Abstract
Francisella tularensis, the causative agent of the zoonotic disease tularemia, has recently gained increased attention due to the emergence of tularemia in geographical areas where the disease has been previously unknown and to the organism's potential as a bioterrorism agent. Although F. tularensis has an extremely broad host range, the bacterial reservoir in nature has not been conclusively identified. In this study, the ability of virulent F. tularensis strains to survive and replicate in the amoeba Acanthamoeba castellanii was explored. We observe that A. castellanii trophozoites rapidly encyst in response to F. tularensis infection and that this rapid encystment phenotype is caused by factor(s) secreted by amoebae and/or F. tularensis into the coculture medium. Further, our results indicate that in contrast to the live vaccine strain LVS, virulent strains of F. tularensis can survive in A. castellanii cysts for at least 3 weeks postinfection and that the induction of rapid amoeba encystment is essential for survival. In addition, our data indicate that pathogenic F. tularensis strains block lysosomal fusion in A. castellanii. Taken together, these data suggest that interactions between F. tularensis strains and amoebae may play a role in the environmental persistence of F. tularensis.
Francisella tularensis is the etiological agent of the zoonotic disease tularemia, also known as rabbit fever (35, 53). Strains belonging to F. tularensis subsp. tularensis and F. tularensis subsp. holarctica, which are both prevalent in the Northern Hemisphere, cause the majority of reported cases of tularemia (36). Subspecies tularensis is highly contagious, with an infectious dose of 1 to 10 bacteria, and is associated with more severe disease (21). Though described more than a century ago as a disease common among hunters and trappers, tularemia has recently been reported in areas with no previous known risk (20, 25, 31, 42). F. tularensis infects a broad range of wildlife species (36), and a number of arthropods, such as ticks and flies, are known to be vectors (36, 49). Humans are usually infected either through an insect bite or by inhalation of aerosolized bacteria (49). Tularemia can be fatal in up to 30% of untreated cases (36, 49), with the mortality rate reaching 90% in pneumonic infections, as described in early studies conducted with vaccinated human volunteers (44-46, 49). Due to its highly infectious nature and its potential for use as a bioterrorism agent, F. tularensis has been classified as a class A biothreat pathogen by the Centers for Disease Control and Prevention (CDC), which has mandated that human tularemia be a reportable disease since 2000 (15, 37). In addition, the absence of a licensed vaccine for prophylaxis (36) makes understanding the virulence mechanisms used by this pathogen imperative for the development of efficacious measures to prevent or treat human disease.
Though F. tularensis has been isolated from more than 250 wildlife species (21), the acute nature of the infections and the resultant high mortality rates in these hosts indicate that the bacterial reservoir(s) in nature have yet to be identified. Tularemia outbreaks involving F. tularensis subsp. holarctica have often been linked to water sources (6, 40), and a positive PCR field test was reported for Francisella during such an outbreak in Norway (5). Abd et al. reported that the F. tularensis live vaccine strain LVS is able to survive and replicate in the amoeba Acanthamoeba castellanii (1), suggesting a potential link between amoeba-Francisella interactions and environmental persistence. A. castellanii, a free-living environmental amoeba, is known to serve as a reservoir for a number of pathogenic microorganisms (24). However, to date, interactions of virulent F. tularensis subspecies tularensis strains with amoebae have not been documented. The ability of several human intracellular pathogens, including Legionella pneumophila and Mycobacterium avium, to infect and survive within amoebae has been well characterized (10, 12). In addition to playing a role in environmental survival and dissemination, growth in A. castellanii has been shown to enhance the ability of L. pneumophila and M. avium to survive and replicate in host macrophages (10, 12) and to enhance the virulence of both species in mice (7, 12). Since F. tularensis species are facultative intracellular pathogens that primarily survive in macrophages, probing the Francisella-amoeba interaction may provide insights into Francisella pathogenesis, as well as environmental survival. In this study, we investigated the ability of virulent type A strains of F. tularensis to survive in A. castellanii with a focus on understanding the role of Francisella-amoeba interactions in environmental persistence.
MATERIALS AND METHODS
Strains and growth conditions.
F. tularensis subsp. holarctica strain LVS and F. tularensis subsp. novicida strain U112 (NOV) were obtained from the CDC. F. tularensis subsp. tularensis strain Schu S4 (SCHU) was obtained from the Rocky Mountain Laboratories, Hamilton, MT. F. tularensis subsp. tularensis clinical strains 1 through 10 were obtained from the Departments of Public Health in Utah and New Mexico (Table 1). All F. tularensis strains used in this study were grown on modified Mueller-Hinton agar (Difco) supplemented with 0.025% ferric pyrophosphate (Sigma), 0.02% IsoVitaleX (Becton Dickinson), 0.1% glucose, and 0.025% calf serum (Invitrogen-Gibco) at 37°C with 5% CO2 or modified Mueller-Hinton broth supplemented as described above with aeration at 37°C.
TABLE 1.
F. tularensis strains used in this study
| Strain | Designation | Subtype | Region | Origin | Year isolated |
|---|---|---|---|---|---|
| F. tularensis subsp. holarctica LVS | LVSb | B | Europe | Sheep | 1949 |
| F. tularensis subsp. novicida U112 | NOV | NA | Utah | Human | 1951 |
| F. tularensis subsp. tularensis strains | |||||
| Schu S4 | SCHUc | A | Ohio | Human lesion | 1941 |
| 70102163 | Ft-1 | Aa | Utah | Human blood | 2001 |
| 79101574 | Ft-2 | Aa | Utah | Human | 1991 |
| 1365 | Ft-3 | Aa | New Mexico | Human | 1997 |
| AS1284 | Ft-4 | Aa | New Mexico | Rodent | 2003 |
| 79400960 | Ft-5 | Aa | Utah | Human | 1990 |
| 80700069 | Ft-6 | Aa | Utah | Human lesion | 2007 |
| 80502541 | Ft-7 | Aa | Utah | Human lesion | 2005 |
| 1385 | Ft-8 | Aa | New Mexico | Rabbit | 2001 |
| 1773a | Ft-9 | Aa | New Mexico | Human | 1999 |
| AS2058 | Ft-10 | Aa | New Mexico | Rabbit | 2002 |
Subtyping was done using primers in IS100 elements unique to type A strains (Victoria Lao, Patrick Chain, and Emilio Garcia, unpublished results).
LVS was produced as a vaccine in 1961 by passaging a virulent F. tularensis subsp. holarctica strain isolated in 1949 (17).
Laboratory-passaged strain.
Cell lines and culture conditions.
A. castellanii (ATCC 30234) amoebae were grown axenically in PYG broth (12) to 90% confluence at 23°C in the dark in 75-cm2 tissue culture flasks (Falcon). The amoebae were harvested before use by rapping the flask sharply to bring them into suspension, and the number of viable cells was determined as described previously (12). To induce amoeba encystment, A. castellanii was suspended in high-salt (HS) buffer for 3 days as described previously (4, 33).
Entry and adherence assays.
