Abstract
The fhl1 gene encoding four-and-a-half LIM protein-1 (FHL1) and its spliced isoform, SLIMMER, is mutated in reducing body myopathy, X-linked myopathy with postural muscle atrophy, scapuloperoneal myopathy, and rigid spine syndrome. In this study we have identified a novel function for SLIMMER in delaying skeletal muscle apoptosis via an interaction with the proapoptotic protein Siva-1. Siva-1 was identified as a SLIMMER-specific-interacting protein using yeast two-hybrid screening, direct-binding studies, and glutathione S-transferase pulldown analysis of murine skeletal muscle lysates. In C2C12 skeletal myoblasts, SLIMMER and Siva co-localized in the nucleus; however, both proteins exhibited redistribution to the cytoplasm following the differentiation of mononucleated myoblasts to multinucleated myotubes. In sections of mature skeletal muscle from wild type mice, SLIMMER and Siva-1 co-localized at the Z-line. SLIMMER and Siva-1 were also enriched in Pax-7-positive satellite cells, muscle stem cells that facilitate repair and regeneration. Significantly, SLIMMER delayed Siva-1-dependent apoptosis in C2C12 myoblasts. In skeletal muscle sections from the mdx mouse model of Duchenne muscular dystrophy, SLIMMER and Siva-1 co-localized in the nucleus of apoptotic myofibers. Therefore, SLIMMER may protect skeletal muscle from apoptosis.
Normal postnatal skeletal muscle is a dynamic tissue, which can regenerate following injury. The satellite cell is the resident muscle stem cell that facilitates the regenerative capacity of muscle (reviewed in Ref. 1, 2). Upon injury or postnatal growth of skeletal muscle (hypertrophy), satellite cells activate, proliferate, and fuse to form new fibers or fuse with existing fibers to repair muscle damage (1, 2).
Accelerated myofiber apoptosis occurs with human aging (3, 4), possibly resulting from oxidative stress/damage, increased mitochondrial dysfunction, and/or activation of the BCL-2 family apoptotic pathway (3, 5, 6). The satellite cell population diminishes with age due to a reduction in self-renewing capacity (1, 2) and/or increased apoptosis (7) leading to a reduced capacity for muscle repair/regeneration. For example, satellite cells from aged rats exhibit increased apoptosis in response to unloading (8), decreased BCL-2 expression, and increased BAX expression (9). Targeted deletion of the transcription factor Pax-7 (a satellite cell marker) in mice compromises muscle regeneration due to increased satellite cell apoptosis (reviewed in Ref. 2). Myofiber apoptosis also occurs in many human myopathies including Duchenne muscular dystrophy (DMD)3 (10, 11).
LIM domain-containing proteins regulate skeletal muscle development and cytoarchitecture and, when mutated, cause human myopathy. The LIM domain comprises a double zinc-finger motif, which mediates protein-protein interactions (12) and functions as a molecular scaffold/adaptor linking signaling and structural complexes with gene transcription. The four-and- a-half LIM (FHL) family of proteins is expressed from five mammalian genes (FHL1–4 and ACT). FHL proteins contain an N-terminal half-LIM motif or single zinc-finger followed by four complete LIM domains (13). FHL1 (SLIM/SLIM1/KyoT/KyoT1/FHL1A) and the splice isoforms KyoT2 (FHL1C) and SLIMMER (FHL1B/KyoT3) are all encoded by the fhl1 gene and are highly expressed in skeletal muscle (14–16). KyoT2 potentially plays a role in embryonic myogenesis by functioning as a potent transcriptional co-repressor of Notch/RBP-Jκ signaling (16). FHL1 is important for the maintenance of muscle mass. Skeletal muscle-specific FHL1 transgenic mice exhibit skeletal muscle hypertrophy and increased muscle strength by co- activating calcineurin/NFATc1-mediated transcription (17). Recently, 14 different mutations in the fhl1 gene have been identified in four distinct human skeletal muscle diseases, reducing body myopathy (RBM) (18–20), X-linked dominant scapuloperoneal disease (21), rigid spine syndrome (22), and X-linked myopathy with postural muscle atrophy (XMPMA) (23), which lead to muscle wasting and in severe cases respiratory and cardiac failure.
We previously identified SLIMMER as a FHL1 splice isoform of unknown function (15). In this study, we have identified the proapoptotic protein Siva-1 (24) as a SLIMMER-specific binding partner. Significantly we demonstrate that SLIMMER slows the onset of Siva-1-induced apoptosis of myoblasts, identifying SLIMMER as a novel regulator of myoblast cell survival.
EXPERIMENTAL PROCEDURES
Materials
The following cells and materials were used: C2C12 and COS-1 cells (American Type Culture Collection), Opti-MEM (Invitrogen), Lipofectamine 2000, To-Pro-3, SlowFade Gold anti-fade reagent, and Escherichia coli TOP10 (Invitrogen), restriction and DNA-modifying enzymes (Promega, MBI Fermentas, or New England Biolabs), DNA oligonucleotides (Geneworks or Micromon), Big Dye version 3.1 sequencing terminators (Applied Systems), In situ death detection kit, TMR red (Roche Applied Science). All other reagents were from Sigma-Aldrich or BDH Chemicals unless indicated.
Plasmids and Cloning
Oligonucleotides are listed in supplemental Table 1. The plasmids pCGN (25), pCGN-β-gal (26), pCGN-FHL1, and pCGN-SLIMMER (15) were described previously. The yeast-2-hybrid constructs pGBKT7, pGBKT7-p53, pGADT7-SV40T, and pACT2 human skeletal muscle library were from Clontech. Yeast baits were PCR-amplified and cloned into pGBKT7 (EcoRI site). Murine KyoT2 sequence was from pEFBOS-myc-KyoT2 (16), a gift from Dr. Tasuku Honjo (Kyoto University, Japan). pET-30a(+) (Novagen), pET-30a(+)-(LIM3+ER) was generated by direct cloning from pGBKT7-(LIM3+ER) into the EcoRI site. pACT2-Siva-1 was isolated from the Y2H library. Siva-1 was PCR-amplified from pACT2-Siva-1 and cloned into pGEX-5x-1 (Amersham Biosciences) (EcoRI site) and into pEFBOS-FLAG (27) (MluI site) (from Dr. Tracey Wilson, Walter and Eliza Hall Institute).
pTrio12R-HEN was generated by replacing the GST cassette in pTrio12R-HGN (26, 28) with eGFP (henceforth GFP). This vector encodes a tricistronic mRNA using internal ribosome reentry sequences to direct expression of separate HA- and GFP-tagged proteins and G418 resistance. pTrio12R-HEN retains the XbaI site of the parental pCGN plasmid, facilitating direct cloning of β-gal and SLIMMER sequences to generate pTrio12R-HA-β-gal-EN and pTrio12R- HA-SLIMMER-EN. pTrio12R-HA-SLIMMER-GFP-Siva-1-N was cloned by adding the Siva-1 sequence from pEFBOS-FLAG-Siva-1 into pTrio12R-HA-SLIMMER-EN (MluI site). Likewise pTrio12R-HA-β-gal-GFP-Siva-1-N was cloned in the same manner.
Sequence size upstream of an internal ribosome reentry sequence can alter the expression level of downstream cistrons, so the larger β-gal mRNA sequence was C-terminally truncated to equalize expression of GFP-Siva-1 between β-gal and SLIMMER vectors. Truncation was achieved by EcoRV (internal β-gal site) and SmaI (3′ to β-gal) digestion and religation of the major fragment.
Antibodies
The following antibodies were used: mouse anti-HA.11 clone 16B12 (1:5000 WB, 1:1000 IF, Covance), mouse anti-polyhistidine clone HIS-1 (1:3000 WB, Sigma- Aldrich), goat anti-GST polyclonal (1:1000 WB, Amersham Biosciences), mouse anti-FLAG clone M2 (1:5000 WB, 3 μl immunoprecipitation, Sigma-Aldrich), rabbit anti-FLAG (1:500 IF, Sigma Aldrich), mouse anti-myc (3 μl of non-immune antibody solution, Invitrogen), goat anti-SLIMMER (1:50 IF-myoblasts/myotubes, 1:100 IF, skeletal muscle sections, 1:1000 WB, Abcam raised to the SLIMMER unique sequence 231KRTVSRVSHPVSKARK246 (15), rabbit Siva (recognizes both Siva-1 and Siva-2, 1:10–25 IF, 1:500 WB, Santa Cruz Biotechnology M-175), goat Siva (Santa Cruz Biotechnology C-20) mouse anti-β-tubulin (1:1000 WB, Zymed Laboratories), mouse anti-GFP clones 7.1/13.1 (1:1000 WB, Roche), mouse anti-skeletal muscle α-actinin (1:1000 IF, Sigma-Aldrich), mouse anti-Pax-7 (1:20 IF, R&D Systems), and mouse anti-pan actin Ab-5 clone ACTN05 (1:3333 dilution, NeoMarkers). Also used were horseradish peroxidase-conjugated secondary antibodies (1:10,000 WB, Chemicon) and Alexa-Fluor-conjugated 488, 594, and 647 secondary antibodies (1:600 IF, Molecular Probes).
