Abstract
Maspin is a serpin that has multiple effects on cell behavior, including inhibition of migration. How maspin mediates these diverse effects remains unclear, as it is devoid of protease inhibitory activity. We have previously shown that maspin rapidly inhibits the migration of vascular smooth muscle cells (VSMC), suggesting the involvement of direct interactions with cell surface proteins. Here, using immunofluorescence microscopy, we demonstrate that maspin binds specifically to the surface of VSMC in the dedifferentiated, but not the differentiated, phenotype. Ligand blotting of VSMC lysates revealed the presence of several maspin-binding proteins, with a protein of 150 kDa differentially expressed between the two VSMC phenotypes. Western blotting suggested that this protein was the β1 integrin subunit, and subsequently both α3β1 and α5β1, but not αvβ3, were shown to associate with maspin by coimmunoprecipitation. Specific binding of these integrins was also observed using maspin-affinity chromatography, using HT1080 cell lysates. Direct binding of maspin to α5β1 was confirmed using a recombinant α5β1-Fc fusion protein. Using conformation-dependent anti-β1 antibodies, maspin binding to VSMC was found to lead to a decrease in the activation status of the integrin. The functional involvement of α5β1 in mediating the effect of maspin was established by the inhibition of migration of CHO cells overexpressing human α5 integrin, but not those lacking α5 expression. Our observations suggest that maspin engages in specific interactions with a limited number of integrins on VSMC, leading to their inactivation, and that these interactions are responsible for the effects of maspin in the pericellular environment.
Maspin is a member of the serpin family of serine protease inhibitors (SERPINB5).2 It was originally identified as a gene down-regulated in invasive breast cancer and proposed as a class II tumor suppressor (1), and has since been shown to have many effects on cellular behavior that are consistent with this activity. It has been shown to decrease the proliferation, migration, and metastasis of tumor cells in vivo (1, 2) and their invasion in vitro (3, 4), and to increase apoptosis of endothelial cells (5) and inhibit angiogenesis (6). However, the cellular effects of maspin are not restricted to tumor cells, and we have demonstrated that maspin can inhibit the migration of vascular smooth muscle cells (7).
VSMC migration is a key event in the development of atherosclerosis (8), and contributes significantly to restenosis after angioplasty (9) and transplant arteriosclerosis (10). VSMC are not terminally differentiated and acquire migratory capacity as part of a phenotypic switch from a contractile, quiescent state to a dedifferentiated phenotype, characterized by proliferation and increased extracellular matrix synthesis, in addition to motility (11). This allows VSMC to respond to environmental cues following vascular injury. The phenotypic plasticity of VSMC is regulated by an array of signals, among which integrin-mediated association with surrounding extracellular matrix and changes in the expression of matrix-degrading proteases are prominent (12–14).
How maspin mediates its various cellular effects is unclear. Maspin has been reported to be an inhibitor of plasminogen activation (3, 15, 16), but we have shown that maspin is unable to inhibit either uPA- or tPA-catalyzed plasminogen activation under conditions in which the serpin PAI-1 was completely inhibitory (7). The anti-proteolytic inhibitory mechanism of serpins is dependent on characteristics of the reactive center loop (RCL) allowing it to adopt the necessary canonical conformation and rearrangements subsequent to protease binding (17). The RCL of maspin does not have the required characteristics (7, 18), and the conclusion that maspin is a non-inhibitory serpin is fully supported by its crystal structure (19, 20).
Another confounding factor in understanding the mechanisms underlying the cellular effects of maspin is that, in common with the serpin PAI-2, it lacks an authentic secretion signal sequence. Nevertheless it has been shown to enter secretory vesicles (21) and is found extracellularly, in the cytoplasm and also in the nucleus (21, 22). Cytoplasmic and nuclear binding proteins for maspin have been identified (23–25), and may be responsible for its effects on proliferation and apoptosis. How secreted, extracellular maspin exerts its effects is unclear, but a function as a cell signaling ligand has been proposed (26–28). However, the characteristics of the maspin inhibitory effect on VSMC migration point to a more direct effect of maspin.
To determine the mechanism of the maspin effect on VSMC migration, we have now attempted to identify maspin-binding proteins on the surface of these cells. In this report we provide biochemical, cellular, and functional evidence that the effect of maspin on cell migration is mediated by specific binding to cell adhesion receptors of the integrin family. We find that maspin binds specifically to β1 integrins on the surface of dedifferentiated VSMC, which leads to a reduction in the activation status of the integrin, and that the binding of maspin to α5β1 is sufficient for its inhibitory effects on cell migration and may represent a more general mechanism underlying its diverse biological effects.
EXPERIMENTAL PROCEDURES
Cells, Antibodies, and Proteins
Primary aortic smooth muscle cells, media, and supplements were from TCS Cell Works (Buckingham, UK). These cells were routinely maintained in the dedifferentiated state in Medium 231 supplemented with 4.9% fetal bovine serum, FGF2 (2 ng/ml), EGF (0.5 ng/ml), heparin (5 ng/ml), insulin (5 μg/ml), and BSA (0.2 μg/ml). When necessary these cells were differentiated by culturing in Medium 231 supplemented with 1% fetal bovine serum and heparin (30 μg/ml) for 7 days. α5CHO (clone 17) and B2CHO cell lines were as previously detailed (29) and HT1080 were obtained from ATCC. These cells were maintained in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% (v/v) fetal calf serum, and 0.1 mm non-essential amino acids. All cell culture reagents including ECM components were from Invitrogen (Paisley, UK).
Primary antibodies to maspin were goat polyclonal (R&D Systems, Abingdon, UK) and mouse monoclonal G167-70 (BD Biosciences, NJ). Monoclonal antibodies to β1 integrin subunit for Western blotting and immunoprecipitation were from AbD Serotec (Oxford, UK) and for immunocytochemistry from Fedor Beditchevski (University of Birmingham). α3 and α5 monoclonal antibodies were from Millipore (Danvers, MA). αv antibodies for immunocytochemistry (L230) were a gift from Celltech (Slough, UK), for Western blotting and immunoprecipitation from R&D Systems, and function blocking (MAB1980) from Millipore. Monoclonal antibodies 12G10 and mAb13 were from Martin Humphries (University of Manchester, UK). Polyclonal anti-β3 integrin was kindly provided by Barry Coller (Rockefeller University, New York). The monoclonal antibodies to uPA (clone 5) and uPAR (R4) were kindly provided by Gunilla Høyer-Hansen (Finsen Laboratory, Copenhagen). Polyclonal anti-GAPDH was from Cell Signaling Technology (Danvers, MA). Secondary antibodies were from Dako (Ely, UK) and Invitrogen.