A. castellanii entry and adherence assays were carried out as described previously (10, 13) in 24-well tissue culture plates (Costar). Briefly, A. castellanii was seeded into plates at a concentration of 2 × 105 cells per well and allowed to adhere overnight at 23°C. The amoebae were washed with HS buffer (4) and incubated in 1 ml of HS buffer for 1 h at 37°C with 5% CO2 prior to infection. F. tularensis overnight cultures were added to amoeba trophozoites at a multiplicity of infection (MOI) of 10. After coincubation for 30 min, the cells were washed once and incubated in HS buffer plus 100 μg gentamicin per ml for 2 h at 37°C and 5% CO2. The amoebae were then washed once to remove gentamicin and lysed by incubation in 1% saponin (Sigma) for 5 min followed by vigorous pipetting. Dilutions were plated on supplemented Mueller-Hinton agar to determine viable CFU counts. Adherence assays were carried out in a similar manner, except that after the amoebae were infected with the bacteria for 30 min, they were immediately washed three times with HS buffer prior to lysis. The percent entry was calculated as follows: (CFUintracellular/CFUinoculum) × 100. Saponin (1%) had no effect on the viability of F. tularensis strains, and all strains used displayed comparable levels of killing by gentamicin.
Intracellular survival assays.
Intracellular survival assays were carried out in a manner similar to that of the entry assays with the following modifications: after gentamicin treatment, fresh HS buffer was added and the amoebae were incubated at 37°C with 5% CO2, lysed, and plated at different times postinfection, indicated below. Survival is expressed as the percentage of CFU present intracellularly at each time point (Tx) compared to that at time zero (2.5 h), i.e., percent survival = (CFU Tx/CFU T0) × 100.
Long-term survival assays.
Amoebae were infected with F. tularensis strains in six-well plates (Costar) as described above. After gentamicin treatment and washing, 3 ml of fresh HS buffer was added to each well and the plates incubated at 37°C with 5% CO2 for up to 3 weeks. To recover bacteria, plates were spun down for 5 min at 100 × g and the HS buffer in each well was replaced with 3 ml PYG medium supplemented with IsoVitaleX. The plates were reincubated at 37°C with 5% CO2 until the wells became turbid or for 72 h (whichever occurred first). The cultures were then plated on Mueller-Hinton agar, and samples were gram stained to confirm the presence of F. tularensis. To inhibit amoeba encystment, A. castellanii trophozoites were treated with 25 μg/ml cycloheximide concurrently with gentamicin treatment for 2 h and then washed before the addition of HS buffer. To inhibit amoeba excystment, A. castellanii cysts were treated with 25 μg/ml cycloheximide in HS buffer for 1 h prior to the addition of PYG medium for bacterial recovery.
Cytotoxicity assays.
A. castellanii was seeded in 96-well plates at a concentration of 5 × 104 cells per well and allowed to adhere overnight at 23°C. The medium was replaced with HS buffer, and the amoebae were incubated at 37°C with 5% CO2 for 1 h prior to infection. F. tularensis overnight cultures were added to cells at MOIs of 10. After coincubation for 1 h at 37°C and 5% CO2, the medium was replaced with fresh HS buffer. Cell death was quantified colorimetrically using the Cyto-Tox96 lactate dehydrogenase release kit (Promega) according to the manufacturer's recommendations.
Fractionation and HPLC analyses.
Amoeba-Francisella cocultures were collected at 6 h postinfection, size fractionated, and concentrated by using amplicon filters (Millipore) with 100 kDa, 30 kDa, and 3 kDa pore sizes sequentially. Twenty-five-ml aliquots of F. tularensis-A. castellanii coculture media were loaded into the filter with the largest pore size and centrifuged. The eluate was then loaded into the filter with the next-smaller pore size, and the sequence was repeated until the last eluate was filtered through the smallest-pore-size filter. Centrifugation was done at 740 × g for 5 to 60 min according to the manufacturer's instructions. Total protein concentrations were determined separately for each eluted fraction. Amounts of 150 and 250 μg of total protein from each fraction were then diluted with running buffer and loaded separately on SuperDex 200 10/300 high-pressure liquid chromatography (HPLC) columns (GE Healthcare) and subfractioned according to the manufacturer's recommendations. The subfractions were either concentrated or diluted with phosphate-buffered saline to 1-ml final volumes.
Protein identification.
The proteins present in F. tularensis-amoeba coculture subfractions were identified commercially by ProtTech, Inc. (Norristown, PA) using Nano liquid chromatography-tandem mass spectrometry (LC-MS-MS) peptide-sequencing technology. The proteins were first denatured by the addition of 8 M urea, followed by reduction of the Cys residues in the solution with 20 mM dithiothreitol, and then alkylated with 20 mM iodoacetamide. The samples were then diluted to 2 M urea with 100 mM ammonium bicarbonate (pH 8.5), and the proteins digested by the addition of sequencing-grade modified trypsin (Promega, Madison, WI). The resulting peptide mixture was desalted with a PepClean spin column (Pierce, Rockford, IL) and analyzed using an LC-MS-MS system, in which an HPLC reverse-phase C18 column with a 75-μm inner diameter was coupled on-line with an ion trap mass spectrometer (Thermo, Palo Alto, CA). The mass spectrometric data acquired from the LC-MS-MS analyses were used to search against the recently established nonredundant protein database RefSeq from GenBank (http://www.ncbi.nlm.nih.gov/RefSeq) using ProtTech's ProtQuest software package. Except where specified, all other chemicals used were purchased from Sigma (St. Louis, MO).
Microscopy.
To evaluate amoeba encystment, A. castellanii was infected with overnight cultures of F. tularensis strains at an MOI of 20 in triplicate. The infection was allowed to proceed for 2 h, and the infected amoebae were examined using a Nikon TE300 light microscope with differential interference contrast options attached to a digital screen. The morphologies of the infected amoebae were noted and quantified by counting the number of amoeba trophozoites and cysts present in three random fields per infected well. Transmission electron microscopy (TEM) was used to examine the ultrastructure of A. castellanii amoebae infected with F. tularensis strains. For TEM, A. castellanii was infected at an MOI of 10 for 10 min at 37°C with 5% CO2, washed twice with HS buffer, and incubated in fresh HS buffer at 37°C and 5% CO2. After incubation for durations indicated below, the amoebae were suspended in medium with a rubber policeman, pelleted by centrifugation for 2 min at 740 × g at 25°C, fixed, and prepared for electron microscopy as previously described (12, 19). To track endosomal vacuoles, A. castellanii trophozoites were preloaded with SPI-Mark 10-nm colloidal gold unconjugated particles (SPI Supplies) overnight before infection. The samples were suspended in 2% glutaraldehyde for 1 h, treated with 1% OsO4 for 2 h, and then postfixed with 0.5% uranyl acetate at 4°C overnight. The cells were embedded and sectioned as described previously (19). Immunofluorescence microscopy (IF) was performed as described previously (18). Briefly, A. castellanii lysosomes were preloaded with LysoTracker red DND-99 (Invitrogen-Molecular Probes) for 30 min before infection, infected for different durations, and then washed with HS buffer and fixed with 4% paraformaldehyde in 100 mM phosphate buffer (pH 7.4). The amoebae were permeabilized with 95% ethanol, blocked with 3% bovine serum albumin, and stained with a chicken anti-F. tularensis polyclonal antibody. To stain infected mature amoeba cysts, the cysts were washed with HS buffer, centrifuged onto slides by using a Cytospin centrifuge (Thermo Fisher) for 3 min at 1,500 × g, fixed for 30 min with 4% paraformaldehyde, and then permeabilized for 30 to 60 min with 95% ethanol. The amoebae were visualized by fluorescent microscopy using a Zeiss AxioVert microscope after staining with Alexa Fluor-coupled secondary antibodies (Invitrogen-Molecular Probes). z-stacks of slices (1 μm thick) were captured using Zeiss Axiovision software, and reconstructed three-dimensional images were assembled by use of Volocity software (Improvision version 5.1; Perkin-Elmer).