Generation of Rabbit Polyclonal Antibodies
A unique peptide sequence of SLIMMER 284YRKNRSLAAPRGPG297 (human GenBankTM accession number AAC72886.1) and Siva-1 82ARGQMLIGPDGRL94 (human GenBank accession number NP_006418) were synthesized (Mimotopes), and each was conjugated to diphtheria toxoid by a linking N-terminal cysteine. The conjugated peptide was injected subcutaneously into New Zealand White rabbits. SLIMMER and Siva-1 antibodies were purified by affinity chromatography using their respective immunizing peptide-coupled thiopropyl-Sepharose resin (Mimotopes) and eluted in 0.1 m glycine HCl, pH 2.5.
Yeast Two-hybrid Analysis
The Matchmaker 3 GAL4-based yeast two-hybrid system was used. The yeast strain AH109, was transformed with pGBKT7-(LIM3+ER) encoding the GAL4 DNA-binding domain (GAL4DBD) fused with the LIM3 plus extra region (LIM3+ER) of SLIMMER. Yeast expressing pGBKT7-(LIM3+ER) were transformed with pACT2 human skeletal muscle cDNA library fused to the GAL4 activation domain (GAL4AD) following the manufacturer's instructions (Clontech). Plasmids from positive clones were extracted and characterized. Y187 yeast were transformed with pACT2-Siva-1, mated with AH109 yeast expressing bait plasmids of the LIM and other domains from FHL1, SLIMMER, and KyoT2, and then plated onto selective media according to the manufacturer's instructions (Clontech).
Culturing of C2C12 and COS-1 Cells
Murine C2C12 myoblasts were maintained in growth media (Dulbecco's modified Eagle's medium supplemented with 20% fetal calf serum, 2 mm l-glutamine, 100 units/ml penicillin, and 0.1% streptomycin). COS-1 cells were maintained in media (Dulbecco's modified Eagle's medium supplemented with 10% fetal or newborn calf serum, 2 mm l-glutamine, 100 units/ml penicillin, and 0.1% streptomycin). Following transfection, cells were grown in media without antibiotics.
Western Blot Analysis
Cells were washed twice in PBS and scraped into HEPES lysis buffer (10 mm HEPES, pH 8, 10 mm KCl, 0.1 mm EDTA, 0.2% Nonidet P-40, Roche Complete mini protease mixture inhibitor tablet) or Triton X-100 lysis buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, Roche Complete mini protease mixture inhibitor tablet). Samples were rocked for 1 h at 4 °C and then pelleted for 10 min at 16,000 × g at 4 °C. Protein concentration was determined using the Bio-Rad DC protein assay kit. 12.5–100 μg of lysate was analyzed by SDS-PAGE and Western blotting. Lysis buffers and amounts of lysate are indicated in the figure legends of Figs. 1–8 and supplemental Fig. 1.
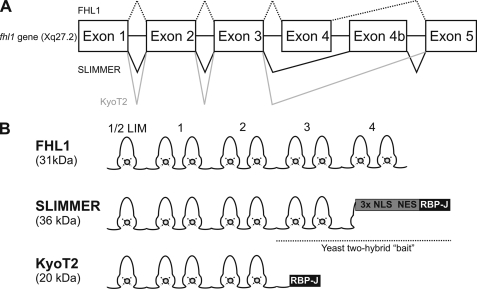
FIGURE 1.
FHL1, SLIMMER, and KyoT2 domain structure. A, fhl1 gene exon structure with alternative splicing indicated. It generates FHL1, SLIMMER, and KyoT2 mRNAs. B, protein structure of FHL1 and alternative splice isoforms. The SLIMMER bait used in yeast two-hybrid studies comprised C-terminal LIM domain 3 followed by the SLIMMER-specific region including the nuclear localization signals (NLS), nuclear export sequences (NES), and the RBP-J-binding domain.
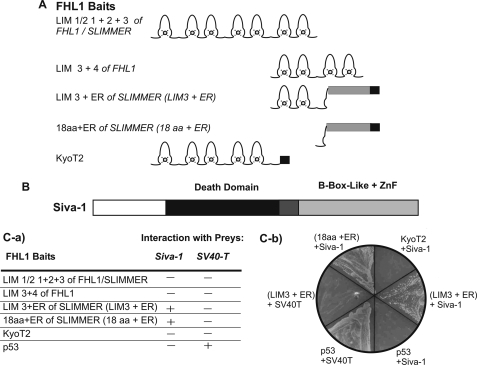
FIGURE 2.
Siva-1 binds SLIMMER in a yeast two-hybrid assay. AH109 yeast expressing the GAL4-DNA-binding domain (GAL4DBD) fused to the indicated LIM domains of FHL1, SLIMMER, KyoT2, or p53 (control) “baits” (A) were mated with Y187 yeast expressing GAL4-activation domain (GAL4AD) fused to Siva-1 or SV40-T (control) “preys” (B). C, transformants were plated onto media lacking tryptophan and leucine (−W−L), to select for diploid yeast expressing both bait and prey constructs and then recultured on media lacking tryptophan, leucine, adenine, and histidine and assessed for growth and LacZ activity (−4+X-α-gal). C-a, table summarizing the interaction of FHL1 bait proteins with either Siva-1 or SV40-T negative control. The presence of an interaction (+) was assessed by growth on minimal media lacking tryptophan, leucine, adenine, and histidine. No interaction is indicated by a minus sign (−). C-b, representative image of yeast growth on minimal media (lacking tryptophan, leucine, adenine, and histidine). Yeast expressing different combinations of bait and prey plasmids are indicated.
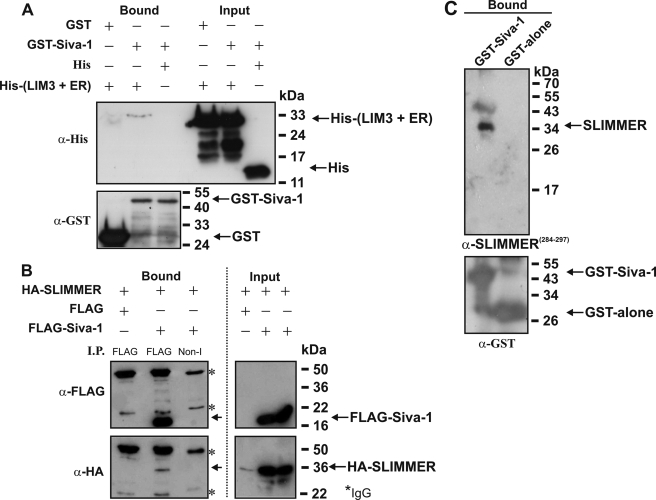
FIGURE 3.
Siva-1 binds SLIMMER. A, GST-Siva-1 and His-tagged LIM domain 3 + ER of SLIMMER (His-(LIM3 + ER)) were co-expressed in E. coli. GST-proteins (5 μg) and associated proteins were bound to glutathione-Sepharose, washed extensively with Tris-buffered saline with 1% Triton X-100, and eluted with SDS-PAGE reducing buffer. Eluted proteins (Bound) and whole-cell extracts (Input) were immunoblotted for recombinant GST (lower panel) or His protein (upper panel) expression. Western blots are representative of three experiments. B, HA-SLIMMER and FLAG-Siva-1 or FLAG-vector were co-transfected into COS-1 cells, and lysates (∼1 mg) prepared in Tris lysis buffer with 1% Triton X-100 were immunoprecipitated (I.P.) with non-immune (Non-I) or anti-FLAG antibodies, washed in Tris-buffered saline without detergent, and immunoblotted with anti-FLAG to confirm FLAG-Siva-1 immunoprecipitation (upper left). Co-immunoprecipitation of HA-SLIMMER specifically with FLAG-Siva-1 was confirmed by immunoblotting with anti-HA (lower left). Protein expression shown by anti-FLAG (upper right) or anti-HA (lower right) immunoblots. Asterisks indicate IgG heavy and light chains. Blots are representative of three independent experiments. Input lanes were loaded with ∼25 μg of protein. C, GST-Siva-1 pulldown of endogenous SLIMMER from skeletal muscle lysates. GST-Siva-1 or GST-alone was expressed in E. coli and ∼500 μg of lysate, purified using glutathione-Sepharose. GST-Siva-1 or GST-alone bound to glutathione-Sepharose was incubated with murine skeletal muscle lysate, the Sepharose beads were washed extensively in Tris-buffered saline without detergent, and bound protein was eluted with SDS-PAGE reducing buffer. Samples were separated on 12.5% SDS-PAGE and immunoblotted with antibodies specific for SLIMMER (α-SLIMMER284–297) (upper panel) or the GST-tag (lower panel).
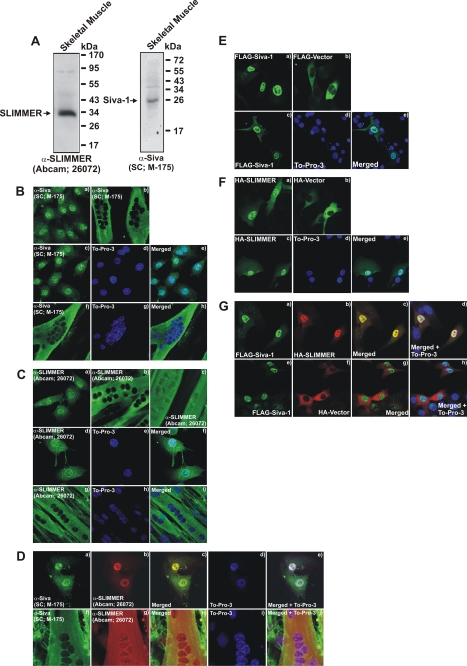
FIGURE 4.
