Canadian Institute of Health Research (to A. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Tables 1–5.
- EMT
- epithelial-to-mesenchymal transition
- MDCK
- Madin-Darby canine kidney
- DMEM
- Dulbecco's modified minimum essential medium
- FBS
- fetal bovine serum
- HA
- hemagglutinin
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- ChIP
- chromatin immunoprecipitation
- ChAP
- chromatin affinity purification
- DAPI
- 4′,6-diamidino-2-phenylindole
- G6PDH
- glyceraldehyde-3-phosphate dehydrogenase
- TBS
- Tris-buffered saline.
REFERENCES
- 1.Christofori G. (2006) Nature 441, 444–450 [DOI] [PubMed] [Google Scholar]
- 2.Thiery J. P. (2002) Nat. Rev. Cancer 2, 442–454 [DOI] [PubMed] [Google Scholar]
- 3.Grünert S., Jechlinger M., Beug H. (2003) Nat. Rev. Mol. Cell Biol. 4, 657–665 [DOI] [PubMed] [Google Scholar]
- 4.Peinado H., Olmeda D., Cano A. (2007) Nat. Rev. Cancer 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 5.Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., García De Herreros A. (2000) Nat. Cell Biol. 2, 84–89 [DOI] [PubMed] [Google Scholar]
- 6.Cano A., Pérez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., del Barrio M. G., Portillo F., Nieto M. A. (2000) Nat. Cell Biol. 2, 76–83 [DOI] [PubMed] [Google Scholar]
- 7.Poser I., Domínguez D., de Herreros A. G., Varnai A., Buettner R., Bosserhoff A. K. (2001) J. Biol. Chem. 276, 24661–24666 [DOI] [PubMed] [Google Scholar]
- 8.Yokoyama K., Kamata N., Hayashi E., Hoteiya T., Ueda N., Fujimoto R., Nagayama M. (2001) Oral Oncol. 37, 65–71 [DOI] [PubMed] [Google Scholar]
- 9.Hajra K. M., Chen D. Y., Fearon E. R. (2002) Cancer Res. 62, 1613–1618 [PubMed] [Google Scholar]
- 10.Nieto M. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 155–166 [DOI] [PubMed] [Google Scholar]
- 11.Michl P., Ramjaun A. R., Pardo O. E., Warne P. H., Wagner M., Poulsom R., D'Arrigo C., Ryder K., Menke A., Gress T., Downward J. (2005) Cancer Cell 7, 521–532 [DOI] [PubMed] [Google Scholar]
- 12.Nepveu A. (2001) Gene 270, 1–15 [DOI] [PubMed] [Google Scholar]
- 13.Sansregret L., Nepveu A. (2008) Gene 412, 84–94 [DOI] [PubMed] [Google Scholar]
- 14.Aleksic T., Bechtel M., Krndija D., von Wichert G., Knobel B., Giehl K., Gress T. M., Michl P. (2007) Oncogene 26, 5939–5949 [DOI] [PubMed] [Google Scholar]
- 15.Coqueret O., Bérubé G., Nepveu A. (1996) J. Biol. Chem. 271, 24862–24868 [DOI] [PubMed] [Google Scholar]
- 16.Coqueret O., Martin N., Bérubé G., Rabbat M., Litchfield D. W., Nepveu A. (1998) J. Biol. Chem. 273, 2561–2566 [DOI] [PubMed] [Google Scholar]
- 17.Michl P., Knobel B., Downward J. (2006) J. Biol. Chem. 281, 15138–15144 [DOI] [PubMed] [Google Scholar]
- 18.Santaguida M., Ding Q., Bérubé G., Truscott M., Whyte P., Nepveu A. (2001) J. Biol. Chem. 276, 45780–45790 [DOI] [PubMed] [Google Scholar]
- 19.Coqueret O., Bérubé G., Nepveu A. (1998) EMBO J. 17, 4680–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S., Aufiero B., Schiltz R. L., Walsh M. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7166–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goulet B., Baruch A., Moon N. S., Poirier M., Sansregret L. L., Erickson A., Bogyo M., Nepveu A. (2004) Mol. Cell 14, 207–219 [DOI] [PubMed] [Google Scholar]
- 22.Goulet B., Sansregret L., Leduy L., Bogyo M., Weber E., Chauhan S. S., Nepveu A. (2007) Mol. Cancer Res. 5, 899–907 [DOI] [PubMed] [Google Scholar]
- 23.Goulet B., Nepveu A. (2004) Cell Cycle 3, 986–989 [PubMed] [Google Scholar]
- 24.Truscott M., Denault J. B., Goulet B., Leduy L., Salvesen G. S., Nepveu A. (2007) J. Biol. Chem. 282, 30216–30226 [DOI] [PubMed] [Google Scholar]
- 25.Moon N. S., Premdas P., Truscott M., Leduy L., Bérubé G., Nepveu A. (2001) Mol. Cell. Biol. 21, 6332–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goulet B., Truscott M., Nepveu A. (2006) Biol. Chem. 387, 1285–1293 [DOI] [PubMed] [Google Scholar]
- 27.Moon N. S., Bérubé G., Nepveu A. (2000) J. Biol. Chem. 275, 31325–31334 [DOI] [PubMed] [Google Scholar]
