Abstract
Estrogen receptor related receptor (ERR)α was one of the first identified (1988) orphan nuclear receptors. Many of the orphan receptors identified after ERRα were deorphanized in a timely manner and appreciated as key transcriptional regulators of metabolic pathways. ERRα, however, remains an orphan. Nevertheless, recent studies have defined regulatory mechanisms and transcriptional targets of ERRα, allowing this receptor to join ranks with other nuclear receptors that control metabolism. Notably, mice lacking ERRα show defects when challenged with stressors that require a ‘shift of gears’ in energy metabolism, such as exposure to cold, cardiac overload or infection. These findings establish the importance of ERRα for adaptive energy metabolism, and suggest that strategies targeting ERRα may be useful in fighting metabolic diseases.
Introduction
The nuclear receptor (NR) superfamily comprises a large number of ligand-regulated transcription factors that play important roles in development, reproduction and metabolism [1]. Cloning of cDNAs encoding nuclear receptors for known hormones, e.g. steroid hormones, was soon followed by the identification of structurally related proteins that had no known ligands and were classified as orphan receptors [2]. Estrogen receptor related receptors (ERR) α and β were the first orphan receptors described [3], followed by more than 30 other family members. The subsequent identification of ligands for many of these orphan receptors offered insights into their physiologic functions, and provided necessary experimental tools. For example, the recognition of lipids and known antidiabetic drugs as ligands of peroxisome proliferator activated receptors (PPARs), oxysterols as ligands of the liver X receptor, and bile acids as ligands of the farnesoid X receptor, enabled the elucidation of the role of these receptors in lipid, cholesterol and bile acid metabolism, respectively [4,5].
Even though ERRα was the first orphan NR identified, its role in metabolism has been appreciated only recently. Most initial studies focused on the roles of ERRα in estrogen signaling, cancer, bone differentiation and development. Such studies have been covered in other reviews [6-10] and will not be discussed here. This review focuses on the role of ERRα in regulating diverse aspects of oxidative metabolism, and the physiologic contexts where this role becomes important.
The ERR family
ERRα is one of three members of the ERR family. ERRα, ERRβ and ERRγ (NR3B1–3, also known as ERR1–3) possess the typical structural features of NRs: a central zinc finger DNA-binding domain that is highly conserved among the three members and enables recognition of DNA sites bearing TNAAGGTCA sequences (also known as ERR response elements or ERREs); a conserved C-terminal domain with a putative ligand binding domain (LBD) and interaction surfaces for coactivators and corepressors; and a less conserved N-terminal region [6]. Despite their similarity to estrogen receptors, ERRs are not activated by estrogens or estrogen-like molecules. Instead, ERRs attain active LBD conformations in the absence of ligand, consistent with the notion that these receptors are constitutively active [11-13]. For ERRα in particular, screening and structure-guided design efforts have identified synthetic inverse agonists that inhibit ERRα function, but no agonists, suggesting that ERRα may stay an orphan [14-17].
Regulation of ERRα: the roles of PGC-1α and PGC-1β
If small molecule ligands are not required to activate ERRα, how is ERRα function controlled? Insights into the regulation of ERRα came from studies of the transcriptional coactivators (Box 1) PGC-1α and PGC-1β, which integrate nutrient and energy state signals and control the expression of genes important for mitochondrial biogenesis, oxidative metabolism, oxidative stress responses and, in the case of PGC-1α, gluconeogenesis [18,19]. The ability of PGC-1 coactivators to induce target genes relies on their recruitment at target genomic regulatory sites via physical interactions with nuclear receptors and other transcription factors [18,19]. In the case of nuclear receptors, PGC-1 amphipathic helices that bear LxxLL-type motifs bind a hydrophobic groove in the receptors' LBD. Given the conserved nature of the LBD across the NR family, it is not surprising that PGC-1α and PGC-1β interact with ERRα. However, several observations suggest that the interactions of PGC-1α/β with ERRα are of unique importance for ERRα and PGC-1 function.
Box 1: Coactivators and corepressors.
Coregulators interact with DNA-binding transcription factors and assist them in the regulation of transcription of target genes. Coregulators that promote the activation of gene transcription are referred to as coactivators. Conversely, corepressors assist transcription factors to repress gene expression. Coactivators and corepressors do not bind DNA by themselves.
First, PGC-1α has a protein interaction surface that is dedicated specifically to the ERRs. PGC-1α harbors three LxxLL motifs, two of which, (L2 and L3), serve as NR interaction sites [20-24]. The L2 site bears a consensus LxxLL motif, similar to ones found in other receptor coactivators, and interacts with most NRs, including the three ERRs. The L3 site is an ERR-dedicated site, with an atypical LLxYL motif that interacts potently and specifically with the ERRs [23-25]. The ERR-specific L3 site suggests that ERRs are not in competition with other NRs for binding PGC-1α, and may have the ‘undivided attention’ of this coactivator. Moreover, the ERR-specific site has enabled the construction of PGC-1α mutants that interact only with ERRs, and that have been used as powerful tools to dissect the role of ERRs versus other NRs downstream of PGC-1α [24,26].
