Abstract
Fission 1 (Fis1) is an evolutionarily conserved, type II integral membrane protein implicated in maintaining the proper morphology of mitochondria and peroxisomes. A concave surface on the cytosolic domain of Fis1 from Saccharomyces cerevisiae is implicated in binding other fission proteins, yet structural studies reveal that this surface is sterically occluded by its N-terminal arm. Here we address the question of whether the N-terminal arm of yeast Fis1 exists in a dynamic equilibrium that would allow access to this functionally important surface. NMR measurements sensitive to dynamics occurring on a wide range of time scales (picoseconds to minutes) were used to assess whether the Fis1 arm is dynamic. Hydrogen–deuterium exchange experiments revealed that the Fis1 arm, α-helix 6, and proximal loops were not protected from solvent exchange, consistent with motions on the second to minute time scale. An engineered cysteine, I85C, located on the concave surface that lies underneath the Fis1 arm, was readily modified by a fluorescent probe, revealing more solvent accessibility of this position than would be predicted from the structure. Chemical denaturation, NMR chemical shift perturbation, and residual dipolar coupling experiments support the idea that the dynamic equilibrium can be shifted on the basis of changing pH and temperature, with the changes primarily localizing to the Fis1 arm and proximal regions. The data as a whole are consistent with the Fis1 arm adopting a primarily “closed” conformational state able to undergo dynamic excursions that reveal the concave surface and therefore may be important for binding other fission factors and for Fis1 function.
Fission 1 (Fis1) is a type II integral membrane protein important in maintaining mitochondrial morphology, presumably by mediating mitochondrial fission of the outer membrane. This process is important in the regulation of cell fate and homeostasis in humans, worms, and budding yeast (1–10). Fis1 is also involved in the regulation of peroxisome morphology in human and yeast cells (11, 12). This molecule is conserved throughout species that contain these organelles, including plants, fungi, and animals (13, 14). How Fis1 acts in these processes is beginning to be understood, especially for its role in mitochondrial morphology in budding yeast. Fis1 appears to be uniformly localized on the surface of the mitochondria by a C-terminal transmembrane anchor (15). On the basis of genetic and cytological data, Fis1 is proposed to recruit mitochondrial division protein l (Mdv1), CCR4-associated factor 4 (Caf4), dynamin-related protein l (Dnm1), and possibly other proteins to mitochondria to assemble a protein machinery that causes scission of the outer membrane in a GTP-dependent manner (15–22). In this process, Mdv1 is suggested to act as an adapter protein important in the activation of the protein machinery, with Caf4 acting in a somewhat redundant fashion that appears to be essential for establishing polarity (21, 22). Dnm1 is a dynamin-related large GTPase that is essential for scission (23–25). Both Fis1 and Dnm1 have orthologs in many species ranging from plants to mammals; however, Mdv1 and Caf4 orthologs have been identified only in fungi (13, 26).
High-resolution three-dimensional structures of Fis1 from human, mouse, and yeast are consistent with its proposed role in recruiting other fission factors (27–30). Fis1 is composed of an antiparallel array of six α-helices that adopt a tetratricopeptide repeat-like fold proposed to mediate protein–protein interactions (31, 32). Although no universal mechanism for how TPR1 motifs recognize other proteins has emerged (33–35), the Fis1 structures reveal a concave surface created by α-helices 2,4, and 6 (Figure 1). This concave surface is lined with evolutionarily conserved residues that are not involved in conferring the TPR fold, which suggest an important role for this surface in mediating interactions with other proteins. Indeed, a temperature sensitive allele of Fis1 that affects mitochondrial morphology consists of three conservative point mutants that lie on α-helix 4, the central helix of this concave surface (26). Structures of Fis1 reveal that access to this important surface is blocked by an N-terminal region, or arm, which is tucked into this concave surface (29).
Figure 1.
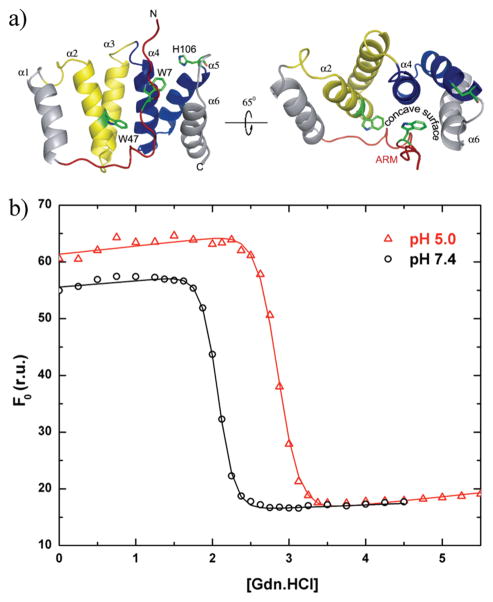
Thermodynamic stability of Fis1ΔTM, which is lower at pH 7.4 than at pH 5.0 as measured by equilibrium chemical denaturation experiments. (a) Ribbon model depicting the NMR-derived structure of the cytosolic domain of Fis1 atpH 5.5 and 32°C, which displays a 16-residue N-terminal region (the Fis1 arm) draped over a concave surface created by α-helices 2, 4, and 6 [PDB entry 1y8m (29)]. The structure is color-coded in this and other figures to highlight the Fis1 arm (red) and the first (yellow) and second (blue) tetratricopeptide repeats. (b) Chemical denaturation experiments in which the unfolding reaction was followed as a function of increasing GdnHCl concentration by measuring the intrinsic tryptophan fluorescence of Trp7 and Trp47 at 325 nm upon excitation at 295 nm. The resulting data were fit to a two-state model using the method of Santoro and Bolen (56). The free energy of unfolding (ΔG)equals 12.1 ± 0.9 kcal/mol at pH 5.0 and 10.4 ± 1.1 kcal/mol at pH 7.4 (1.7 kcal/mol lower). Note the differences and similarities in the baselines for the folded and unfolded states, respectively.
The importance of the Fis1 arm in regulating mitochondrial fission in yeast has been underscored by studies that display altered mitochondrial morphologies upon expression of constructs in which the Fis1 arm has been partially or completely deleted (ΔN-Fis1) (29). These studies suggest that the Fis1 arm is indispensable. However, this phenotype can be rescued upon overexpression of Mdv1, suggesting the altered morphology could be a result of misregulation of the fission complex (26, 36), as it has been shown that ΔN-Fis1 retains binding to %. both Mdv1 and Dnm1 in vitro (37). The Fis1 arm is also important in binding fission factors. Mdv1 binds Fis1 more tightly in the presence of the arm and is shown to have direct interactions with the Fis1 arm in a structure of the cocomplex (35, 37). By contrast, binding between Fis1 and the GTPase Dnm1 in biochemical pull-down experiments is at least 100-fold stronger when the Fis1 arm (residues 1–16) is deleted. When single residues in Δ N-Fis1 were replaced with glutamic acid residues, those on the concave surface disrupted Dnm1 binding (37). Interestingly, Dnm1 is able to compete with the Fis1 arm for the concave surface at high Dnm1 concentrations (37). These data support the possibility that the Fis1 arm may stabilize interactions with Mdv1 (35) but may inhibit interactions with Dnm1 by blocking access to the concave surface (37)
Thus, the Fis1 arm appears to block access to a functionally important surface, raising the question of how Dnm1 gains access to this surface. One possibility is that the Fis1 arm exists in a dynamic equilibrium between closed and open conformational states, which we test here with structural, dynamic, and thermo-dynamic experiments with budding yeast Fis1 lacking its C-terminal transmembrane domain. We find that the Fis1 arm lies predominantly in a closed conformation but also find evidence for a conformational change that is consistent with a dynamic equilibrium involving the Fis1 arm and α-helix 6. This conformational change could be readily modulated by other fission factors to regulate access to this functionally important surface on Fis1.
MATERIALS AND METHODS
Protein Expression and Purification
A gene encoding budding yeast Fis1ΔTM lacking the 29 C-terminal residues was subcloned into the pET38b expression vector using standard methods (38). Escherichia coli cells were transformed with this vector and grown at 37°C in Luria broth medium containing kanamycin (30 μg/mL) for selection to an an OD600 of ≈0.7. Protein expression was induced at 20°C by addition of IPTG to a final concentration of 0.4 mM. The cells were grown for 16–18 h and harvested by centrifugation; the resulting cell pellet was resuspended in 20 mM Tris-HCl, 500 mM NaCl, pH 8.0 buffer containing protease inhibitors (Roche) and pepstatin A(10 μg/mL). The cells were then lysed by either three passes through a French press or sonication. The lysate was separated from cellular debris by centrifugation at 30000g for 35 min. The supernatant was acidified to pH 5.0 by the addition of 50 mM acetic acid (HOAc). This acidified lysate was separated from acid-precipitated debris by centrifugation at 30000g for 35 min. Fis1ΔTM remained soluble and was thoroughly precipitated out of solution via addition of a saturated solution of (NH4)2SO4 (4 M) under continuous stirring in an ice–water bath. The precipitated Fis1ΔTM was collected by centrifugation at 30000 rcf for 45 min and gently resuspended in 50 mM NaOAc (pH 4.6). This suspension was further clarified by centrifugation at 30000g for 30 min. The supernatant was diluted 5-fold with 50 mM NaOAc (pH 4.6), loaded onto an anionic exchange chromatography column (SP Sepharose XL, GE Healthcare), and eluted with a 0 to 500 mM NaCl gradient in 50 mM NaOAc buffer (pH 4.6). The resulting chromatographic fractions containing Fis1ΔTM were concentrated and further purified by size exclusion chromatography (Superdex 75, GE Healthcare). Sample purity was checked by standard Coomassie-stained SDS–PAGE and was always greater than 95%
NMR Chemical Shift Perturbations as a Function of pH
NMR experiments at 298 K were performed at 18.8 T on a Varian Inova800 spectrometer outfitted with a HCN cold-probe to determine pH-dependent chemical shift changes. Two-dimensional 1H-15N heteronuclear single-quantum correlation (HSQC) experiments were conducted at pH 5.0 and 7.4 using WATERGATE for solvent suppression (39). Uniformly 15N-labeled Fis1ΔTM samples were prepared by extensive buffer exchange in either 25 mM d4-NaOAc (pH 5.0) or 25 mM d11-Tris-HCl (pH 7.4). Both samples contained 50 mM NaCl, 10 mM EDTA, 1 mM DTT, and 10% 2H2O. The protein concentration in the samples used for these experiments ranged from 0.2 to 1.1 mM for both pH values for evaluation of potential protein concentration effects, which were not observed. The pH of all these samples was precisely adjusted at ±0.1 unit of the desired value with no correction for 10% D2O. HSQC spectra were collected at a rate of four scans per increment, 1280 (t2)×256 (t1) complex points with acquisition times of 64 ms (1H) and 71 ms (15N). The carrier frequencies were centered on the chemical shift of water in 1H and in the center of the amide region at 117.5 ppm in 15N. NMR data processing was conducted with NMRPipe (40), and data were subsequently analyzed with NMRView (41, 42). Average chemical shift differences between pH values were determined for each residue as Δ = [(ΔδH)2 + (¼ Δ δN)2]½, where Δ is the total chemical shift change, Δ δH is the chemical shift change in 1H, and Δ δN is the chemical shift change in 15N expressed in parts per million.