Recombinant maspin, expressed in Saccharomyces cerevisiae, was as previously described (7). Recombinant α5β1-Fc integrin, which is the ectodomain of α5β1 integrin expressed as a fusion protein with mutated human γ1 Fc domains to promote heterodimer formation, was kindly provided by Martin Humphries (University of Manchester) (30).
Immunoprecipitation, Western, and Ligand Blotting
Cell lysates were prepared by washing subconfluent monolayers twice with PBS prior to harvesting in PBS containing complete EDTA-free inhibitors (Roche Applied Science, Burgess Hill, UK). Cells were lysed at 5 × 107 cell/ml in 50 mm Hepes pH 7.8, 150 mm NaCl containing 1% (v/v) Triton X-100 and protease inhibitors. After 30 min on ice, insoluble material was pelleted by centrifugation at 2000 × g for 5 min at 4 °C. The protein content of the soluble fractions was measured with a BCA protein assay kit according to the manufacturer's instructions (Thermo Scientific, Rockford, IL). The preparation of biotinylated samples was performed as detailed previously (31).
Immunoprecipitation was carried out according to a modification of a method described previously (31). Briefly, 100-μl aliquots of VSMC lysate precleared with protein G-Sepharose were incubated with recombinant maspin (0, 1, or 5 μg maspin to 200 μg of cellular protein) for 12 h at 4 °C. 5 μg of anti-maspin or anti-integrin was added, and a further incubation performed for 12 h at 4 °C. 50 μl of 50% (v/v) protein G-Sepharose was added to each sample, and immune complexes allowed to bind for at least 1 h at 4 °C. The beads were washed four times with lysis buffer, and adsorbed material eluted in non-reducing Laemmli sample buffer.
For Western and ligand blotting, samples separated by SDS-PAGE on a 10% resolving gel were transferred to PVDF (polyvinylidene difluoride) membranes (Bio-Rad). Protein bands were detected by incubation with the appropriate primary antibody followed by horseradish peroxidase (HRP)-conjugated secondary antibodies (0.65 μg/ml). Biotinylated samples were visualized with HRP-conjugated streptavidin (0.1 μg/ml). For ligand blotting, the antibody incubations were preceded by an incubation with 1 μg/ml ligand: maspin or α5β1-Fc.
Immunofluorescence Microscopy
Subconfluent monolayers were fixed with 4% formaldehyde, followed by blocking with 1% BSA (bovine serum albumin) (w/v)/PBS. Protein and antibody incubations were performed in 1% BSA (w/v)/PBS for 30 min at 37 °C; between steps samples were washed with 0.1% Tween 20 (v/v)/PBS. The concentrations of primary antibodies used are given in the respective figure legends. Secondary antibody was Alexa Fluor 488 goat anti-mouse IgG at 2 μg/ml. Slides were mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Cells were visualized and images captured with a CCD upright microscope (Carl Zeiss Ltd, Hertfordshire, UK). The same exposure conditions were used for all slides and images were analyzed using Axiovision 4.5 software. To quantify β1 integrin in the experiments using conformation-specific antibodies, the mean fluorescence intensity of outlined cells was measured using Andor iQ software (Andor, Belfast, UK). The mean intensity was multiplied by the total number of pixels to obtain the integrated intensity and then divided by the area of the cell to calculate fluorescence intensity/μm2.
Affinity Chromatography
A maspin affinity column was made using a 1-ml HiTrapTM NHS HP column from GE Healthcare (Amersham Biosciences). 1 mg of recombinant maspin was coupled to the column according to the product guidelines. A lysate prepared from 6 × 107 HT1080 cells was applied to the column. After incubation for 15 min at 25 °C, the column was washed with PBS to remove unbound protein. Eluates were collected by sequential application of 3 ml of PBS containing 0.5 m NaCl, 3 ml PBS containing 1 m NaCl and 3 ml of wash buffer (0.1 m sodium acetate pH 4.0, 0.5 m NaCl). Fractions were collected (1 ml) and analyzed by SDS-PAGE and Western blotting.
Plasmids and Transfection
The full-length maspin cDNA was a gift from Dr. Margaret Worrall (Department of Biochemistry, University College Dublin, Ireland). The maspin coding region was subcloned into the mammalian expression vector pcDNA3.2-V5 using the Gateway system (Invitrogen) to generate a maspin plasmid referred to as pcDNA3.2-Maspin. Transfection of CHO cells was performed using FuGENE 6 reagent (Roche Applied Science) according to the manufacturer's instructions; optimized at 1 μg of DNA per 2 × 105 cells.
Cell Migration Assays
Cell migration was determined using time lapse video microscopy as detailed previously (7). Briefly, 24-well plates were coated with 5 μg/ml fibronectin, collagen I or laminin in PBS overnight at 4 °C. Wells blocked for 30 min at 37 °C with 1% BSA (w/v)/PBS, prior to seeding cells at a density of 7500 cells/ml/well. After 16 h, cells were put in serum-free conditions. After 24 h, 100 nm recombinant maspin or 5 μg/ml function-blocking antibody was added as required, and cells were placed in a humidified chamber (37 °C, 5% CO2) and migration recorded by computerized time-lapse video microscopy with a CCD inverted microscope (Carl Zeiss Ltd). Images were acquired every 10 min for 17 h. 10 randomly selected cells were tracked per movie, quantified using Axiovision software, and migration expressed in μm/h.
Taqman Real-time PCR
Total RNA was extracted from subconfluent monolayers using the High Pure RNA Isolation kit (Roche Applied Science) according to the manufacturer's instructions. For each sample, 1 μg of total RNA was reverse-transcribed using 10 μg/ml random hexamers and superscript II reverse transcriptase (Invitrogen, Paisley, UK) according to the manufacturer's instructions. Integrin expression was quantified by use of specific primers designed for use with universal library probes (Universal Probe Library Assay Design Centre, Roche Applied Science). To control against amplification of genomic DNA, primers spanned intron/exon boundaries. 18S rRNA was used as a reference gene to normalize differences in the amounts of total RNA. Taqman reactions were performed using the 7500 Fast Real-Time PCR System (Applied Biosystems, Warrington, UK), using the manufacturer's protocol. Each reaction was performed in 25 μl and contained the equivalent of 5 ng of reverse-transcribed RNA (1 ng RNA for 18S), 50% TaqMan® Master Mix (Applied Biosystems). Results were analyzed using the standard curve method.