- 28.Harada R., Vadnais C., Sansregret L., Leduy L., Bérubé G., Robert F., Nepveu A. (2008) Nucleic Acids Res. 36, 189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truscott M., Raynal L., Premdas P., Goulet B., Leduy L., Bérubé G., Nepveu A. (2003) Mol. Cell. Biol. 23, 3013–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santaguida M., Nepveu A. (2005) J. Biol. Chem. 280, 32712–32721 [DOI] [PubMed] [Google Scholar]
- 31.Li S., Moy L., Pittman N., Shue G., Aufiero B., Neufeld E. J., LeLeiko N. S., Walsh M. J. (1999) J. Biol. Chem. 274, 7803–7815 [DOI] [PubMed] [Google Scholar]
- 32.Nishio H., Walsh M. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11257–11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Truscott M., Harada R., Vadnais C., Robert F., Nepveu A. (2008) Mol. Cell. Biol. 28, 3127–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Wijnen A. J., Cooper C., Odgren P., Aziz F., De Luca A., Shakoori R. A., Giordano A., Quesenberry P. J., Lian J. B., Stein G. S., Stein J. L. (1997) J. Cell. Biochem. 66, 512–523 [DOI] [PubMed] [Google Scholar]
- 35.van Wijnen A. J., van Gurp M. F., de Ridder M. C., Tufarelli C., Last T. J., Birnbaum M., Vaughan P. S., Giordano A., Krek W., Neufeld E. J., Stein J. L., Stein G. S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11516–11521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hackett A. J., Smith H. S., Springer E. L., Owens R. B., Nelson-Rees W. A., Riggs J. L., Gardner M. B. (1977) J. Natl. Cancer Inst. 58, 1795–1806 [DOI] [PubMed] [Google Scholar]
- 37.Mazzoni E., Adam A., Bal de Kier Joffe E., Aguirre-Ghiso J. A. (2003) Mol. Cancer Res. 1, 776–787 [PubMed] [Google Scholar]
- 38.Sansregret L., Goulet B., Harada R., Wilson B., Leduy L., Bertoglio J., Nepveu A. (2006) Mol. Cell. Biol. 26, 2441–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szulc J., Wiznerowicz M., Sauvain M. O., Trono D., Aebischer P. (2006) Nat. Methods 3, 109–116 [DOI] [PubMed] [Google Scholar]
- 40.Weinmann A. S., Farnham P. J. (2002) Methods 26, 37–47 [DOI] [PubMed] [Google Scholar]
- 41.Lee K. A., Bindereif A., Green M. R. (1988) Gene Anal. Tech. 5, 22–31 [DOI] [PubMed] [Google Scholar]
- 42.Dankort D., Jeyabalan N., Jones N., Dumont D. J., Muller W. J. (2001) J. Biol. Chem. 276, 38921–38928 [DOI] [PubMed] [Google Scholar]
- 43.Defilippi P., Olivo C., Venturino M., Dolce L., Silengo L., Tarone G. (1999) Microsc. Res. Tech. 47, 67–78 [DOI] [PubMed] [Google Scholar]
- 44.Schmitz A. A., Govek E. E., Böttner B., Van Aelst L. (2000) Exp. Cell Res. 261, 1–12 [DOI] [PubMed] [Google Scholar]
- 45.Yamazaki D., Kurisu S., Takenawa T. (2005) Cancer Sci. 96, 379–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goulet B., Watson P., Poirier M., Leduy L., Bérubé G., Meterissian S., Jolicoeur P., Nepveu A. (2002) Cancer Res. 62, 6625–6633 [PubMed] [Google Scholar]
- 47.Royal I., Park M. (1995) J. Biol. Chem. 270, 27780–27787 [DOI] [PubMed] [Google Scholar]
- 48.Peinado H., Marin F., Cubillo E., Stark H. J., Fusenig N., Nieto M. A., Cano A. (2004) J. Cell Sci. 117, 2827–2839 [DOI] [PubMed] [Google Scholar]
- 49.Ripka S., König A., Buchholz M., Wagner M., Sipos B., Klöppel G., Downward J., Gress T., Michl P. (2007) Carcinogenesis 28, 1178–1187 [DOI] [PubMed] [Google Scholar]
- 50.Cavallaro U., Christofori G. (2004) Ann. N.Y. Acad. Sci. 1014, 58–66 [DOI] [PubMed] [Google Scholar]
- 51.Hazan R. B., Qiao R., Keren R., Badano I., Suyama K. (2004) Ann. N.Y. Acad. Sci. 1014, 155–163 [DOI] [PubMed] [Google Scholar]
- 52.Bachelder R. E., Yoon S. O., Franci C., de Herreros A. G., Mercurio A. M. (2005) J. Cell Biol. 168, 29–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barberà M. J., Puig I., Domínguez D., Julien-Grille S., Guaita-Esteruelas S., Peiró S., Baulida J., Francí C., Dedhar S., Larue L., García de Herreros A. (2004) Oncogene 23, 7345–7354 [DOI] [PubMed] [Google Scholar]
- 54.Julien S., Puig I., Caretti E., Bonaventure J., Nelles L., van Roy F., Dargemont C., de Herreros A. G., Bellacosa A., Larue L. (2007) Oncogene 26, 7445–7456 [DOI] [PubMed] [Google Scholar]
- 55.Vallin J., Thuret R., Giacomello E., Faraldo M. M., Thiery J. P., Broders F. (2001) J. Biol. Chem. 276, 30350–30358 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.