Statistical analyses.
The means and standard deviations of the results from triplicate samples were calculated for representative experiments. All experiments were repeated at least three times, unless otherwise noted. Significance was determined by analysis of variance using the paired Student t test. P values of <0.05 were considered significant.
RESULTS
F. tularensis strains enter and replicate in A. castellanii amoebae with different efficiencies.
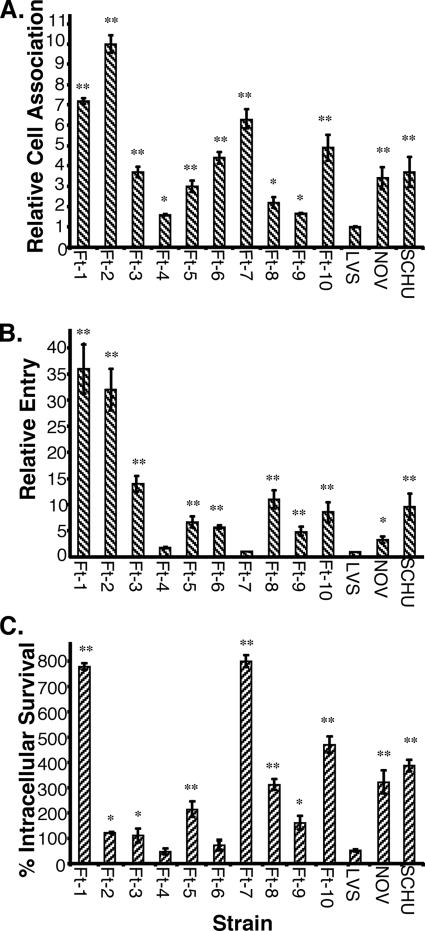
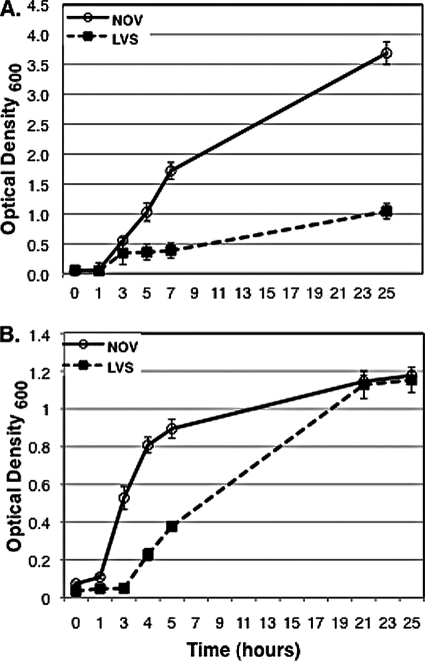
To determine if pathogenic F. tularensis strains enter and survive in A. castellanii, we infected amoebae with 10 clinical isolates of F. tularensis subsp. tularensis strains chosen from geographically and temporally separate tularemia outbreaks (Ft-1 to Ft-10, Table 1) and with the laboratory strain F. tularensis subsp. tularensis Schu S4 (SCHU). For purposes of comparison, we also infected A. castellanii with the commonly used laboratory strains of F. tularensis subsp. novicida (NOV) and F. tularensis subsp. holarctica strain (LVS). Our results demonstrate that F. tularensis strains associate with, enter, and survive in A. castellanii with various efficiencies (Fig. 1). LVS was significantly less efficient at adherence, entry, and survival in A. castellanii than the other strains tested (P ≤ 0.02). We therefore chose to represent the results of infection experiments relative to those for LVS. F. tularensis strains varied in their ability to adhere to A. castellanii amoebae, with the least efficient strains, Ft-4 and Ft-9, adhering at a rate 1.5 to 2 times higher than that of LVS (P = 0.05) and the most efficient strains, Ft-1 and Ft-2, adhering at a rate 7 to 10 times higher than that of LVS (P = 0.02) (Fig. 1A). With the exception of strains Ft-4 and Ft-7, which entered at rates comparable to that of LVS, all the strains tested consistently entered A. castellanii at a rate 5 to 50 times higher than that of LVS (P ≤ 0.05) (Fig. 1B). We also examined the ability of F. tularensis strains to survive and replicate in A. castellanii at 24 h postinfection (Fig. 1C). For LVS, only half of the CFU at time zero (2.5 h) were recovered after 24 h. These data are significantly different from the data described by Abd et al. (1); this can be attributed to their use of a rich medium, which we observed allowed for replication of LVS extracellularly (Fig. 2A). Strains Ft-4 and Ft-6 were recovered at a rate similar to that of LVS, whereas the rest of the strains varied widely in their ability to survive and replicate. Ft-3, Ft-9, and Ft-2 were recovered at rates 1.2 to 1.5 times higher than that of LVS after 24 h (P = 0.05). Ft-5, Ft-8, Ft-10, SCHU, and NOV were recovered at rates that were two to five times higher (P = 0.02), and Ft-1 and Ft-7 were recovered at rates that were seven to eight times higher than that of LVS after 24 h (P = 0.01). With the exception of Ft-7, all other strains tested correlated in their ability to attach to, enter, and survive in A. castellanii. Though Ft-7 showed a relatively high rate of immediate attachment to the amoebae, the numbers of CFU recovered at time zero were consistently low. Interestingly, by 24 h postinfection, the Ft-7 CFU counts were approximately seven times higher than the CFU counts recovered at time zero. These data suggest that either Ft-7 enters at a low rate but replicates very efficiently or that the strain is killed early in the infection but is able to recover and replicate after a lag phase. We were unable to assess intracellular growth past 24 h by CFU counts since the majority of the amoebae were encysted by then and were extremely resistant to lysis, both chemical and mechanical, consistent with previous reports (28-30).
FIG. 1.
Cell association (A), entry (B), and intracellular survival (C) of F. tularensis strains in A. castellanii amoebae. Rates of cell association (A) and entry (B) of LVS were arbitrarily set at 1. The intracellular survival rate (C) is represented as the ratio of numbers of CFU recovered at 24 h to those recovered at time zero for each strain. Data points and error bars represent the means and standard deviations, respectively, of the results of assays done in triplicate from representative experiments (**, P ≤ 0.02; *, P ≤ 0.05).
FIG. 2.
Growth curves of F. tularensis strains in PYG medium (A) and Mueller-Hinton broth (B) at an optical density of 600 nm over a 24-h time period. Data points and error bars represent the means and standard deviations, respectively, of the results of assays done in triplicate from a representative experiment.
Pathogenic F. tularensis strains are present in spacious vacuoles at 30 min postinfection.