Statistical Analysis
Data are presented as mean ± S.D. Significance was judged using the Student's t test.
RESULTS
Maspin Binds to the Surface of VSMC
We have previously shown that maspin can dynamically regulate the migration of VSMC, inhibiting migration at low concentrations and over a short time scale (7). We hypothesized that this was due to the binding of maspin to cell surface proteins involved in cell-matrix interactions, with integrins being potential candidates. In the present study we set out to identify maspin-binding cell surface proteins, using VSMC cultured to generate two phenotypic states. A differentiated or contractile, non-migratory phenotype, and a de-differentiated, synthetic phenotype that both migrates and proliferates and is thought to resemble cells responding to vascular injury.
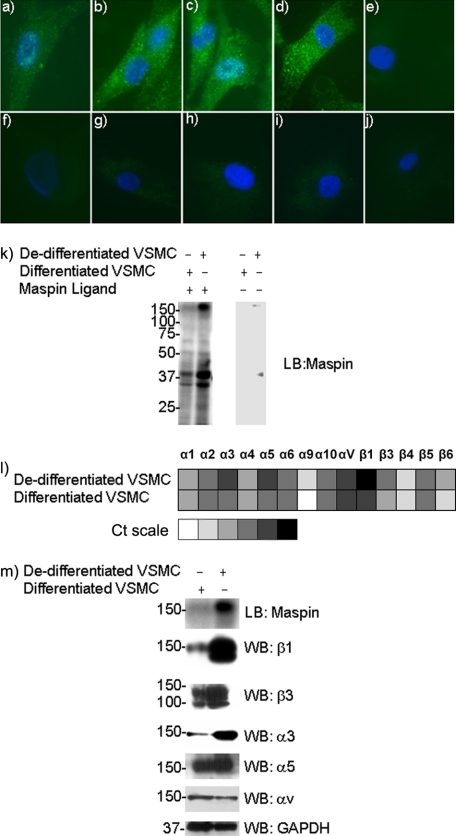
To investigate the binding of exogenous maspin to the surface of VSMC, cells were incubated with varying concentrations of recombinant maspin, which was subsequently detected by immunofluorescence microscopy using a maspin specific antibody. Maspin was found to bind de-differentiated VSMC in a concentration-dependent manner and displayed a punctate pattern of staining on the cell surface (Fig. 1, panels a–e). By contrast, differentiated VSMC displayed no binding of maspin (Fig. 1, panels f–j).
FIGURE 1.
Maspin binding to VSMC. Panels a–j, maspin binding to VSMC detected by immunofluorescence microscopy. Purified recombinant maspin (0 nm) (panels a and f), 40 nm (panels b and g), 80 nm (panels c and h), and 250 nm (panels d and i) was incubated with either de-differentiated (panels a–e) or differentiated (panels f–j) VSMC. Isotype-matched IgG (2 μg/ml) controls are shown for cells incubated with 250 nm maspin (panels e and j). Anti-maspin (2 μg/ml) was used for detection. Secondary antibodies were Alexa Fluor 488-labeled (green), and nuclei were counterstained with DAPI (blue). k, ligand blot analysis of maspin binding to VSMC proteins. Whole cell lysates of both differentiated and de-differentiated VSMC were blotted onto PVDF membranes, incubated with recombinant maspin and probed with an anti-maspin antibody. Control blots in the absence of maspin incubation are also shown. l, quantitative real-time PCR (TaqMan) analysis of integrin gene expression in VSMC. Relative expression levels are shown as a “heatmap” based on Ct values, ranging from undetectable expression (40 cycles), through very low (35–39 cycles), low (30–34 cycles), moderate (25–29 cycles), high (20–24 cycles) to very high expression (<20 cycles). m, Western blot analysis of integrin expression in VSMC. Whole cell lysates of both differentiated and de-differentiated VSMC were blotted onto PVDF membranes and probed with anti-integrin antibodies. Ligand blot for maspin binding is shown for comparison, and GAPDH is used as a loading control (10 μg of cellular protein per lane).
To identify candidate maspin binding proteins we used ligand or far Western blotting. Whole cell lysates were separated by SDS-PAGE, and blots incubated with maspin, prior to detection with a maspin specific antibody. VSMC of both phenotypes showed a similar pattern of maspin-binding proteins, although the intensity of the bands detected varied (Fig. 1k). Three major protein bands were observed at 35, 40, and 150 kDa. The intensity of the two lower molecular weight bands was only slightly higher in the de-differentiated cells. However, the 150-kDa band showed a large increase under these conditions and has a mobility consistent with an integrin subunit.
As a first step toward determining the role of integrins in binding maspin, we characterized integrin expression in VSMC by Taqman real time PCR (Fig. 1l), and those most highly expressed confirmed at the protein level by Western blot (Fig. 1m). Comparing the levels of expression in the two VSMC phenotypes it could be seen that α3, α5, and β1 subunits were up-regulated in de-differentiated VSMC, with little change in αv or β3 subunits, raising the possibility that these integrins are involved in the binding of maspin to the surface of VSMC.
α3β1 and α5β1 Bind to a Maspin Affinity Chromatography Column
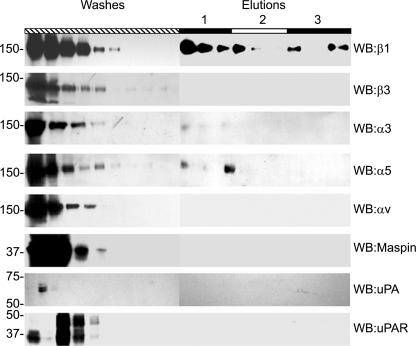
To directly investigate the binding of maspin to cellular proteins, we employed affinity chromatography using maspin as a ligand. Primary VSMC presented limitations regarding the large number of cells needed for these experiments; therefore HT-1080 cells were used as an alternative source of material. We have previously shown that maspin inhibits the migration of these cells (7), and they have a similar integrin profile to VSMC (data not shown). HT-1080 cell lysates were applied to the maspin affinity column and specific binding and elution of integrin subunits determined by Western blot. We found that the integrin subunits α3, α5, and β1 bound specifically to the maspin column, whereas αv and β3 were only detected in the flow through (Fig. 2). This provides direct evidence that that the α3β1 and α5β1 integrin heterodimers may be involved in the binding of maspin to the surface of VSMC. Neither uPA and uPAR, which have been implicated as maspin-binding proteins, nor endogenous maspin bound to the affinity column.
FIGURE 2.