Second, ERRα, which is a weak activator of transcription in cells with low PGC-1 activity and where other NRs are transcriptionally active, is transformed to a potent transcriptional activator when PGC-1α or PGC-1β is introduced into the system [23,24,27]. The striking dependency of ERRα on PGC-1 coactivators likely reflects the much higher affinity of ERRα for PGC-1 over other widely used NR coactivators [13,25], and suggests that ERRα activity is primarily controlled by interaction with ‘protein ligands,’ such as PGC-1α and PGC-1β, rather than small molecule ligands.
Third, PGC-1α regulates not only the activity but also the expression levels of ERRα [24]. This results from ERRα binding to conserved ERREs in its own promoter, inducing transcription in a positive autoregulatory loop upon activation by PGC-1α [28-30]. Consistent with the ability of PGC-1 coactivators to induce ERRα expression, ERRα mRNA levels are highest in tissues that have high levels of PGC-1α or PGC-1β, such as brown adipose tissue, heart, kidney, small intestine and skeletal muscle [19,24,31-34]. Moreover, ERRα mRNA levels are increased in response to physiologic signals that induce PGC-1α expression, such as fasting in liver [34], cold exposure in brown adipose tissue and muscle [24] and exercise in skeletal muscle [35].
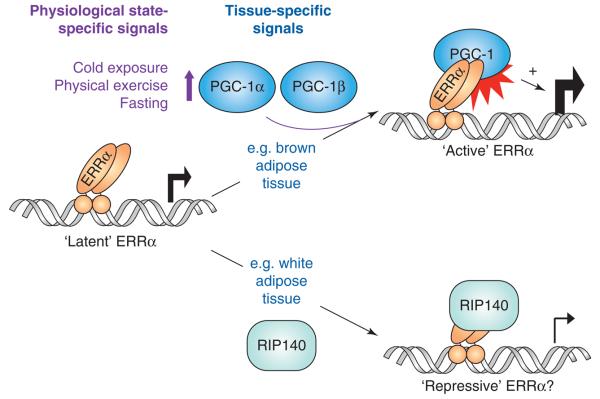
In summary, ERRα is activated by interactions with PGC-1 coactivators and, therefore, by tissue- and physiological state-specific signals that determine PGC-1α/β levels and activity (Figure 1). Additional signals are likely to impact ERRα function via other mechanisms, including ERRα phosphorylation and sumoylation [36-39]. Importantly, ERRα also interacts with the corepressor (Box 1) RIP140, which can suppress ERRα-dependent transcription and is thus likely to antagonize PGC-1α/β function at ERRα target genes [40] (Figure 1).
Figure 1.
ERRα is regulated by protein-protein interactions with coregulators. The ability of ERRα to affect gene expression is controlled by its association with specific coregulators, such as the coactivators PGC-1α and PGC-1β and the corepressor RIP140. Consequently, ERRα activity is regulated by the tissue- and physiologic state-specific signals that regulate levels and activity of PGC-1α/β and RIP140. For example, ERRα is a positive regulator of the TCA cycle gene IDH3A in brown adipose tissue, which expresses high levels of PGC-1α/β, but a negative regulator of the OxPhos gene SDHB in white adipocytes, which express high levels of RIP140 [44,54]. Gene-specific factors, such as the type of ERRE sequence and the proteins binding in the vicinity of the ERRE, may also contribute to the ability of ERRα to interact with PGC-1α/β versus RIP140.
Cellular function of ERRα: regulation of energy metabolism genes
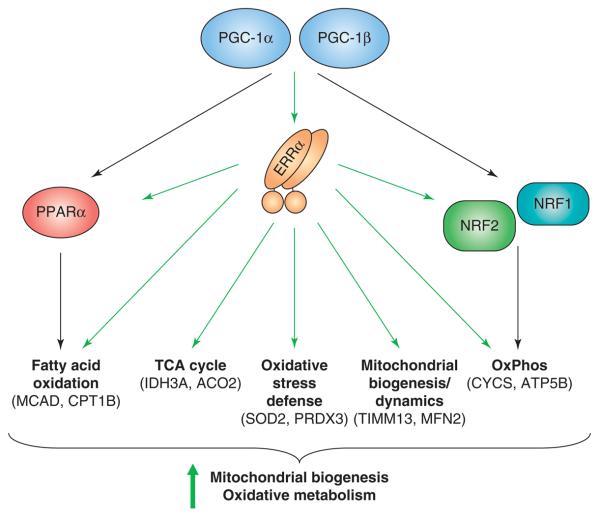
The finding that ERRα is activated by PGC-1α/β has given insights into the cellular and physiologic context of ERRα function and suggested a role for ERRα in PGC-1 signaling. Indeed, several studies have established not only that ERRα, like PGC-1α/β, regulates the expression of energy metabolism genes, but also that ERRα is a key downstream effector of PGC-1α/β. First, whereas PGC-1α mutants that lack the ability to interact with all NRs (including ERRs) fail to induce expression of energy metabolism genes, PGC-1α mutants that allow interaction with ERRs but not other NRs are as efficient as wild-type PGC-1α in enhancing gene expression [26,41]. Second, inhibition of ERRα activity by either siRNA or an ERRα inverse agonist impairs the ability of PGC-1α to induce expression of energy metabolism genes and enhance mitochondrial biogenesis and oxidative capacity [26,29]. Conversely, overexpression of ERRα in primary cardiomyocytes, which express endogenous PGC-1α/β, induces many PGC-1 target genes involved in substrate utilization and energy production [42]. Interestingly, while ERRα overexpression in cells lacking endogenous PGC-1α and PGC-1β fails to induce target gene expression, consistent with a requirement for PGC-1 co-activators (Figure 1), overexpression of a chimeric, constitutively active form of ERRα bearing the VP16 activation domain bypasses this requirement and induces mitochondrial gene expression and biogenesis [26]. Collectively, these and later studies have established that ERRα activated by PGC-1α or PGC-1β induces genes with roles in lipid transport [42,43], fatty acid oxidation [24,26,29,41-44], TCA cycle [24,41,44-46], oxidative phosphorylation [26,29,41,42,44-46], mitochondrial biogenesis [26,44], mitochondrial dynamics [35,47] and oxidative stress defense [45] (Figure 2 and Table 1). Accordingly, ERRα, depending on the cell type, has been shown to promote lipid uptake and oxidation, mitochondrial biogenesis, respiratory capacity and ROS production, and to inhibit glucose oxidation [26,29,42,43,46,48].