To characterize the ultrastructure of pathogenic F. tularensis vacuoles in the amoebae, we infected A. castellanii trophozoites with LVS, NOV, SCHU, and Ft-1 and examined them by TEM. By 30 min postinfection, the majority of vacuoles containing NOV, SCHU, and Ft-1 were significantly more spacious (P ≤ 0.02) (Fig. 3A and Table 2) than vacuoles containing LVS. To determine if F. tularensis strains disrupt the endosomal pathway, we preloaded amoeba lysosomes with colloidal gold particles prior to bacterial infection, which allowed us to assess the frequency of lysosomal fusion (Fig. 3B). We were able to confirm a statistically significant correlation between tight vacuole formation and the presence of gold particles, and we found that by 2 h postinfection, 82% of bacterial tight vacuoles formed by all four strains colocalized with gold particles (P = 0.02) (Table 2). This number rose to 88% at 24 h postinfection (P = 0.01), suggesting that tight vacuoles are lysosomal in nature while spacious ones are not. At 24 h postinfection, over 90% of amoebae infected with NOV, SCHU, and Ft-1 contained bacteria present in spacious vacuoles, compared to 57% of amoeba infected with LVS (Table 2). By this stage, the majority of A. castellanii infected with NOV, SCHU, and Ft-1 were encysted and intact bacteria were observed within the double-wall layers of the amoeba cysts. An “early” or “young” cyst containing bacteria is shown in Fig. 3C. Trophozoites still present at this point contained bacteria in spacious vacuoles, and multiple vacuoles containing bacteria were observed within the same trophozoite (Fig. 3D). The bacterial structure appears typical of that described for other pleiomorphic bacteria. Similar to what has been previously described with LVS, strains NOV, SCHU, and Ft-1 recruit mitochondria and membrane structures suggestive of the endoplasmic reticulum to the bacterial vacuoles (Fig. 3E), which display intact phagosomal membranes (Fig. 3F).
FIG. 3.
Transmission electron micrographs of A. castellanii amoebae infected with F. tularensis strains. Ft-1 (A) is present in spacious vacuoles, while LVS (B) is present in tight vacuoles at 2 h postinfection; long black arrows point to spacious vacuoles, and short white arrows to tight vacuoles. (B) LVS tight vacuole fused with colloidal-gold-labeled lysosomes at 2 h postinfection; short white arrows point to gold particles. (C) Early cyst of A. castellanii amoeba containing SCHU bacteria at 24 h postinfection. (D) A. castellanii trophozoite with multiple SCHU vacuoles at 24 h postinfection. (E and F) Bacterial vacuoles shown in panel D were enlarged to show mitochondrion and endoplasmic reticulum (white arrows) recruitment to the phagosome (E) and intact phagosomal membranes (F). b, bacteria; m, mitochondria.
TABLE 2.
Quantification of F. tularensis bacteria in infected A. castellanii amoebae
| Time | % of tight vacuolesa |
% of tight vacuole fusionb | P valuee | % of lysosomal fusionc |
P valuef | ||||
|---|---|---|---|---|---|---|---|---|---|
| LVS | NOV | SCHU | Ft-1 | LVS | NOV | ||||
| 30 min | NDd | ND | ND | ND | ND | ND | 48 ± 3 | 26 ± 2 | 0.02 |
| 2 h | 40 ± 3 | 10 ± 3 | 8 ± 2 | 9 ± 3 | 82 ± 2 | 0.02 | 89 ± 4 | 43 ± 3 | 0.01 |
| 24 h | 46 ± 2 | 9 ± 2 | 8 ± 3 | 10 ± 3 | 88 ± 4 | 0.01 | ND | ND | ND |
Percentage of tight bacterial vacuoles in cells containing at least one bacterial vacuole. Results are the means ± standard deviations of two counts of 50 cells in different sections of two separate preparations.
Percentage of colocalization of colloidal gold particles with bacterial vacuoles from all F. tularensis strains processed. Results are the means ± standard deviations of two counts of 50 cells containing at least one bacterial vacuole in different sections of two separate preparations.
Percentage of colocalization of LysoTracker red with bacterial vacuoles containing at least one bacterium by IF. Results are the means ± standard deviations of two counts of 25 cells from four separate preparations.
ND, not done (sample too small).
P values indicate the significance of the percentage of fusion of F. tularensis tight vacuoles with colloidal gold particles.
P values indicate the significance of the percentage of colocalization of NOV and LVS vacuoles with LysoTracker red.
LVS colocalizes with the lysosomal marker LysoTracker red.
To confirm our observations that LVS-containing tight vacuoles are lysosomal in nature, we preloaded amoeba lysosomes with the lysosomal marker LysoTracker red and infected them with LVS and NOV strains expressing green fluorescent protein (GFP). By 30 min postinfection, 26% of NOV bacteria colocalized with LysoTracker red, compared to 48% of LVS bacteria (P = 0.02) (Fig. 4 and Table 2). By 2 h postinfection, 43% of NOV bacteria colocalized with LysoTracker red, compared with 89% of LVS bacteria (P = 0.01) (Table 2). These data confirm that LVS bacteria reside predominantly in lysosomal vacuoles while NOV bacteria appear able to block lysosomal fusion in A. castellanii.
FIG. 4.
Immunofluorescence z-stack projections of A. castellanii trophozoites infected with LVS expressing GFP for 2 h (a, b, and c) and NOV stained with an anti-F. tularensis antibody (green) (f, g, and h) after 2 h and 30 min, respectively. At 2 h postinfection, the majority of LVS bacteria (a) colocalize with LysoTracker red (c), which appears diffuse within the trophozoite (b). The majority of NOV bacteria (f) do not colocalize with LysoTracker red (h), which appears localized (g). Enlarged cross sections of panels c and h represent trophozoites infected with LVS (d) and NOV (i). These were rotated by 90° using Volocity software to confirm the presence of the bacteria intracellularly (e and j).
NOV is present in A. castellanii cysts at 7 days postinfection.
To confirm that the structures observed by TEM within the double walls of amoeba cysts were bacteria, we infected A. castellanii with either LVS and NOV strains or LVS and NOV strains expressing GFP and incubated them for various times after infection. At the time points indicated below, amoeba cysts were either fixed and visualized directly (GFP-expressing strains) or stained with a chicken anti-F. tularensis polyclonal antibody prior to processing for IF and visualization. A. castellanii cysts stained positive for NOV but not LVS at 24 h and 7 days postinfection (Fig. 5). A. castellanii cysts infected with GFP-expressing NOV bacteria were only positive for NOV after 24 h (data not shown). Since GFP is not expressed at 7 days postinfection, the NOV bacteria present in A. castellanii cysts may either have lost the plasmid or be metabolically inactive.
FIG. 5.
Nomarski DC z-stack projections of uninfected A. castellanii cysts (e and f) and cysts infected with NOV (a and b) and stained with an anti-NOV antibody (b and f). Enlarged cross sections of panels b and f (c and g) were rotated by 90° using Volocity software to confirm the presence (d) or absence (h) of intracellular bacteria.
Recovery of viable F. tularensis bacteria from A. castellanii cysts 21 days postinfection.
Demonstrating the presence of viable F. tularensis bacteria in A. castellanii cysts by intracellular growth assays was not possible due to increased resistance of amoeba cysts to lysis, as previously reported (28-30). To circumvent this problem, we designed an experiment based on our observations that all the F. tularensis strains we tested replicate vigorously in PYG medium (Fig. 2A) but not in HS buffer (data not shown). This allowed us to determine whether virulent F. tularensis strains were able to survive longer than 24 h in A. castellanii. A. castellanii trophozoites were infected with NOV, LVS, SCHU, Ft-1, and Ft-7. After gentamicin treatment, fresh HS buffer was added to each well and the amoebae were allowed to incubate for various durations. Weekly, infected cysts were spun down and the buffer replaced with PYG. The presence of the rich PYG medium enabled the amoebae to excyst and the bacteria present in the cysts to be released into the medium, where they could replicate. As shown in Table 3, viable NOV, Ft-1, and Ft-7 bacteria were recovered up to 3 weeks postinfection. We observed that about 80% of the amoebae excysted by 24 h after the addition of PYG broth, consistent with previous data describing the kinetics of A. castellanii excystment (28). We did not observe bacterial turbidity in the wells with infected amoebae, nor were we able to recover bacterial CFU before 36 to 48 h after the replacement of HS buffer with PYG broth. These data suggest that F. tularensis bacteria were present intracellularly and not just associated with the outer surface of the cysts. To confirm these results, we blocked the excystment of F. tularensis-infected cysts by using cycloheximide as previously described (9) and did not recover any bacteria upon the addition of PYG (Table 3). Not unexpectedly, we were only able to recover viable LVS bacteria up to 3 days postinfection, consistent with the results of our CFU assays demonstrating that the LVS strain is attenuated in amoeba infections.