Maspin affinity chromatography. HT1080 cell lysate was applied to a maspin affinity column, after incubation the column was washed extensively with PBS to remove unbound protein (Washes). Specifically bound proteins were then eluted by sequential application of 3 ml of PBS containing 0.5 m NaCl, 1.0 m NaCl, and acetate buffer pH 4.0, and collected as 1-ml fractions (Elutions, labeled 1–3, respectively). 30 μl of each fraction was separated by SDS-PAGE prior to Western blotting with primary antibodies to β1 (10 μg/ml), β3 (10 μg/ml), α3 (1:1000 antiserum), α5 (1:1000 antiserum), αv (10 μg/ml), maspin (1 μg/ml), uPA (10 μg/ml), or uPAR (10 μg/ml).
VSMC α3β1 and α5β1 Coimmunoprecipitate with Maspin
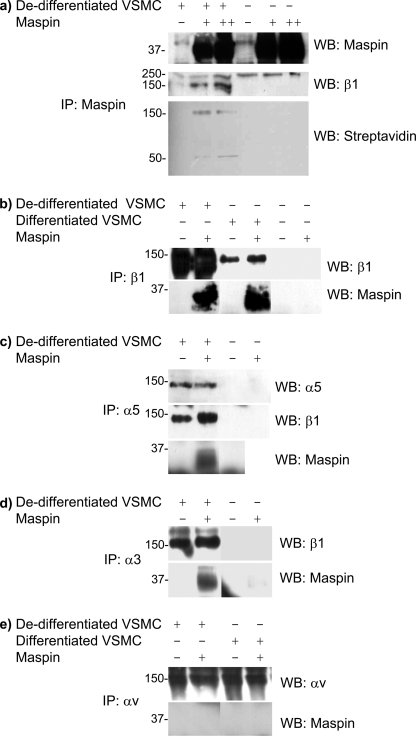
To give further support for the involvement of the α3β1 and α5β1 integrin heterodimers in the binding of maspin to the surface of VSMC, we determined whether these proteins could be coimmunoprecipitated. Proteins on the surface of de-differentiated VSMC were labeled with biotin, and cell lysates incubated with recombinant maspin, before immunoprecipitation with a maspin antibody. Maspin was efficiently immunoprecipitated from both cell lysates and controls under these conditions (Fig. 3a). First, streptavidin was used to visualize biotinylated proteins associated with maspin. Two specific bands at 50 and 150 kDa were detected, the latter coinciding with the maspin-binding protein detected by ligand blot (Fig. 1k) and precipitated at a low concentration of maspin. Western blotting for β1 integrin confirmed the presence of this integrin subunit in the coimmunoprecipitate with maspin and that it had a mobility indistinguishable from the biotinylated protein (Fig. 3a).
FIGURE 3.
Coimmunoprecipitation of maspin and VSMC integrins. Lysates of differentiated and de-differentiated VSMC were incubated for 14 h at 4 °C with recombinant maspin, and subsequently immunoprecipitated using antibodies to either maspin or various integrin subunits. a, cell lysates were incubated with 0, 1, or 5 μg maspin per 200 μg of cellular protein, samples immunoprecipitated with anti-maspin goat mAb and Western blots performed with anti-maspin mouse mAb, anti-β1 mouse mAb. Cell surface proteins were also biotinylated and detected by streptavidin after immunoprecipitation. b–e, lysates were incubated with 1 μg of maspin (chosen as optimal amount), samples immunoprecipitated with anti-integrin antibodies to β1 (b), α5 (c), α3 (d), or αv (e), and Western blots performed with antibodies to either maspin or various integrins, as indicated.
To investigate the involvement of other integrin subunits, similar experiments were performed on lysates of cells that had not been modified by biotin and by immunoprecipitation with integrin rather than maspin antibodies. Consistent with the observations with biotinylated cells, the β1 integrin antibody was able to coimmunoprecipitate maspin, and this was observed in VSMC of both phenotypes (Fig. 3b). Using an antibody to α5, both maspin and the β1 subunit could be coimmunoprecipitated (Fig. 3c). Similarly, an antibody to α3 immunoprecipitated both maspin and the β1 subunit (Fig. 3d). However, antibodies to either αv (Fig. 3e) or β3 (data not shown) failed to coimmunoprecipitate maspin with the relevant integrin. In differentiated VSMC maspin was not immunoprecipitated by either α3 or α5 antibodies (data not shown), most likely due to the lower levels of these integrin subunits under these conditions. These data are therefore consistent with the observations made with the maspin affinity column, and suggest that maspin interacts specifically with α3β1 and α5β1 on the surface of de-differentiated VSMC.
Maspin Interacts with the Extracellular Domain of α5β1
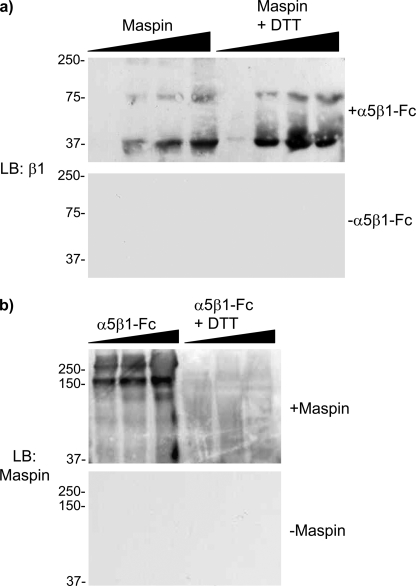
To demonstrate a direct interaction between maspin and α5β1 we have made use of an α5β1-Fc fusion protein, containing the entire extracellular part of the integrin heterodimer, using a ligand blot approach. Maspin was run on SDS-PAGE, blotted, incubated with α5β1-Fc, and probed with an antibody to the β1 integrin subunit. Concentration-dependent binding of α5β1-Fc to immobilized maspin was detected as a band at 37 kDa, with a minor band at 75 kDa representing a dimerized form that also bound the integrin (Fig. 4b). Reduction with dithiothreitol did not affect the interaction with the integrin as, despite the presence of 8 cysteine residues, maspin has no disulfide bonds.
FIGURE 4.
Interaction of purified maspin and α5β1-Fc fusion proteins. a, recombinant maspin (10, 50, 100, and 250 pmol), in the presence or absence of reduction with DTT, was run on SDS-PAGE, and blots subsequently incubated with α5β1-Fc fusion protein (1 μg/ml) and probed with an anti-β1 antibody. b, α5β1-Fc (5, 10, and 30 pmol), in the presence or absence of reduction with DTT, was run on SDS-PAGE, and blots subsequently incubated with maspin (1 μg/ml) and probed with an anti-maspin antibody. Each panel also shows control blots, in which incubation with either α5β1-Fc or maspin, respectively, has been omitted.