SLIMMER and Siva-1 co-localize in C2C12 skeletal myoblasts and myotubes. A, Siva and SLIMMER are expressed in murine skeletal muscle lysates. The gastrocnemius muscle was dissected from wild type mice, the tissue was homogenized, and protein was extracted using 1% Nonidet P-40 lysis buffer. 50 μg of Nonidet P-40-soluble lysates was separated on 12.5% SDS-PAGE and immunoblotted with a goat polyclonal SLIMMER antibody (Abcam 26072) or rabbit polyclonal Siva antibody (Santa Cruz Biotechnology M-175) as indicated. B and C, localization of endogenous Siva and SLIMMER in undifferentiated C2C12 myoblasts or differentiated myotubes. Myoblasts were plated onto fibronectin-coated coverslips, cultured for 24 h, fixed and stained with either a rabbit polyclonal Siva-specific antibody (B-a) or a goat polyclonal SLIMMER-specific antibody (C-a), and co-stained with To-Pro-3 iodide to detect nuclei (B, c–e, Siva) (C, d–f, SLIMMER) as indicated. Alternatively, myoblasts were cultured for a further 24 h until confluent and switched to low serum media to induce differentiation to multinucleated myotubes. Myotubes were also fixed and stained with either a rabbit polyclonal Siva-specific antibody (B-b) or a goat polyclonal SLIMMER-specific antibody (C, b and c) and co-stained with To-Pro-3 iodide to detect nuclei (B, f–h, Siva; C, g–i, SLIMMER) as indicated. D, endogenous Siva and SLIMMER co-localize in myoblasts and myotubes. Myoblasts (D, a–e) and myotubes (D, f–j) were prepared as described in B and C and co-stained with rabbit polyclonal Siva (D, a and f), goat polyclonal SLIMMER (D, b and g) antibodies, and the nuclear stain To-Pro-3 iodide (D, d and i) as indicated. E–G, C2C12 myoblasts were transiently transfected with FLAG-Siva-1 (E, a and c–e), FLAG-vector (E-b), HA-SLIMMER (F, a and c–e), or HA-vector (F-b) as indicated and stained with anti-FLAG (E) or anti-HA (F) antibodies, respectively. A subset of cells were also co-stained with To-Pro-3 iodide to detect nuclei (E, c–e, and F, c–e). G, FLAG-Siva-1 co-localizes with HA-SLIMMER in myoblasts. C2C12 myoblasts were co-transfected with FLAG-Siva-1 and either HA-SLIMMER (G, a–d) or HA-vector (G, e–h). Cells were co-stained with anti-FLAG and anti-HA antibodies and To-Pro-3 to detect nuclei. All cells were viewed using laser-scanning confocal microscopy.
FIGURE 5.
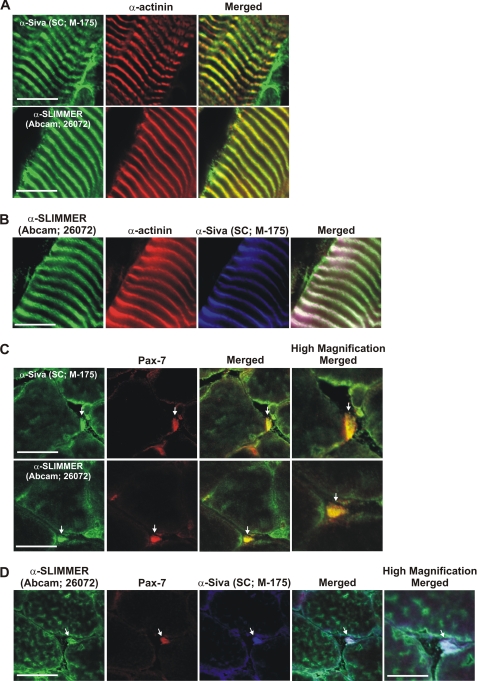
SLIMMER and Siva-1 co-localize at the Z-line in mature skeletal muscle and are enriched in satellite cells. Immunohistochemistry of longitudinal (A and B) and transverse (C and D) cryosections from wild type mouse gastrocnemius muscle. A and B, longitudinal sections were co-stained with rabbit anti-Siva antibody (Santa Cruz Biotechnology M-175), goat anti-SLIMMER (Abcam 26072), and mouse α-actinin antibodies as indicated. C and D, transverse sections were co-stained with rabbit anti-Siva antibody (Santa Cruz Biotechnology M-175), goat anti-SLIMMER (Abcam 26072), and mouse Pax-7 antibodies as indicated. Arrows indicate satellite cells, shown in high magnification images. Sections were viewed using laser-scanning confocal microscopy. Scale bar equals 20 μm (5 μm in high magnification images).
FIGURE 6.
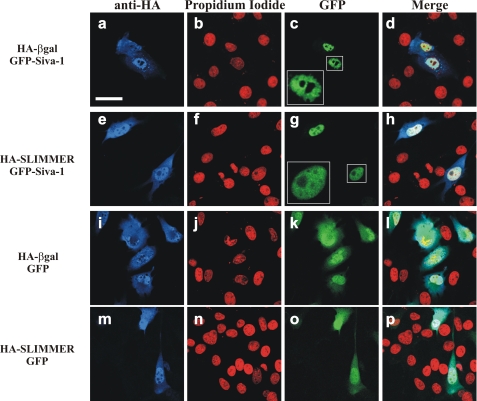
SLIMMER alters Siva-1-induced nuclear morphology in C2C12 myoblasts. C2C12 myoblasts were co-transfected with HA and GFP constructs as indicated. Myoblasts were harvested following 24 h post-transfection, stained with anti-HA (blue) and propidium iodide (red), and then imaged by confocal microscopy. Expressed GFP and GFP-Siva-1 proteins are naturally fluorescent (green). To demonstrate nuclear morphology, high magnification images of nuclei from GFP-Siva-1 transfected cells are shown in the boxed insets. Merge panels show the merged images of the green, red, and blue color channels. Scale bar, 40 μm.
FIGURE 7.
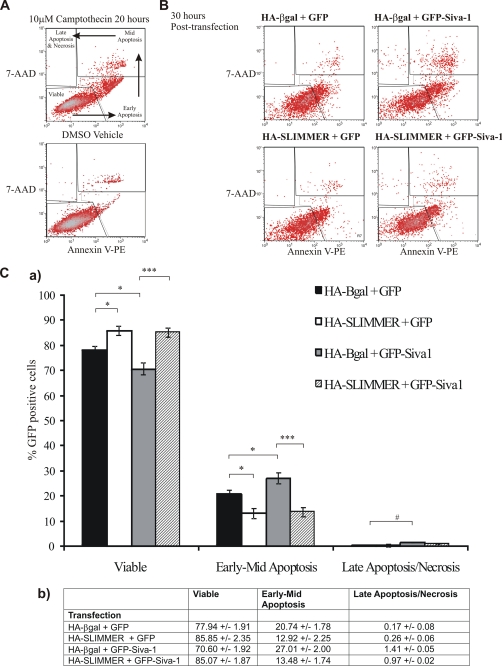
SLIMMER delays Siva-1-dependent apoptosis in C2C12 myoblasts. A, C2C12 myoblasts were treated for 20 h with 10 μm camptothecin, and both adherent and detached myoblasts were analyzed by flow cytometry for phosphatidylserine exposure by annexin V-PE labeling (horizontal axis) and permeability transition by 7AAD uptake (vertical axis). Scatter plot is shown. The apoptotic myoblast subpopulation were defined by Annexin V labeling and 7AAD permeability: viable cells (annexin V-PE- and 7AAD-negative), early apoptotic (annexin V-PE-positive, 7AAD-negative), mid-apoptotic (annexin V-PE- and 7AAD-positive), and late apoptotic/necrotic (annexin V-PE-negative and 7AAD-positive). B, C2C12 myoblasts were transfected with 25 μg of HA-SLIMMER plus GFP-Siva-1 constructs or control vectors as indicated. GFP gating was established by comparison with HA-β-gal + GST-transfected myoblasts (data not shown). Representative scatter plots are shown. Five independent experiments were performed. C-a, the percentage of the transfected population representing viable or apoptotic cells was determined from the five flow cytometry experiments outlined in B, and shown here as a bar graph. Black bars, HA-β-gal + GFP co-expressing myoblasts; white bars, HA-SLIMMER + GFP; gray bars, HA-β-gal + GFP-Siva-1; striped bars, HA-SLIMMER + GFP-Siva-1. Error bars represent S.E. *, p < 0.05; ***, p < 0.0005; #, p = 0.05. C-b, table shows the data for the graph presented in C-a as a percentage of the GFP-positive cells. Data represents the mean ± S.E.