TABLE 3.
Survival of F. tularensis strains in A. castellanii
| Time (days) | Result after indicated treatmenta |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LVS |
NOV |
SCHU |
Ft-1 |
Ft-7 |
UI (− C) | ||||||
| − C | + C | − C | + C | − C | + C | − C | + C | − C | + C | ||
| 3 | + | NG | + | NG | + | NG | + | NG | + | NG | NG |
| 7 | NG | ND | + | NG | + | NG | + | NG | + | NG | NG |
| 14 | NG | ND | + | NG | + | ND | + | ND | + | ND | NG |
| 21 | NG | ND | + | ND | NG | ND | + | ND | + | ND | NG |
A. castellanii cysts were treated with (+ C) or without (− C) 25 μg/ml cycloheximide prior to the addition of HS buffer to prevent excystment. UI, uninfected A. castellanii control; +, turbidity was observed in experimental wells 48 to 72 h after replacement of buffer with rich medium (F. tularensis growth was confirmed by plating for viable CFU and gram staining; in all cases where turbidity was observed, >108 CFU/ml were present); NG, no growth after replacement of buffer with rich medium and plating for viable CFU; ND, not done.
F. tularensis subsp. novicida and virulent F. tularensis strains induce the rapid encystment of A. castellanii.
In contrast to infection with LVS (Fig. 6B), our initial observations of A. castellanii infected with NOV and SCHU showed that a large number of amoeba trophozoites began encysting within 2 h of infection (Fig. 6C and E). Although A. castellanii is known to encyst in response to starvation, desiccation, and other adverse environmental conditions (24) and can be induced to artificially encyst in the laboratory with ∼3 days of growth in HS buffer (4) (also known as “encystment buffer”) (33), the presence of NOV and SCHU rapidly accelerated this natural phenomenon. The rapid encystment phenotype (REP) we observed also occurred in response to F. tularensis strains even when the amoebae were grown xenically in the presence of heat-killed Escherichia coli as a food source, as previously described (48). In addition, REP was not associated with an increase in cytotoxicity, confirming that the amoebae were encysted and not dead (data not shown). We investigated the ability of F. tularensis clinical isolates (Ft-1 to Ft-10) to cause the rapid encystment of A. castellanii trophozoites compared with that of NOV, LVS, and SCHU. F. tularensis clinical isolates varied in their ability to cause REP (Fig. 6). Furthermore, Ft-3 (Fig. 6D), Ft-4, and Ft-9 did not cause REP, while Schu (Fig. 6E), Ft-2 (Fig. 6F), Ft-7 (Fig. 6G), and Ft-10 (Fig. 6H) caused the highest REP levels. Interestingly, out of the 10 clinical strains tested, 5 strains (50%) were able to induce a significantly higher level of A. castellanii encystment than was observed in uninfected trophozoites or trophozoites infected with LVS (P ≤ 0.03) (Fig. 7). NOV, however, caused the highest levels of REP observed, a phenomenon that may be explained by the rapid growth of NOV compared to the growth of type A F. tularensis strains. It is interesting to note that the same five strains that induced higher levels of encystment (Ft-1, Ft-2, Ft-7, Ft-8, and Ft-10) were associated with higher rates of attachment, entry, and survival in A. castellanii than all other strains tested (Table 4).
FIG. 6.
Rapid encystment of A. castellanii in response to infection with virulent F. tularensis strains, LVS, and NOV. Shown are uninfected A. castellanii trophozoites (A); A. castellanii trophozoites infected with LVS (B) and Ft-3 (D) for 2 h; and A. castellanii infected with NOV (C), Schu (E), Ft-2 (F), Ft-7 (G), and Ft-10 (H) for 2 h. Arrows point to early cysts, and asterisks indicate trophozoites.
FIG. 7.
Quantification of the encystment of A. castellanii at 2 h postinfection with F. tularensis strains. Data points and error bars represent the means and standard deviations, respectively, of three random field counts of assays done in triplicate from representative experiments (**, P ≤ 0.02; *, P = 0.05).
TABLE 4.
Survival of F. tularensis strains in A. castellanii in the presence and absence of encystment
| Time (days) | Result(s) after indicated treatmenta |
|||||||
|---|---|---|---|---|---|---|---|---|
| SCHU |
Ft-1 |
NOV |
LVS |
|||||
| − C | + C | − C | + C | − C | + C | R | + R | |
| 3 | +, E | NG, NE | +, E | NG, NE | +, E | NG, NE | + | +, E |
| 8 | +, E | NG, NE | +, E | NG, NE | +, E | NG, NE | NG | +, E |
| 14 | +, E | ND | +, E | ND | +, E | ND | NG | NG |
A. castellanii trophozoites were treated with (+ C) or without (− C) 25 μg/ml cycloheximide to prevent encystment or A. castellanii amoebae were infected with LVS in the presence (+ R) or absence (− R) of the REP fractions from NOV-A. castellanii or SCHU-A. castellanii cocultures. +, turbidity was observed in experimental wells 48 to 72 h after replacement of buffer with rich medium (F. tularensis growth was confirmed by plating for viable CFU and gram staining; in all cases where turbidity was observed, >108 CFU/ml were present); E, at least 50% of A. castellanii amoebae present were encysted; NG; no growth after replacement of buffer with rich medium and plating for viable CFU; NE, less than 10% of A. castellanii amoebae present were encysted; ND, not done.
A protein fraction isolated from F. tularensis-A. castellanii cocultures is responsible for the rapid encystment of A. castellanii.
To investigate whether the rapid amoeba encystment we observed was a result of direct contact between the bacterium and amoeba or mediated by soluble factor(s), we used a transwell culture system to physically separate the amoeba monolayer from the bacteria by using an insert with a 0.2-μm filter. The inability of F. tularensis strains to pass through the filter was confirmed by plating for viable CFU (data not shown). In the absence of cell-cell contact using the transwell system, A. castellanii trophozoites still induced REP in response to the same bacterial strains (data not shown). These data suggest that REP may be caused by factor(s) secreted by F. tularensis strains in response to A. castellanii and/or factor(s) secreted by A. castellanii in response to bacterial infection. Since spent bacterial culture medium did not confer REP (data not shown), we concluded that cross talk occurring between F. tularensis and A. castellanii is necessary for induction of the encystment phenotype. To determine whether the soluble factors in REP were proteinaceous in nature, cocultures of bacteria and amoebae were boiled or subjected to a 30-min treatment with proteinase K. Both of these treatments resulted in abrogation of REP (data not shown), suggesting that the factor(s) responsible are proteins. The coculture media from A. castellanii infected with LVS, NOV, SCHU, Ft-1, Ft-2, Ft-7, and Ft-8 were then fractionated and analyzed by HPLC. Four different-size fractions, ≥100 kDa, <100 kDa to ≥30 kDa, <30 kDa to ≥3 kDa, and <3 kDa, were obtained for each strain. The addition of these four fractions separately to naïve A. castellanii trophozoites demonstrated that the fraction of ≥3 kDa to <30 kDa from NOV, SCHU, Ft-1, Ft-7, and Ft-8 infections induces REP (Fig. 8). The fraction of <100 kDa to >30 kDa from NOV was also able to confer REP, suggesting that there may be slight size differences between the factor(s) produced by type A strains and NOV. The addition of the fraction of ≥3 kDa to <30 kDa from the LVS-A. castellanii coculture did not result in REP, and the results resembled those for cultures treated with other fractions or medium alone.