When α5β1-Fc was blotted and incubated with maspin an interaction was again detected, but only under non-reducing conditions indicating that maintenance of the integrin ectodomain structure was essential for binding and making a nonspecific interaction between maspin and α5β1-Fc unlikely. A major band was observed at 150 kDa representing the β1 integrin subunit, and a minor band at >250 kDa representing the heterodimer. These data demonstrate that maspin interacts specifically with the extracellular domain of α5β1 and that this interaction likely involves the β chain.
Inhibition of CHO Migration by Maspin Requires the α5 Integrin Subunit
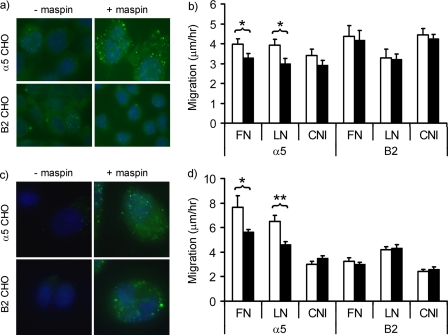
To obtain direct evidence for the role of α5β1 in the binding of maspin and its effects on cell migration, we have made use of CHO cells either stably transfected with the human α5 subunit (32) or the B2 clone which is essentially α5-null (33). Maspin was found to bind specifically to α5CHO cells with a punctate distribution (Fig. 5a), as previously observed on VSMC. However, no binding of maspin to B2CHO cells was detected (Fig. 5c), consistent with the presence of α5β1 being sufficient for maspin binding to the cell surface.
FIGURE 5.
α5β1-dependent inhibition of CHO cell migration. a, binding of exogenous maspin to α5CHO and B2CHO cells detected by immunofluorescence microscopy. Cells were incubated in the absence or presence of maspin (40 nm) and binding detected and visualized as detailed in Fig. 1. Maspin binding to the cell surface is detected as green and nuclei are counterstained blue. b, effect of exogenous maspin on the migration of α5CHO and B2CHO cells on fibronectin (FN), laminin (LN), or collagen I (CNI) was determined by time-lapse video microscopy over a period of 17 h. Experiments were performed both in the presence (closed bars) and absence (open bars) of recombinant maspin (100 nm). c, binding of endogenously expressed maspin to α5CHO and B2CHO cells transfected with either pcDNA3.2 (−mapsin) or pcDNA3.2-Maspin (+maspin). d, migration of α5CHO and B2CHO cells transfected with pcDNA3.2 (open bars) or pcDNA3.2-Maspin (closed bars). Data shown represent the means and S.E. of three independent experiments. *, p < 0.05; **, p < 0.005.
To determine the effect of maspin binding to α5β1 on cell migration using this model, migration of α5CHO and B2CHO cells was assessed in the presence and absence of exogenously added maspin (Fig. 5b). The addition of 100 nm maspin, previously determined as optimal (7), significantly reduced the migration of α5CHO cells on both fibronectin and laminin, but not on collagen. By contrast, maspin had no effect on the migration of B2CHO cells on any of the three matrices. Similar effects were also observed with endogenously expressed maspin, as transfection of α5CHO and B2CHO with wild-type maspin aused a significant reduction in the migration of α5CHO on fibronectin and laminin, while having no effect on the migration of B2CHO (Fig. 5d). Unexpectedly, both cell types displayed a punctate, cell surface staining pattern when maspin was endogenously expressed (Fig. 5c). As this was not observed in the presence of exogenously added maspin, it is possible that secreted maspin is primarily associated with the matrix underlying the cells, as has been previously observed (34), and therefore apparently unaffected by the presence or absence of α5β1. Overall, these data demonstrate that maspin binds specifically to α5β1 and that this is sufficient for maspin to exert its inhibitory effect on cell migration.
Maspin Binding to α5β1 on VSMC Alters Integrin Activation Status
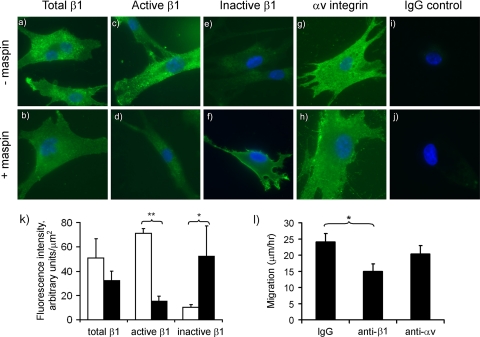
To begin to understand the mechanisms involved in the effect of maspin on α5β1 function, we investigated whether maspin influenced the activation state of β1 integrins on VSMC using two conformation-dependent antibodies in immunofluorescence microscopy. These antibodies, 12G10 (35) and mAb13 (36), preferentially recognize “active” and “inactive” conformations of β1, respectively (37, 38). Total β1 integrin on the surface of VSMC, determined using a non-conformation-dependent antibody, was unchanged in the presence of maspin both in terms of its cell surface distribution (Fig. 6, panels a and b), and its abundance, as determined by semi-quantitative image analysis (Fig. 6k). However, using the conformation-dependent antibodies, clear differences in staining intensities were observed in the absence and presence of maspin. There was a ∼5-fold reduction in the detection of the active conformation of β1 (Fig. 6, panels c and d), and this was accompanied by a concomitant 5-fold increase in the detection of the inactive conformation (Fig. 6, panels e and f). Similar results were also obtained with a 5-fold higher concentration of maspin and when the incubation time was increased to 24 h. Therefore, binding of maspin appears to result in a redistribution of the balance between active and inactive conformations of β1 integrin, possibly as a consequence of the direct binding of maspin, leading to an overall reduction in β1 activity. Consistent with this as the functional mechanism, antibody-mediated blocking of β1 function led to a 40% inhibition of VMSC migration (Fig. 6l), in contrast to which blocking of αvβ3 function had no significant effect.
FIGURE 6.