NMR Chemical Shift Assignment
Sequential backbone assignments of the Fis1Δ TM protein resonances at pH 5.0 and 7.4 and at 25°C were completed from the analysis of standard double- and triple-resonance three-dimensional experiments, including HNCA, HNCACB, HNCOCA, CBCACONH, 15N-edited NOESY, HMQC-NOESY-HSQC, and HNN (43). These data sets were collected on a 0.8–1.3 mM Fis1Δ TM sample uniformly labeled with 13Cand15N at 14.2 T on a Bruker AVANCE600 instrument equipped with a TXI cryoprobe. The chemical shift assignments at pH 5.0 were verified by comparison with assignments at pH 5.5 that were kindly provided by M. Suzuki and R. J. Youle (29).
Fluorescence-Monitored Chemical Denaturations
To determine the thermodynamic stability of Fis1Δ TM at pH 5.0 and 7.4, equilibrium chemical denaturation experiments were performed by following the intrinsic fluorescence of Trp7 and Trp47 in Fis1Δ TM as a function of an increasing concentration of GdnHCl at 25°C in a PTI model A1010 fluorimeter (Photon Technology International). Fis1ΔTM protein samples were prepared by extensive dialysis in either 25 mM NaOAc (pH 5.0) or 25 mM Tris-HCl (pH 7.4). Both samples also contained 50 mM NaCl, 10 mM EDTA, and 2 mM DTT. Protein samples at pH 5.0 and 7.4 were prepared using an automatic titrator (Microlab 500 series, Hamilton) at a concentration of 3 μ M. The GdnHCl concentration ranged from 0 to 5.5 M for the pH 5.0 samples and from 0 to 4.5 M for the pH 7.4 samples. The excitation wavelength was 295 nm, and the emission spectrum were collected between 305 and 450 nm at a scan rate of 1 nm/s for all the samples with slit widths at 1 and 5 nm for excitation and emission, respectively.
NMR-Detected Hydrogen–Deuterium Exchange
A 0.8 mM 15N-labeled Fis1ΔTM sample was prepared by extensive dialysis in 25 mM d11-Tris-HCl, 50 mM NaCl, 10 mM EDTA, and 2 mM DTT (pH 7.4). After dialysis, the sample was completely lyophilized and the hydrogen–deuterium exchange reaction was started by rapidly dissolving the lyophilized protein into 100% 2H2O. The pD of the sample was confirmed after the experiment was concluded. A series of 21 two-dimensional 1H–15N HSQC experiments were conducted at 298 K and 14.2 T as a function of increasing time on a Bruker AVANCE600 spectrometer. HSQC spectra were collected at a rate of eight scans per increment, 1280 (t2) × 128 (t1) complex points with 64 ms (1H) and 37 ms (15N) acquisition times. To follow the H–D exchange reaction, we assumed that the cross-peak volume (or intensity) was proportional to the proton occupancy of the corresponding amide group. For each point in the volume (or intensity) decay, the time was assigned to be the middle of the acquisition time for each HSQC spectrum, considering “zero time” the time when the protein sample was dissolved in 100% 2H2O. Cross-peak intensities were fitted to the single-exponential decay function I = I∞ +I0 exp(−kext) to determine the exchange rate values, kex. I is the intensity of the cross-peak at each time point (in seconds). I0 is the amplitude of the exchange decay curve. I∞ is the intensity of the cross-peak at the infinity time point. Cross-peak volumes were fitted in a similar manner and gave similar results.
NMR Spin Relaxation Measurements
All spin relaxation experiments were conducted at 14.2 T on a Bruker Avance600 spectrometer at 298 K with a TXI cryoprobe. The 15N spin–lattice relaxation rate constant R1, the spin–spin relaxation rate constant R2, and the steady-state {1H}–15N NOE were measured using inversion recovery, Carr–Purcell–Meiboom–Gill (CPMG), and steady-state NOE pulse sequences (44, 45). Each R1 spectrum was acquired with 1280 (t2)× 220 (t1) complex data points, spectral widths of 10000 Hz in 1H and 1702 Hz in 15N, and 16 scans per t1 point. The recycle delay was 1.2 s. Eight R1 delay times were selected to fully define the exponential decay curve (7, 119, 252, 420, 623, 840, 1246, and 2996 ms). For the R2 experiments, the acquisition parameters were identical to those of the R1 experiments except for nine delay times of 16.5, 33,49.5, 66,82.5,99,115.5,165, and 280.5 ms. The recycle delay for these experiments was 1.5 s. Acquisition order was randomized to eliminate the effect of sample or field inhomogeneity over time. For each R1 and R2 analysis, at least two time points were recorded in duplicate for error analysis. For the hetero-nuclear NOE measurements, spectral widths were identical to those of the R1 and R2 experiments. A saturation time of 4 s was used for all NOE experiments. The 15N R1 and R2 relaxation rate constants were determined by fitting individual R1 and R2 decay curves to a single exponential using CURVEFIT (46). R1 and R2 uncertainties were estimated from Monte Carlo simulations of the data as described previously (45, 47, 48). NOE values were obtained from the ratio of the intensities of experiments conducted with and without proton saturation. The uncertainties of the NOEs were set to the standard deviation between intensities of duplicate experiments and determined by error propagation. R1, R2, and ssNOE data were analyzed using FastModelFree (49) implementation of ModelFree 4.01 (50).
NMR Saturation Transfer Experiments
All saturation transfer NMR experiments were conducted at 18.8 T on a Varian INOVA800 spectrometer at 298 K equipped with an HCN probe. These experiments measure magnetization transfer from water to amide protons and are corrected for the apparent amide proton T1 relaxation rate constant (T1app). T1 apparent experiments and saturation transfer experiments were performed with standard pulse sequences as previously described (51)with 1664 (t2) × 200 (t1) complex data points, spectral widths of 12,800 Hz in 1H and 2500 Hz in 15N, and 16 scans per t1 point. The recycle delay was 1.2 s. For the T1app experiment, T1 delay times of 1, 5, 10, 15, 20, 25, 30, 40, 50, and 2000 ms were used. Acquisition order was randomized to eliminate the effect of sample or field in homogeneity over time. Duplicates for two experiments were collected for error analysis. Individual T1 decay curves were fit to a single exponential as described previously using the data analysis software IGOR PRO. Uncertainties were estimated from the data fits.
Saturation transfer experiments were performed in an interleaved manner, alternating an on-resonance and off-resonance saturation pulse train for 1s using a string of Gaussian-3 pulses. Saturation ratios were obtained from the ratio of the intensities of experiments recorded with and without water saturation. The uncertainties for the saturation transfer experiments were estimated as a 5% error in peak height intensities, which was then used in standard error propagation to estimate the reported uncertainties in kex and kex′ values. The true uncertainty in kex and kex′ is likely to be higher on the basis of the assumption of an average intrinsic R1 value (see eq 1), which has previously been estimated to introduce uncertainties of up to 25% (52). In saturation transfer experiments, the exchange rate constant, kex, can be estimated from the ratio of amide intensities with and without water presaturation from the following equation if the saturation time is short with respect to R1:
| (1) |
where M0 and Mps are equilibrium intensities of the amide proton resonances in the absence and presence of water presaturation, respectively. Here the proton R1 is the intrinsic R1 in the absence of chemical exchange and is estimated to be 5.56 s−1, which is the mean proton R1 value of well-structured helical residues in α2–α5 that define the core of this TPR-like protein. The measured values of kex are more appropriately an apparent exchange rate constant because of NOE contributions from spin diffusion and coincidental Hα protons that resonate close to the water frequency (and are also saturated during the experiment). To correct for these contributions to the exchange rate constant, we also measured apparent kex values at pH 5.0 and used eq 2 to determine a corrected exchange rate constant, kex′:
| (2) |
Given eq 1, the slowest exchange rate constants to which saturation transfer experiments are sensitive are limited by the intrinsic R1 values. In practice with an estimated average proton R1 of 5.56 s−1, rate constants much smaller than ~1s−1 will not be accurately measured due to signal loss during the experiment. The fastest exchange rate constants to which these experiments are sensitive are on the order of 100 s−1, as faster exchange rates result in signal losses during coherence transfer given a 1JNH of ~ 95 Hz.
