Abstract
Replication protein A (RPA), a highly conserved single-stranded DNA-binding protein in eukaryotes, is a stable complex comprising three subunits termed RPA1, RPA2, and RPA3. RPA is required for multiple processes in DNA metabolism such as replication, repair, and homologous recombination in yeast (Saccharomyces cerevisiae) and human. Most eukaryotic organisms, including fungi, insects, and vertebrates, have only a single RPA gene that encodes each RPA subunit. Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), however, possess multiple copies of an RPA gene. Rice has three paralogs each of RPA1 and RPA2, and one for RPA3. Previous studies have established their biochemical interactions in vitro and in vivo, but little is known about their exact function in rice. We examined the function of OsRPA1a in rice using a T-DNA insertional mutant. The osrpa1a mutants had a normal phenotype during vegetative growth but were sterile at the reproductive stage. Cytological examination confirmed that no embryo sac formed in female meiocytes and that abnormal chromosomal fragmentation occurred in male meiocytes after anaphase I. Compared with wild type, the osrpa1a mutant showed no visible defects in mitosis and chromosome pairing and synapsis during meiosis. In addition, the osrpa1a mutant was hypersensitive to ultraviolet-C irradiation and the DNA-damaging agents mitomycin C and methyl methanesulfonate. Thus, our data suggest that OsRPA1a plays an essential role in DNA repair but may not participate in, or at least is dispensable for, DNA replication and homologous recombination in rice.
In a population of organisms, it is crucial to maintain the integrity of genome among individuals as well as shuffle genetic information at the population level. To maintain such genetic integrity, cells have evolved elaborate mechanisms such as base excision repair (BER; Hegde et al., 2008), nucleotide excision repair (NER; Shuck et al., 2008), homologous recombination (HR; Li and Heyer, 2008) repair, and nonhomologous end joining (Weterings and Chen, 2008) pathways to repair diverse types of DNA damage. To allow for variation, however, organisms utilize meiosis to shuffle genetic material so as to increase genetic diversity in populations and in the species.
DNA double-strand break (DSB) repair is particularly important in maintaining the integrity of genome among individuals and shuffling genetic information among population, because DSBs are generated not only in meiotic cells but also from the action of certain endogenous or exogenous DNA-damaging agents and during repair of other kinds of DNA lesions by NER or BER (West et al., 2004; Bleuyard et al., 2006). The past decade has witnessed an explosion in understanding of this complex process by using yeast (Saccharomyces cerevisiae) as a model organism (Aylon and Kupiec, 2004). Cells can repair DSBs by the relatively inaccurate process of rejoining the two broken ends directly (i.e. nonhomologous end joining) or much more accurately by HR (Bleuyard et al., 2006; Wyman and Kanaar, 2006). These two pathways appear to compete for DSBs, but the balance between them differs widely among species, between different cell types of a single species, and during different cell cycle phases of a single cell type (Shrivastav et al., 2008). According to the current general model for meiotic DSB repair (Bishop and Zickler, 2004; Ma, 2006; San Filippo et al., 2008), when DSBs occur the MRN complex (composed of Mre11, Rad50, and NBS1) resects the DSBs to generate 5′→3′ single-stranded DNA (ssDNA) ends. Subsequently, the replication protein A (RPA) protein complex binds to the ssDNA ends to protect them from attack by endogenous exonucleases; then, in concert with catalysis by Rad52, Rad55, and Rad57, the recombinase Rad51 displaces RPA, resulting in the generation of a Rad51 nucleoprotein filament that in turn catalyzes the search and invasion into the recombination partner with the help of proteins belonging to the RAD52 epistasis group to form a D loop that accompanies DNA synthesis. Thereafter, at least two competing mechanisms may come into play. One is the DSB repair pathway, in which the capture of the second DSB end and additional DNA synthesis result in an intermediate that harbors two Holliday junctions. The subsequent resolution of Holliday junctions results in the formation of crossovers. Alternatively, in the synthesis-dependent strand annealing pathway, the D loop dissociates and the invading single strand with newly synthesized DNA reanneals with the other DSB end, followed by gap-filling DNA synthesis and ligation, forming only noncrossover products (Ma, 2006; San Filippo et al., 2008).
RPA is comprised of three subunits of RPA1, 2, and 3, alternatively termed as RPA70, 32, and 14, respectively, according to their apparent Mrs (Wold, 1997; Iftode et al., 1999). RPA is an essential protein in various DNA metabolism pathways such as DNA replication, repair, and HR (Wold, 1997; Iftode et al., 1999). In these pathways, the most basic function of RPA is binding to ssDNA to protect it from exonucleases, and its general roles in DNA metabolism depend on its interactions with other proteins in various pathways (Wold, 1997; Iftode et al., 1999). For example, in human NER pathway, RPA binds to damaged DNA and interacts with xeroderma pigmentosum damage-recognition protein, XPA, in the damage recognition step, and then the endonucleases XPG and ERCC1/XPF are recruited to the RPA-XPA-damaged DNA complex in the excision step (He et al., 1995). Interactions of RPA with those proteins are critical in this process (Wold, 1997; Iftode et al., 1999). A great deal of protein dynamics research has indicated that the interactions between RPA and other DNA-metabolism proteins are choreographed on the ssDNA to recruit the required protein present at the proper time (Fanning et al., 2006).
Human, animals, and fungi have single copy for each subunit of RPA (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), however, have multiple genes for most RPA subunits (Ishibashi et al., 2006; Shultz et al., 2007). Most of them have not unveiled exact function up to now. To elucidate the molecular basis of meiosis in rice, we performed a large-scale screen for sterile mutants using our T-DNA insertion mutant library (Wu et al., 2003). Previously, we reported the cloning of OsPAIR3, a novel gene required for homologous chromosome pairing and synapsis in rice (Yuan et al., 2009). Here we report the characterization of another sterile mutant with a T-DNA insertion in OsRPA1a. Our results indicate that OsRPA1a is essential for DNA repair but may play redundant roles in DNA replication and recombination in rice.
RESULTS
Identification of the osrpa1a Mutant
The osrpa1a mutant was identified by screening a rice mutant library (Wu et al., 2003) for sterile mutants. The osrpa1a mutant was not distinguishable from the wild type in plant morphology, but it was sterile (Fig. 1, A–C). However, partial mutants occasionally produced one or two seeds.
Figure 1.
Phenotypic characterization of the osrpa1a mutant. A, A wild-type plant (left) and osrpa1a mutant (right) at maturity. B and C, Spikelets from wild-type (B) and osrpa1a mutant (C) plants are morphologically identical. D and E, Viability of mature pollen grains of wild type (D) and the osrpa1a mutant (E) as assessed by I2-KI staining. Bars = 100 μm. F and G, Structure of mature embryo sacs from wild type (F) and osrpa1a mutant (G). AN, Antipodals; PN, polar nucleus; SY, synergids. Bars = 50 μm. [See online article for color version of this figure.]