Maspin alters β1 integrin conformation on VSMC. The cell surface expression of integrins on VSMC was determined in the absence (panels a, c, e, g, and i). and presence (panels b, d, f, h, and j) of incubation with maspin (100 nm) for 1 h at 37 °C. Antibodies were to total β1 (1:50 dilution), active β1 (12G10, 5 μg/ml), inactive β1 (mAb13, 5 μg/ml), αv integrin (L230, 10 μg/ml), and nonspecific IgG control (5 μg/ml). Integrins detected by Alexa Fluor 488 fluorescence (green) and nuclei are shown counterstained with DAPI (blue). k, images taken from randomly chosen cell fields were analyzed for Alexa Fluor 488 staining intensity in four independent experiments. Data are shown for total β1, active β1 (12G10) and inactive β1 (mAb13) in the absence (open bars) and presence of maspin (closed bars). The intensity of αv staining, used here as control non-maspin-binding integrin, was unchanged. Data shown are mean ± S.D. (n = 4). *, p < 0.05; **, p < 0.0005. l, effect of function blocking antibodies on VSMC migration. Cells were incubated with either anti-β1 mAb13, anti-αv MAB1980, or control IgG (all at 5 μg/ml) and migration assessed by time-lapse video microscopy (7). Data shown are mean ± S.D. (n = 3). *, p < 0.01.
DISCUSSION
Maspin has been shown to influence many aspects of cell behavior, including migration, invasion, proliferation, and apoptosis. Although these effects have been proposed to involve both intracellular and extracellular activities of maspin, the mechanisms responsible for these activities remain unclear. We have previously demonstrated that the inhibitory effect of maspin on cell migration is an extracellular activity of maspin, and furthermore that it does not involve protease inhibition by maspin (7). These observations suggested that maspin may exert its effect by acting as a ligand for cell surface receptors, rather than acting as a classical serpin. Consistent with this notion, we demonstrate here that maspin binds to cell surface integrins, in particular α5β1, that this binding causes inactivating conformational changes in the integrin and leads to the inhibitory effect of maspin on cell migration.
Our previous study focused on VSMC (7), the migration of these cells being a major contributory factor in the development of atherosclerosis and other fibroproliferative vascular diseases. Here we find that these cells bind exogenously added maspin, with a punctate pattern observed on the cell surface. This binding was only observed on VSMC displaying the de-differentiated or synthetic phenotype. In contrast to the differentiated or contractile phenotype, these cells are motile and resemble the cells present in fibroproliferative vascular lesions. Using ligand blotting, maspin was found to bind to a limited number of proteins in lysates of both cell types. One of these proteins, with a molecular weight of 150 kDa, and subsequently demonstrated to be the β1 integrin subunit, was detected almost exclusively in the de-differentiated cells. Consistent with this observation, expression of β1 integrin was shown to be up-regulated on de-differentiation of VSMC. Further evidence for the specific binding of maspin to β1 integrin comes from the retention of this protein on a maspin affinity column, coimmunoprecipitation of maspin and β1 integrin, and the binding of maspin to purified recombinant α5β1. Western blot analysis of cellular proteins binding to maspin by affinity chromatography also indentified α3 and α5 integrin subunits, both of which are partners for the β1 subunit in integrin heterodimers. Both α3β1 and α5β1 are major integrins in VSMC. However, αvβ3, another major integrin in VSMC, did not bind to the maspin affinity column as neither αv or β3 subunits were detected by Western blot. Therefore, maspin appears to be capable of binding to multiple β1 integrins.
The role of maspin binding to α5β1 in the inhibition of cell migration was demonstrated in a model system consisting of two CHO cell lines; B2CHO which are essentially devoid of α5 and α5CHO stably transfected with the human α5 subunit. Maspin had no effect on the migration of B2CHO cells on a variety of extracellular matrix substrates, whereas the migration of α5CHO cells on both fibronectin and laminin was inhibited by maspin. This inhibition of migration was to a similar extent to that previously observed on VSMC (7), and was observed both when cells were treated with exogenous maspin and when cells were transfected with maspin. Attempts to directly demonstrate a role for α5β1 in the inhibition of VSMC migration by maspin were unsuccessful, as we were unable to efficiently silence α5 expression in these cells.3
Integrins have complex roles in regulating cell motility and migration. This involves direct engagement of extracellular matrix ligands to mediate cell adhesion, which can be modulated by “inside-out” signals to alter the affinity for these ligands, and also to transduce “outside-in” signals to the cytoplasm. The effect of maspin on VSMC migration is rapid (7), which is consistent with maspin having a direct mechanism of action, rather than a mechanism involving downstream signal transduction. Consistent with this we found that maspin led to a change in the activation status of β1 integrins in VSMC. This activation status is dependent on the conformation of the integrin heterodimer (39, 40), which can be detected by conformation-specific monoclonal antibodies (35, 36). The binding of maspin to α5β1 caused an increase in the proportion of β1 integrin in the inactive, low ligand-binding affinity conformation, with a concomitant decrease in the active, high ligand binding affinity conformation. The reduction in binding of the 12G10 antibody could possibly be due to simple steric hindrance of antibody binding by maspin, rather than any change in integrin conformation, but a conformational inactivation of the integrin is strongly supported by the observed increase in mAb13 binding. Consistent with this, antibody-mediated blocking of β1 integrin function also inhibited VSMC migration. Therefore maspin binding to α5β1, and potentially other β1 integrins, is likely to affect cell migration directly, making this effect distinct from the previously reported increased expression of integrins, including α5β1, in breast cancer cells on prolonged treatment with maspin (26).
The observed inhibitory effect of maspin on the migration of α5CHO cells on laminin is perhaps surprising, as laminin is not a ligand for α5β1 and no effect was observed with B2CHO cells lacking α5β1. This makes it possible that maspin binding to α5β1 can also indirectly influence the behavior of other integrins. This may also be related to the observation that B2CHO appeared to bind maspin when it was expressed by the cells but not when it was added exogenously, despite these cells lacking α5 expression.
Despite the inhibitory effect of maspin on the migration of both VSMC and α5CHO cells, we have found it to have no significant effect on the adhesion of these cells to a variety of extracellular matrix substrates.3 This is in contrast to the maspin-induced increase in cell adhesion observed in other cell types (26, 41–43). Whether these differences are cell-type related or methodological is unresolved but, in support of our conclusions here, one of these studies implicated β1 integrins in the pro-adhesive effect as maspin and β1 integrin colocalised on the cell surface and could be coimmunoprecipitated (43). Maspin has previously been reported to bind to collagens I and III (44). However we found that maspin had no effect on cell migration on collagen, consistent with its specific interactions with non-collagen-binding α5β1 and α3β1 integrins.