FIGURE 8.
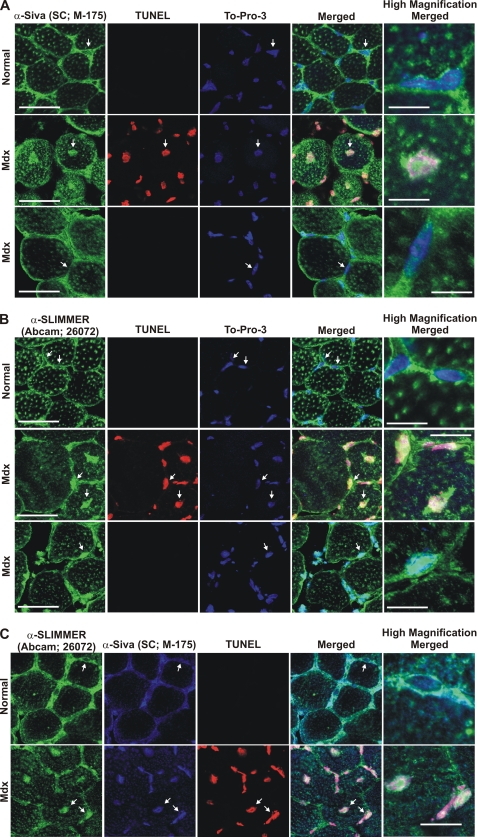
Siva and SLIMMER co-localize in apoptotic myonuclei in skeletal muscle from dystrophic mdx mice. Transverse gastrocnemius skeletal muscle sections from 4-week-old dystrophic mdx mice (Mdx) or normal control mice (Normal) were co-stained with either rabbit polyclonal Siva antibody (Santa Cruz Biotechnology M-175) (A) or goat polyclonal SLIMMER antibody (Abcam 26072) (green) (B) followed by TUNEL staining to detect apoptotic nuclei (red) and the nuclear stain To-Pro-3 (blue). Alternatively, transverse sections were co-stained with both the Siva- and SLIMMER-specific antibodies followed by TUNEL staining (C). In A and B, for skeletal muscle from mdx mice, two areas of muscle fibers are shown: apoptotic (TUNEL-positive, middle panel) and a neighboring non-apoptotic region (TUNEL-negative, bottom panel). Arrows indicate the myonuclei represented in the high magnification images. Scale bar, 20 μm (5 μm in high magnification images).
Transfection of Plasmid DNA
Transfection quality plasmid DNA was prepared using Qiagen midiprep kits and transfected using Lipofectamine 2000 according to the manufacturer's protocol.
Immunofluorescence Labeling of C2C12 Cells
Immunofluorescence staining of C2C12 myoblasts and myotubes was performed as described (26).
Microscopy
Sections and slides were mounted using SlowFade Gold reagent and viewed using laser-scanning confocal microscopy (Olympus Fluoroview 500 or Leica TCS NT, Monash Microimaging).
In Vitro Direct Protein Interaction
The E. coli strain BL21 DE3 Gold Codon Plus RP (Stratagene) were co-transformed with pGEX-5×1 (GST-tag), pGEX-5×1-Siva-1 and pET-30a(+), or pET-30a(+)-(LIM3+ER) (HIS-tag). Growth, extraction, GST pulldown, and immunoblotting were performed as described previously (28).
Co-immunoprecipitation from COS-1 Cells
The protocol was performed as described previously (26) with minor modifications as follows. 16 μg of plasmid DNA was transfected with cells being harvested 24 h later. Lysates were extracted for 1 h and pelleted for 10 min. Precleared lysates were immunoprecipitated with 3 μg of antibody as indicated together with 60 μl of protein A-Sepharose for 2 h at 4 °C. Bound fraction was washed six times with Tris-saline buffer, and precipitated protein was eluted with SDS-PAGE reducing buffer.
GST Pulldown from Skeletal Muscle Lysates
Recombinant GST or GST-Siva-1 were produced by auto-induction of E. coli BL21 DE3 pLysS (Novagen) transformed with pGEX-5×1 (Amersham Biosciences) or pGEX-5×1-Siva-1 as described (28). Muscle lysates were prepared from 4-week-old male C57BL/6 mice, and GST pulldown was performed with 400 μl of muscle lysates (∼500 μg total protein) as described previously (17).
Preparation of Mouse Tissue for Western Blot or Sectioning
Mice were humanely sacrificed by CO2 inhalation and cervical dislocation following the guidelines of the National Health and Medical Research Council, Monash University, animal ethics number BAM/2001/22 and SOBSB/B/2007/46. Tissue extracts were prepared in 1% Nonidet P-40 extraction buffer and analyzed by SDS-PAGE and Western blotting as described (17). Protein concentration was determined using a Bio-Rad DC protein assay kit, and lysates were analyzed by SDS-PAGE and Western blotting. Frozen longitudinal and transverse sections of mouse gastrocnemius muscle were prepared, fixed, stained, and viewed as described (17, 26, 29).
Flow Cytometry Analysis of Apoptosis
C2C12 myoblasts (6.25 × 105) were seeded in 10-cm dishes for 24 h and then treated with 10 μm camptothecin (or DMSO vehicle) for 20 h or, alternatively, transfected with 25 μg of pTrio12R-based plasmid DNA using Lipofectamine 2000 and analyzed 30 h later. Cell media containing floating cells were collected and pooled with dislodged cells from a single PBS rinse and adherent cells that were detached by incubation with 2 ml of trypsin for 5 min. The cell pellet, collected by centrifugation at 1,200 rpm 15 min at 4 °C, was resuspended in 2 ml of PBS(+). Cells were counted using a hemocytometer, and aliquots of 2 × 106 cells were resuspended in 0.5 ml of annexin V binding buffer (Roche Applied Science) plus 1% bovine serum albumin (filtered), 0.5 μl of annexin V-PE (Molecular Probes), and 2.5 μl of 1 mg/ml 7AAD (Sigma-Aldrich) and incubated for 2 min at room temperature. The labeled cells were pelleted, resuspended in 0.5 ml of PBS(+), and analyzed immediately on a FACSCalibur (BD Biosciences) flow cytometer. Initial forward and side scatter gating was set to encompass all cells except for undissociated large cell clusters; when applicable, GFP fluorescent cells were gated for analysis with GFP gating established by comparison to GST-transfected cells. Appropriate nonlabeled, single, double, and triple labeled controls were used to calibrate and compensate channel bleed-through. The protocol for fluorescence-activated cell sorter analysis of apoptosis in GFP-positive cells using 7AAD and annexin V-PE was adapted from a previously published source (30).
Myofiber Apoptosis
mdx mice, the mouse model of DMD, were obtained from the Animal Resource Centre, Western Australia. Myofiber apoptosis was examined in the gastrocnemius muscle from 4-week-old mdx mice (C57BL/6) versus age- and sex-matched normal control C57BL/6 mice as described previously (31). Frozen, transverse sections of gastrocnemius muscle (8 μm) were fixed in 4% paraformaldehyde for 20 min, washed extensively with PBS, and permeabilized in 0.1% Triton X-100, 0.1% sodium citrate for 2 min on ice. Sections were washed extensively with PBS and blocked (1% bovine serum albumin, 5% horse serum in PBS) for 15 min. Sections were stained with primary antibodies overnight at 4 °C and secondary antibodies for 1 h at room temperature as listed under “Antibodies.” For detection of myonuclei, sections were pretreated with RNase A (10 mg/ml) overnight at 4 °C and stained with To-Pro-3 (1:100) for 5 min. Apoptotic nuclei were detected using TUNEL staining (in situ cell death detection kit, TMR red; Roche Applied Science) according to manufacturer's instructions.
Statistical Analysis
All statistical analysis was performed using the unpaired Student's t test, and p values of <0.05 were considered significant.
RESULTS
FHL1 and the Splice Isoforms KyoT2 and SLIMMER Domain Structure
The fhl1 gene contains five exons (1–5) and encodes three different isoforms (FHL1, SLIMMER, and KyoT2) by alternate mRNA splicing (15) (Fig. 1). FHL1 contains an N-terminal half-LIM domain followed by four complete LIM domains derived from exons 1–5. KyoT2 contains the N-terminal two and one-half LIM domains identical to FHL1; however, alternate splicing and removal of exon 4 leads to a frameshift and a novel C terminus encoding an RBP-J-binding site (16). SLIMMER is generated by inclusion of an alternative exon 4b, leading to a 200-bp insert and frameshift after LIM domain 3. SLIMMER contains the FHL1 N-terminal three-and-a-half LIM domains, followed by a unique C terminus with bipartite nuclear localization signals, a leucine-rich nuclear export sequence, and an RBP-J-binding site (15, 16).
Identification of the Proapoptotic Protein Siva-1 as a Novel SLIMMER Binding Partner
Proteins that bind to and are regulated by FHL1 and KyoT2 have been identified; however, SLIMMER-specific binding partners have not been reported, and the specific function of this spliced isoform is unknown. To identify SLIMMER-interacting proteins, a yeast two-hybrid screen was performed. To this end a truncated SLIMMER bait comprising LIM domain 3 followed by the nuclear import and export sequences and the RBP-J-binding domain, designated SLIMMER (LIM3+ER) (Figs. 1B and 2A), was expressed in AH109 yeast and co-transformed with a pACT2 human skeletal muscle library. After screening and sequence analysis, one putative interacting clone was identified as the full-length coding sequence for the proapoptotic protein Siva-1 (Fig. 2B).