To characterize the osrpa1a sterility phenotype, we examined male and female gamete fertility in mutant and wild-type plant. Staining of mutant pollen with iodium potassium iodide solution, which indicates pollen viability, showed that only approximately 54.6% of the pollen could be stained (n = 1,051, Fig. 1E). The embryo sacs from the mutant were empty (n > 100, Fig. 1G), whereas wild-type sacs had antipodals, polar nuclei, and synergids (Fig. 1F). Thus, we speculated that the osrpa1a mutant was partly male sterile and completely female sterile. Reciprocal crosses between osrpa1a and wild-type plants confirmed this speculation. No seeds were obtained from 253 spikelets in three independent panicles using osrpa1a as maternal recipients. When using a wild-type plant as a maternal recipient pollinated with osrpa1a pollen, the seed setting rate was 31.6% and 34.0% in two independent panicles.
Isolation and Characterization of the OsRPA1a Gene
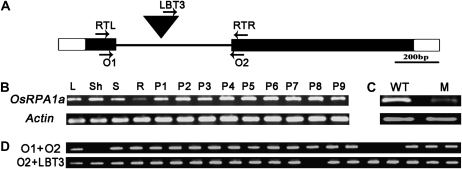
A BLASTn search of the T-DNA flanking sequence isolated by thermal asymmetric interlaced PCR (Zhang et al., 2007) against the rice genomic sequence database (http://rice.plantbiology.msu.edu) showed that the T-DNA was inserted into the intron of OsRPA1a (LOC_Os02g53680; Fig. 2A), which encodes RPA1a, one of the isoforms of RPA1 subunit in rice (Ishibashi et al., 2006).
Figure 2.
Analysis of T-DNA tagging of OsRPA1a and the expression of OsRPA1a. A, Structure of OsRPA1a and the position of the T-DNA insertion (inverted filled triangle). The intron (line), open reading frame (black box), and 5′ and 3′ untranslated regions (white boxes) are indicated. The positions of the primers (RTL and RTR used in B and C; O1, O2, and LBT3 used in D) are indicated by arrows. B, Spatial and temporal expression pattern of OsRPA1a in leaf (L), sheath (Sh), stem (S), root (R) from a booting plant and panicles at various developmental stages according to their length (P1–P9). P1: approximately 0.5 cm; P2: approximately 1.0 cm; P3: approximately 2.0 cm; P4: approximately 3.5 cm; P5: approximately 4.5 cm; P6: approximately 11 cm; P7: approximately 16.5 cm; P8: approximately 19 cm; P9: approximately 22 cm. C, RT-PCR analysis of OsRPA1a expression in panicles from wild type (WT) and osrpa1a mutant (M). Actin is used as the reference for the mRNA level in B and C. D, Genotyping of the OsRPA1a T-DNA-tagging progeny.
We examined the expression of OsRPA1a transcripts in various tissues by reverse transcription (RT)-PCR. OsRPA1a expression was high in all stages of the developing panicles, and moderately high in leaf, sheath, and stem, but low in root (Fig. 2B). We also investigated the OsRPA1a expression pattern by searching the chip database of the Collection of Rice Expression Profiles (http://crep.ncpgr.cn/) and RiceGE (http://signal.salk.edu/cgi-bin/RiceGE?JOB=EXPR&TYPE). The chip data were consistent with our RT-PCR results and showed that OsRPA1a was expressed in seeds, callus, endosperm, shoot apical meristem, stigma, ovary, hull, and so on (data not shown). RT-PCR showed that OsRPA1a expression was dramatically lower in the mutant panicle compared to wild type (Fig. 2C).
PCR genotyping analysis of the T1 generation showed that all three sterile plants were homozygous for the T-DNA insertion in OsRPA1a and that the remaining 17 fertile plants were heterozygous or homozygous for OsRPA1a (Fig. 2D). In the T2 generation, all 83 homozygous plants from the progeny of three independent heterozygotes were sterile, whereas 197 heterozygous plants and 93 homozygous wild-type plants showed normal fertility. These results indicated that the sterile phenotype was cosegregated with the T-DNA insertion.
Genetic Complementation and Examination of OsRPA1a by RNA Interference
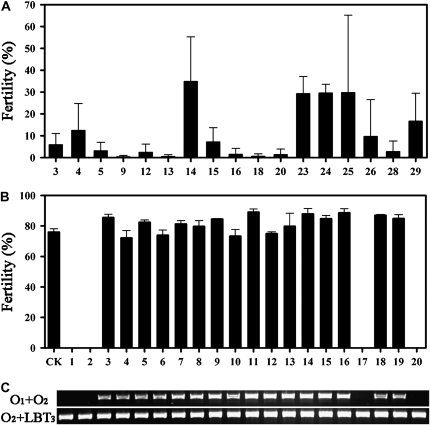
To confirm that osrpa1a corresponded to the sterile phenotype, a genetic complementation experiment was performed. Construct pC2301-RPA1a, containing the entire OsRPA1a open reading frame, 2.7-kb upstream sequences, and a 3.6-kb downstream region, and an empty pCAMBIA2301 vector were introduced into a osrpa1a mutant background by Agrobacterium tumefaciens-mediated transformation (Wu et al., 2003). PC2301-RPA1a restored fertility in 17 of 26 independent transgenic plants (Fig. 3A), whereas a sterility phenotype similar to that of osrpa1a was observed in all four plants regenerated with empty vector and six negative transgenic plants with pC2301-RPA1a. In the progeny of the rescued plants, segregation of OsRPA1a coincided with normal fertility (Fig. 3, B and C). We also generated transgenic plants in which OsRPA1a expression was suppressed by RNA interference and showed that the sterile phenotype was attributable to reduced OsRPA1a expression (Supplemental Fig. S1). Taken together, these results confirmed that the sterility of mutants was caused by the loss of OsRPA1a function.
Figure 3.
Transgenic complementation of the osrpa1a mutants. A, Fertility of the 17 complemented osrpa1a lines of the T0 generation. B, Fertility of the progenies of a single-copy insertion complemented osrpa1a line of the T1 generation. A wild-type plant was used as control (CK). The average fertility (mean ± se) was based on three panicles selected randomly from each plant in A and B. C, Genotyping of the plants used to obtain the data shown in B.