Maspin is considered to be primarily an intracellular serpin that can also enter secretory vesicles (21), and there is considerable controversy regarding its site of action. The inhibitory effect of maspin on α5CHO cell migration was observed both when purified maspin was added to the cells and when the cells were transfected with maspin. As maspin was also observed to bind to the cell surface, these effects must be due to an extracellular activity of maspin. The interaction of maspin with the purified recombinant α5β1-Fc is important in this respect, as it demonstrates that maspin binds to the extracellular region of the integrin rather than to the cytoplasmic tail, which is not present in this fusion protein. Therefore, the integrin-mediated effect of maspin is not dependent on an intracellular mechanism involving an interaction between non-secreted maspin and the intracellular tail of the integrin. The extracellular regions of integrins are known to interact with a variety of proteins, in addition to their principal extracellular matrix ligands. These interactions can affect integrin function (45), and we have demonstrated here that maspin can similarly act as an integrin-associated protein.
Although our interest in maspin specifically relates to its inhibitory effect on VSMC migration, the integrin-mediated mechanism is not restricted to these cells as it is also observed on CHO cells. Therefore, the binding of secreted, extracellular maspin to β1 integrins may be a more general mechanism for its biological effects, which include the inhibition of cancer cell invasion and metastasis. In addition, the maspin knock-out is embryonically lethal in mice, which has been attributed to an impaired ability of endodermal cells to interact with their surrounding extracellular matrix (42). The β1 integrin knock-out displays very similar characteristics (46), and thus the defect in maspin-null mice has been speculated to be due to maspin interactions with β1 integrin (42).
How maspin interacts with β1 integrins has yet to be determined. It has been reported that the reactive center loop, central to the function of protease inhibitory serpins, is involved in the extracellular effects of maspin on cell adhesion and migration (4, 41, 47), as well as to its intracellular effects on apoptosis (25, 48). However, we find that mutation of the reactive center loop of maspin has no effect on its ability to inhibit cell migration.4 This suggests that maspin contains multiple functional epitopes, a notion also supported by other studies (43, 44). Another non-inhibitory serpin, pigment epithelium-derived factor (SERPINF1), which displays neuroprotective, anti-angiogenic and tumor suppressive activities has similarly been demonstrated to affect these processes using different functional epitopes, and by interacting with different putative receptors (49–51). Identification of the structural epitope of maspin that interacts with β1 integrins will lead to a further understanding the activity of this multifunctional, but non-inhibitory, serpin and inform further studies on potential maspin-based therapeutics.
Acknowledgments
We thank Dr. Fedor Berditchevski (University of Birmingham), Prof. Barry Coller (Rockefeller University), Prof. Martin Humphries (University of Manchester), Dr. Gunilla Høyer-Hansen (Finsen Laboratory, Copenhagen, Denmark), Prof. Rudy Juliano (University of North Carolina), Prof. Yoshikazu Takada (UC Davis School of Medicine), Dr. Margaret Worrall (University College Dublin, Ireland) for their kind gifts of various cell lines and reagents.
This work was supported by grants from the British Heart Foundation (FS/05/063), Norfolk and Waveney Big C Appeal, Biotechnology and Biological Sciences Research Council, and by the EU Framework Programme 6 Cancerdegradome Project LSHC-CT-2003-503297.
R. Bass and V. Ellis, unpublished experiments.
L. Wagstaff, V. Ellis, and R. Bass, manuscript in preparation.
- SERPIN
- serine protease inhibitors
- BSA
- bovine serum albumin
- PBS
- phosphate-buffered saline
- PVDF
- polyvinylidene difluoride
- DTT
- dithiothreitol
- VSMC
- vascular smooth muscle cells.
REFERENCES
- 1.Zou Z., Anisowicz A., Hendrix M. J., Thor A., Neveu M., Sheng S., Rafidi K., Seftor E., Sager R. (1994) Science 263, 526–529 [DOI] [PubMed] [Google Scholar]
- 2.Shi H. Y., Zhang W., Liang R., Abraham S., Kittrell F. S., Medina D., Zhang M. (2001) Cancer Res. 61, 6945–6951 [PubMed] [Google Scholar]
- 3.Biliran H., Jr., Sheng S. (2001) Cancer Res. 61, 8676–8682 [PubMed] [Google Scholar]
- 4.Sheng S., Carey J., Seftor E. A., Dias L., Hendrix M. J., Sager R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11669–11674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z., Shi H. Y., Zhang M. (2005) Oncogene 24, 2008–2019 [DOI] [PubMed] [Google Scholar]
- 6.Zhang M., Volpert O., Shi Y. H., Bouck N. (2000) Nat. Med. 6, 196–199 [DOI] [PubMed] [Google Scholar]
- 7.Bass R., Fernández A. M., Ellis V. (2002) J. Biol. Chem. 277, 46845–46848 [DOI] [PubMed] [Google Scholar]
- 8.Ross R. (1993) Nature 362, 801–809 [DOI] [PubMed] [Google Scholar]
- 9.Schwartz R. S., Holmes D. R., Jr., Topol E. J. (1992) J. Am. Coll. Cardiol. 20, 1284–1293 [DOI] [PubMed] [Google Scholar]
- 10.Häyry P., Isoniemi H., Yilmaz S., Mennander A., Lemström K., Räisänen-Sokolowski A., Koskinen P., Ustinov J., Lautenschlager I., Taskinen E., Krogerus L., Aho P., Paavonen T. (1993) Immunol. Rev. 134, 33–81 [DOI] [PubMed] [Google Scholar]
- 11.Owens G. K. (1995) Physiol. Rev. 75, 487–517 [DOI] [PubMed] [Google Scholar]