Siva-1 was originally identified as a CD27-binding protein (24). Siva-1 contains a death domain homology region followed by two cysteine-rich domains in the C terminus, a B-box-like zinc-finger domain, and a zinc-finger domain (Fig. 2B). Siva-1 functions as a proapoptotic protein in many cellular contexts, as follows. Siva-1 expression is activated during DNA damage response (32) and is also regulated by transcription factors, including p53 (32, 33) and E2F1 (34). Siva-1 is critical for p53-dependent neuronal cell death (35) and also induces apoptosis of T lymphocytes via a caspase-dependent mitochondrial pathway (36). Bcl-XL binds to and is regulated by Siva-1 (37). Yeast transformed individually with the SLIMMER (LIM3+ER) bait or the Siva-1 prey did not autonomously activate the reporter genes when grown on minimal media (minus amino acids histidine, leucine, adenine, and tryptophan) and did not activate reporter genes when co-transformed with the control prey (SV40-T) and bait (p53) proteins, respectively (Fig. 2C-b). These control studies validate the SLIMMER-Siva-1 interaction in yeast.
Siva-1 Binds SLIMMER and Does Not Interact with FHL1 and KyoT2
We next examined the specificity of the SLIMMER-Siva-1 interaction and determined whether Siva-1 also binds the splice isoforms FHL1 and KyoT2. Yeast two-hybrid baits for specific domains from FHL1, SLIMMER, or KyoT2 were cloned into the pGBKT7 vector (Fig. 2A), transformed into AH109 yeast, and mated with Y187 yeast transformed with the Siva-1 prey plasmid. Diploid yeast were assayed for growth on minimal media (Fig. 2C). A bait containing the LIM domains common to both FHL1 and SLIMMER (LIM-½ 1+2+3 FHL1/SLIMMER) did not bind Siva-1, as indicated by the absence of growth on minimal media (Fig. 2C). No interaction was observed between the C-terminal LIM domains 3 and 4 of FHL1 (LIM 3+4 FHL1), suggesting that Siva-1 does not bind FHL1 or its LIM domain 3. The LIM3+ER construct, which comprises the SLIMMER-specific nuclear import and export sequences and the RBP-J-binding domain common to SLIMMER and KyoT2, was able to bind Siva-1. However, it is unlikely that the RBP-J-binding domain of SLIMMER mediates this interaction, as this domain is also present in KyoT2, which did not bind Siva-1 (Fig. 2C). It is also unlikely that Siva-1 binds to LIM domain 3 of SLIMMER, as deletion of this domain (18aa+ER) did not impair binding to Siva-1. Therefore Siva-1 most likely binds the unique region of SLIMMER, which contains the nuclear import and export domains, and thus represents the first SLIMMER-specific binding partner identified to date.
SLIMMER and Siva-1 Complex in Muscle Cells
We determined whether SLIMMER and Siva-1 bind directly, without the cooperation or requirement for additional proteins, by assessing the binding of purified GST-Siva-1 to the His-SLIMMER truncation mutant containing the “LIM 3+ER” region (His-LIM3+ER) with the predicted Siva-1-binding site. Recombinant His-(LIM3+ER) and GST-Siva-1 were co-expressed in E. coli, and bacterial lysates were incubated with glutathione-Sepharose, washed extensively, and immunoblotted with either anti-His or anti-GST antibodies (Fig. 3A, bound fractions). His-(LIM3+ER) bound GST-Siva-1 but not GST. In control studies the HIS-tag alone did not interact with GST-Siva-1. GST antibody immunoblot analysis confirmed binding of recombinant GST-Siva-1 or GST to the glutathione-Sepharose beads. An anti-His immunoblot revealed equal input of His-(LIM3+ER) and HIS-tag alone (Fig. 3A, input fractions).
To demonstrate an interaction between full-length SLIMMER and Siva-1 in mammalian cells, COS-1 cells were co-transfected with FLAG-Siva-1 and HA-SLIMMER. We reproducibly detected increased protein expression of HA-SLIMMER in COS-1 cells when it was co-expressed with FLAG-Siva-1 relative to that detected when transfected with FLAG-vector alone, suggesting that the complex between Siva-1 and SLIMMER stabilizes SLIMMER protein expression (see Fig. 3B, input lanes). Lysates were immunoprecipitated with a FLAG antibody to pull down Siva-1 and immunoblotted with anti-HA to detect SLIMMER. HA-SLIMMER was co-immunoprecipitated by FLAG-Siva-1 but not FLAG tag alone (Fig. 3B, lower panel). In control studies FLAG-Siva-1 immunoprecipitation was confirmed by immunoblotting with anti-FLAG antibody (Fig. 3B, upper panel). HA-SLIMMER and FLAG-Siva-1 were not immunoprecipitated by a control non-immune antibody. Expression of recombinant proteins was confirmed in input lysates. Therefore SLIMMER and Siva-1 can complex in mammalian cells.
To demonstrate an interaction between endogenous SLIMMER and Siva-1 we undertook co-immunoprecipitation experiments of murine skeletal muscle lysates using a series of rabbit and goat polyclonal SLIMMER and Siva antibodies. We generated a rabbit polyclonal anti-peptide antibody specific to the unique SLIMMER sequence 284YRKNRSLAAPRGPG297 (human GenBank accession number AAC72886) (supplemental Fig. 1A), conserved between human and mouse SLIMMER isoforms (murine GenBank accession number NP_001070829, labeled four-and-a-half LIM domains 1 isoform 1) but not shared by FHL1 or KyoT2 (38) (supplemental Fig. 1). This SLIMMER antibody, but not preimmune serum, detected recombinant HA-SLIMMER expressed in COS-1 cells and a 36-kDa immunoreactive polypeptide in C2C12 myoblasts. We also occasionally detected a 50–55-kDa immunoreactive polypeptide using both our SLIMMER antibody (supplemental Fig. 1B, a and b) and the commercial (Abcam 26072) antibodies (Fig. 4A). We also generated a Siva-1-specific anti-peptide antibody to amino acids 82ARGQMLIGPDGRL94 (human GenBank accession number NP_006418) (anti-Siva-1(82–94); data not shown). This antibody demonstrated similar results in immunoblots to commercially available Siva antibodies (Santa Cruz Biotechnology, rabbit M-175 or goat C-20) (not shown). Therefore, we have presented results here using only the commercial antibody. Unfortunately we were unable to immunoprecipitate endogenous SLIMMER or Siva-1 using their respective antibodies, either commercial or our antibodies (data not shown). Therefore, to further confirm the interaction between SLIMMER and Siva-1, a GST pulldown assay was undertaken. GST-Siva-1 or GST produced in E. coli was purified by coupling to glutathione-Sepharose beads and incubated with gastrocnemius muscle lysates from wild type mice. Bound proteins were immunoblotted with the SLIMMER anti-peptide antibody or anti-GST antibody. A ∼36-kDa polypeptide corresponding to endogenous SLIMMER was pulled down by GST-Siva-1 but not GST (Fig. 3C, upper panel). Immunoblot analysis with anti-GST confirmed relatively equal binding of the recombinant GST-Siva-1 or GST to the glutathione-Sepharose (Fig. 3C, lower panel). Hence endogenous skeletal muscle-derived SLIMMER binds GST-Siva-1, further confirming an interaction between these proteins.
SLIMMER and Siva-1 Co-localize in Myoblasts and Differentiated Myotubes
To further investigate the association between Siva-1 and SLIMMER, we analyzed their subcellular localization in myoblasts and multinucleated myotubes. Epitope-tagged Siva-1 localizes predominantly to the nucleus (36), with some staining also detected within the cytosol and plasma membrane (39). The subcellular localization of Siva-1 in skeletal muscle or myoblasts has not been reported, although Northern blot analysis has revealed that Siva-1 is expressed in skeletal muscle (40). We demonstrated a ∼26-kDa immunoreactive polypeptide in skeletal muscle lysates using anti-Siva antibodies (Santa Cruz M-175) (Fig. 4A) and using our own anti-Siva-1(82–94) antibody (not shown), indicating that Siva-1 is expressed in muscle. In myoblasts endogenous Siva-1 localized predominantly to the nucleus (Fig. 4B, a and c), as shown by its co-localization with the nuclear stain To-Pro-3 (Fig. 4B, c–e). FLAG-Siva-1 when expressed in myoblasts also showed a predominantly nuclear distribution (Fig. 4E, a and c–e). Interestingly, following the differentiation of myoblasts to form multinucleated myotubes, which involves the exit of myoblasts from the cell cycle and myoblast fusion, Siva-1 was detected exclusively in the cytoplasm (Fig. 4B, b and f–h).