Figure 2.
ERRα regulates genes important for mitochondrial biogenesis and oxidative metabolism. ERRα acts as a downstream effector of PGC-1α and PGC-1β in the regulation of expression of genes with roles in fatty acid oxidation, the TCA cycle, oxidative stress responses, mitochondrial biogenesis and dynamics, and oxidative phosphorylation (OxPhos). It acts both directly on genes encoding mitochondrial structural components and enzymes, as well as indirectly by enhancing expression of other transcriptional factors, such as PPARD, NRF1 and NRF2/GABP, that regulate these pathways.
Table 1.
ERRα target genes in metabolism
| Metabolic pathway |
Gene | Regulated by ERRα in | Direct target in | Promoter study? |
|---|---|---|---|---|
| Fatty acid oxidation |
Acadm (Mcad) | HeLa [24], SAOS2 [26], C2C12 [29], MDA-MB-231 [15], HL-1 atrial myocytes [49], soleus muscle [42], BAT [44], heart [52] |
BAT [44], heart [49] | Yes [23] |
| Cpt1b | MEFs [45], BAT [44] | BAT [44] | – | |
| TCA cycle | Aco2 | Macrophages [46]; BAT [44] | BAT [44], macrophages [46] | – |
| Idh3a | SAOS2 [26], HepG2 [41,56], macrophages [46]; BAT [44] | BAT [44], macrophages [46] | – | |
| Sdha | Macrophages [46] | Macrophages [46] | – | |
| Sdhd | Macrophages [46] | Heart [49], macrophages [46] | – | |
| OxPhos | Atp5b | SAOS2 [26], HepG2 [41,56], macrophages [46]; liver [56], BAT [44] |
BAT [44], heart [49], macrophages [46] | Yes [26] |
| Atp5 g3 | Macrophages [46] | Macrophages [46] | – | |
| Coq7 | Macrophages [46] | Macrophages [46] | – | |
| Cox8a | Macrophages [46] | Macrophages [46] | – | |
| Cycs | SAOS2 [26], HepG2 [41,56], MEFs [45], macrophages [46]; BAT [44], heart [52] |
BAT [44], heart [49], macrophages [46] | Yes [26,49] | |
| Ndufs7 | Macrophages [46] | Macrophages [46] | – | |
| Mitochondrial transport |
Timm13a | Macrophages [46] | Macrophages [46] | – |
| Slc25a4 (Ant1) | HepG2 [41], heart [52] | Heart [49], macrophages [46] | – | |
| Mitochondrial dynamics |
Mfn2 | SAOS2 [35] | SAOS2 [35], HeLa [47] | Yes [47] |
| Regulator of metabolism |
Esrra | C2C12 [29], MDA-MB-231 [15], | Heart [49] | Yes [28,29] |
| Ppara | HepG2 [41], BAT [44], heart [52] | L6 myoblasts [42] | Yes [42] | |
| Gabpa (Nrf2A) | C2C12 [29], FAO hepatoma [29], MEFs [45] | Heart [49], macrophages [46] | Yes [29] | |
| Nrip1 (Rip140) | 3T3-L1 [66] | 3T3-L1 [66] | Yes [66] | |
| Lipid transport | Apoa4 | Intestine [43] | Caco-2 [43] | Yes [43] |
| Gluco/Glycero- neogenesis |
Pck1 (Pepck)* | H4IIE hepatoma cells [56]; liver [56] | H4IIE [56] | Yes [34,56] |
ERRα targets shown here are ones with roles in metabolism, regulated by endogenous ERRα (i.e. genes whose expression is reduced, as determined by quantitative RT–PCR, in cells when ERRα is inhibited by siRNA or the inverse agonist XCT790, cells derived from ERRα null mice, or tissues of ERRα null mice) and directly recognized by ERRα (i.e. direct ERRα targets, as determined by quantitative ChIP). Target genes whose ERRE-containing regulatory regions have been tested for conferring ERRα/PGC-1 regulation to reporter genes in transient transfections are indicated by ‘yes’ under ‘promoter study’. Notably, some targets are regulated by ERRα in a cell-type specific manner. For example, Acadm is not regulated by ERRα in macrophages [46] and Gabpa is not regulated by ERRα in SAOS2 [26] and macrophages [46].