NMR Residual Dipolar Coupling Experiments
We investigated several different media to introduce a degree of molecular alignment required to measure RDCs before finding that a stretched polyacrylamide gel gave robust residual dipolar couplings that ranged from −18 to 15 Hz. Two-dimensional IPAP1H–15N HSQC spectra were acquired with echo–antiecho detection to attenuate natural abundance signals from gels (53), on a 300 μM 15N-labeled Fis1ΔTM sample, in a 5% stretched acrylamide gel (pH 7.4 or 5.0) at 14.2 T on a Bruker Avance600 spectrometer. Spectra were acquired with 400 complex points in t1 (0.120 s acquisition time) with a 15N spectral width of 1643 Hz. Residual dipolar couplings were extracted from the IPAP data sets as described previously (54). We first determined an alignment tensor using model 1 from the structural ensemble 1Y8M and FitAlign with RDC values for a subset of residues that showed no pH dependence on their chemical shifts and lie in the core of the structure (helices 2–5). From this alignment tensor, we used euler_pdb to rotate the coordinates of 1Y8M into this frame and then calculated the predicted RDC values from this aligned structure using calc_rdip2. These programs were courtesy of J. R. Tolman.
Cysteine Modification Assay
WT or I85C-Fis1ΔTM was allowed to react with a 5-fold molar excess of AlexaFluor 488 C5-maleimide (Invitrogen) in 100 mM phosphate buffer (pH 7.4) and 50 mM NaCl. Aliquots were taken out at the indicated time points, and the reaction was stopped upon addition of 50% BME and SDS–PAGE loading buffer. To control for background fluorescence, we prepared a negative control with protein in BME and loading buffer, to which AlexaFluor 488 C5-maleimide was added. Samples (0.5 μg of protein) were run on 15% SDS–PAGE gels to separate the unreacted dye from the conjugated protein. The gel was scanned on a Typhoon 9400 imager, and the intensity of fluorescence was measured for each band. The data were subtracted for background using the negative control and fitted using either a mono- or double-exponential fit. Mono: y = yo + A exp(−τ/x). Double: y = yo1 + A1 exp(−τ1/x) + yo2 + A2 exp(−τ2/x), where y is the intensity at time t, x is the time in minutes, yo is the initial intensity, A1 and A2 are the max intensities, and τ is the rate constant in inverse minutes. Fis1ΔTM contains two native Cys residues, C79 and C87, located on helix 4 and mostly buried. The figures are representative of three independent measurements which have been averaged to give the rate constants and standard error of the mean in the table shown. We estimated an uncertainty of 10% in the densitometry measurements (from the variability in loading the gel and the measurement itself). Given the first data were collected at 10 s, the calculated upper limit of τ we can reliably measure is ~10 s−1.
RESULTS
To address the question of whether the N-terminal Fis1 arm existed in a dynamic equilibrium, we first isolated recombinantly expressed Fis1ΔTM from bacteria. We used a qualitative solubility screen (55) at a protein concentration of 10 mg/mL to identify buffer conditions suitable for dynamics measurements by NMR spectroscopy. We found that the molecule was most soluble under slightly acidic conditions (> 25 mg/mL at pH 5.0) but precipitated at concentrations of > 10 mg/mL near pH 7.4 in the absence of salt. Such pH dependence on solubility is not unusual especially near the isoelectric point of a protein, which is estimated to be ~7.7 for Fis1ΔTM and may account for the observed solubility differences. However, we wondered if this solubility difference might arise from a change in conformation and therefore measured the free energy of unfolding that could be sensitive to differences in conformation. We monitored the intrinsic tryptophan fluorescence of the two tryptophans in Fis1ΔTM, Trp7 and Trp47 (Figure 1a), as a function of increasing concentration of guanidinium hydrochloride (GdnHCl) at pH 5.0 and 7.4, which represent the end points in pH titration experiments (Figure 1 of the Supporting Information). The resulting data at pH 5.0 and 7.4 demonstrated a cooperative unfolding transition that was well-fitted to a two-state unfolding mechanism. Using the method of Santoro and Bolen (56), the estimated free energy of unfolding (ΔG°)at pH 5.0 equals 12.1 ± 0.9 kcal/mol and at pH 7.4 equals 10.4 ± 1.1 kcal/mol (Figure 1b). Thus, Fis1ΔTM is less stable at pH 7.4 than at pH 5.0 by ~1–2 kcal/mol given the experimental uncertainty in this measurement. In thermal unfolding experiments, the midpoint temperature decreased 8.3 °C from 77.3 °C at pH 5.0 to 69.3 °C at pH 7.4 (data not shown). A decrease in the free energy of unfolding could arise from pH-dependent differences in either the unfolded or the folded conformational state. However, from the denaturation curve, we noted that the intrinsic fluorescence of the fully unfolded conformational states (>3.5 M GdnHCl) did not exhibit a pH dependence (Figure 1b). In contrast, the intrinsic tryptophan fluorescence of the folded conformational states (<1.5 M GdnHCl) was greater at pH 5.0 than at pH 7.4, consistent with more solvent protection of the Trp residues at pH 5.0 than at pH 7.4. Trp7 is located in the Fis1 arm and Trp47 in α-helix 2 that lies under the Fis1 arm (Figure 1a). Thus, upon protonation, it appears that Fis1ΔTM experiences an increase in its thermodynamic stability of ~1–2 kcal/mol, and changes in the intrinsic tryptophan fluorescence may detect a conformational change.
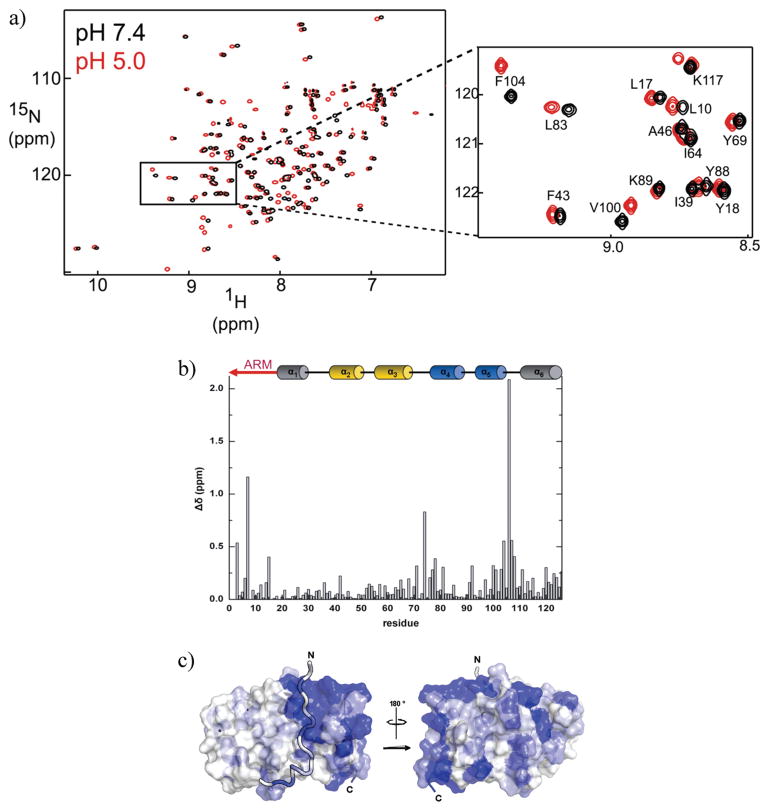
A structure of Fis1ΔTM was determined at pH 5.5 (29), and given the pH dependence noted above, we asked whether Fis1ΔTM adopted a different conformation at pH 7.4. We collected 1H–15N HSQC spectra as a function of increasing pH from pH 3 to 8. Spectra collected at pH 7.4 and 5.0 were chosen as being representative of the end points of the pH titration. At pH 7.4, only a subset of cross-peaks differed in either proton and nitrogen chemical shifts from those at pH 5.0, with a handful of cross-peaks with significant decreases in volume at pH 7.4 (Figure 2). To assess these differences, we calculated the average chemical shift difference, Δδ, using both the 1H and 15N chemical shifts. These differences ranged from 0 to >2 ppm with only 16% of the residues displaying a pH-induced difference of > 0.25 ppm (Figure 2b). To interpret these differences in light of the secondary and tertiary structure of Fis1ΔTM [calculated from data recorded at pH 5.5 and 32 °C(29)], we assigned the 1H and 15N chemical shifts to their respective residues at pH 5.0 and 7.4 using standard triple-resonance methods. More than 95% of the cross-peaks in the 1H–15N HSQC spectrum were assigned at pH 5.0 and 7.4.
Figure 2.
NMR chemical shifts differ at pH 7.4 and 5.0 for a subset of residues. (a) 1H–15N HSQC spectra of a 1.0 mM sample of Fis1ΔTM at pH 7.4 (black) and pH 5.0 (red) and 25 °C recorded at 18.8 T. (b) Observed change in chemical shift, Δδ, between pH 7.4 and 5.0 plotted as a function of residue number, where Δδ = {(1HδpH7.4 − 1HδpH5.0)2 + [¼(15NδpH7.4 − 15NδpH5.0)]2}1/2. (c) Mapping of the observed chemical shift changes onto a surface representation of the pH 5.5 structure of Fis1ΔTM [PDB entry 1y8m (29)]. This figure was made with PYMOL (81) using a color gradient (82) to display average chemical shift differences from 0.25 to 2.0 ppm (white to dark blue, respectively).