Female Gametophyte Development in osrpa1a Is Disrupted at the Megasporocyte Meiosis Stage
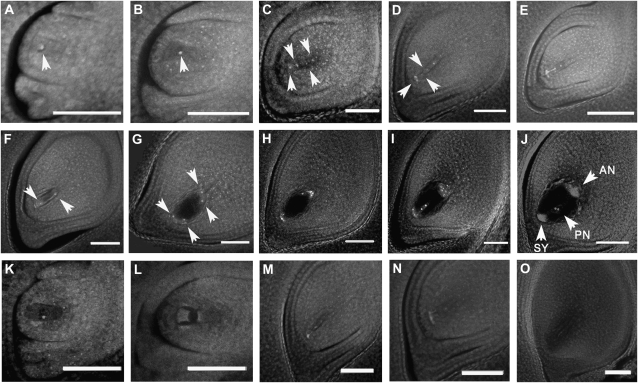
To characterize the cytological course of the osrpa1a mutation, we monitored the formation and development of the embryo sac in osrpa1a mutants in comparison with wild type. Embryo sac formation and development in wild type was divided into eight stages as described (Liu et al., 1997; Yuan et al., 2009). The wild-type archespore was generated from one of the subepidermal cells in the nucellus at the archesporial cell formation stage (Fig. 4A) and developed into the megaspore mother cell at the megasporocyte formation stage (Fig. 4B). The megaspore mother cell then underwent meiotic division to form a linear tetrad of megaspores at the megasporocyte meiosis stage (Fig. 4C). After meiosis, the three megaspores nearest the micropyle degenerated, and the chalazal megaspore was preserved as the functional megaspore at the functional megaspore formation stage (Fig. 4D). The functional megaspore elongated and enlarged into the uninucleate embryo sac during the mononucleate embryo sac formation stage (Fig. 4E), then initiated three rounds of mitotic division to produce the two- (Fig. 4F), four- (Fig. 4G), and eight-nucleate embryo sac (Fig. 4H) at the embryo sac mitosis stage. Subsequent migration and cellularization of the nuclei occurred during the eight-nucleate embryo sac developing stage (Fig. 4I), resulting in a mature embryo sac with polar nuclei, antipodals, and synergids (Fig. 4J). Compared with the wild type, the megaspore mother cell formed normally prior to meiosis in osrpa1a mutants as it did in wild type (Fig. 4K). The clearly visible nuclei and normal integuments indicated normal initiation of the ovule in osrpa1a mutants. At megasporocyte meiosis stage, however, only one or two or three malformed nuclei were seen in osrpa1a mutants compared with the clear tetrad observed in wild type (Fig. 4, L and M). The aberrant nuclei of the mutants indicated that the nuclei might have begun to degenerate during this stage. Subsequently, only degenerated footprints of nuclei remained in the embryo sac until maturity (Fig. 4, N and O). These observations suggested that the osrpa1a mutation may interfere in normal meiosis in the megasporocyte, resulting in degeneration of abnormal dyad and tetrad with subsequent defects in embryo sac formation.
Figure 4.
Formation and development of the embryo sac in wild-type (A–J) and osrpa1a mutants (K–O). A, Archesporial cell (arrowhead) formation stage. B, Megasporocyte (arrowhead) formation stage. C, Megasporocyte meiosis stage. The megasporocyte undergoes two meiotic nuclear divisions to form a linear tetrad of megaspores (arrowheads). D, Functional megaspore formation stage. Three megaspores nearest the micropyle (arrowheads) degenerate to preserve the chalazal megaspore as the functional megaspore. E, Mononucleate embryo sac formation stage. F to H, Embryo sac mitosis stage. The functional megaspore undergoes three mitotic divisions to yield a two-nucleate (arrowheads) embryo sac (F), four-nucleate (arrowheads) embryo sac (G), and eight-nucleate embryo sac (H), respectively. I, Eight-nucleate embryo sac development stage. J, Mature embryo sac stage. The polar nucleus (PN), synergids (SY), and antipodals (AN) are indicated by arrowheads. K, Megasporocyte formation stage of osrpa1a mutant. In this stage, the megasporocyte forms normally as in wild type. L and M, Megasporocyte meiosis stage. The megasporocyte is likely to undergo its first (L) and second (M) meiotic nuclear division, but the normal dyad and linear tetrad of megaspores do not form. In addition, the dyad and tetrad of megaspores begin to degenerate. N, Functional megaspore formation stage. All of the abnormal megaspores degenerate, and no functional megaspore is formed. O, Mature embryo sac stage. The mature embryo sac is almost empty except for a small footprint of the degenerated nuclei. Bars = 50 μm.
Meiosis in Pollen Mother Cells Is Affected in osrpa1a Mutants
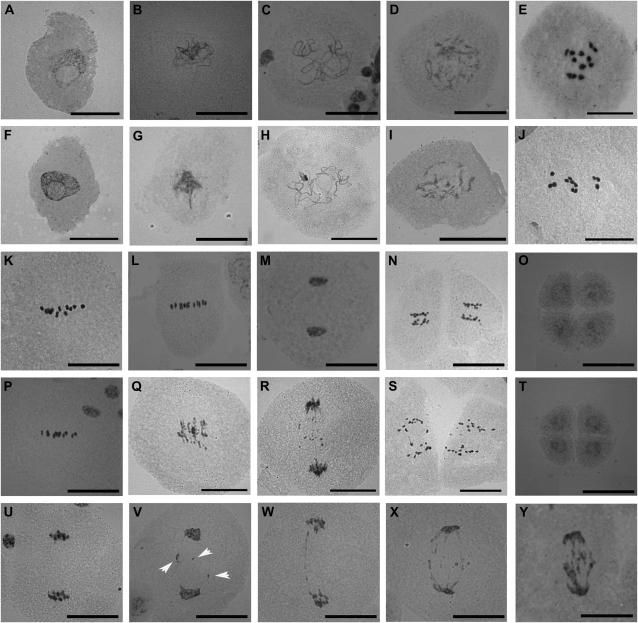
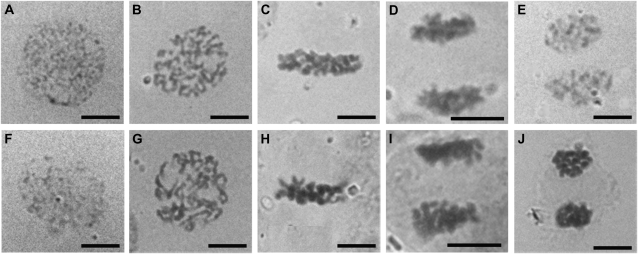
To further investigate the possibility that osrpa1a mutants were defective in meiosis, we examined the meiotic process by staining the pollen mother cells (PMCs) of osrpa1a mutants with carbol fuchsin and compared the results with wild type. In wild type, the chromosome condensed into a thin thread-like structure in the leptotene stage (Fig. 5A), began to pair/juxtapose and synapse at the zygotene stage (Fig. 5B), and finished synapsis to yield a thick thread at the pachytene stage (Fig. 5C). Thereafter, the chromosomes continued condensing, and homologs separated at the diplotene stage except in regions where chiasmata occurred (Fig. 5D), followed by further condensation leading to the formation of 12 bivalents (Fig. 5E). The bivalents aligned at the equatorial plane at metaphase I (Fig. 5K), separated at the metaphase I/anaphase I transition (Fig. 5L), and migrated to opposite poles of the cell at telophase I (Fig. 5M). The second meiotic division (Fig. 5N) then yielded four haploid nuclei (Fig. 5O).