Pepck is repressed by ERRα (all other targets shown here are activated by ERRα).
The effects of ERRα on expression of metabolism genes are mediated by direct and indirect actions at the promoters of these genes. Bioinformatic analyses show a statistically significant enrichment of motifs corresponding to ERREs in genes induced by PGC-1α [29]. The direct action of ERRα, as opposed to other NRs that recognize the same motif, is supported by the detection of endogenous ERRα binding at these sites [35,42-44,48,49]. Furthermore, the functional significance of some of these ERREs has been validated in vitro by showing that these response elements confer ERRα- and PGC-1α-dependent regulation of reporter genes in transfection assays [26,42,43,47-51]. In addition to direct actions, ERRα can enhance mitochondrial gene expression indirectly by inducing the expression of other transcriptional regulators of mitochondrial genes, such as NRF1, the GABPα subunit of GABP/NRF2, and PPARα [29,42] (Figure 2). The ability of ERRα to induce these transcription factors, which may be cell-type specific [24,44], is likely to expand the spectrum of ERRα-dependent genes, as well as increase the magnitude of regulation for genes where ERRα acts both directly and indirectly.
The role of ERRα as a regulator of energy metabolism has been confirmed and broadened by the unbiased identification of genome-wide direct targets of ERRα in the heart, a tissue that expresses high levels of all three ERRs [49]. Using chromatin immunoprecipitation (ChIP) and genomic DNA arrays (ChIP-on-chip), Dufour et al. detected ERRα binding at the promoters of a large number of energy metabolism genes, as well as genes involved in energy sensing, amino acid and carbohydrate metabolism, Ca+2 homeostasis, contractile function and other cellular processes. Strikingly, a comparison of ERRα and ERRγ binding revealed largely similar target genes, suggesting a great overlap in the function of ERRs and implicating ERRγ, and likely ERRβ, in the regulation of cellular energy metabolism [49]. It is too early yet to tell the extent to which all targets identified by ChIP-on-chip are positively regulated by ERRα and ERRγ. The expression of many of them is not decreased in the hearts of ERRα null mice, which may be due to compensation by ERRβ and/or ERRγ [49,52]. It will be interesting to combine genome-wide ChIP and expression analyses in a cell system where there is no compensation by other ERRs, to elucidate the positive and negatively regulated ERRα targets.
Most of the studies have focused on the ability of ERRα to activate energy metabolism genes. However, ERRα can also act as a repressor [53]. At least two distinct modes of repression by ERRα have been observed in the context of metabolism. First, ERRα can act as a downstream effector of the corepressor RIP140 [40,54]. RIP140 acts as the antithesis of PGC-1α, suppressing many of the energy metabolism genes that PGC-1α activates [55] (Figure 1). In fact, the ability of RIP140 to repress expression of Glut4 and SDHB (identified as direct ERR targets [49]) relies on endogenous ERRα [54]. Thus, ERRα may function as a sensor of energy demand signals by interpreting the relative levels and activity of PGC-1s versus RIP140, and accordingly acting as an activator or repressor, possibly in a gene-specific manner (Figure 1). A different mode of repression has been shown in the case of the gluconeogenic/glyceroneogenic gene PEPCK, where ERRα binding at a complex response element inhibits PGC-1α recruitment and PEPCK induction [56]. It is not clear why ERRα at the PEPCK promoter does not recruit PGC-1α, as at other ERREs. One possibility is that the sequence of the PEPCK ERRE and/or the proximity to other elements directs ERRα binding as a monomer, which cannot recruit PGC-1α [36,57].
Physiological function of ERRα: lessons from mouse models
Mice genetically engineered to lack ERRα have been instrumental for understanding the physiological role of ERRα in the regulation of energy metabolism. Initial characterization of ERRα null mice showed them to be viable and fertile, with no gross anatomical alterations except for a small but significant decrease in adipose mass [58]. Gene expression analyses revealed modest decreases in the expression of some ERRα metabolic targets, but also upregulation of others and no evident organ malfunction [42,43,58]. ERRα null mice are not only lean but also resistant to high fat diet-induced obesity, a phenotype that does not appear congruent with the role of ERRα in promoting mitochondrial oxidative function. Given the multitude of transcription factors that regulate energy metabolism genes [59] and the fact that ERRα function is context (tissue- and signal-) dependent, the apparent lack of phenotype, other than the resistance to diet-induced obesity, may not be so surprising. Still, the cellular role of ERRα downstream of PGC-1α and PGC-1β suggested an important role for ERRα in metabolic adaptations to signals of increased energy demands. Indeed, three studies in the last year have shown roles for ERRα in the adaptation to stressful states that require increased use of energy stores.