Endogenous SLIMMER (Fig. 4C, a and d–f) and HA-SLIMMER (Fig. 4F, a and c–e) localized to the nucleus in myoblasts as we reported previously (15). Following the differentiation of myoblasts to myotubes, SLIMMER, like Siva-1, exhibited a prominent cytoplasmic distribution (Fig. 4C, b and g–i). However, some myotubes retained nuclear SLIMMER staining (Fig. 4C-c). The ability of SLIMMER to shuttle in and out of the nucleus is conferred by its bipartite nuclear localization signal and leucine-rich nuclear export sequence (15). Endogenous Siva-1 and SLIMMER co-localized in the nucleus of myoblasts (Fig. 4D, a–e) and the cytoplasm of myotubes (Fig. 4D, f–j), consistent with complex formation between these species. FLAG-Siva-1 also co-localized with HA-SLIMMER (Fig. 4G, a–d) in the nucleus of myoblasts but not HA-vector (Fig. 4G, e–h).
SLIMMER and Siva-1 Co-localize at the Z-line and Subsarcolemma of Mature Skeletal Muscle and in Satellite Cells
To define the localization of endogenous Siva-1 and SLIMMER in mature muscle, murine gastrocnemius sections were co-stained with commercial goat anti-SLIMMER (Abcam 26072) (15), and rabbit anti-Siva (Santa Cruz Biotechnology, M-175) (Fig. 5). The sarcomeric Z-line was identified by co-staining with a mouse monoclonal α-actinin antibody (41). In longitudinal sections, both SLIMMER and Siva-1 co-localized with the Z-line marker, α-actinin (Fig. 5, A and B). In transverse sections, SLIMMER and Siva-1 also co-localized at the myofiber periphery, most likely at the sarcolemma or subsarcolemma (Fig. 8C, upper panel). To assess whether SLIMMER and Siva-1 were also present in satellite cells, transverse muscle sections were co-stained with the satellite cell marker Pax-7 (42). Both SLIMMER and Siva-1 staining was detected in Pax-7-positive satellite cells (Fig. 5, C and D (see arrows)). Similar results were also obtained with the rabbit anti-Siva-1(82–94) antibody (data not shown). Therefore SLIMMER and Siva-1 co-localize in skeletal muscle at the Z-line and subsarcolemma and are co-expressed in satellite cells.
SLIMMER Regulates Siva-1-induced Apoptosis
Siva-1 functions as a proapoptotic protein in many cell types (35, 36, 43–45). However, the role that Siva-1 plays in myoblast apoptosis has not been examined. To investigate this role, HA-SLIMMER or HA-β-gal (vector control) were co-expressed in myoblasts with GFP or GFP-Siva-1 (we achieved a 20% transfection efficiency of the dual expression pTrio12R plasmids into myoblasts), and 24 h post-transfection, cells were co-stained with anti-HA and propidium iodide (to label the nuclei) and examined (Fig. 6). In some cells expression of GFP-Siva-1 in C2C12 myoblasts resulted in nuclei with an irregular or ragged appearance (Fig. 6, a–d), not evident in vector controls (Fig. 6, i–l), reminiscent of the early morphological signs of apoptosis (46); this was reduced following co-expression with HA-SLIMMER (Fig. 6, e–h) but not HA-β-gal control (Fig. 6, a–d). In control studies HA and GFP immunoblot analysis revealed equivalent expression of recombinant proteins under these conditions (supplemental Fig. 2).
Myoblast apoptosis was further analyzed by flow cytometry for membrane exposure of phosphatidylserine by annexin V-PE labeling and permeability transition by 7-AAD uptake, with transfected cells gated by the presence of GFP fluorescence. C2C12 myoblasts were maintained under subconfluent, proliferative conditions in order to prevent the complication of normal phosphatidylserine exposure as part of the differentiation program (47). As a positive control and to establish adequate gating for the cell populations, C2C12 myoblasts were treated for 20 h with 10 μm camptothecin, a DNA-damaging agent that induces apoptosis in many cells including myoblasts (48) via the mitochondrial apoptotic pathway in which Siva is implicated (35). Camptothecin-treated C2C12 myoblasts showed a typical apoptotic shift from a double-negative proliferative subpopulation, above that detected in DMSO treated cells, to an early apoptotic annexin V-positive subpopulation within the time frame observed (Fig. 7A, upper panel). However, later stages of apoptosis, as indicated by the uptake of 7-AAD to form a double-positive subpopulation, and also loss of annexin V-staining in the late apoptotic and necrotic cells correlating with a 7AAD-only positive subpopulation, were also evident (Fig. 7A, upper panel) (49). 30 h post-transfection, all C2C12 myoblasts including control HA-β-gal-GFP-vector cells exhibited apoptotic subpopulations ((Fig. 7, B (top left) and C). GFP-Siva-1 expression increased the proportion of cells gating in the annexin V-positive and the annexin V/7-AAD-double positive regions (Fig. 7B, top right). Co-expression of HA-SLIMMER with GFP-Siva-1 reduced the proportion of cells gating in the annexin V-positive and the annexin V/7-AAD-double positive regions (Fig. 7B, bottom right).
The proportion of transfected myoblasts gating into each region, representing viable cells (double-negative), early-mid apoptosis (annexin V-positive), and late apoptosis/necrosis (7-AAD-positive, annexin V-negative), was quantified (Fig. 7, C-a, and raw values in C-b). GFP-Siva-1 increased the proportion of myoblasts undergoing early-to-mid- apoptosis compared with GFP-vector controls (*, p < 0.05). Furthermore co-expression of HA-SLIMMER reduced both basal (*, p < 0.05) and GFP-Siva-1-mediated apoptosis (***, p < 0.0005). Therefore SLIMMER delays the onset of Siva-1-mediated apoptosis.
Siva-1 and SLIMMER Co-localize in Apoptotic Myofiber Nuclei in DMD Muscle
DMD (reviewed in Ref. 50) is associated with rapid muscle degeneration and early death (51). Myofiber apoptosis is prominent in myopathies/dystrophies including DMD (10, 11). In the mdx mouse model of DMD myofiber apoptosis precedes necrosis and peaks at 4 weeks of age (31). To determine whether Siva-1 and SLIMMER associate with myofiber apoptosis in a muscle disease model, the localization of both proteins was examined in transverse skeletal muscle sections from 4-week-old mdx mice. In skeletal muscle from normal mice, Siva-1 staining was confined to the sarcolemma (Fig. 8A, top panel) and was not present in myofiber nuclei (Fig. 8A, see arrows and high magnification image). No TUNEL staining was evident in wild type muscle as described (11). Significantly Siva-1 was detected in apoptotic myofiber nuclei in mdx muscle where it co-localized with TUNEL staining. Siva-1 nuclear localization was most apparent in myofibers with centralized nuclei, a marker of degenerating/regenerating myofibers (52) (Fig. 8A, middle panel, see arrows), but was absent from the nuclei of neighboring non-apoptotic myofibers (TUNEL-negative) (Fig. 8A, bottom panel), revealing a correlation between Siva-1 nuclear localization and myofiber apoptosis. SLIMMER was confined to the sarcolemma in skeletal muscle from normal mice where it co-localized with Siva and was not evident in myonuclei (Fig. 8, B (top panel) and C). SLIMMER was detected in apoptotic myonuclei in mdx muscle (Fig. 8B, middle panel), where it co-localized with Siva-1 (Fig. 8C, lower panel). However, nuclear SLIMMER was also present in the neighboring non-apoptotic nuclei in mdx muscle (Fig. 8B, bottom panel).
DISCUSSION
In this study, we have identified the proapoptotic protein Siva-1 as a SLIMMER-specific binding partner and have demonstrated that SLIMMER inhibits Siva-1-induced apoptosis. Evidence that Siva-1 and SLIMMER complex was shown by yeast two-hybrid interaction, direct-binding studies, co-immunoprecipitation of recombinant proteins from intact cells, GST-Siva-1 pulldown of endogenous SLIMMER from skeletal muscle lysates, and co-localization of Siva-1 and SLIMMER in the nucleus of normal myoblasts and the apoptotic myonuclei of diseased, dystrophic muscle. Therefore these studies identify a unique binding partner for the FHL1 isoform SLIMMER, which acts as a regulator of apoptosis induced by Siva-1.
Despite their related domain structure and amino acid similarity, the splice variants of the fhl1 gene, FHL1, SLIMMER, and KyoT2, exhibit distinct subcellular localization and unique functions in skeletal muscle. KyoT2 localizes to the nucleus and cytoplasm and represses RBP-J-mediated transcriptional activity by competing with the coactivators EBNA2 and Notch1 for binding to RBP-J (16, 53, 54). SLIMMER has also recently been shown to inhibit the activity of RBP-J (55), a highly conserved transcription factor critical for embryonic development of a number of structures including somites (56), from which skeletal muscle progenitors are derived (57). SLIMMER shares three common LIM domains with FHL1; therefore SLIMMER will share common transcriptional targets in the nucleus with FHL1, but via its unique C terminus, it binds Siva-1 and regulates myofiber apoptosis. SLIMMER localizes to the nuclei of myoblasts and the cytoplasm of myotubes. We have reported here that SLIMMER nuclear and cytoplasmic localization correlated in general with that of its binding partner, Siva-1, in both undifferentiated myoblasts and multinucleated myotubes and also in both normal skeletal muscle and in the centralized nuclei of myofibers in a mouse model of Duchenne muscular dystrophy.