Figure 5.
Cytological analysis of PMC meiosis in wild type (A–E, K–O) and osrpa1a mutants (F–J, P–Y). A and F, Leptotene. B and G, Zygotene. C and H, Pachytene. D and I, Diplotene. E and J, Diakinesis. K and P, Metaphase I. L and Q, Metaphase I/anaphase I transition. M and R, Telophase I. N and S, Anaphase II. O and T, Four newly formed nuclei (pollen tetrad). U to Y, PMCs at telophase I show different levels of chromosome fragmentation (the unconspicuous chromosome fragmentation in V are indicated by arrowheads). Bars = 30 μm.
Chromosome dynamics in osrpa1a mutants were phenotypically indistinguishable from wild type in prophase I (Fig. 5, F–J) and metaphase I (Fig. 5P). The chromosomes condensed, paired/juxtaposed, and pairing correctly to yield 12 bivalents (Fig. 5J) as in wild type. However, osrpa1a exhibited clearly chromosome fragmentation at the metaphase I/anaphase I transition when homologous chromosomes begin to separate from each other (Fig. 5Q), and this abnormality persisted to the end of meiosis (Fig. 5, R and S), suggesting the existence of unrepaired DNA breakage in these PMCs. Although the number of chromosome fragments was difficult to quantify, visual estimation indicated that the degree of fragmentation varied greatly (Fig. 5, U–Y). About 11.7% (n = 222) of osrpa1a meiocytes showed no fragmentation, similar to wild type at telophase I (Fig. 5U), whereas the remaining osrpa1a samples showed varying degrees of fragmentation at telophase I (Fig. 5, V–Y), indicating different levels of unrepaired DNA breaks among the cells examined. Despite chromosome fragmentation during homologous chromosome and sister chromatid segregation in osrpa1a (Fig. 5, Q–S and V–Y), cytokinesis of meiocytes (n > 200) occurred normally and resulted in only dyads at telophase I (Fig. 5, R and V–Y) and tetrads at the end of meiosis (Fig. 5T) rather than irregular triads or polyads. This cytological analysis suggested that OsRPA1a is essential for ensuring chromosome integrity but is dispensable for normal cytokinesis during meiosis.
The osrpa1a Mutants Are Hypersensitive to DNA Mutagens
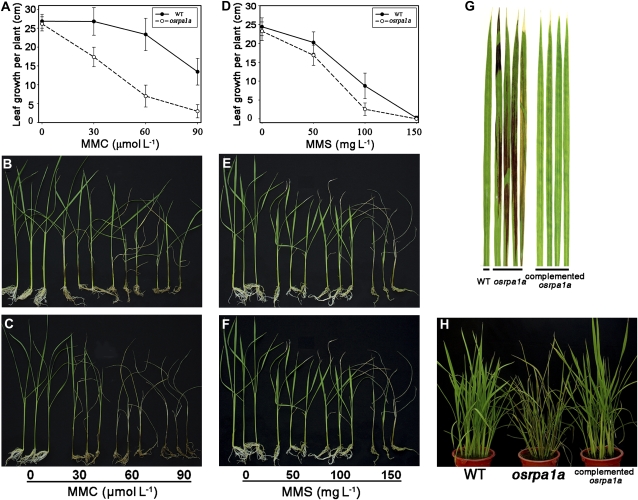
Many mutants of genes related to DNA metabolism are hypersensitive to DNA mutagens such as mitomycin C (MMC), methyl methanesulfonate (MMS), and UV irradiation (Garcia et al., 2003; Bleuyard and White, 2004; Bleuyard et al., 2005; Waterworth et al., 2007). In yeast and human, RPA is involved extensively in various DNA repair pathways (Wold, 1997; Iftode et al., 1999). Considering that OsRPA1a was expressed not only in panicles but also in vegetative organs in rice (Fig. 2B), we speculated that OsRPA1a may also be involved in DNA repair pathways in vegetative tissues. To address this possibility, we tested the effect of MMC, MMS, and UV-C irradiation on seedling growth in progenies from a T-DNA insertion heterozygous plant. When 15-d-old seedlings were grown in culture solutions containing various concentrations of MMC for 20 d, the osrpa1a mutant plants grew significantly slower than wild type (P < 0.01; Fig. 6, A–C). In addition, the mutant seedlings could not survive at 90 μmol L−1 MMC (Fig. 6C) whereas none of the wild type died at this concentration (Fig. 6B). No difference was observed between osrpa1a and wild-type seedlings grown in culture solution without MMC (P = 0.35; Fig. 6, A–C). Similar although milder effects were also observed when mutant plants were exposed to MMS in comparison with wild-type plants (Fig. 6, D–F).
Figure 6.
osrpa1a mutants are hypersensitive to MMC, MMS, and UV-C irradiation. A and D, Leaf growth per plant of wild-type (WT) and osrpa1a mutant plants during MMC (A) or MMS (D) treatment. The average leaf growth (mean ± se) was based on 12 independent seedlings. B and E, Phenotypes of wild-type growth in culture solution plus MMC (B) or MMS (E) at different concentrations. C and F, Hypersensitivity of osrpa1a mutants to MMC (C) and MMS (F). G and H, Hypersensitivity of osrpa1a mutants to UV-C irradiation. [See online article for color version of this figure.]
We also exposed osrpa1a and wild-type plants to UV-C irradiation. Upon exposure for 3 h, the mutants showed necrotic lesions on leaves 2 d later (data not shown), and 6 h of UV-C irradiation resulted in more extensive necrosis (Fig. 6, G and H). Wild-type plants exhibited no necrosis when irradiated with UV-C for up to 6 h. The complemented osrpa1a plants were also examined for their sensitivity to MMS and UV-C irradiation. For these plants, the results were similar to those for wild type (Fig. 6, G and H; data not shown). These data clearly demonstrated that plants lacking OsRPA1a were hypersensitive to DNA mutagens, indicating that OsRPA1a played an essential role in DNA repair pathways in somatic cells.
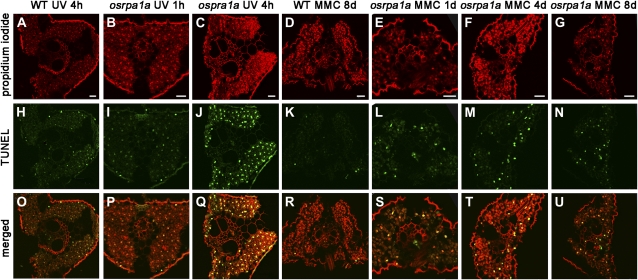
To examine if the necrotic lesions (Fig. 6G) and the slow growth (Fig. 6, A–C) of osrpa1a under DNA mutagens were due to the unrepaired DNA damage, terminal deoxyribonucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay was carried out in the leaves of osrpa1a and wild type. It showed that the TUNEL signal could be detected after 4 h of UV-C irradiation in wild type (Fig. 7H). However, clear TUNEL signal appeared in osrpa1a leaves after 1 h of UV-C irradiation (Fig. 7I) and became stronger after 4 h of irradiation (Fig. 7J). When 20-d-old seedlings were grown in culture solutions containing 30 μmol L−1 MMC, the TUNEL-positive nuclei were detected in osrpa1a leaves 1 d later (Fig. 7L) and lasted to 8 d (Fig. 7, M and N), while little positive TUNEL signal was found in wild-type leaves until 8 d (Fig. 7K). These observations demonstrated that treatment with UV-C or MMC caused more DNA damage in osrpa1a than wild type, indicating that OsRPA1a plays an essential role in DNA repair pathway in somatic cells.