In the first study, ERRα null mice were shown to be deficient in maintaining their core body temperature when exposed to cold [44]. Adaptive thermogenesis in rodents relies on heat production in brown adipose tissue (BAT), which is rich in mitochondria and expresses high levels of PGC-1D, PGC-1β and ERRα. Brown adipocytes of ERRα null mice have decreased expression of FAO, TCA cycle and OxPhos genes, decreased mitochondrial mass and oxidative capacity, and increased lipid accumulation. Despite the normal induction of genes important for thermogenesis, such as uncoupling protein 1 and deiodinase 2, ERRα null mice become hypothermic when exposed to even mild cold (13°C) temperatures, reflecting their decreased capacity for respiration, and thereby, for the generation of energy necessary for thermogenesis [44].
In the second study, ERRα was shown to be important in the heart for the bioenergetic and functional adaptation to cardiac pressure overload [52]. In the absence of stress, hearts of ERRα null mice show deregulated expression of several energy metabolism genes but normal myofibrillar structure, mitochondrial morphology and mass, and contractile function. When challenged with increased workload imposed by chronic pressure overload, ERRα null hearts develop signs of heart failure, such as chamber dilatation, left ventricular systolic dysfunction, wall thinning and fibrosis. At the bioenergetic level, ERRα null hearts have a decreased capacity for ATP synthesis and exhibit defects in their high energy phosphate reserves after E-adrenergic stimulation or ischemia/reperfusion [52]. At the gene expression level, the role of ERRα in maintaining expression of energy metabolism genes becomes accentuated in chronically overloaded hearts, suggesting that defects in expression of these genes underlies the energetic defect and the development of contractile dysfunction [52].
The third study using ERRα mice reveals a key role for ERRα in IFN-γ stimulated oxidative metabolism in macrophages [46]. IFN-γ activates the antimicrobial actions of macrophages by inducing the expression of a wide range of genes. It also induces the expression of PGC-1β, ERRα and genes important for mitochondrial metabolism, and enhances respiration. ERRα is required for the IFN-γ induced metabolic response. ERRα null macrophages show compromised mitochondrial gene induction, an inability to increase mitochondrial respiration and mitochondrial ROS, and decreased clearance of the intracellular bacterial pathogen Listeria monocytogenes in response to IFN-γ. As a result, ERRα null mice, and wild-type mice whose bone marrow-derived cells are ERRα null, have impaired survival when infected with L monocytogenes [46].
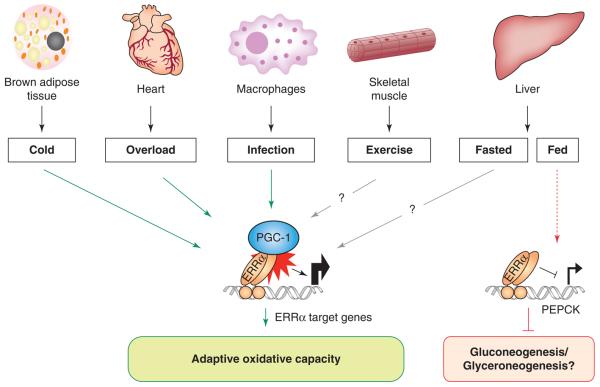
The common theme in all three studies is that ERRα null mice are unable to mount proper responses to stressors that require shifts in energy production or utilization (Figure 3). Although small, the bioenergetic defects are significant and compromise organismal survival. By extension, it can be hypothesized that ERRα null mice show defects in other states of high energy demands, e.g. physical exercise. Indeed, ERRα null mice show decreased physical activity and decreased capacity for exercise when running on treadmills (J.A.V. and A.K., unpublished data). It will be interesting to test other paradigms, e.g. whether small defects seen in intestinal lipid absorption of ERRα null mice [43] are accentuated in states requiring increased nutrient absorption.
Figure 3.
ERRα function in vivo is important for adaptive metabolic responses under conditions of increased energetic demand. Mice lacking ERRα show compromised responses to stresses imposed by exposure to cold, cardiac overload, and bacterial infection. In all three cases, a decreased capacity for oxidative metabolism leads to reduced fitness [44,46,52]. A role for ERRα in skeletal muscle is supported by findings that ERRα null mice have a reduced capacity for exercise (Villena and Kralli, unpublished data). ERRα may have additional roles in other states, as indicated by the increased expression of the gluconeogenic/glyceroneogenic enzyme PEPCK in livers of fed mice [56].
Do these findings mean that ERRα function is not important for basal energy metabolism? And why are ERRα null mice lean and resistant to high fat diet-induced obesity? Although the answers to these questions are not yet clear, the following observations give some insights. First, increased expression of other regulators, as seen with ERRJ and PGC-1α in heart [42,52], may compensate for the lack of ERRα and mask ERRα function in basal energy metabolism. Overcompensation by ERRγ/PGC-1α or differences in genes targeted by ERRα and ERRγ may also contribute to the increased expression of some ERR targets and the lean phenotype in ERRα null mice. Second, the role of ERRα in specific tissues or under particular physiological states may promote energy intake and storage. Such a role is consistent with the notion of ERRα as a factor that enables the organism to cope with energetically stressful states, e.g. by ensuring availability of energy stores. In line with this role, ERRα null mice show defects in intestinal lipid absorption and decreased lipid stores in WAT [43,58]. ERRα may act with PGC-1α to promote transcription of genes with roles in lipid transport [42,43] in the intestine, where ERRα and PGC-1α are highly expressed, and/or act with the corepressor RIP140 to suppress energy expenditure genes [54] in WAT, where RIP140 levels are high. Finally, it is possible that effects of ERRα on development and differentiation, e.g. by ERRα promoting adipogenesis [60], might underlie some of the phenotypes in the ERRα null mice.