As shown here, overexpression of Siva-1 induces apoptosis of C2C12 myoblasts. At 24 h post-transfection Siva-1-expressing cells developed a ragged, nuclei morphology reminiscent of an early morphological sign of myoblast apoptosis (10, 11, 58). During the latter stages, apoptosis was assessed using the markers of phosphatidylserine exposure and 7AAD uptake. Overexpression of Siva-1 increased the level of apoptosis above GFP-vector control-transfected cells. Significantly, co-expression of SLIMMER reduced the induction of apoptosis by Siva-1. This modest reduction in Siva-1-mediated apoptosis induced by HA-SLIMMER expression was consistent with the modest reduction in apoptosis also observed by co-expression of the human papillomavirus-16 E7 protein and GFP-Siva-1 in HaCaT (human immortalized keratinocytes) (43). These observations suggest that SLIMMER acts an anti-apoptotic protein in myoblasts. To further confirm the effect of SLIMMER on Siva-1-induced apoptosis, we have also attempted small, interfering RNA (siRNA)-mediated knockdown of SLIMMER using oligonucleotides targeted to the unique region of SLIMMER (data not shown). Unfortunately, preliminary reverse transcription-PCR studies indicate the siRNA oligonucleotides also knock down FHL1 mRNA, suggesting that the oligonucleotides can target prespliced mRNA (data not shown).
Although the precise molecular mechanisms by which Siva proteins induce apoptosis are poorly understood, emerging evidence suggests that this may occur through modulation of the BCL-2 pathway. Siva-1 binds to BCL-2 and Bcl-XL, but not BAX, and sensitizes MCF7 and MDA-MB-231 breast cancer cells to UV-mediated apoptosis (37, 59). Interestingly the BCL-2/Bax intrinsic apoptotic pathway is also implicated in skeletal muscle apoptosis, but a direct role for Siva-1 in muscle apoptosis has not been reported to date (46). In T lymphocytes Siva-1 and Siva-2 promote apoptosis via a caspase-dependent mitochondrial pathway, attenuated by overexpression of Bcl-2 or Bcl-XL (36). The precise mechanism by which SLIMMER regulates Siva-1-dependent apoptosis is currently unknown and will be the focus of future studies.
Expression of Siva-1 is altered in a number of pathological processes including acute ischemic injury (45) and Coxsackievirus infection (44) and during chemotherapy resistance in cancer (32, 61). Interestingly, we have also shown here that Siva-1 nuclear localization in apoptotic muscle fibers from dystrophic mdx mice correlates with TUNEL-positive apoptotic muscle fibers. Siva-1 staining has also been detected in TUNEL-positive cells present in rat kidneys following induction of acute ischemic injury (45). Therefore our data suggest that the recruitment of Siva-1 to nuclei may play a role in cellular apoptosis. Indeed, a recent study has shown Siva-1 is sequestered at the cytoplasm/plasma membrane through binding the lysophosphatidic acid-2 receptor (LPA2) (39, 62). Dissociation of the LPA2·Siva-1 complex leads to translocation of Siva-1 to the nucleus and subsequent activation of its proapoptotic function.
Recently, 14 different mutations in the fhl1 gene have been identified in patients with four diseases associated with muscle wasting: RBM (18–20), X-linked myopathy with postural muscle atrophy (23), scapuloperoneal disease (21), and rigid spine syndrome (22). Clinically the most severe of these, RBM, ranges from early childhood-onset muscle weakness followed rapidly by death due to respiratory failure to adult-onset disease with slower but relentless progression. Interestingly, myofiber apoptosis has been implicated as a possible cause of skeletal muscle degeneration in a fatal form of RBM (60). Several of the mutations in the fhl1 gene affect highly conserved cysteine and histidine zinc-binding residues in the second LIM domain common to FHL1, SLIMMER, and KyoT2. Mutation of these residues leads to destabilization of the second LIM domain and ultimately aggregation of the misfolded FHL1/SLIMMER protein into insoluble, nonfunctional inclusions (17, 18). It is possible under these conditions that SLIMMER will be trapped in inclusion bodies allowing unopposed Siva-1 induced apoptosis, which may contribute to the myopathy.
In this study we have shown a unique function of the SLIMMER splice variant of the fhl1 gene. SLIMMER functions to impede apoptosis by interacting directly with the proapoptotic protein Siva-1. It is interesting to speculate that deregulation of this interaction may control the survival of myofibers and satellite cells and hence may contribute to muscle apoptosis and impair regeneration.
Supplementary Material
Acknowledgments
We thank Monash Micro Imaging and Dr. R. Gurung, Department of Biochemistry, Monash University, Australia, for technical assistance.
This work was supported by Grant 9284272 from the National Health and Medical Research Council, Australia.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Table 1.
- DMD
- Duchenne muscular dystrophy
- FHL
- four-and-a-half LIM
- SLIM
- skeletal muscle LIM
- SLIMMER
- SLIM1 with extra regions
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- β-gal
- β-galactosidase
- RBM
- reducing body myopathy
- ER
- extra region
- 7ADD
- 7-amino-actinomycin D
- HA
- hemagglutinin
- PBS
- phosphate-buffered saline
- TUNEL
- terminal dUTP nick-end labeling
- RBP-J
- recombination signal binding protein for immunoglobulin kappa J region
- WB
- Western blot
- IF
- immunofluorescence.
REFERENCES
- 1.Le Grand F., Rudnicki M. A. (2007) Curr. Opin. Cell Biol. 19, 628–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryson-Richardson R. J., Currie P. D. (2008) Nat. Rev. 9, 632–646 [DOI] [PubMed] [Google Scholar]
- 3.Marzetti E., Lawler J. M., Hiona A., Manini T., Seo A. Y., Leeuwenburgh C. (2008) Free Radic. Biol. Med. 44, 160–168 [DOI] [PubMed] [Google Scholar]
- 4.Whitman S. A., Wacker M. J., Richmond S. R., Godard M. P. (2005) Pflugers Arch. 450, 437–446 [DOI] [PubMed] [Google Scholar]
- 5.Dirks A. J., Leeuwenburgh C. (2005) Sports Med. 35, 473–483 [DOI] [PubMed] [Google Scholar]
- 6.Pollack M., Leeuwenburgh C. (2001) J. Gerontol. A Biol. Med. Sci. 56, B475–B482 [DOI] [PubMed] [Google Scholar]
- 7.Jejurikar S. S., Kuzon W. M., Jr. (2003) Apoptosis 8, 573–578 [DOI] [PubMed] [Google Scholar]
- 8.Siu P. M., Pistilli E. E., Butler D. C., Alway S. E. (2005) Am. J. Physiol. Cell Physiol. 288, C338–C349 [DOI] [PubMed] [Google Scholar]
- 9.Jejurikar S. S., Henkelman E. A., Cederna P. S., Marcelo C. L., Urbanchek M. G., Kuzon W. M., Jr. (2006) Exp. Gerontol. 41, 828–836 [DOI] [PubMed] [Google Scholar]
- 10.Sandri M., Minetti C., Pedemonte M., Carraro U. (1998) Lab. Invest. 78, 1005–1016 [PubMed] [Google Scholar]
- 11.Sandri M., El Meslemani A. H., Sandri C., Schjerling P., Vissing K., Andersen J. L., Rossini K., Carraro U., Angelini C. (2001) J. Neuropathol. Exp. Neurol. 60, 302–312 [DOI] [PubMed] [Google Scholar]
- 12.Kadrmas J. L., Beckerle M. C. (2004) Nat. Rev. Mol. Cell Biol. 5, 920–931 [DOI] [PubMed] [Google Scholar]
- 13.Fimia G. M., De Cesare D., Sassone-Corsi P. (2000) Mol. Cell. Biol. 20, 8613–8622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greene W. K., Baker E., Rabbitts T. H., Kees U. R. (1999) Gene 232, 203–207 [DOI] [PubMed] [Google Scholar]
- 15.Brown S., McGrath M. J., Ooms L. M., Gurung R., Maimone M. M., Mitchell C. A. (1999) J. Biol. Chem. 274, 27083–27091 [DOI] [PubMed] [Google Scholar]