The most perturbed chemical shift difference (>2.0 ppm) arose from His106, the sole histidine residue at the end of α-helix 5 whose pKa value we determined is 6.04 ± 0.06 from long-range HMQC experiments (Figure 1 of the Supporting Information). In general, the largest differences in average chemical shift (>0.25 ppm) cluster primarily into a single region of the structure involving residues 4–15, 71–81, and 99–111 (Figure 2b). This cluster corresponds to the N-terminal Fis1 arm and regions that lie within a few angstroms of the arm (Figure 2c). We also noted that cross-peak intensities were substantially decreased at pH 7.4 for residues K3 and T9 in the Fis1 arm, E73 and S74 in the loop between α-helices 3 and 4, L103 in α-helix 5, and K111 in the loop between α-helices 5 and 6. pH-dependent differences in chemical shifts can arise from a variety of effects, including salt, buffer, and/or conformational differences, but we found the observed chemical shift differences between spectra acquired at pH 5.0 and 7.4 to be independent of salt and buffer concentration (data not shown).
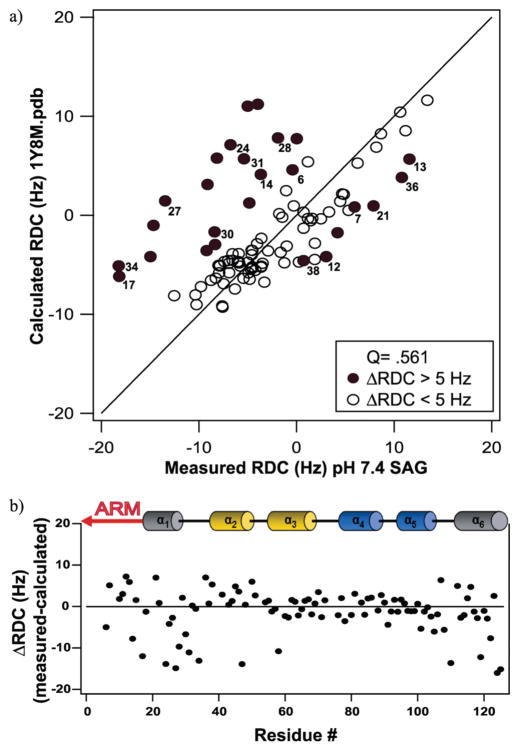
To assess these chemical shift differences further, we measured residual dipolar couplings (RDCs) from the backbone amide of Fis1ΔTM at both pH 5.0 and 7.4. RDCs can provide both high-resolution structural information and motional information about a wide range of time scales (57). We were able to reliably measure RDCs using IPAP-based experiments for 73% of Fis1ΔTM residues at pH 7.4 and 83% of the residues at pH 5.0 where the level of solvent exchange is reduced and chemical shift dispersion is improved. We compared our experimental RDC measurements at physiological pH with those predicted from the reported structure of Fis1ΔTM determined at pH 5.5 using FitAlign (Figure 3). To eliminate differences in RDC values that derive from differences in alignment tensors, we first generated an alignment tensor using a subset of Fis1ΔTM residues that exhibited little, if any, change in chemical shift (Δδ < 0.1 ppm) between pH 5.0 and 7.4. We then rotated model 1 from the reported structure into a coordinate axis system consistent with this alignment tensor using euler_pdb. The predicted RDC values from this adjusted structure were calculated using calc_rdip2. This allowed us to compare differences in RDC values that do not arise purely from a difference in the alignment tensor. The RDC values we measured at pH 7.4 did not compare favorably with those predicted with a Q factor of 0.561. Interestingly, the largest differences in the predicted and measured RDC values at pH 7.4 localized to the Fis1 arm, helix 1, the loop between helices 1 and 2, and helix 6. By contrast, the largest differences at pH 5.0 were distributed throughout the sequence (Figure 2 of the Supporting Information). Taken together, these data are consistent with a difference in the structure or dynamics of the arm and proximal regions of Fis1ΔTM at pH 7.4 compared to more acidic conditions.
Figure 3.
Residual dipolar coupling analysis of Fis1ΔTM at 14.2 T, 25 °C, and pH 7.4. (a) RDC values calculated from modified PDB files plotted vs measured values at pH 7.4. The reported Q value shows a poor correlation with the previously reported structure. (b) The differences between measured and calculated RDC values are plotted as a function of residue number to highlight the structural regions that give rise to the discrepancies in Q values with the reported structure. The largest differences between the structure and measured RDC values at pH 7.4 lie in the arm, helix 1, and helix 6.
To address whether these conformational differences in Fis1ΔTM involved a dynamic equilibrium, we used three different NMR-based probes that are sensitive to motions on different time scales. We first conducted R1, R2, and steady-state hetero-nuclear NOE experiments at pH 7.4 (Figure 3 of the Supporting Information). R1 and steady-state NOE values are sensitive to motions on the nanosecond to picosecond time scale, while R2 is also sensitive to slower motions on the microsecond to millisecond time scales. The resulting R1, R2, and steady-state NOE data were analyzed using the FASTModelFree (49) implementation of ModelFree 4.01 (50) which fits spin relaxation data to the Lipari–Szabo formalism assuming completely isotropic motion (58). At pH 7.4, the majority of Fis1ΔTM residues were described well by model 1 with an average rotational correlation time (τm) of 8.25 ± 0.32 ns, which compares favorably with the value of 8.3 ± 0.57 ns estimated from analysis of R2/R1 values using Quadric_Diffusion (59). The ModelFree analysis also provides a measure of the internal motional restriction of the amide bond vector in the form of a generalized squared order parameter S2, which is formally bounded between 0 and 1 for completely isotropic and restricted motion, respectively. For Fis1ΔTM at pH 7.4, S2 values appear uniformly distributed for residues that lie within the helical regions of Fis1ΔTM (mean S2= 0.95 ± 0.05 at pH 7.4) with slightly lower values for some residues in loop regions. Data collected and analyzed identically at pH 5.0 showed few differences from data collected at pH 7.4 (data not shown). We interpret these data to suggest that the backbone of Fis1ΔTM is well-ordered on the millisecond to picosecond time scale.
Fis1ΔTM may be dynamic on a time scale slower than milliseconds to picoseconds, so we measured amide proton exchange rates by saturation transfer NMR. These experiments can be sensitive to motions on the millisecond to minute time scale via detection of exchange of amide protons with solvent protons. In saturation transfer experiments, the ratio of amide intensities with and without water presaturation measured at different pH values can be used to estimate a corrected exchange rate constant at pH 7.4, kex′, which is corrected for spurious effects that arise from NOEs and coincidental chemical shifts (52). For Fis1ΔTM, these experiments are sensitive to motions only on the time scale of ~1–100 s−1 (see Materials and Methods) At pH 7.4, the corrected exchange rate constants for certain amide protons of Fis1ΔTM residues in loops are higher than those of the majority of amide protons that show exchange rates below the limits of detection (Figure 4 of the Supporting Information). In the Fis1 arm residues, we observe only a few amide protons with significant exchange rate constants (K3, D5, W7, L10, and I11) and negligible exchange with solvent for most residues in the arm.
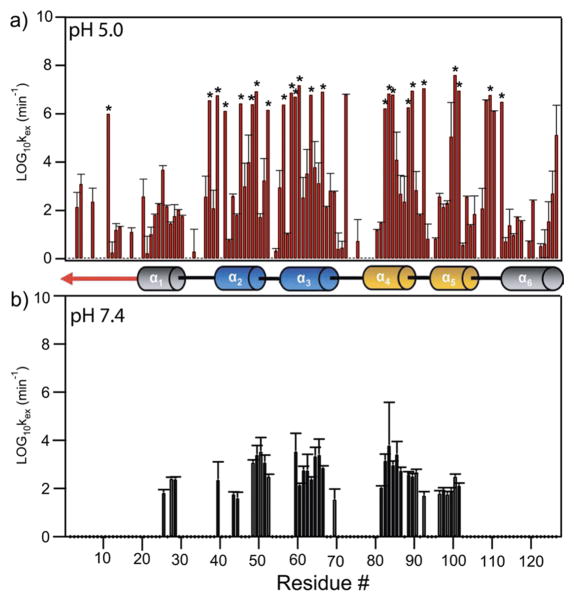
Given that the majority of Fis1ΔTM backbone residues exchange more slowly with solvent than 1 s−1, we used hydrogen–deuterium exchange experiments detected by NMR spectroscopy. In these experiments, a sample lyophilized in H2O is suspended in D2O and the exchange of backbone amide protons with solvent deuterons is monitored by collecting 1H–15N HSQC spectra as a function of time. Amide protons buried in the hydrophobic core or hydrogen-bonded in secondary structure will not readily exchange with solvent deuterons unless a dynamic event allows for such exchange. Such dynamic events can be either small internal fluctuations or local unfolding events that are related to the global unfolding of the protein. Regardless of the exact mechanism, these experiments can provide a measure of protein dynamics on a time scale of minutes and longer. HDX data were collected at pH 5.0 and 7.4 and were well-fitted to a single exponential, resulting in determination of the rate constant for solvent exchange (kex) for each nonoverlapped residue. At pH 5.0, backbone amides in helices 1–5 of Fis1ΔTM were more protected from solvent exchange than loop regions, with notable exceptions involving the loop between helices 5 and 6. Many residues showed little exchange with solvent over a 7 day collection period and corresponded to residues in the core TPR helices of Fis1 with the notable exception of Lys11 in the Fis1 arm (denoted with an asterisk in Figure 4a). Other residues in the Fis1 arm were protected from solvent exchange to the same degree as residues in helices 1 and 5. By contrast, at pH 7.4, backbone amides in the Fis1 arm and α-helix 6 were not protected at all. The marked decrease in the degree of solvent protection at pH 7.4 for residues in the Fis1 arm, helix 1, the loop between helices 5 and 6, and helix 6 is not solely an effect in the increase in the level of base-catalyzed exchange as these data have been corrected for such effects. Thus, these data suggest these residues, including the Fis1 arm and proximal loop residues, undergo motions on the minute time scale not observed at a more acidic pH.