FIG. 8.
Quantification of the encystment of naïve A. castellanii trophozoites 4 h after the addition of fractions from F. tularensis-A. castellanii cocultures. Data points and error bars represent the means and standard deviations, respectively, of three random field counts of assays done in triplicate from representative experiments (*, P ≤ 0.02).
REP fractions from NOV and SCHU contain proteins that mediate A. castellanii encystment.
To identify the proteins that may be responsible for the observed REP, we had the REP fractions from NOV and SCHU analyzed commercially by LC-MS-MS. In parallel, we also analyzed fractions of the same size isolated from uninfected A. castellanii trophozoites, A. castellanii cysts, and laboratory-grown NOV. Interestingly, we identified the same proteins in REP fractions from NOV and SCHU. LC-MS-MS analyses revealed four unique A. castellanii proteins that were only present in the A. castellanii-F. tularensis REP fraction and one protein (subtilisin-like serine proteinase) that was present in both the REP fraction and the A. castellanii cyst fraction (Table 5). We were able to infer that the subtilisin serine proteinase is present in the REP fractions at four times the amount it was present in the A. castellanii cyst fraction because the number of peptides sequenced by LC-MS-MS from each protein can be used as an indication for the relative abundance of proteins in a sample. This serine proteinase shows 97% identity and 98% homology at the C-terminal region to a previously identified subtilisin family serine proteinase that has recently been shown to mediate the encystation of A. castellanii (33, 34). Interestingly, the REP fraction also contained ubiquitin and a ubiquitin fusion protein. The proteasome/ubiquitin system has been shown to be involved in the encystment of amoebae, a process requiring extensive protein degradation (22). The other two proteins identified in the fractions were actin and the α-chain of profilin II. Profilin is an actin-binding protein involved in the dynamic turnover and restructuring of the actin cytoskeleton (27, 51). The protein is normally found in association with monomeric actin in Acanthamoeba (27). We also identified a large protein which is predicted to be a chaperone belonging to the DnaK/HSP70 family. Considering that the size of this protein is larger than 30 kDa, it is unlikely that it is actually secreted; most probably it was released into the medium as a result of amoeba lysis during encystment. In addition to A. castellanii proteins, we also identified seven F. tularensis proteins that were only present in the REP fraction and not in laboratory-grown F. tularensis. Since most of the genes encoding these are annotated in the databases as encoding “hypothetical proteins,” we are not able to speculate on their function. We are currently constructing mutations in the genes encoding these proteins and expressing them in vitro to identify their role in inducing amoeba encystment.
TABLE 5.
Identification of A. castellanii proteins in the REP fraction
| Protein | Locus | Size (kDa) | No. of peptidesa |
|---|---|---|---|
| Ubiquitin | P49634 | 15.4 | 4 |
| Ubiquitin fusion protein | CAA53293 | 194 | 4 |
| Profilin II, α-chain | P19984 | 21.9 | 2 |
| Actin | P02578 | 29.8 | 2 |
| Encystation-mediating serine protease | ABY63398 | 32.1 | 16 |
| Subtilisin-like serine protease | AAF91465 | 16.5 | 7 |
| DnaK molecular chaperone/HSP70 family protein | AAU94654 | 37.5 | 2 |
Number of peptides sequenced by Nano LC-MS-MS.
DISCUSSION
Francisella tularensis was first identified as the cause of a plague-like outbreak in ground squirrels in 1911 (32). There has been a rising interest in F. tularensis (49) in recent years, due in large part to the recognition of F. tularensis as a potential bioterrorism agent (47). F. tularensis exhibits an extremely broad host range, and the bacterium is known to infect hundreds of wildlife species (41), which facilitates human infections. However, the acute nature of the infections in vertebrate and invertebrate hosts identified so far suggests that the reservoir(s) of F. tularensis in the environment have not been identified (36). Previous studies have correlated tularemia outbreaks with aquatic environments (5, 6) and suggested that environmental amoebae may serve as bacterial reservoirs in nature (40). So far, only two such studies have examined the survival of F. tularensis strains in the amoeba Acanthamoeba castellanii (1, 43). Though these studies concluded that F. tularensis LVS and NOV are able to survive in A. castellanii for weeks postinfection, the authors used a rich medium that allowed for the replication of the bacteria extracellularly for all survival experiments, making it impossible to discern true long-term intracellular survival. In addition, the authors did not characterize the interaction of virulent F. tularensis strains with amoebae, leaving unanswered the question of whether amoebae can serve as environmental reservoirs for F. tularensis strains that are pathogenic for humans.
In the present study, we conducted a detailed characterization of the interaction of multiple F. tularensis strains with the amoeba A. castellanii and have demonstrated for the first time the ability of fully virulent strains to enter and survive in amoebae. To ensure that we were able to quantify long-term survival without confounding factors, all our experiments were performed in an HS buffer that supports the survival of A. castellanii but does not allow for the growth of F. tularensis (data not shown). In addition to the interactions of the most commonly used laboratory strains, LVS, SCHU, and NOV, with A. castellanii, those of 10 clinical strains isolated from human, rodent, and lagomorph outbreaks in New Mexico and Utah were also examined. To maximize our chances of obtaining disparate isolates, we chose strains that were isolated from geographically separate outbreaks over a 10-year period. Since their isolation, these strains have not been manipulated in the laboratory and have remained largely uncharacterized. Our results demonstrate for the first time that fully virulent F. tularensis strains can associate with and enter A. castellanii, albeit with disparate efficiencies. Further, we have demonstrated that long-term survival of pathogenic F. tularensis isolates in amoebae is dependent on the induction of amoeba encystment. Not surprisingly, we observed that LVS is the least efficient at both association and entry, consistent with the nonpathogenic nature of this isolate. However, F. tularensis strains in general replicated at much lower rates in A. castellanii than other amoeba-resistant bacteria, such as M. avium and L. pneumophila (10, 11). The variation in the ability of clinical F. tularensis strains to associate with and survive in A. castellanii suggests that more than one environmental host may exist for F. tularensis.
To assess downstream events after bacterial entry, we examined the ultrastructure of F. tularensis-infected amoebae by TEM. We observed that F. tularensis strains were associated with two types of bacterial vacuoles: tight vacuoles, which directly conform to the bacterial shape, and spacious vacuoles, usually associated with multiple bacteria. Quantification of the type of bacterial vacuoles associated with each strain revealed that LVS was enclosed in tight vacuoles at a rate four times higher than the rates for NOV, SCHU, and Ft-1. In addition, though we were unable to calculate the frequency of lysosomal fusion with individual strains due to the low numbers of trophozoites present after 24 h of infection, we found that 88% of tight vacuoles colocalized with colloidal gold particles. These data suggest that spacious vacuoles may not be lysosomal in nature and that enclosure within these vacuoles may provide a survival advantage to NOV, SCHU, and Ft-1. This would be similar to what has been described for survival of Salmonella in macrophages (26). We were able to calculate the frequency of lysosomal fusion using IF and found that by 2 h postinfection, 89% of LVS vacuoles colocalize with lysosomes, compared to 43% of NOV vacuoles, consistent with our TEM observations.