- 16.Taniguchi Y., Furukawa T., Tun T., Han H., Honjo T. (1998) Mol. Cell. Biol. 18, 644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cowling B. S., McGrath M. J., Nguyen M. A., Cottle D. L., Kee A. J., Brown S., Schessl J., Zou Y., Joya J., Bönnemann C. G., Hardeman E. C., Mitchell C. A. (2008) J. Cell Biol. 183, 1033–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schessl J., Zou Y., McGrath M. J., Cowling B. S., Maiti B., Chin S. S., Sewry C., Battini R., Hu Y., Cottle D. L., Rosenblatt M., Spruce L., Ganguly A., Kirschner J., Judkins A. R., Golden J. A., Goebel H. H., Muntoni F., Flanigan K. M., Mitchell C. A., Bönnemann C. G. (2008) J. Clin. Invest. 118, 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schessl J., Taratuto A. L., Sewry C., Battini R., Chin S. S., Maiti B., Dubrovsky A. L., Erro M. G., Espada G., Robertella M., Saccoliti M., Olmos P., Bridges L. R., Standring P., Hu Y., Zou Y., Swoboda K. J., Scavina M., Goebel H. H., Mitchell C. A., Flanigan K. M., Muntoni F., Bönnemann C. G. (2009) Brain 132, 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shalaby S., Hayashi Y. K., Nonaka I., Noguchi S., Nishino I. (2009) Neurology 72, 375–376 [DOI] [PubMed] [Google Scholar]
- 21.Quinzii C. M., Vu T. H., Min K. C., Tanji K., Barral S., Grewal R. P., Kattah A., Camaño P., Otaegui D., Kunimatsu T., Blake D. M., Wilhelmsen K. C., Rowland L. P., Hays A. P., Bonilla E., Hirano M. (2008) Am. J. Hum. Genet. 82, 208–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shalaby S., Hayashi Y. K., Goto K., Ogawa M., Nonaka I., Noguchi S., Nishino I. (2008) Neuromuscul. Disord. 18, 959–961 [DOI] [PubMed] [Google Scholar]
- 23.Windpassinger C., Schoser B., Straub V., Hochmeister S., Noor A., Lohberger B., Farra N., Petek E., Schwarzbraun T., Ofner L., W. N., Wagner K., H., Vincent J. B., Quasthoff S. (2008) Am. J. Hum. Genet. 82, 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasad K. V., Ao Z., Yoon Y., Wu M. X., Rizk M., Jacquot S., Schlossman S. F. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6346–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka M., Herr W. (1990) Cell 60, 375–386 [DOI] [PubMed] [Google Scholar]
- 26.McGrath M. J., Cottle D. L., Nguyen M. A., Coghill I. D., Robinson P. A., Holdsworth M., Cowling B. S., Hardeman E. C., Mitchell C. A., Brown S. (2006) J. Biol. Chem. 281, 7666–7683 [DOI] [PubMed] [Google Scholar]
- 27.Mizushima S., Nagata S. (1990) Nucleic Acids Res. 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cottle D. L., McGrath M. J., Cowling B. S., Coghill I. D., Brown S., Mitchell C. A. (2007) J. Cell Sci. 120, 1423–1435 [DOI] [PubMed] [Google Scholar]
- 29.Coghill I. D., Brown S., Cottle D. L., McGrath M. J., Robinson P. A., Nandurkar H. H., Dyson J. M., Mitchell C. A. (2003) J. Biol. Chem. 278, 24139–24152 [DOI] [PubMed] [Google Scholar]
- 30.Ljungberg K., Whitmore A. C., Fluet M. E., Moran T. P., Shabman R. S., Collier M. L., Kraus A. A., Thompson J. M., Montefiori D. C., Beard C., Johnston R. E. (2007) J. Virol. 81, 13412–13423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tidball J. G., Albrecht D. E., Lokensgard B. E., Spencer M. J. (1995) J. Cell Sci. 108, 2197–2204 [DOI] [PubMed] [Google Scholar]
- 32.Qin L. F., Lee T. K., Ng I. O. (2002) Life Sci. 70, 1677–1690 [DOI] [PubMed] [Google Scholar]
- 33.Daoud S. S., Munson P. J., Reinhold W., Young L., Prabhu V. V., Yu Q., LaRose J., Kohn K. W., Weinstein J. N., Pommier Y. (2003) Cancer Res. 63, 2782–2793 [PubMed] [Google Scholar]
- 34.Fortin A., MacLaurin J. G., Arbour N., Cregan S. P., Kushwaha N., Callaghan S. M., Park D. S., Albert P. R., Slack R. S. (2004) J. Biol. Chem. 279, 28706–28714 [DOI] [PubMed] [Google Scholar]
- 35.Jacobs S. B., Basak S., Murray J. I., Pathak N., Attardi L. D. (2007) Cell Death Differ. 14, 1374–1385 [DOI] [PubMed] [Google Scholar]
- 36.Py B., Slomianny C., Auberger P., Petit P. X., Benichou S. (2004) J. Immunol. 172, 4008–4017 [DOI] [PubMed] [Google Scholar]
- 37.Xue L., Chu F., Cheng Y., Sun X., Borthakur A., Ramarao M., Pandey P., Wu M., Schlossman S. F., Prasad K. V. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6925–6930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgan M. J., Madgwick A. J. (1999) Biochem. Biophys. Res. Commun. 255, 245–250 [DOI] [PubMed] [Google Scholar]
- 39.Lin F. T., Lai Y. J., Makarova N., Tigyi G., Lin W. C. (2007) J. Biol. Chem. 282, 37759–37769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon Y., Ao Z., Cheng Y., Schlossman S. F., Prasad K. V. (1999) Oncogene 18, 7174–7179 [DOI] [PubMed] [Google Scholar]
- 41.Lane B. P., Elias J., Drummond E. (1977) J. Histochem. Cytochem. 25, 69–72 [DOI] [PubMed] [Google Scholar]
- 42.Seale P., Sabourin L. A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M. A. (2000) Cell 102, 777–786 [DOI] [PubMed] [Google Scholar]
- 43.Severino A., Abbruzzese C., Manente L., Valderas A. A., Mattarocci S., Federico A., Starace G., Chersi A., Mileo A. M., Paggi M. G. (2007) J. Cell. Physiol. 212, 118–125 [DOI] [PubMed] [Google Scholar]
- 44.Henke A., Launhardt H., Klement K., Stelzner A., Zell R., Munder T. (2000) J. Virol. 74, 4284–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Padanilam B. J., Lewington A. J., Hammerman M. R. (1998) Kidney Int. 54, 1967–1975 [DOI] [PubMed] [Google Scholar]
- 46.Sandri M., Carraro U. (1999) Int. J. Biochem. Cell Biol. 31, 1373–1390 [DOI] [PubMed] [Google Scholar]
- 47.van den Eijnde S. M., van den Hoff M. J., Reutelingsperger C. P., van Heerde W. L., Henfling M. E., Vermeij-Keers C., Schutte B., Borgers M., Ramaekers F. C. (2001) J. Cell Sci. 114, 3631–3642 [DOI] [PubMed] [Google Scholar]
- 48.Arsic N., Zacchigna S., Zentilin L., Ramirez-Correa G., Pattarini L., Salvi A., Sinagra G., Giacca M. (2004) Mol. Ther. 10, 844–854 [DOI] [PubMed] [Google Scholar]
- 49.Raj G. V., Sekula J. A., Guo R., Madden J. F., Daaka Y. (2004) Prostate 61, 105–113 [DOI] [PubMed] [Google Scholar]
- 50.Deconinck N., Dan B. (2007) Pediatric Neurology 36, 1–7 [DOI] [PubMed] [Google Scholar]
- 51.A., Rudnicki M. A. (2008) Mol. Diagn. Ther. 12, 99–108 [DOI] [PubMed] [Google Scholar]
- 52.Briguet A., Courdier-Fruh I., Foster M., Meier T., Magyar J. P. (2004) Neuromuscul. Disord. 14, 675–682 [DOI] [PubMed] [Google Scholar]
- 53.Qin H., Du D., Zhu Y., Li J., Feng L., Liang Y., Han H. (2005) FEBS Lett. 579, 1220–1226 [DOI] [PubMed] [Google Scholar]
- 54.Qin H., Wang J., Liang Y., Taniguchi Y., Tanigaki K., Han H. (2004) Nucleic Acids Res. 32, 1492–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang L., Zhang H. W., Liang J., Niu X. L., Zhang S. Z., Feng L., Liang Y. M., Han H. (2008) Biochim. Biophys. Acta 1779, 805–810 [DOI] [PubMed] [Google Scholar]
- 56.Oka C., Nakano T., Wakeham A., de la Pompa J. L., Mori C., Sakai T., Okazaki S., Kawaichi M., Shiota K., Mak T. W., Honjo T. (1995) Development 121, 3291–3301 [DOI] [PubMed] [Google Scholar]
- 57.Buckingham M., Bajard L., Chang T., Daubas P., Hadchouel J., Meilhac S., Montarras D., Rocancourt D., Relaix F. (2003) J. Anat. 202, 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandri M., Massimino M. L., Cantini M., Giurisato E., Sandri C., Arslan P., Carraro U. (1998) Neurosci. Lett. 252, 123–126 [DOI] [PubMed] [Google Scholar]
- 59.Chu F., Borthakur A., Sun X., Barkinge J., Gudi R., Hawkins S., Prasad K. V. (2004) Apoptosis 9, 83–95 [DOI] [PubMed] [Google Scholar]
- 60.Ikezoe K., Nakagawa M., Osoegawa M., Kira J., Nonaka I. (2004) Acta Neuropathol. 107, 439–442 [DOI] [PubMed] [Google Scholar]
- 61.Chu F., Barkinge J., Hawkins S., Gudi R., Salgia R., Kanteti P. V. (2005) Cancer Res. 65, 5301–5309 [DOI] [PubMed] [Google Scholar]
- 62.E, S., Lai Y. J., Tsukahara R., Chen C. S., Fujiwara Y., Yue J., Yu J. H., Guo H., Kihara A., Tigyi G., Lin F. T. (2009) J. Biol. Chem. 284, 14558–14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.