Figure 4.
NMR-derived hydrogen–deuterium exchange data of Fis1ΔTM at 14.2 T, 25 °C, and pH 7.4 and 5.0 as a function of residue number. The rate constants for exchange of the amide backbone with solvent deuterons, log10 kex, are derived from a series of HSQC experiments conducted with a 15N-labeled sample after dissolution in D2O. Note the N-terminal region and helix 6 are not protected from solvent exchange at pH 7.4 but are at pH 5.0. Residues marked with an asterisk (38, 40, 42, 46, 49, 50, 53, 57, 59–61, 64, 67, 83–85,89,90,92,100,101,109, and 112) showed “super protection”.
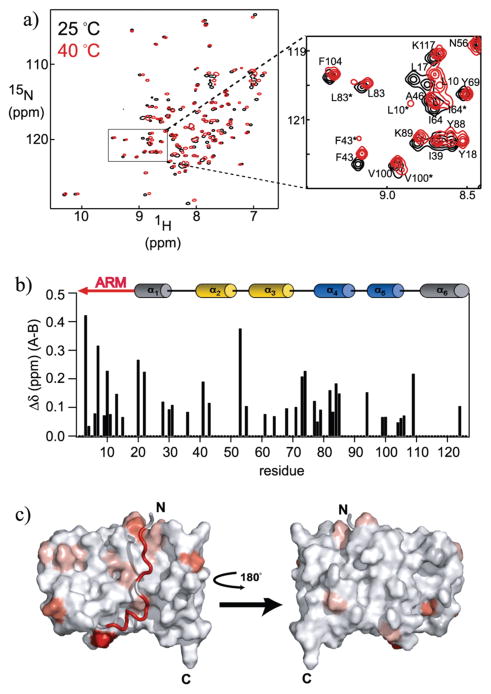
The NMR data taken as a whole suggest a slow opening rate for the Fis1 arm that could be consistent with a small population of Fis1ΔTM molecules with the concave surface exposed to solvent. This model would predict that other perturbations to this conformational equilibrium, such as temperature, would result in displacement of the Fis1 arm that might be more readily detected than in the experiments described herein. To test this idea, we monitored the 1H and 15N chemical shifts of Fis1ΔTM at 40 °C (Figure 5a). We found the most significant perturbations of chemical shifts for only a subset of residues. We assigned the chemical shifts of 40 °C spectra using standard methods and calculated the average chemical shift difference, Δδ, using both the 1H and 15N chemical shifts (Figure 5 of the Supporting Information). While differences in temperature-induced chemical shifts can arise from both conformational and hydrogen bond length differences, we found the largest difference involved the Fis1 arm and proximal regions (Figure 5 of the Supporting Information). In addition to temperature-induced chemical shift changes, we noted new additional cross-peaks for a subset of residues at 40 °C. These additional cross-peaks were observed for only a subset of residues (Figure 5a, inset). Additional cross-peaks in NMR spectra can arise from either conformational or sample heterogeneity. HPLC analysis and MALDI-TOF mass spectrometry demonstrated that NMR samples were >95% homogeneous. Furthermore, these cross-peaks can disappear when the temperature is decreased to 25 °C. Thus, the observed changes in relative cross-peak intensities are consistent with the existence of major and minor conformations in solution. Consistent with this interpretation, we found when the sample temperature was increased to 50 °C that cross-peak intensities of the minor species increased while cross-peak intensities of the major species concomitantly decreased (data not shown). We conclude that a minor species of Fis1ΔTM is in slow exchange on the NMR time scale with a major species. We calculated the chemical shift difference between the major and minor species, Δδ, and mapped these differences onto a structural model of Fis1ΔTM (Figure 5b,c). The minor species appears to involve many residues with a subset that cluster in the Fis1 arm and proximal regions (Figure 5c). We interpret these data to support the idea of a dynamic equilibrium involving the Fis1 arm. However, we note that not all residues in the Fis1 arm exhibited this behavior, consistent with a dynamic process that modulates residues nonuniformly and suggests more complex dynamics than a simple opening reaction involving the Fis1 arm.
Figure 5.
NMR chemical shifts differ at 40 and 25 °C at pH 7.4 for a subset of residues. (a) 1H–15N HSQC spectra of a 1.0 mM sample of Fis1ΔTM at 25 (black) and 40 °C (red) and at pH 7.4 recorded at 14.2 T. For a subset of residues, a minor conformation (B) in slow exchange with a major conformation (A) appears and is indicated with an asterisk. (b) Observed change in chemical shift, Δδ, between conformations A and B at 40 °C plotted as a function of residue number, where Δδ = {(1HδpH7.4−1HδpH5.0)2+[¼(15NδpH7.4− 15NδpH5.0)]2}1/2. A minor conformation was observed for residues 3,4, 6,7,9–11,13,15,20,22,28,30,31,36,41,43,53,55,61,64,68,71,73, 74,77–79, 82–85,94,99,100,104–106,109, and 124. (c) Mapping of the observed chemical shift changes between conformations A and B onto a surface representation of the pH 5.5 structure of Fis1ΔTM [PDB entry 1 y8m (29)]. This figure was made with PYMOL (81) using a color gradient (82) to display average chemical shift differences from 0 to 0.43 ppm (from white to red, respectively). The residues for which no chemical shift data are available are colored light gray.
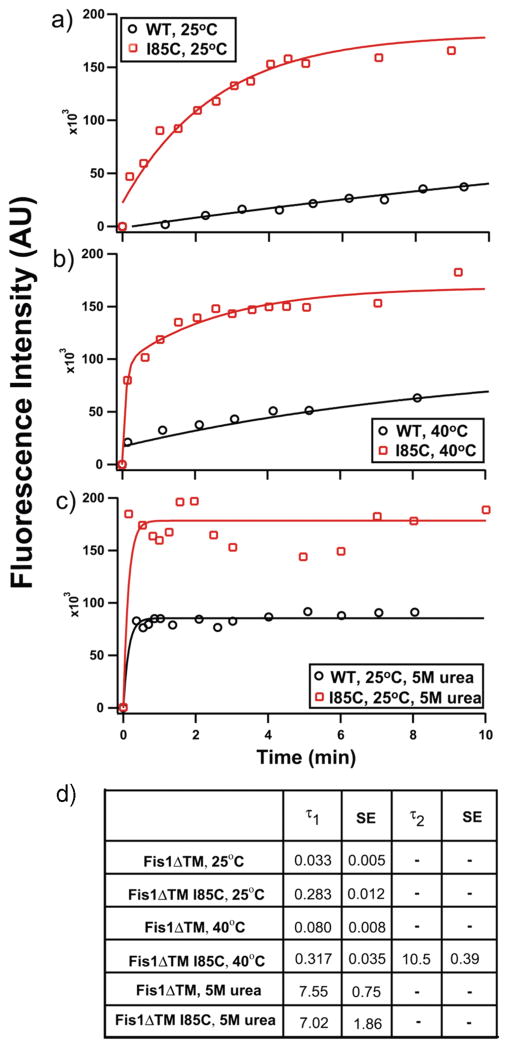
If the Fis1 arm were involved in a dynamic equilibrium, we would expect that we could detect this equilibrium using a cysteine modification experiment used as a measure of solvent accessibility to a specific side chain. From our data given above, we predicted that a cysteine probe placed at the interface of the concave surface and the N-terminal arm would exhibit solvent accessibility if the arm were undergoing dynamic excursions away from the concave surface. To test this prediction, we replaced isoleucine 85 with cysteine (I85C) and monitored the covalent modification over time with the fluorescent cysteine-reactive dye AlexaFluor Maleimide 488 (Figure 6a). We chose this position as Ile85 lies in the middle of the concave surface on α-helix 4, directly below the Fis1 arm.
Figure 6.
Chemical modification of a buried residue may be consistent with Fis1 arm dynamics. Modification of an engineered cysteine residue by AlexaFluor Malemide 488 was monitored as a function of time. I85C lies on α-helix 4 beneath the Fis1 arm. Fis1ΔTM also contains two native Cys residues, C79 and C87, located on the opposite side of helix 4 and are mostly buried: (a) wild type (black circles) and I85C (red squares) Fis1ΔTM at 25 °C, (b) wild-type (black circles) and I85C (red squares) Fis1ΔTM at 40 °C, and (c) unfolded wild-type (black circles) and unfolded I85C (red squares) Fis1ΔTM. (d) τ1 and τ2 values from single- or double-exponential fits. The figures are representative of three independent measurements which have been averaged to give the rate constants and standard error of the mean in the table shown.
Wild-type Fis1ΔTM has two native cysteines (residues 79 and 87), also located on helix 4, pointing toward the opposite surface from Ile85. These cysteines appear to be solvent inaccessible in the published structure, and indeed, we saw a slow rate of modification (τ = 0.033 ± 0.005) upon addition of AlexaFluor Maleimide 488 (Figure 6a). In contrast, I85C-Fis1ΔTM exhibited a rate of modification almost 10-fold faster (τ = 0.283 ± 0.012) (Figure 6a) than that of the wild type. Replacement of Ile85 itself could perturb the structure of the protein, allowing all cysteines in I85C to become more accessible to chemical modification. However, 15N-labeled I85C-Fis1ΔTM exhibited very similar backbone amide 1H and 15N chemical shifts, with most Δδ being <0.1 ppm when compared to wild-type Fis1ΔTM, indicating no large structural changes had occurred upon replacement of Ile85 with Cys (Figure 6 of the Supporting Information). Further, we also noted that the amplitudes of the modification curves are consistent with the increased amplitude of I85C arising primarily from modification of the non-native cysteine.