Even though the majority of A. castellanii trophozoites encyst within 24 h after infection, we have demonstrated for the first time the ability of virulent F. tularensis strains to survive in amoebal cysts for up to 3 weeks postinfection. Surprisingly, viable SCHU bacteria were only recovered up to 2 weeks postinfection. This observation may be explained by the fact that the SCHU strain has been propagated under laboratory conditions for almost 70 years since its initial isolation from a clinical case (8, 16), and some loss of virulence is to be expected. This extended propagation may account for the inability of SCHU to survive past 2 weeks when compared with the survival of the other type A strains (e.g., Ft-1 and Ft-7) that have been only minimally manipulated in the laboratory. Unlike Mycobacterium and Legionella spp., the F. tularensis strains examined do not replicate to a high degree in A. castellanii cysts but appear to survive by inducing amoeba encystment. It is still possible that these F. tularensis strains may indeed grow to large numbers in amoeba trophozoites when abundant nutrients are present, and in fact, many amoeba-resistant microorganism are known to be endosymbiotic or lytic in a given amoeba depending on environmental conditions (23, 24). Another plausible hypothesis is that amoeba-resistant microorganisms have developed multiple approaches to environmental survival. Our data from using the eukaryotic protein synthesis inhibitor cycloheximide suggest that the ability to cause amoeba encystment is necessary for the survival of the F. tularensis strains tested in A. castellanii. Alternatively, cycloheximide may be acting by preventing the synthesis of an amoebal protein or by blocking a protein-mediated process the bacteria need to survive upon internalization. Considering the drought resistance and hardiness of amoeba cysts, cyst formation could enable intracellular F. tularensis bacteria to survive desiccation and food shortage in the environment. F. tularensis could then be transmitted orally to animals that drink from water contaminated with amoeba cysts. Infected animals would then complete the environmental cycle by fecal shedding of Francisella in or near aquatic environments, which are prime amoeba habitats (2, 3).
Though amoeba encystment in response to bacterial infection has been reported previously, this usually occurs in the presence of a high ratio of bacteria to amoebae (52). To explore the cause of the rapid encystment of A. castellanii trophozoites in response to F. tularensis infection, we verified that the amoebae were actually encysted and not dead by conducting cytotoxicity assays comparing infected and uninfected A. castellanii amoebae (data not shown). We also confirmed that the amoebae were not simply encysting in response to overwhelming numbers of bacteria or lack of nutrients by reducing the MOI and/or growing the amoeba xenically in the presence of heat-killed E. coli as a food source (48). At MOIs of 1 and 5, we were not able to recover viable organisms following F. tularensis infections but we still obtained high levels of amoeba encystment. Interestingly, it has been observed that A. castellanii does not usually undergo encystment in response to low levels of bacterial replication (14, 54) (Ling Yan, personal communication). These data suggest that the encystment was specific to F. tularensis and not simply due to the presence of high numbers of bacteria or low nutrient levels. In addition, using a transwell culture system, we demonstrated that encystment does not require direct bacterium-cell contact, as REP still occurred even though the bacteria were physically separated from the amoeba trophozoites.
Quantification of encystment showed that F. tularensis strains varied in their ability to cause REP. NOV showed the highest level of REP, most likely because of its rapid rate of growth compared to the growth rates of LVS and virulent F. tularensis strains. Along with NOV, strains SCHU, Ft-1, Ft-2, Ft-7, Ft-8, and Ft-10 caused very high rates of REP, while LVS, Ft-3, Ft-4, and Ft-6 did not. Since all virulent F. tularensis strains and LVS showed similar rates of growth, the failure of LVS, Ft-3, Ft-4, and Ft-6 to induce rapid encystment in amoebae cannot be attributed to the number of CFU present. It is interesting to note that F. tularensis strains causing the highest levels of encystment were also the strains showing the highest levels of attachment, entry, and replication in A. castellanii.
The induction of REP appears to involve proteins produced as a result of F. tularensis-A. castellanii cross talk. The results of our experiments show that the addition of coculture media from A. castellanii and F. tularensis strains that cause REP confer REP on naïve amoeba trophozoites, while spent culture media from the same F. tularensis strains alone does not. These data also suggest that soluble factor(s) secreted into the coculture medium by the bacteria and/or the amoebae mediate the phenotype. Consequently, we conducted some preliminary analyses of the coculture media in order to narrow down the size and nature of the fraction responsible for inducing the phenotype. We found the factors responsible for inducing encystment to be between 3 and 30 kDa and concluded that the active component(s) of the fraction that induces REP are likely to be protein(s), since boiling or proteinase K treatment abrogated the activity of the fraction, as evidenced by the loss of REP. LC-MS-MS analyses of the REP-inducing fraction revealed that the same proteins are present in REP-inducing fractions from NOV and Ft-1. We identified a 16.5-kDa subtilisin-like serine proteinase that was present in REP fractions at four times the quantity it was present in media from A. castellanii cysts. This subtilisin-like serine proteinase shows 97% identity and 98% homology at the C terminus containing the peptidase S8 region (pfam0082) to a previously identified 33-kDa encystment-mediating serine proteinase (EMSP) that has recently been shown to mediate the encystation of A. castellanii (33, 34). This suggests that our protein may be a cleavage product of EMSP. Subtilases have been associated with autophagosomes (39, 50), and EMSP small interfering RNA-treated A. castellanii amoebae show defects in substance degradation and autophagy maturation during encystation (34). The abundance of EMSP-like protein in the REP fraction suggests that autophagy may be activated in A. castellanii in response to F. tularensis infection. The REP fraction also contained ubiquitin and a ubiquitin fusion protein. The proteasome/ubiquitin system is responsible for degradation of proteins (22, 38). It is likely that this system is used to degrade proteins present in trophozoites that will not be required in the emerging cysts. The other two proteins identified in the fractions were actin and the α-chain of profilin II. Profilin is an actin-binding protein involved in the dynamic turnover and restructuring of the actin cytoskeleton (27, 51). This protein is normally found in association with monomeric actin in members of the genus Acanthamoeba (27). The presence of both of these proteins in the REP fraction is likely due to the cytoskeletal rearrangements that occur upon encystment and not to their secretion.
In addition to A. castellanii proteins, we also identified seven F. tularensis proteins that were only present in the REP-inducing fraction and not in the medium of laboratory-grown F. tularensis. We are currently characterizing these proteins to identify the role they play in the induction of amoeba encystment. Previous studies have shown that interactions of a number of intracellular bacterial pathogens with amoebae result in an enhancement of the pathogen's ability to enter and survive in mammalian cells (7, 11, 12). Since it is likely that cross talk between bacteria and amoebae results in amoeba encystment, this process could also result in the upregulation of bacterial virulence factors that may be required for subsequent entry and survival in mammalian cells, in addition to enhancing the ability of F. tularensis bacteria to persist in the environment.
Acknowledgments
We thank Stan Falkow and Lucy Tompkins for their feedback and helpful suggestions. We thank Nafisa Ghori for help with the electron microscopy and Upi Singh, Paul Henderson, Jeffrey Cirillo, and Emilio Garcia for technical advice. We thank Brent Segelke, Matt Coleman, and Manuel Amieva for critical reading of the manuscript. We also thank Jean Celli for the Schu S4 strain and Karin Elkins for the U112 and LVS strains.