Concluding remarks
In the last five years, extraordinary progress has been made in our understanding of ERRα function. In response to activation by PGC-1α and/or PGC-1β, ERRα regulates genes important for mitochondrial biogenesis and function. Mice lacking ERRα show tissue- and signal-specific defects in mitochondrial biogenesis and respiratory capacity, leading to cold sensitivity, cardiac malfunction and susceptibility to infection. Mitochondrial dysfunction and/or decreased PGC-1α/β levels have been associated with insulin resistance, type 2 diabetes, cardiovascular disease and neurodegenerative disorders [61-65]. The contribution of ERRα to these disorders is not yet clear. Neither is it known whether increases in ERRα activity could have any harmful effects. Nevertheless, the expectation is that enhancing ERRα activity should enhance mitochondrial oxidative metabolism, as well as the oxidative stress defense system, and benefit patients suffering from such diseases. The emerging evidence that ERRγ shares the ability to regulate energy metabolism genes further raises expectations for the therapeutic potential of ERR agonists. It is clearly essential to understand the specific roles and contributions of each ERR to mitochondrial function in different tissues and physiological states.
Acknowledgements
We thank Ben Hock and Ulrich Müller for discussions and comments, and NIDDK for funding (DK064951 and DK074660 to A.K.).
Glossary
- ACO2
aconitase 2
- ATP5B
ATP synthase, H+ transporting, mitochondrial F1 complex, beta subunit precursor
- ChIP
chromatin immunoprecipitation
- CPT1b
carnitine palmitoyltransferase 1b (muscle)
- CYCS
cytochrome c, somatic
- ERR
estrogen-receptor related receptor
- ERRE
ERR response element
- FAO
fatty acid oxidation
- GABP
GA binding protein
- GLUT4
glucose transporter 4 (insulin responsive)
- IDH3A
isocitrate dehydrogenase 3, alpha subunit
- LBD
ligand binding domain
- MCAD
medium-chain acyl-CoA dehydrogenase
- MFN
mitofusin
- NR
nuclear receptor
- NRF
nuclear respiratory factor
- OxPhos
oxidative phosphorylation
- PEPCK
phosphoenol pyruvate carboxykinase
- PGC
PAR gamma coactivator
- PPAR
peroxisome proliferator activated receptor
- PRDX3
peroxiredoxin 3
- RIP140
receptor-interacting protein 140 (also known as nuclear receptor interacting protein 1 or NRIP1)
- SDHB
succinate dehydrogenase complex, subunit B
- SOD
superoxide dismutase
- TCA
tricarboxylic acid
- TIM
translocase of inner mitochondrial membrane
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Malley B. The steroid receptor superfamily: more excitement predicted for the future. Mol. Endocrinol. 1990;4:363–369. doi: 10.1210/mend-4-3-363. [DOI] [PubMed] [Google Scholar]
- 3.Giguere V, et al. Identification of a new class of steroid hormone receptors. Nature. 1988;331:91–94. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- 4.Kliewer SA, et al. Orphan nuclear receptors: shifting endocrinology into reverse. Science. 1999;284:757–760. doi: 10.1126/science.284.5415.757. [DOI] [PubMed] [Google Scholar]
- 5.Chawla A, et al. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 6.Giguere V. To ERR in the estrogen pathway. Trends Endocrinol. Metab. 2002;13:220–225. doi: 10.1016/s1043-2760(02)00592-1. [DOI] [PubMed] [Google Scholar]
- 7.Bonnelye E, Aubin JE. Estrogen receptor-related receptor alpha: a mediator of estrogen response in bone. J. Clin. Endocrinol. Metab. 2005;90:3115–3121. doi: 10.1210/jc.2004-2168. [DOI] [PubMed] [Google Scholar]
- 8.Bardet PL, et al. Studying non-mammalian models? Not a fool's ERRand! Trends Endocrinol. Metab. 2006;17:166–171. doi: 10.1016/j.tem.2006.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ariazi EA, Jordan VC. Estrogen-related receptors as emerging targets in cancer and metabolic disorders. Curr. Top. Med. Chem. 2006;6:203–215. doi: 10.2174/1568026610606030203. [DOI] [PubMed] [Google Scholar]
- 10.Stein RA, McDonnell DP. Estrogen-related receptor alpha as a therapeutic target in cancer. Endocr. Relat. Cancer. 2006;13(Suppl 1):S25–S32. doi: 10.1677/erc.1.01292. [DOI] [PubMed] [Google Scholar]
- 11.Greschik H, et al. Structural and functional evidence for ligand-independent transcriptional activation by the estrogen-related receptor 3. Mol. Cell. 2002;9:303–313. doi: 10.1016/s1097-2765(02)00444-6. [DOI] [PubMed] [Google Scholar]
- 12.Horard B, Vanacker JM. Estrogen receptor-related receptors: orphan receptors desperately seeking a ligand. J. Mol. Endocrinol. 2003;31:349–357. doi: 10.1677/jme.0.0310349. [DOI] [PubMed] [Google Scholar]
- 13.Kallen J, et al. Evidence for ligand-independent transcriptional activation of the human estrogen-related receptor alpha (ERRalpha): crystal structure of ERRalpha ligand binding domain in complex with peroxisome proliferator-activated receptor coactivator-1alpha. J. Biol. Chem. 2004;279:49330–49337. doi: 10.1074/jbc.M407999200. [DOI] [PubMed] [Google Scholar]