To test whether temperature affects a dynamic equilibrium involving the Fis1 arm, we measured chemical modification of I85C at 40 °C (Figure 6b). The rate of cysteine modification for wild-type Fis1ΔTM increased slightly at 40 °C compared to that at 25 °C[τ = 0.080 ± 0.008 (Figure 6b)]. In contrast, the rate of cysteine modification for I85C could not be fit to a single exponential (Figure 6b). Instead, these data required fitting to a double-exponential equation with the following rate constants: τ1 = 0.317 ± 0.035, and τ2 = 10.5 ± 0.39. To determine whether these rate constants arose from a misfolding of the I85C variant at 40 °C, we collected a 1H–15N HSQC spectrum of 15N-labeled I85C-Fis1ΔTM at 40 °C and found minor changes in chemical shift, indicating the protein was still folded at this temperature (Figure 6 of the Supporting Information).
DISCUSSION
The concave surface of Fis1ΔTM, occluded by the N-terminal arm in both NMR and X-ray structures (29,35), is thought to be displaced for Fis1 to be able to recruit the essential GTPase Dnm1 to sites of membrane scission (26,37). We therefore tested the hypothesis that the Fis1 arm may exist in a dynamic equilibrium between an open and closed conformation that might allow access to this concave surface. We interpret our data to support a model in which the N-terminal arm and α-helix 6 are predominantly structured as seen in the high-resolution structures but exhibit dynamic excursions away from this mean structure. These conformational excursions could lead to a loss of protection from amide proton exchange with solvent as measured by HDX (Figure 4) and that may be detected by RDC measurements that indicated significant differences in the structure of the Fis1 arm and proximal regions when compared to the reported structures (Figure 3). The NMR data as a whole indicate that the Fis1 arm and α-helix 6 may be dynamic on a time scale slower than 1 s−1 (from saturation transfer experiments) but faster than ~0.7 × 10−3 s−1 at pH 7.4 (given the lag time between dissolution in D2O and collection of the first HDX data point). These data could be consistent with a small population of conformational states in which the Fis1 arm is not occluding access to the concave surface. Interestingly, proline isomerization occurs on a very similar time scale (60). There are two prolines in the N-terminal arm of yeast Fis1 (P8 and P16) that are conserved among Fis1 from fungal species. The structure of yeast Fis1 at acidic pH shows both prolines in the cis conformation (29). Thus, it is tempting to speculate that the slow interconversion of proline isomers may govern when the arm is occluding access to the concave surface.
Our chemical modification experiments with I85C are also consistent with a dynamic equilibrium involving the Fis1 arm. We found that Cys85 that lies under the N-terminal arm is readily modified by a fluorescent probe. In these experiments, we cannot eliminate the possibility that replacing Ile85 with Cys itself perturbs the Fis1 arm equilibrium. However, we found few differences in chemical shifts between the wild type and I85C-Fis1ΔTM. To address the innate limitations in these chemical modification experiments, we measured the modification rate at two temperatures. Measuring the modification rate of the I85C protein at 25 and 40 °C allows us to observe a perturbation in the dynamic equilibrium. We could fit our 40 °C data only to a double exponential, with one rate constant that corresponded very closely to that of I85C at 25 °C (τ40C = 0.317 ± 0.035 vs τ25C = 0.283 ± 0.012), while the other corresponded closely to the rate of modification of completely accessible cysteines in an unfolded protein (τ2 = 10.5 ± 0.039 vs τunfolded = 7.02 ± 1.86) (Figure 6c). This suggests that I85C exists in two populations: one that is as folded like the wild type and one that is unfolded. However, our NMR data of I85C at 40 °C show no evidence of an unfolded population, arguing that the second rate constant derives primarily from Cys85 being freely accessible to the fluorescent dye at 40 °C as would occur upon displacement of the Fis1 arm. Consistent with this interpretation, the chemical shift of the native Cys79 in the I85C mutant did not change, though Cys87 was not readily assignable. These residues serve as useful controls as they also lie on helix 4, though on the opposite side of the concave surface. Interestingly, perturbing the equilibrium of conformational states involving the Fis1 arm at temperatures even higher than those reported here results in formation of Fis1ΔTM homodimers. We have also found that mutagenesis of residues that would be expected to displace the Fis1 arm from the concave surface (by introducing charged residues in the arm itself or concave surface) results in homodimer formation (unpublished data). Thus, the Fis1 arm appears to occlude a concave surface that is important for homodimerization and binding to other fission factors.
Previous analysis of backbone 13C′ spin relaxation data has been reported for Fis1ΔTM and is largely consistent with our analysis here, with the exception that Ile85 was found to undergo chemical exchange on a microsecond to millisecond time scale that is not sensitive to detection by 15N spin relaxation methods (61). While other residues were not reported to be involved in chemical exchange, this observation may be consistent with the idea of a flexible Fis1 arm that could give rise to such selective exchange, consistent with our observations. Dynamic excursions away from the mean structure involving the Fis1 arm are consistent with biochemical pull-down experiments that detected binding only between Fis1 and Dnm1 after Western blot analysis with extended exposure times, which is contrasted by robust binding observed upon removal of the Fis1 arm (37).
If the role of Fis1 is to recruit Dnm1 to the mitochondrial surface but the Fis1 arm prevents this interaction, then the question becomes how this apparent autoinhibition is relieved. A likely candidate is Mdv1, an adaptor protein essential for fission. However, several lines of evidence suggest that Mdv1 does not play this role. First, the altered mitochondrial morphology caused by ΔN-Fis1 (29) is rescued by Mdv1 overexpression (26), which argues against Mdv1 acting to displace the Fis1 arm. Second, Mdv1 is thought to act late in the fission process after a Fis1 –Dnm1 complex has formed (36). Third, we saw no evidence of an increased level of Dnm1 binding in the presence of increasing amounts of Mdv1ΔC in biochemical pull-down experiments. However, this result could be due to the presence of large fusion constructs necessary for these experiments (37). Finally, the crystal structure of Fis1ΔTM in complex with a fragment of Mdv1 determined to high-resolution shows that the Fis1 arm in a closed conformation similar to the structure in the absence of Mdv1 (35). In this cocomplex, the N-terminal residues of the Fis1 arm are making intimate contacts with Mdv1 that would appear to favor the Fis1 arm in a closed conformation, consistent with biochemical pull-down data that showed a slight decrease in the level of binding upon removal of the Fis1 arm (37). These data suggest that Mdv1 does not displace the Fis1 arm to expose the conserved, concave surface.
Is it possible that conformational substates of Fis1ΔTM that are sparsely populated are important in allowing access to the conserved, concave surface important in Dnm1 binding? Support for such a possibility can be found in other systems. For example, the Db1 homology domain of guanine nucleotide exchange factor Vav1 is also autoinhibited by an N-terminal helix (arm) that sterically occludes access to the active site (62). In this system, the inhibitory helix (arm) also adopts a primarily closed conformation but exhibits brief, dynamic excursions to a conformation that allows phosphorylation and relief of autoinhibition (63, 64). Thus, conformational substates that are fleetingly populated, and therefore invisible by most forms of spectroscopy that detect only the ensemble average, may be important in regulation of a protein’s activity. This view of protein structure consisting of a dynamic population of conformational substates is not new (65) but has been demonstrated computationally and experimentally only recently (66–75) in large part due to the development of new NMR methods (76–78) that can detect these so-called “invisible states” if the time scale of motion is suitable (79, 80).
These considerations support the idea that a small fraction of Fis1ΔTM molecules with the Fis1 arm in an open conformation might be sufficient to relieve the apparent autoinhibition. We note that the presence of the transmembrane domain and the mitochondrial membrane that are absent in our experiments may shift this conformational equilibrium further toward an open conformation. Whether autoinhibition is relieved entirely by intrinsic dynamics of Fis1 or requires other mechanisms, such as catalyzed prolyl isomerization or phosphorylation, remains to be determined. In summary, the dynamic nature of yeast Fis1ΔTM demonstrated by these experiments is consistent with a model in which the Fis1 arm regulates access to a concave surface important in binding to Dnm1, a GTPase essential for mitochondrial and peroxisomal fission.
Supplementary Material
Acknowledgments
We thank Kevin R. MacKenzie, Joel R. Tolman, and Evangelos Moudrianakis for helpful suggestions and reading earlier drafts of the manuscript. We also thank Joel R. Tolman for the software used in analyzing RDC data. We are grateful to Joel R. Tolman and James T. Stivers for suggesting the cysteine modification experiments and Ira Ropson for use of the autotitrator. We appreciate Motoshi Suzuki and Richard J. Youle for kindly providing chemical shift assignments of Fis1 used to confirm our assignments.
Footnotes
Abbreviations: TPR, tetratricopeptide repeat fold; TM, transmembrane domain; Tris, tris(hydroxymethyl)aminomethane; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; GdnHCl, guanidinium hydrochloride; HMQC, heteronuclear multiple-quantum coherence; HSQC, heteronuclear single-quantum coherence; NOE, nuclear Overhauser enhancement; HDX, hydrogen–deuterium exchange; RDC, residual dipolar couplings; PDB, Protein Data Bank.