- 12.Galis Z. S., Khatri J. J. (2002) Circ. Res. 90, 251–262 [PubMed] [Google Scholar]
- 13.Dollery C. M., Libby P. (2006) Cardiovasc. Res 69, 625–635 [DOI] [PubMed] [Google Scholar]
- 14.Newby A. C. (2006) Cardiovasc. Res. 69, 614–624 [DOI] [PubMed] [Google Scholar]
- 15.McGowen R., Biliran H., Jr., Sager R., Sheng S. (2000) Cancer Res. 60, 4771–4778 [PubMed] [Google Scholar]
- 16.Sheng S., Truong B., Fredrickson D., Wu R., Pardee A. B., Sager R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huntington J. A., Read R. J., Carrell R. W. (2000) Nature 407, 923–926 [DOI] [PubMed] [Google Scholar]
- 18.Pemberton P. A., Wong D. T., Gibson H. L., Kiefer M. C., Fitzpatrick P. A., Sager R., Barr P. J. (1995) J. Biol. Chem. 270, 15832–15837 [DOI] [PubMed] [Google Scholar]
- 19.Al-Ayyoubi M., Gettins P. G., Volz K. (2004) J. Biol. Chem. 279, 55540–55544 [DOI] [PubMed] [Google Scholar]
- 20.Law R. H., Irving J. A., Buckle A. M., Ruzyla K., Buzza M., Bashtannyk-Puhalovich T. A., Beddoe T. C., Nguyen K., Worrall D. M., Bottomley S. P., Bird P. I., Rossjohn J., Whisstock J. C. (2005) J. Biol. Chem. 280, 22356–22364 [DOI] [PubMed] [Google Scholar]
- 21.Pemberton P. A., Tipton A. R., Pavloff N., Smith J., Erickson J. R., Mouchabeck Z. M., Kiefer M. C. (1997) J. Histochem. Cytochem. 45, 1697–1706 [DOI] [PubMed] [Google Scholar]
- 22.Smith S. L., Watson S. G., Ratschiller D., Gugger M., Betticher D. C., Heighway J. (2003) Oncogene 22, 8677–8687 [DOI] [PubMed] [Google Scholar]
- 23.Bailey C. M., Khalkhali-Ellis Z., Kondo S., Margaryan N. V., Seftor R. E., Wheaton W. W., Amir S., Pins M. R., Schutte B. C., Hendrix M. J. (2005) J. Biol. Chem. 280, 34210–34217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin S., Li X., Meng Y., Finley R. L., Jr., Sakr W., Yang H., Reddy N., Sheng S. (2005) J. Biol. Chem. 280, 34985–34996 [DOI] [PubMed] [Google Scholar]
- 25.Li X., Yin S., Meng Y., Sakr W., Sheng S. (2006) Cancer Res. 66, 9323–9329 [DOI] [PubMed] [Google Scholar]
- 26.Seftor R. E., Seftor E. A., Sheng S., Pemberton P. A., Sager R., Hendrix M. J. (1998) Cancer Res. 58, 5681–5685 [PubMed] [Google Scholar]
- 27.Odero-Marah V. A., Khalkhali-Ellis Z., Chunthapong J., Amir S., Seftor R. E., Seftor E. A., Hendrix M. J. (2003) Cancer Biol. Ther. 2, 398–403 [DOI] [PubMed] [Google Scholar]
- 28.Shi H. Y., Stafford L. J., Liu Z., Liu M., Zhang M. (2007) Cell Motil. Cytoskeleton 64, 338–346 [DOI] [PubMed] [Google Scholar]
- 29.Bass R., Ellis V. (2009) Thromb. Haemost. 101, 954–962 [PubMed] [Google Scholar]
- 30.Coe A. P., Askari J. A., Kline A. D., Robinson M. K., Kirby H., Stephens P. E., Humphries M. J. (2001) J. Biol. Chem. 276, 35854–35866 [DOI] [PubMed] [Google Scholar]
- 31.Bass R., Werner F., Odintsova E., Sugiura T., Berditchevski F., Ellis V. (2005) J. Biol. Chem. 280, 14811–14818 [DOI] [PubMed] [Google Scholar]
- 32.Sechler J. L., Corbett S. A., Schwarzbauer J. E. (1997) Mol. Biol. Cell 8, 2563–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schreiner C. L., Bauer J. S., Danilov Y. N., Hussein S., Sczekan M. M., Juliano R. L. (1989) J. Cell Biol. 109, 3157–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khalkhali-Ellis Z., Hendrix M. J. (2007) Cancer Res. 67, 3535–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mould A. P., Garratt A. N., Askari J. A., Akiyama S. K., Humphries M. J. (1995) FEBS Lett. 363, 118–122 [DOI] [PubMed] [Google Scholar]
- 36.Akiyama S. K., Yamada S. S., Chen W. T., Yamada K. M. (1989) J. Cell Biol. 109, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Humphries J. D., Schofield N. R., Mostafavi-Pour Z., Green L. J., Garratt A. N., Mould A. P., Humphries M. J. (2005) J. Biol. Chem. 280, 10234–10243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mould A. P., Akiyama S. K., Humphries M. J. (1996) J. Biol. Chem. 271, 20365–20374 [DOI] [PubMed] [Google Scholar]
- 39.Luo B. H., Springer T. A. (2006) Curr. Opin. Cell Biol. 18, 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Askari J. A., Buckley P. A., Mould A. P., Humphries M. J. (2009) J. Cell Sci. 122, 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ngamkitidechakul C., Warejcka D. J., Burke J. M., O'Brien W. J., Twining S. S. (2003) J. Biol. Chem. 278, 31796–31806 [DOI] [PubMed] [Google Scholar]
- 42.Gao F., Shi H. Y., Daughty C., Cella N., Zhang M. (2004) Development 131, 1479–1489 [DOI] [PubMed] [Google Scholar]
- 43.Cella N., Contreras A., Latha K., Rosen J. M., Zhang M. (2006) FASEB J. 20, 1510–1512 [DOI] [PubMed] [Google Scholar]
- 44.Blacque O. E., Worrall D. M. (2002) J. Biol. Chem. 277, 10783–10788 [DOI] [PubMed] [Google Scholar]
- 45.Brown E. J. (2002) Curr. Opin. Cell Biol. 14, 603–607 [DOI] [PubMed] [Google Scholar]
- 46.Stephens L. E., Sutherland A. E., Klimanskaya I. V., Andrieux A., Meneses J., Pedersen R. A., Damsky C. H. (1995) Genes Dev. 9, 1883–1895 [DOI] [PubMed] [Google Scholar]
- 47.Sheng S., Pemberton P. A., Sager R. (1994) J. Biol. Chem. 269, 30988–30993 [PubMed] [Google Scholar]
- 48.Jiang N., Meng Y., Zhang S., Mensah-Osman E., Sheng S. (2002) Oncogene 21, 4089–4098 [DOI] [PubMed] [Google Scholar]
- 49.Becerra S. P., Sagasti A., Spinella P., Notario V. (1995) J. Biol. Chem. 270, 25992–25999 [DOI] [PubMed] [Google Scholar]
- 50.Filleur S., Volz K., Nelius T., Mirochnik Y., Huang H., Zaichuk T. A., Aymerich M. S., Becerra S. P., Yap R., Veliceasa D., Shroff E. H., Volpert O. V. (2005) Cancer Res. 65, 5144–5152 [DOI] [PubMed] [Google Scholar]
- 51.Filleur S., Nelius T., de Riese W., Kennedy R. C. (2009) J. Cell. Biochem. 106, 769–775 [DOI] [PubMed] [Google Scholar]