Figure 7.
Detection of nuclear DNA fragmentation in leaves by in situ TUNEL assay. A to G, Cross sections stained with propidium iodide. H to N, TUNEL results of the corresponding sections of A to G. O to U, Merged images of propidium iodide and TUNEL signals. Bars = 20 μm. WT, Wild type. [See online article for color version of this figure.]
Mitosis Is Unaffected in osrpa1a Mutants
Human RPA interacts with several proteins involved in DNA replication pathway; these proteins include DNA polymerase α, simian virus 40 large tumor antigen, and Epstein-Barr virus EBNA-1 (Wold, 1997; Iftode et al., 1999). Mutation in Arabidopsis MRE11, a gene involved in meiotic DNA repair, results in mitotic chromosomal fragmentation at anaphase (Puizina et al., 2004). RT-PCR assays and the expression pattern obtained from chip databases revealed that OsRPA1a was expressed universally, with rather high expression in proliferating tissues such as callus and shoot apical meristem (http://crep.ncpgr.cn/; http://signal.salk.edu/cgi-bin/RiceGE?JOB=EXPR&TYPE). We hypothesized that OsRPA1a might also be involved in DNA replication and that OsRPA1a mutations may impact mitosis in rice. Because there was no distinguishable difference in phenotype between osrpa1a and wild type at the vegetative stage, we further explored whether chromosome behavior was abnormal during mitosis by examining chromosome behavior in anther wall cells from osrpa1a and wild-type plants. The osrpa1a mutants displayed a mitotic phenotype identical to that of wild type (Fig. 8). The nucleus migrated to the center of cells at preprophase in both wild-type and mutant plants (Fig. 8, A and F). Subsequent chromatin condensation resulted in normal chromosome formation at prophase (Fig. 8, B and G), followed by further condensation and alignment at the equatorial plane at metaphase (Fig. 8, C and H). Thereafter, sister chromatids were pulled apart to opposite ends of the cell at anaphase (Fig. 8, D and I), each set of separated sister chromatids unfolded back into chromatin at telophase (Fig. 8, E and J), and finally the nuclear envelope formed to yield two new daughter cells. To verify the results from anther wall cells, we monitored mitosis in root tip cells and found no difference between osrpa1a and wild type (data not shown).
Figure 8.
Mitotic chromosome behavior is identical in wild type (A–E) and osrpa1a mutants (F–J). A and F, Preprophase. B and G, Prophase. C and H, Metaphase. D and I, Anaphase. E and J, Telophase. Bars = 5 μm.
DISCUSSION
Fertility in osrpa1a Is Higher in Male Gametes Than Female Gametes
Reciprocal crosses between osrpa1a and wild-type plants confirmed that osrpa1a male gametophytes were partially fertile whereas female gametophytes were completely sterile. Cytological analysis also revealed that the partial male fertility remained (Fig. 1E) and that no functional embryo sac was formed in osrpa1a mutants. We did find that one or two seeds were occasionally produced in osrpa1a plants (fertility <0.1%), indicating that functional embryo sacs formed very infrequently. Our RT-PCR analysis did in fact detect residual OsRPA1a expression in osrpa1a (Fig. 2C), suggesting that low levels of functional OsRPA1a may be sufficient for partial male gametophyte development but only limited embryo sac formations, and indicating differential dependence on OsRPA1a between male and female meiosis. Another explanation is likely that other proteins exhibit partial functional redundancy with OsRPA1a for chromosome repair, resulting in a low level of normal male gametophyte production and partial male fertility. The relatively higher level of male fertility in osrpa1a may reflect different meiotic checkpoint systems required for normal male and female gametogenesis.
Distinctive Chromosome Behavior of osrpa1a Mutants and the Origin of Chromosome Fragmentation in osrpa1a Mutants
In Arabidopsis, quite a large number of mutants with defect in meiotic DNA repair have been identified (Ma, 2006; Mercier and Grelon, 2008). Among these mutants, atrad50, atmre11, atrad51, atrad51c, and atxrcc3, which have defects in the repairing of AtSPO11-induced DSB, exhibited similar meiotic chromosome behaviors like asynaptic homologs and/or chromosome fragmentation (Bleuyard et al., 2004a; Bleuyard and White, 2004; Li et al., 2004, 2005; Puizina et al., 2004). The meiotic chromosome fragmentation could be suppressed by mutation in AtSPO11-1 (Bleuyard et al., 2004b; Li et al., 2004, 2005; Puizina et al., 2004), which provides a direct evidence that the origin of the DNA breaks in these mutants is the unrepaired DSBs generated by AtSPO11. However, the atspo11-1 mutation only partially rescues meiotic chromosome fragmentation in atspo11-1/atatr, atspo11-1/atatm double mutants and the atspo11-1/atatr/atatm triple mutant (Culligan and Britt, 2008). In addition, atspo11-1 cannot prevent chromosome fragmentation in atmei1 and atxir1 (Grelon et al., 2003; Dean et al., 2009). These experiments suggest that DNA breakage in PMCs can also be generated in an AtSPO11-independent manner. Thus, the origin of chromosome fragmentation in osrpa1a still needs to be elucidated by employing the osspo11/osrpa1a double mutant.
The chromosome behavior during meiotic process is distinct in osrpa1a compared to these mutants. Homologous chromosome pairing seemed normal to form bivalents properly at diakinesis (Fig. 5J), but chromosome fragmentation occurred after anaphase I (Fig. 5, Q–S, V–Y). Moreover, the chromosome fragmentations are quite mild in osrpa1a mutant (Fig. 5, U–Y), indicating that whether the generation of DSB is SPO11 dependent or not, DNA damage could be partially repaired in osrpa1a mutants. Given that the osrpa1a mutant used in this research is not a null one (Fig. 2C) and there are multiple RPA copies in rice, we speculate that this DNA damage could be repaired by the residual OsRPA1a or other RPA paralogs in osrpa1a, leading to some unrepaired DNA damage that could not be detected at prophase I in this experiment. The other possible role of OsRPA1a in rice, suggested by the function of coordination of the loading of RAD51/DMC1 from its otholog in yeast and human, may be conferred by another family member in osrpa1a mutants.