This work was supported by National Institutes of Health (NIH) Grant RO1GM067180 and American Cancer Society Award IRG-58-005-41. NMR instrumentation was secured in part with National Science Foundation Grant DBI-0216077 and NIH Grant S10-RR020922. S.C. was a recipient of a postdoctoral fellowship from the Spanish Government. A. C. M. was supported in part by HHMI summer undergraduate research fellowships.
SUPPORTING INFORMATION AVAILABLE
pKa determination of His106 of Fis1 by long-range HMQC experiments at 25 °C, NMR residual dipolar coupling analysis of Fis1 at pH 5.0, NMR spin relaxation analysis of Fis1 at pH 7.4, NMR saturation transfer experiments of Fis1 at pH 7.4, NMR chemical shift analysis at 25 and 40 °C, HSQC spectra of I85C mutant at 25 and 40 °C, and a table of residual dipolar coupling values at pH 5.0 and 7.4. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Chan DC. Mitochondria: Dynamic Organelles in Disease, Aging, and Development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Martinou JC, Youle RJ. Which Came First, the Cytochrome c Release or the Mitochondrial Fission? Cell Death Differ. 2006;13:1291–1295. doi: 10.1038/sj.cdd.4401985. [DOI] [PubMed] [Google Scholar]
- 3.Jagasia R, Grote P, Westermann B, Conradt B. DRP-1-Mediated Mitochondrial Fragmentation during EGL-1-Induced Cell Death in C. elegans. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- 4.Delivani P, Adrain C, Taylor RC, Duriez PJ, Martin SJ. Role for CED-9 and Egl-1 as Regulators of Mitochondrial Fission and Fusion Dynamics. Mol Cell. 2006;21:761–773. doi: 10.1016/j.molcel.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 5.Fannjiang Y, Cheng WC, Lee SJ, Qi B, Pevsner J, McCaffery JM, Hill RB, Basanez G, Hardwick JM. Mitochondrial Fission Proteins Regulate Programmed Cell Death in Yeast. Genes Dev. 2004;18:2785–2797. doi: 10.1101/gad.1247904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Roles of the Mammalian Mitochondrial Fission and Fusion Mediators Fis1, Drp1, and Opa1 in Apoptosis. Mol Biol Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu T, Fox RJ, Burwell LS, Yoon Y. Regulation of Mitochondrial Fission and Apoptosis by the Mitochondrial Outer Membrane Protein hFis1. J Cell Sci. 2005;118:4141–4151. doi: 10.1242/jcs.02537. [DOI] [PubMed] [Google Scholar]
- 8.Heath-Engel HM, Shore GC. Mitochondrial Membrane Dynamics, Cristae Remodeling and Apoptosis. Biochim Biophys Acta. 2006;1763:549–560. doi: 10.1016/j.bbamcr.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Stojanovski D, Koutsopoulos OS, Okamoto K, Ryan MT. Levels of Human Fis1 at the Mitochondrial Outer Membrane Regulate Mitochondrial Morphology. J Cell Sci. 2004;117:1201–1210. doi: 10.1242/jcs.01058. [DOI] [PubMed] [Google Scholar]
- 10.James DI, Parone PA, Mattenberger Y, Martinou JC. HFis1, a Novel Component of the Mammalian Mitochondrial Fission Machinery. J Biol Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 11.Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. A Role for Fis1 in both Mitochondrial and Peroxisomal Fission in Mammalian Cells. Mol Biol Cell. 2005;16:5077–5086. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schrader M. Shared Components of Mitochondrial and Peroxisomal Division. Biochim Biophys Acta. 2006;1763:531–541. doi: 10.1016/j.bbamcr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Shaw JM, Nunnari J. Mitochondrial Dynamics and Division in Budding Yeast. Trends Cell Biol. 2002;12:178–184. doi: 10.1016/s0962-8924(01)02246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Logan DC. Plant Mitochondrial Dynamics. Biochim Biophys Acta. 2006;1763:430–441. doi: 10.1016/j.bbamcr.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-Mediated Mitochondrial Fission is a Multi-Step Process Requiring the Novel Integral Membrane Component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tieu Q, Nunnari J. Mdv1p is a WD Repeat Protein that Interacts with the Dynamin-Related GTPase, Dnm1p, to Trigger Mitochondrial Division. J Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tieu Q, Okreglak V, Naylor K, Nunnari J. The WD Repeat Protein, Mdv1p, Functions as a Molecular Adaptor by Interacting with Dnm1p and Fis1p during Mitochondrial Fission. J Cell Biol. 2002;158:445–452. doi: 10.1083/jcb.200205031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerveny KL, McCaffery JM, Jensen RE. Division of Mitochondria Requires a Novel DMN1-Interacting Protein, Net2p. Mol Biol Cell. 2001;12:309–321. doi: 10.1091/mbc.12.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerveny KL, Jensen RE. The WD-Repeats of Net2p Interact with Dnm1p and Fis1p to Regulate Division of Mitochondria. Mol Biol Cell. 2003;14:4126–4139. doi: 10.1091/mbc.E03-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakobs S, Martini N, Schauss AC, Egner A, Westermann B, Hell SW. Spatial and Temporal Dynamics of Budding Yeast Mitochondria Lacking the Division Component Fis1p. J Cell Sci. 2003;116:2005–2014. doi: 10.1242/jcs.00423. [DOI] [PubMed] [Google Scholar]
- 21.Griffin EE, Graumann J, Chan DC. The WD40 Protein Caf4p is a Component of the Mitochondrial Fission Machinery and Recruits Dnm1p to Mitochondria. J Cell Biol. 2005;170:237–248. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schauss AC, Bewersdorf J, Jakobs S. Fis1p andCaf4p, but Not Mdv1p, Determine the Polar Localization of Dnm1p Clusters on the Mitochondrial Surface. J Cell Sci. 2006;119:3098–3106. doi: 10.1242/jcs.03026. [DOI] [PubMed] [Google Scholar]
- 23.Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, Shaw JM. The Dynamin-Related GTPase, Dnm1p, Controls Mitochondrial Morphology in Yeast. J Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sesaki H, Jensen RE. Division Versus Fusion: Dnm1p and Fzo1p Antagonistically Regulate Mitochondrial Shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The Dynamin-Related GTPase Dnm1 Regulates Mitochondrial Fission in Yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karren MA, Coonrod EM, Anderson TK, Shaw JM. The Role of Fis1p-Mdv1p Interactions in Mitochondrial Fission Complex Assembly. J Cell Biol. 2005;171:291–301. doi: 10.1083/jcb.200506158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki M, Jeong SY, Karbowski M, Youle RJ, Tjandra N. The Solution Structure of Human Mitochondria Fission Protein Fis1 Reveals a Novel TPR-Like Helix Bundle. J Mol Biol. 2003;334:445–458. doi: 10.1016/j.jmb.2003.09.064. [DOI] [PubMed] [Google Scholar]
- 28.Dohm JA, Lee SJ, Hardwick JM, Hill RB, Gittis AG. Cytosolic Domain of the Human Mitochondrial Fission Protein Fis1 Adopts a TPR Fold. Proteins. 2004;54:153–156. doi: 10.1002/prot.10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki M, Neutzner A, Tjandra N, Youle RJ. Novel Structure of the N Terminus in Yeast Fis1 Correlates with a Specialized Function in Mitochondrial Fission. J Biol Chem. 2005;280:21444–21452. doi: 10.1074/jbc.M414092200. [DOI] [PubMed] [Google Scholar]
- 30.Ohashi W, Hirota H, Yamazaki T, Koshiba S, Hamada T, Yoshida M, Yokoyama S. Protein Data Bank; 2002. Solution Structure of RSGI RUH-001, a Fis1p-Like and CGI-135 Homologous Domain from a Mouse cDNA. http://www.rcsb.org/pdb/explore.do?structure-Id=1IYG. [Google Scholar]
- 31.Blatch GL, Lassle M. The Tetratricopeptide Repeat: A Structural Motif Mediating Protein-Protein Interactions. BioEssays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 32.D’Andrea LD, Regan L. TPR Proteins: The Versatile Helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 33.Cortajarena AL, Regan L. Ligand Binding by TPR Domains. Protein Sci. 2006;15:1193–1198. doi: 10.1110/ps.062092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cliff MJ, Williams MA, Brooke-Smith J, Barford D, Ladbury JE. Molecular Recognition Via Coupled Folding and Binding in a TPR Domain. J Mol Biol. 2005;346:717–732. doi: 10.1016/j.jmb.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Chan DC. Structural Basis for Recruitment of Mitochondrial Fission Complexes by Fis1. Proc Natl Acad Sci U S A. 2007;104:18526–18530. doi: 10.1073/pnas.0706441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naylor K, Ingerman E, Okreglak V, Marino M, Hinshaw JE, Nunnari J. Mdv1 Interacts with Assembled Dnm1 to Promote Mitochondrial Division. J Biol Chem. 2006;281:2177–2183. doi: 10.1074/jbc.M507943200. [DOI] [PubMed] [Google Scholar]
- 37.Wells RC, Picton LK, Williams SC, Tan FJ, Hill RB. Direct Binding of the Dynamin-like GTPase, Dnm1, to Mitochondrial Dynamics Protein Fis1 is Negatively Regulated by the Fis1 N-Terminal Arm. J Biol Chem. 2007;282:33769–33775. doi: 10.1074/jbc.M700807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor Laboratory Press; Plainview, NY: 2001. [Google Scholar]
- 39.Plotto M, Saudek V, Sklenar V. Gradient Tailored Excitation for Single-Quantum NMR Spectroscopy of Aqueous Solutions. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 40.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A Multidimensional Spectral Processing System Based on UNIX Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 41.Johnson BA, Blevins RA. NMR View: A Computer Program for the Visualization and Analysis of NMR Data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 42.Johnson BA. using NMR View to Visualize and Analyze the NMR Spectra of Macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 43.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ. Protein NMR Spectroscopy. Academic Press; San Diego: 1996. [Google Scholar]
- 44.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. Comparison of the Backbone Dynamics of a Folded and an Unfolded SH3 Domain Existing in Equilibrium in Aqueous Buffer. Biochemistry. 1995;34:868–878. doi: 10.1021/bi00003a021. [DOI] [PubMed] [Google Scholar]
- 45.Skelton NJ, Palmer AG, Akke M, Kordel J, Rance M, Chazin WJ. Practical Aspects of Two-Dimensional Proton-Detected 15N Spin Relaxation Measurements. J Magn Reson, Ser B. 1993;102:253–264. [Google Scholar]
- 46.Palmer AG. CurveFit: Programs for Linear and Non-Linear Least Squares Fitting. 2003 http://www.palmer.hs.columbia.edu/software/curvefit.html.