- 14.Busch BB, et al. Identification of a selective inverse agonist for the orphan nuclear receptor estrogen-related receptor alpha. J. Med. Chem. 2004;47:5593–5596. doi: 10.1021/jm049334f. [DOI] [PubMed] [Google Scholar]
- 15.Willy PJ, et al. Regulation of PPARgamma coactivator 1alpha (PGC-1alpha) signaling by an estrogen-related receptor alpha (ERRalpha) ligand. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8912–8917. doi: 10.1073/pnas.0401420101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyatt SM, et al. On the intractability of estrogen-related receptor alpha as a target for activation by small molecules. J. Med. Chem. 2007;50:6722–6724. doi: 10.1021/jm7012387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kallen J, et al. Crystal structure of human estrogen-related receptor alpha in complex with a synthetic inverse agonist reveals its novel molecular mechanism. J. Biol. Chem. 2007;282:23231–23239. doi: 10.1074/jbc.M703337200. [DOI] [PubMed] [Google Scholar]
- 18.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 20.Vega RB, et al. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tcherepanova I, et al. Modulation of estrogen receptor-alpha transcriptional activity by the coactivator PGC-1. J. Biol. Chem. 2000;275:16302–16308. doi: 10.1074/jbc.M001364200. [DOI] [PubMed] [Google Scholar]
- 22.Knutti D, et al. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9713–9718. doi: 10.1073/pnas.171184698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huss JM, et al. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J. Biol. Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 24.Schreiber SN, et al. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha) J. Biol. Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 25.Gaillard S, et al. Definition of the molecular basis for estrogen receptor-related receptor-alpha-cofactor interactions. Mol. Endocrinol. 2007;21:62–76. doi: 10.1210/me.2006-0179. [DOI] [PubMed] [Google Scholar]
- 26.Schreiber SN, et al. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamei Y, et al. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laganiere J, et al. A polymorphic autoregulatory hormone response element in the human estrogen-related receptor alpha (ERRalpha) promoter dictates peroxisome proliferator-activated receptor gamma coactivator-1alpha control of ERRalpha expression. J. Biol. Chem. 2004;279:18504–18510. doi: 10.1074/jbc.M313543200. [DOI] [PubMed] [Google Scholar]
- 29.Mootha VK, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu D, et al. Estrogen-related receptor-gamma and peroxisome proliferator-activated receptor-gamma coactivator-1alpha regulate estrogen-related receptor-alpha gene expression via a conserved multi-hormone response element. J. Mol. Endocrinol. 2005;34:473–487. doi: 10.1677/jme.1.01586. [DOI] [PubMed] [Google Scholar]
- 31.Sladek R, et al. The orphan nuclear receptor estrogen-related receptor alpha is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol. Cell. Biol. 1997;17:5400–5409. doi: 10.1128/mcb.17.9.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vega RB, Kelly DP. A role for estrogen-related receptor alpha in the control of mitochondrial fatty acid beta-oxidation during brown adipocyte differentiation. J. Biol. Chem. 1997;272:31693–31699. doi: 10.1074/jbc.272.50.31693. [DOI] [PubMed] [Google Scholar]
- 33.Kressler D, et al. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. J. Biol. Chem. 2002;277:13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- 34.Ichida M, et al. Identification of a specific molecular repressor of the peroxisome proliferator-activated receptor gamma Coactivator-1 alpha (PGC-1alpha) J. Biol. Chem. 2002;277:50991–50995. doi: 10.1074/jbc.M210262200. [DOI] [PubMed] [Google Scholar]
- 35.Cartoni R, et al. Mitofusins 1/2 and ERRalpha expression are increased in human skeletal muscle after physical exercise. J. Physiol. 2005;567:349–358. doi: 10.1113/jphysiol.2005.092031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barry JB, Giguere V. Epidermal growth factor-induced signaling in breast cancer cells results in selective target gene activation by orphan nuclear receptor estrogen-related receptor alpha. Cancer Res. 2005;65:6120–6129. doi: 10.1158/0008-5472.CAN-05-0922. [DOI] [PubMed] [Google Scholar]
- 37.Ariazi EA, et al. Estrogen-related receptor alpha1 transcriptional activities are regulated in part via the ErbB2/HER2 signaling pathway. Mol. Cancer Res. 2007;5:71–85. doi: 10.1158/1541-7786.MCR-06-0227. [DOI] [PubMed] [Google Scholar]
- 38.Vu EH, et al. Phosphorylation-dependent sumoylation of estrogen-related receptor alpha1. Biochemistry. 2007;46:9795–9804. doi: 10.1021/bi700316g. [DOI] [PubMed] [Google Scholar]