OsRPA1a Is Essential for Various DNA Repair Pathways
Homozygous osrpa1a plants are hypersensitive to a wide array of DNA-damaging agents including MMC, MMS, and UV-C irradiation compared with the wild-type plant (Fig. 5). These DNA mutagens can induce different types of DNA lesions. MMS is an alkylating compound that methylates DNA on N7-deoxyguanine and N3-deoxyadenine (Vazquez et al., 2008). In yeast, BER, HR, and DNA damage-tolerance pathways, together with a functional S-phase checkpoint, are essential for cell survival when the DNA template is damaged by MMS (Vazquez et al., 2008). MMC induces interstrand DNA cross-links that can be removed by HR, NER, and translesion synthesis pathways in eukaryotes (Lehoczky et al., 2007). UV irradiation induces DNA lesions such as cyclobutane pyrimidine dimers and pyrimidine (6-4) pyrimidinone dimers (6-4 photoproducts) that can be removed by photoreactivation and NER, BER, mismatch repair, and other DNA repair pathways (Tuteja et al., 2001; Kimura et al., 2004). In Arabidopsis, most of the genes involved in the control of meiotic chromosome dynamics are also required for DNA damage repair in somatic cells (Garcia et al., 2003; Waterworth et al., 2007), but their response spectrums are quite different. The atmre11 mutants are hypersensitive to MMS and x-ray (Bundock and Hooykaas, 2002). The atatm mutants are hypersensitive to MMS and γ-radiation but not to UV-B irradiation (Garcia et al., 2003), whereas atnbs1 mutants have defects in their responses to MMC and MMS but not UV-C and x-rays (Waterworth et al., 2007). The different responses of these mutants indicate that these genes play essential roles in certain DNA repair pathways; moreover, different DNA repair pathways may share some common components, implying that a highly complex network is involved in DNA repair. Thus, the hypersensitivity of the osrpa1a mutants to MMC, MMS, and UV-C irradiation suggests that OsRPA1a is essential for the network, and hence mutations in this gene have a comprehensive effect on DNA repair. This result is consistent with findings that human and yeast RPA is involved in multiple DNA repair pathways (Wold, 1997; Iftode et al., 1999), suggesting that the roles of RPA in DNA repair are likely conserved in eukaryotes.
Functional Diversification of RPA1s in Arabidopsis and Rice
Arabidopsis has five paralogs of RPA1 and two paralogs each of RPA2 and RPA3 (Shultz et al., 2007). The expression patterns and interactions of these genes have not been examined. AtRPA1a is required for interference-sensitive crossover formation but dispensable for meiotic DSB repair (Osman et al., 2009), and the atrpa1b (At5g08020) mutant is indistinguishable from wild type under normal conditions but is hypersensitive to MMS and UV-B (Ishibashi et al., 2005). In addition, mutant plants with one of the AtRPA2 subunits (At2g24490) are smaller than wild type and the transcriptional gene silencing is suppressed in a DNA-methylation-independent manner (Elmayan et al., 2005; Kapoor et al., 2005; Xia et al., 2006), indicating that these individual RPA subunits have different functions in Arabidopsis DNA metabolism.
In rice, there are three paralogs of RPA1 (OsRPA1a, OsRPA1b, and OsRPA1c) and RPA2 (OsRPA2-1, OsRPA2-2, and OsRPA2-3) as well as a single RPA3 (Ishibashi et al., 2006). Yeast two-hybrid assays and pull-down analyses have demonstrated that their specific interactions result in three types of RPA complex: RPA1a-RPA2-2-RPA3 (type A), RPA1b-RPA2-1-RPA3 (type B), and RPA1c-RPA2-3-RPA3 (type C; Ishibashi et al., 2006). Previous studies have shown that OsRPA1a and OsRPA2-1 are expressed highly in proliferating tissues such as suspension cells, root tips, as well as young leaves but weakly in mature leaves (Ishibashi et al., 2001). In this research, RT-PCR analysis indicates that OsRPA1a also have high expression in all stages of the developing panicles (Fig. 2B). The analyses of the osrpa1a mutant showed that OsRPA1a is essential for DNA repair but is dispensable for DNA replication and HR. OsRPA1b is expressed preferentially in proliferating tissues and induces an increase in the intercalary meristem under submergence or treatment with GA. Moreover, OsRPA1b expression precedes that of the DNA replication marker gene histone H3, indicating that it is involved in normal DNA replication and cell proliferation (van der Knaap et al., 1997; Ishibashi et al., 2001). OsRPA1c and OsRPA2-3 expressed specially at panicles and osrpa2-3 mutant exhibited identical chromosome dynamic defects with that of atrpa1a (Y. Chang, Z. Ma, and C. Wu, unpublished data). It seems that each RPA subunit has evolved distinct function(s) between rice and Arabidopsis.
Among the OsRPA1 homologs, OsRPA1a has the highest amino acid sequence identity with AtRPA1a. Moreover, OsRPA1a and AtRPA1a belong to the same phylogenetic clade (Y. Chang, Z. Ma, and C. Wu, unpublished data). Surprisingly, the chromosome defects in osrpa1a and atrpa1a mutants are significantly different. Considering that osrpa2-3 has the same chromosome defects with that of atrpa1a, we deduce that type C (RPA1c-RPA2-3-RPA3) in rice may perform the same function with that of AtRPA1a-containing RPA complex in Arabidopsis and OsRPA1c may be the functional counterpart of AtRPA1a in rice. Meanwhile, there still exists the possibility that OsRPA1a, like AtRPA1a, also participates in coating the D loop and ssDNA tail of the resected second strand from the DSB site to mediate second-end capture through a RAD52-like protein (Osman et al., 2009), and this activity is shared/replaced by another family member in the osrpa1a mutant. Thus, the functional analysis of other subunits of RPA both in rice and Arabidopsis will be a fascinating research in the future.
MATERIALS AND METHODS
Plant Materials
All rice (Oryza sativa) plants used in this study were japonica (O. sativa sp.japonica) cv Zhonghua 11. The osrpa1a T-DNA insertional line was obtained by screening the enhancer trap mutant library (Wu et al., 2003). Plant genotypes were determined by PCR using the T-DNA left border primer LBT3 (5′-ccagtactaaaatccagatcccccgaat-3′) in combination with gene-specific primers on both sides of the insertion: O1 (5′-agcgagtacgtcatcaacga-3′) and O2 (5′-ccgaaacaagaacttccagg-3′). All the plants were cultivated under normal conditions in Wuhan, China (latitude 30.5°N, 15 m above sea level; average daily temperature approximately 28°C).
Vector Construction and Rice Transformation
The transformation recipient for complementary experiment was callus culture that was induced from seeds homozygous for osrpa1a. To generate the complementary vector, a 9.7-kb XbaI-KpnI genomic fragment was isolated from the Nipponbare bacterial artificial chromosome clone OSJNBa0075N09 (kindly provided by R. Wing, University of Arizona) and subcloned into the binary vector pCAMBIA2301, giving pC2301-RPA1a. The empty vector pCAMBIA2301 was used as a negative control.