- 47.Palmer AG, III, Rance M, Wright PE. Intramolecular Motions of a Zinc Finger DNA-Binding Domain from Xfin Characterized by Proton-Detected Natural Abundance 13C Heteronuclear NMR Spectroscopy. J Am Chem Soc. 1991;113:4371–4379. [Google Scholar]
- 48.Hill RB, Bracken C, DeGrado WF, Palmer AG., III Molecular Motions and Protein Folding: Characterization of the Backbone Dynamics and Folding Equilibrium of α2D using 13C NMR Spin Relaxation. J Am Chem Soc. 2000;122:11610–11619. [Google Scholar]
- 49.Cole R, Loria JP. FAST-Modelfree: A Program for Rapid Automated Analysis of Solution NMR Spin-Relaxation Data. J Biomol NMR. 2003;26:203–213. doi: 10.1023/a:1023808801134. [DOI] [PubMed] [Google Scholar]
- 50.Palmer AG. III ModelFree 401. http://biochemistry.hs.columbia.edu/labs/palmer/software/modelfree.html.
- 51.Lyons TA, Ratnaswamy G, Pochapsky TC. Redox-Dependent Dynamics of Putidaredoxin Characterized by Amide Proton Exchange. Protein Sci. 1996;5:627–639. doi: 10.1002/pro.5560050407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spera S, Ikura M, Bax A. Measurement of the Exchange Rates of Rapidly Exchanging Amide Protons: Application to the Study of Calmodulin and its Complex with a Myosin Light Chain Kinase Fragment. J Biomol NMR. 1991;1:155–165. doi: 10.1007/BF01877227. [DOI] [PubMed] [Google Scholar]
- 53.Ottiger M, Delaglio F, Bax A. Measurement of J and Dipolar Couplings from Simplified Two-Dimensional NMR Spectra. J Magn Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 54.Ruan K, Briggman KB, Tolman JR. De Novo Determination of Internuclear Vector Orientations from Residual Dipolar Couplings Measured in Three Independent Alignment Media. J Biomol NMR. 2008;41:61–76. doi: 10.1007/s10858-008-9240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lepre CA, Moore JM. Microdrop Screening: A Rapid Method to Optimize Solvent Conditions for NMR Spectroscopy of Proteins. J Biomol NMR. 1998;12:493–499. doi: 10.1023/a:1008353000679. [DOI] [PubMed] [Google Scholar]
- 56.Santoro MM, Bolen DW. Unfolding Free Energy Changes Determined by the Linear Extrapolation Method. 1 Unfolding of Phenylmethanesulfonyl α-Chymotrypsin using Different Denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 57.Tolman JR, Al-Hashimi HM. NMR Studies of Biomolecular Dynamics and Structural Plasticity using Residual Dipolar Couplings. In: Webb GA, editor. Annual Reports on NMR Spectroscopy. Academic Press; New York: 2003. pp. 105–166. [Google Scholar]
- 58.Lipari G, Szabo A. Model-Free Approach to the Interpretation of Nuclear Magnetic-Resonance Relaxation in Macromolecules. 1 Theory and Range of Validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 59.Palmer AG. III Diffusion Tensor Analysis using the Quadratic Approach. http://biochemistry.hs.columbia.edu/labs/palmer/software/quadric.html.
- 60.Grathwohl C, Wuthrich K. NMR studies of the Rates of Proline Cis-Trans Isomerization in Oligopeptides. Biopolymers. 1981;20:2623–2633. [Google Scholar]
- 61.Tjandra N, Suzuki M, Chang SL. Refinement of Protein Structure Against Non-Redundant Carbonyl 13C NMR Relaxation. J Biomol NMR. 2007;38:243–253. doi: 10.1007/s10858-007-9165-7. [DOI] [PubMed] [Google Scholar]
- 62.Aghazadeh B, Lowry WE, Huang XY, Rosen MK. Structural Basis for Relief of Autoinhibition of the Db1 Homology Domain of Proto-Oncogene Vav by Tyrosine Phosphorylation. Cell. 2000;102:625–633. doi: 10.1016/s0092-8674(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 63.Amarasinghe GK, Rosen MK. Acidic Region Tyrosines Provide Access Points for Allosteric Activation of the Autoinhibited Vav1 Db1 Homology Domain. Biochemistry. 2005;44:15257–15268. doi: 10.1021/bi051126h. [DOI] [PubMed] [Google Scholar]
- 64.Li P, Martins IR, Amarasinghe GK, Rosen MK. Internal Dynamics Control Activation and Activity of the Autoinhibited Vav DH Domain. Nat Struct Mol Biol. 2008;15:613–618. doi: 10.1038/nsmb.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frauenfelder H, Parak F, Young RD. Conformational Substates in Proteins. Annu Rev Biophys Biophys Chem. 1988;17:451–479. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- 66.Frauenfelder H, Sligar SG, Wolynes PG. The Energy Landscapes and Motions of Proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 67.Hilser VJ, Dowdy D, Oas TG, Freire E. The Structural Distribution of Cooperative Interactions in Proteins: Analysis of the Native State Ensemble. Proc Natl Acad Sci U S A. 1998;95:9903–9908. doi: 10.1073/pnas.95.17.9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sadqi M, Casares S, Abril MA, Lopez-Mayorga O, Conejero-Lara F, Freire E. The Native State Conformational Ensemble of the SH3 Domain from α-Spectrin. Biochemistry. 1999;38:8899–8906. doi: 10.1021/bi990413g. [DOI] [PubMed] [Google Scholar]
- 69.Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D. Intrinsic Dynamics of an Enzyme Underlies Catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 70.Whitten ST, Garcia-Moreno EB, Hilser VJ. Local Conformational Fluctuations can Modulate the Coupling between Proton Binding and Global Structural Transitions in Proteins. Proc Natl Acad Sci USA. 2005;102:4282–4287. doi: 10.1073/pnas.0407499102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fitch CA, Whitten ST, Hilser VJ, Garcia-Moreno EB. Molecular Mechanisms of pH-Driven Conformational Transitions of Proteins: Insights from Continuum Electrostatics Calculations of Acid Unfolding. Proteins. 2006;63:113–126. doi: 10.1002/prot.20797. [DOI] [PubMed] [Google Scholar]
- 72.Popovych N, Sun S, Ebright RH, Kalodimos CG. Dynamically Driven Protein Allostery. Nat Struct Mol Biol. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boehr DD, McElheny D, Dyson HJ, Wright PE. The Dynamic Energy Landscape of Dihydrofolate Reductase Catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 74.Casares S, Sadqi M, López-Mayorga O, Martínez JC, Conejero-Lara F. Structural cooperativity in the SH3 domain studied by site-directed mutagenesis and amide hydrogen exchange. FEBS Lett. 2003;539:125–130. doi: 10.1016/s0014-5793(03)00212-6. [DOI] [PubMed] [Google Scholar]
- 75.Vallurupalli P, Hansen DF, Kay LE. Structures of Invisible, Excited Protein States by Relaxation Dispersion NMR Spectroscopy. Proc Natl Acad Sci USA. 2008;105:11766–11771. doi: 10.1073/pnas.0804221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishima R, Torchia DA. Estimating the Time Scale of Chemical Exchange of Proteins from Measurements of Transverse Relaxation Rates in Solution. J Biomol NMR. 1999;14:369–372. doi: 10.1023/a:1008324025406. [DOI] [PubMed] [Google Scholar]
- 77.Palmer AG, III, Kroenke CD, Loria JP. Nuclear Magnetic Resonance Methods for Quantifying Microsecond-to-Millisecond Motions in Biological Macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 78.Korzhnev DM, Kay LE. Probing Invisible, Low-Populated States of Protein Molecules by Relaxation Dispersion NMR Spectroscopy: An Application to Protein Folding. Acc Chem Res. 2008;41:442–451. doi: 10.1021/ar700189y. [DOI] [PubMed] [Google Scholar]
- 79.Thuduppathy GR, Hill RB. Applications of NMR Spin Relaxation Methods for Measuring Biological Motions. Methods Enzymol. 2004;384:243–264. doi: 10.1016/S0076-6879(04)84015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loria JP, Berlow RB, Watt ED. Characterization of Enzyme Motions by Solution NMR Relaxation Dispersion. Acc Chem Res. 2008;41:214–221. doi: 10.1021/ar700132n. [DOI] [PubMed] [Google Scholar]
- 81.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 82.Campbell RL. http://pldserver1.biochem.queensu.ca/~rlc/work/pymol/
- 83.Lipari G, Szabo A. Model-Free Approach to the Interpretation of Nuclear Magnetic-Resonance Relaxation in Macromolecules. 2 Analysis of Experimental Results. J Am Chem Soc. 1982;104:4559–4570. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.