- 39.Tremblay AM, et al. Phosphorylation-Dependent Sumoylation Regulates Estrogen-Related Receptor-{alpha} and -{gamma} Transcriptional Activity through a Synergy Control Motif. Mol. Endocrinol. 2008;22:570–584. doi: 10.1210/me.2007-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castet A, et al. Receptor-interacting protein 140 differentially regulates estrogen receptor-related receptor transactivation depending on target genes. Mol. Endocrinol. 2006;20:1035–1047. doi: 10.1210/me.2005-0227. [DOI] [PubMed] [Google Scholar]
- 41.Gaillard S, et al. Receptor-selective coactivators as tools to define the biology of specific receptor-coactivator pairs. Mol. Cell. 2006;24:797–803. doi: 10.1016/j.molcel.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 42.Huss JM, et al. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrier JC, et al. Estrogen-related receptor alpha (ERRalpha) is a transcriptional regulator of apolipoprotein A-IV and controls lipid handling in the intestine. J. Biol. Chem. 2004;279:52052–52058. doi: 10.1074/jbc.M410337200. [DOI] [PubMed] [Google Scholar]
- 44.Villena JA, et al. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1418–1423. doi: 10.1073/pnas.0607696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rangwala SM, et al. Estrogen-related receptor alpha is essential for the expression of antioxidant protection genes and mitochondrial function. Biochem. Biophys. Res. Commun. 2007;357:231–236. doi: 10.1016/j.bbrc.2007.03.126. [DOI] [PubMed] [Google Scholar]
- 46.Sonoda J, et al. Nuclear receptor ERR alpha and coactivator PGC-1 beta are effectors of IFN-gamma-induced host defense. Genes Dev. 2007;21:1909–1920. doi: 10.1101/gad.1553007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soriano FX, et al. Evidence for a mitochondrial regulatory pathway defined by peroxisome proliferator-activated receptor-gamma coactivator-1 alpha, estrogen-related receptor-alpha, and mitofusin 2. Diabetes. 2006;55:1783–1791. doi: 10.2337/db05-0509. [DOI] [PubMed] [Google Scholar]
- 48.Wende AR, et al. PGC-1alpha coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: a mechanism for transcriptional control of muscle glucose metabolism. Mol. Cell. Biol. 2005;25:10684–10694. doi: 10.1128/MCB.25.24.10684-10694.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dufour CR, et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 50.Araki M, Motojima K. Identification of ERRalpha as a specific partner of PGC-1alpha for the activation of PDK4 gene expression in muscle. FEBS J. 2006;273:1669–1680. doi: 10.1111/j.1742-4658.2006.05183.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. Estrogen-related receptors stimulate pyruvate dehydrogenase kinase isoform 4gene expression. J. Biol. Chem. 2006;281:39897–39906. doi: 10.1074/jbc.M608657200. [DOI] [PubMed] [Google Scholar]
- 52.Huss JM, et al. The nuclear receptor ERRalpha is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007;6:25–37. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 53.Johnston SD, et al. Estrogen-related receptor alpha 1 functionally binds as a monomer to extended half-site sequences including ones contained within estrogen-response elements. Mol. Endocrinol. 1997;11:342–352. doi: 10.1210/mend.11.3.9897. [DOI] [PubMed] [Google Scholar]
- 54.Powelka AM, et al. Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J. Clin. Invest. 2006;116:125–136. doi: 10.1172/JCI26040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christian M, et al. Metabolic regulation by the nuclear receptor corepressor RIP140. Trends Endocrinol. Metab. 2006;17:243–250. doi: 10.1016/j.tem.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 56.Herzog B, et al. Estrogen-related receptor alpha is a repressor of phosphoenolpyruvate carboxykinase gene transcription. J. Biol. Chem. 2006;281:99–106. doi: 10.1074/jbc.M509276200. [DOI] [PubMed] [Google Scholar]
- 57.Barry JB, et al. A single nucleotide in an estrogen-related receptor alpha site can dictate mode of binding and peroxisome proliferator-activated receptor gamma coactivator 1alpha activation of target promoters. Mol. Endocrinol. 2006;20:302–310. doi: 10.1210/me.2005-0313. [DOI] [PubMed] [Google Scholar]
- 58.Luo J, et al. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Mol. Cell. Biol. 2003;23:7947–7956. doi: 10.1128/MCB.23.22.7947-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 60.Ijichi N, et al. Estrogen-related receptor alpha modulates the expression of adipogenesis-related genes during adipocyte differentiation. Biochem. Biophys. Res. Commun. 2007;358:813–818. doi: 10.1016/j.bbrc.2007.04.209. [DOI] [PubMed] [Google Scholar]
- 61.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 62.Patti ME, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 64.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 65.McGill JK, Beal MF. PGC-1alpha, a new therapeutic target in Huntington's disease? Cell. 2006;127:465–468. doi: 10.1016/j.cell.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 66.Nichol D, et al. RIP140 expression is stimulated by estrogen-related receptor alpha during adipogenesis. J. Biol. Chem. 2006;281:32140–32147. doi: 10.1074/jbc.M604803200. [DOI] [PubMed] [Google Scholar]