To generate the OsRPA1a RNAi vector, a 449-bp fragment was amplified with primers RNAi-F (5′-gtgactagtggtaccacgcacttaaggagcgcgag-3′; underlined letters indicate the restriction sites added for subsequent cloning) and RNAi-R (5′-ggggagctcggatccaagctgatacattaagtggcgt-3′) from FL-cDNA clone J033031B02 (http://cdna01.dna.affrc.go.jp/cDNA) and inserted into the pDS1301 vector as described by Chu et al. (2006). The constructed vectors were transferred into Agrobacterium tumefaciens strain EHA105, and rice transformation was performed as described (Wu et al., 2003).
Phenotypic Characterization
To evaluate the viability of mature pollen grains, anthers from mature flowers were dissected in a drop of iodium potassium iodide solution, and images were captured with a Leica DFC480 digital camera system. Photos of spikelets were taken under a stereo microscope (Leica MZFLIII) equipped with a digital camera (Nikon E5400). Embryo sac development was observed by whole-mount eosin B-staining confocal laser-scanning microscopy under a laser-scanning confocal microscope (Leica TCS SP2) as described (Zeng et al., 2007). Images were processed using Adobe Photoshop 7.0.
Meiotic and Mitotic Chromosome Spreads
Meiotic and mitotic chromosome spreads were prepared as described (Ross et al., 1996) with some modifications. Young panicles were fixed with Carnoy's fixative (ethanol:acetic acid, 3:1, v/v) and infiltrated by a vacuum. After fixing at room temperature for 12 to 24 h, they were washed with 75% ethanol for 10 min and stored in 75% ethanol in 4°C.
For PMC meiotic chromosome spreads, anthers were dissected gently with needles, and after transferred to a glass slide together with a drop of improved carbol fuchsin they were staved gently with a needle to release PMCs, and then anther wall debris was carefully removed and a coverslip was added. Finally, the preparation was gently squashed by vertical thumb pressure under a double layer of filter paper and photographed with a Leica DFC480 digital camera system.
Anthers with PMCs at the premeiosis stage were chosen for observation of mitosis in anther wall cells. The anthers were processed according to Ross et al. (1996) except for staining with improved carbol fuchsin instead of 4′,6-diamidino-2-phenylindole. Images were processed using Adobe Photoshop 7.0.
RT-PCR Analysis
Total RNA isolation and RT-PCR were performed as described (Yuan et al., 2009) with gene-specific primers (RTL: 5′-gttgatgacgtactcgctga-3′; RTR: 5′-tggctggactgttggttgga-3′). The PCR conditions were: an initial step of 94°C incubation for 5 min, followed by 32 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 1 min.
MMC, MMS, and UV-C Exposure
Seeds from heterozygous plants (OsRPA1a/osrpa1a) were surface sterilized with 70% ethanol for 1 min and the 0.15% HgCl2 for 15 min. After washing with sterile water seven to eight times, the seeds were germinated on solid one-half Murashige and Skoog medium with 0.3% phytagel. Genotypes were assayed 15 d later. The lengths of each leaf were measured, and the seedlings were regrown in plastic boxes supplied with culture solution plus MMC (Sigma no. 0503) or MMS (Sigma no. 4016) in appropriate concentrations. The length of the leaves was remeasured 18 (for MMS) or 20 (for MMC) d after treatment. Leaf growth per plant (equal to the sum of growth of every leaf on a seedling during the stress treatment) was chosen as the measurement index of sensitivity.
For UV-C irradiation, 70-d-old seedlings were exposed to UV-C in a darkroom. Before and after irradiation, the seedlings were kept in darkroom for 24 h. Images were taken 4 d after irradiation. The irradiance to the top of the seedlings was about 0.29 J m−2 s−1 measured by a UV irradiance meter (model ZQJ-254, Gucun optic instrument factory).
TUNEL Assay
For TUNEL assay, 20-d-old seedlings were treated with 30 μmol L−1 MMC and 70-d-old seedlings were exposed to UV-C. Leaf tissues were collected at different time point after UV-C irradiation (0, 1, 4 h) and MMC treatment (0, 1, 4, 8 d). DNA damage was detected using the DeadEnd Fluorometric TUNEL system (Promega, catalog no. G3250) according to the manufacturer's instructions. The green fluorescence of fluorescein and red fluorescence of propidium iodide was viewed at 520 ± 10 nm and 640 ± 10 nm under a laser-scanning confocal microscope (Leica TCS SP2), respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Suppression of OsRPA1a expression in wild type phenocopies the osrpa1a mutation.
Supplementary Material
Acknowledgments
We thank Dr. Chen Chunli for assistance with preparing chromosome spreads. We also thank Prof. Chris Franklin for helpful comments on the manuscript.
This work was supported by grants from the National Special Key Project of China on Functional Genomics of Major Plants and Animals, the National Natural Science Foundation of China, and the National Program on Key Basic Research Project (grant no. 2007CB108700).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Changyin Wu (cywu@mail.hzau.edu.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Aylon Y, Kupiec M (2004) DSB repair: the yeast paradigm. DNA Repair (Amst) 3 797–815 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, White CI (2004. a) Meiotic defects in the Arabidopsis rad50 mutant point to conservation of the MRX complex function in early stages of meiotic recombination. Chromosoma 113 197–203 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, White CI (2004. b) The atspo11-1 mutation rescues atxrcc3 meiotic chromosome fragmentation. Plant Mol Biol 56 217–224 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, Savigny F, White CI (2005) Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J 41 533–545 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, White CI (2006) Recent advances in understanding of the DNA double-strand break repair machinery of plants. DNA Repair (Amst) 5 1–12 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, White CI (2004) The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J 23 439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Zickler D (2004) Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117 9–15 [DOI] [PubMed] [Google Scholar]
- Bundock P, Hooykaas P (2002) Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 14 2451–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL, et al (2006) Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev 20 1250–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Britt AB (2008) Both ATM and ATR promote the efficient and accurate processing of programmed meiotic double-strand breaks. Plant J 55 629–638 [DOI] [PubMed] [Google Scholar]
- Dean PJ, Siwiec T, Waterworth WM, Schlogelhofer P, Armstrong SJ, West CE (2009) A novel ATM dependant X-ray inducible gene is essential for both plant meiosis and gametogenesis. Plant J 58 791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan T, Proux F, Vaucheret H (2005) Arabidopsis RPA2: a genetic link among transcriptional gene silencing, DNA repair, and DNA replication. Curr Biol 15 1919–1925 [DOI] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR (2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res 34 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Bruchet H, Camescasse D, Granier F, Bouchez D, Tissier A (2003) AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M, Gendrot G, Vezon D, Pelletier G (2003) The Arabidopsis MEI1 gene encodes a protein with five BRCT domains that is involved in meiosis-specific DNA repair events independent of SPO11-induced DSBs. Plant J 35 465–475 [DOI] [PubMed] [Google Scholar]
- He Z, Henricksen LA, Wold MS, Ingles CJ (1995) RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature 374 566–569 [DOI] [PubMed] [Google Scholar]
- Hegde ML, Hazra TK, Mitra S (2008) Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res 18 27–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftode C, Daniely Y, Borowiec JA (1999) Replication protein A (RPA): the eukaryotic SSB. Crit Rev Biochem Mol Biol 34 141–180 [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Kimura S, Furukawa T, Hatanaka M, Hashimoto J, Sakaguchi K (2001) Two types of replication protein A 70 kDa subunit in rice, Oryza sativa: molecular cloning, characterization, and cellular & tissue distribution. Gene 272 335–343 [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Kimura S, Sakaguchi K (2006) A higher plant has three different types of RPA heterotrimeric complex. J Biochem 139 99–104 [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Koga A, Yamamoto T, Uchiyama Y, Mori Y, Hashimoto J, Kimura S, Sakaguchi K (2005) Two types of replication protein A in seed plants. FEBS J 272 3270–3281 [DOI] [PubMed] [Google Scholar]
- Kapoor A, Agarwal M, Andreucci A, Zheng X, Gong Z, Hasegawa PM, Bressan RA, Zhu JK (2005) Mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA-methylation-independent manner in Arabidopsis. Curr Biol 15 1912–1918 [DOI] [PubMed] [Google Scholar]
- Kimura S, Tahira Y, Ishibashi T, Mori Y, Mori T, Hashimoto J, Sakaguchi K (2004) DNA repair in higher plants; photoreactivation is the major DNA repair pathway in non-proliferating cells while excision repair (nucleotide excision repair and base excision repair) is active in proliferating cells. Nucleic Acids Res 32 2760–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehoczky P, McHugh PJ, Chovanec M (2007) DNA interstrand cross-link repair in Saccharomyces cerevisiae. FEMS Microbiol Rev 31 109–133 [DOI] [PubMed] [Google Scholar]
- Li W, Chen C, Markmann-Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA 101 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yang X, Lin Z, Timofejeva L, Xiao R, Makaroff CA, Ma H (2005) The AtRAD51C gene is required for normal meiotic chromosome synapsis and double-stranded break repair in Arabidopsis. Plant Physiol 138 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer WD (2008) Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 18 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XD, Xu XB, Lu YG, Xu SX (1997) The process of embryo sac formation and its stages dividing in rice. Chinese J Rice Sci 11 141–150 [Google Scholar]
- Ma H (2006) A molecular portrait of Arabidopsis meiosis. In C Somerville, E Meyerowitz, J Dangl, M Stitt, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, pp 1–39 [DOI] [PMC free article] [PubMed]
- Mercier R, Grelon M (2008) Meiosis in plants: ten years of gene discovery. Cytogenet Genome Res 120 281–290 [DOI] [PubMed] [Google Scholar]
- Osman K, Sanchez-Moran E, Mann SC, Jones GH, Franklin FC (2009) Replication protein A (AtRPA1a) is required for class I crossover formation but is dispensable for meiotic DNA break repair. EMBO J 28 394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puizina J, Siroky J, Mokros P, Schweizer D, Riha K (2004) Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross KJ, Fransz P, Jones GH (1996) A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res 4 507–516 [DOI] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H (2008) Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77 229–257 [DOI] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA (2008) Regulation of DNA double-strand break repair pathway choice. Cell Res 18 134–147 [DOI] [PubMed] [Google Scholar]
- Shuck SC, Short EA, Turchi JJ (2008) Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res 18 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz RW, Tatineni VM, Hanley-Bowdoin L, Thompson WF (2007) Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol 144 1697–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N, Singh MB, Misra MK, Bhalla PL, Tuteja R (2001) Molecular mechanisms of DNA damage and repair: progress in plants. Crit Rev Biochem Mol Biol 36 337–397 [DOI] [PubMed] [Google Scholar]
- van der Knaap E, Jagoueix S, Kende H (1997) Expression of an ortholog of replication protein A1 (RPA1) is induced by gibberellin in deepwater rice. Proc Natl Acad Sci USA 94 9979–9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez MV, Rojas V, Tercero JA (2008) Multiple pathways cooperate to facilitate DNA replication fork progression through alkylated DNA. DNA Repair (Amst) 7 1693–1704 [DOI] [PubMed] [Google Scholar]
- Waterworth WM, Altun C, Armstrong SJ, Roberts N, Dean PJ, Young K, Weil CF, Bray CM, West CE (2007) NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant J 52 41–52 [DOI] [PubMed] [Google Scholar]
- West CE, Waterworth WM, Sunderland PA, Bray CM (2004) Arabidopsis DNA double-strand break repair pathways. Biochem Soc Trans 32 964–966 [DOI] [PubMed] [Google Scholar]
- Weterings E, Chen DJ (2008) The endless tale of non-homologous end-joining. Cell Res 18 114–124 [DOI] [PubMed] [Google Scholar]
- Wold MS (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 66 61–92 [DOI] [PubMed] [Google Scholar]
- Wu C, Li X, Yuan W, Chen G, Kilian A, Li J, Xu C, Li X, Zhou DX, Wang S, et al (2003) Development of enhancer trap lines for functional analysis of the rice genome. Plant J 35 418–427 [DOI] [PubMed] [Google Scholar]
- Wyman C, Kanaar R (2006) DNA double-strand break repair: All's well that ends well. Annu Rev Genet 40 363–383 [DOI] [PubMed] [Google Scholar]
- Xia R, Wang J, Liu C, Wang Y, Wang Y, Zhai J, Liu J, Hong X, Cao X, Zhu JK, et al (2006) ROR1/RPA2A, a putative replication protein A2, functions in epigenetic gene silencing and in regulation of meristem development in Arabidopsis. Plant Cell 18 85–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Li X, Chang Y, Wen R, Chen G, Zhang Q, Wu C (2009) Mutation of the rice gene PAIR3 results in lack of bivalent formation in meiosis. Plant J 59 303–315 [DOI] [PubMed] [Google Scholar]
- Zeng YX, Hu CY, Lu YG, Li JQ, Liu XD (2007) Diversity of abnormal embryo sacs in indica/japonica hybrids in rice demonstrated by confocal microscopy of ovaries. Plant Breed 126 574–580 [Google Scholar]
- Zhang J, Guo D, Chang Y, You C, Li X, Dai X, Weng Q, Zhang J, Chen G, Li X, et al (2007) Non-random distribution of T-DNA insertions at various levels of the genome hierarchy as revealed by analyzing 13 804 T-DNA flanking sequences from an enhancer-trap mutant library. Plant J 49 947–959 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.