This work was performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under contract DE-AC52-07NA27344 and supported by Laboratory Directed Research and Development grant 06-ERD-057 from Lawrence Livermore National Laboratory and grant AI-65359 from the National Institutes of Health/NIAID to A.R. J.J.M. was supported by NIH training grant GM007276-29, a U.S. Department of Homeland Security fellowship, and a National Science Foundation Graduate Research fellowship.
Footnotes
Published ahead of print on 9 October 2009.
REFERENCES
- 1.Abd, H., T. Johansson, I. Golovliov, G. Sandstrom, and M. Forsman. 2003. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, O. R. 2002. Laboratory and field-based studies of abundances, small-scale patchiness, and diversity of gymnamoebae in soils of varying porosity and organic content: evidence of microbiocoenoses. J. Eukaryot. Microbiol. 49:17-23. [DOI] [PubMed] [Google Scholar]
- 3.Arias Fernandez, M. C., E. Paniagua Crespo, M. Marti Mallen, M. P. Penas Ares, and M. L. Casro Casas. 1989. Marine amoebae from waters of northwest Spain, with comments on a potentially pathogenic euryhaline species. J. Protozool. 36:239-241. [DOI] [PubMed] [Google Scholar]
- 4.Band, R. N., and S. Mohrlok. 1969. The respiratory metabolism of Acanthamoeba rhysodes during encystation. J. Gen. Microbiol. 59:351-358. [Google Scholar]
- 5.Berdal, B. P., R. Mehl, H. Haaheim, M. Loksa, R. Grunow, J. Burans, C. Morgan, and H. Meyer. 2000. Field detection of Francisella tularensis. Scand. J. Infect. Dis. 32:287-291. [DOI] [PubMed] [Google Scholar]
- 6.Berdal, B. P., R. Mehl, N. K. Meidell, A. M. Lorentzen-Styr, and O. Scheel. 1996. Field investigations of tularemia in Norway. FEMS Immunol. Med. Microbiol. 13:191-195. [DOI] [PubMed] [Google Scholar]
- 7.Brieland, J., M. McClain, L. Heath, C. Chrisp, G. Huffnagle, M. LeGendre, M. Hurley, J. Fantone, and C. Engleberg. 1996. Coinoculation with Hartmannella vermiformis enhances replicative Legionella pneumophila lung infection in a murine model of Legionnaires' disease. Infect. Immun. 64:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhuri, R. R., C. P. Ren, L. Desmond, G. A. Vincent, N. J. Silman, J. K. Brehm, M. J. Elmore, M. J. Hudson, M. Forsman, K. E. Isherwood, D. Gurycova, N. P. Minton, R. W. Titball, M. J. Pallen, and R. Vipond. 2007. Genome sequencing shows that European isolates of Francisella tularensis subspecies tularensis are almost identical to US laboratory strain Schu S4. PLoS ONE 2:e352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chisholm, G. E., IV, and M. H. Vaughan. 1979. Isolation and characterization of a cycloheximide-resistant mutant of Acanthamoeba castellanii Neff. J. Bacteriol. 138:280-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cirillo, J. D., S. L. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cirillo, J. D., S. Falkow, and L. S. Tompkins. 1994. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 62:3254-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cirillo, J. D., S. Falkow, L. S. Tompkins, and L. E. Bermudez. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirillo, S. L., L. E. Bermudez, S. H. El-Etr, G. E. Duhamel, and J. D. Cirillo. 2001. Legionella pneumophila entry gene rtxA is involved in virulence. Infect. Immun. 69:508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cirillo, S. L., L. Yan, M. Littman, M. M. Samrakandi, and J. D. Cirillo. 2002. Role of the Legionella pneumophila rtxA gene in amoebae. Microbiology 148:1667-1677. [DOI] [PubMed] [Google Scholar]
- 15.Darling, R. G., C. L. Catlett, K. D. Huebner, and D. G. Jarrett. 2002. Threats in bioterrorism. I: CDC category A agents. Emerg. Med. Clin. North. Am. 20:273-309. [DOI] [PubMed] [Google Scholar]
- 16.Eigelsbach, H. T., W. Braun, and R. D. Herring. 1951. Studies on the variation of Bacterium tularense. J. Bacteriol. 61:557-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eigelsbach, H. T., and C. M. Downs. 1961. Prophylactic effectiveness of live and killed tularemia vaccines. I. Production of vaccine and evaluation in the white mouse and guinea pig. J. Immunol. 87:415-425. [PubMed] [Google Scholar]
- 18.El-Etr, S. H., A. Mueller, L. S. Tompkins, S. Falkow, and D. S. Merrell. 2004. Phosphorylation-independent effects of CagA during interaction between Helicobacter pylori and T84 polarized monolayers. J. Infect. Dis. 190:1516-1523. [DOI] [PubMed] [Google Scholar]
- 19.El-Etr, S. H., L. Yan, and J. D. Cirillo. 2001. Fish monocytes as a model for mycobacterial host-pathogen interactions. Infect. Immun. 69:7310-7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eliasson, H., and E. Back. 2007. Tularaemia in an emergent area in Sweden: an analysis of 234 cases in five years. Scand. J. Infect. Dis. 39:880-889. [DOI] [PubMed] [Google Scholar]
- 21.Ellis, J., P. C. Oyston, M. Green, and R. W. Titball. 2002. Tularemia. Clin. Microbiol. Rev. 15:631-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez, J., G. Bai, U. Frevet, E. J. Corey, and D. Eichinger. 1999. Proteasome-dependent cyst formation and stage-specific ubiquitin mRNA accumulation in Entamoeba invadens. Eur. J. Biochem. 264:897-904. [DOI] [PubMed] [Google Scholar]
- 23.Greub, G., B. La Scola, and D. Raoult. 2003. Parachlamydia acanthamoeba is endosymbiotic or lytic for Acanthamoeba polyphaga depending on the incubation temperature. Ann. N. Y. Acad. Sci. 990:628-634. [DOI] [PubMed] [Google Scholar]
- 24.Greub, G., and D. Raoult. 2004. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17:413-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurcan, S., M. Eskiocak, G. Varol, C. Uzun, M. Tatman-Otkun, N. Sakru, A. Karadenizli, C. Karagol, and M. Otkun. 2006. Tularemia re-emerging in European part of Turkey after 60 years. Jpn. J. Infect. Dis. 59:391-393. [PubMed] [Google Scholar]
- 26.Hernandez, L. D., K. Hueffer, M. R. Wenk, and J. E. Galan. 2004. Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science 304:1805-1807. [DOI] [PubMed] [Google Scholar]
- 27.Kaiser, D. A., V. K. Vinson, D. B. Murphy, and T. D. Pollard. 1999. Profilin is predominantly associated with monomeric actin in Acanthamoeba. J. Cell Sci. 112(Pt. 21):3779-3790. [DOI] [PubMed] [Google Scholar]
- 28.Khunkitti, W., D. Lloyd, J. R. Furr, and A. D. Russell. 1998. Acanthamoeba castellanii: growth, encystment, excystment and biocide susceptibility. J. Infect. 36:43-48. [DOI] [PubMed] [Google Scholar]
- 29.Kilvington, S., W. Heaselgrave, J. M. Lally, K. Ambrus, and H. Powell. 2008. Encystment of Acanthamoeba during incubation in multipurpose contact lens disinfectant solutions and experimental formulations. Eye Contact Lens 34:133-139. [DOI] [PubMed] [Google Scholar]
- 30.Lloyd, D., N. A. Turner, W. Khunkitti, A. C. Hann, J. R. Furr, and A. D. Russell. 2001. Encystation in Acanthamoeba castellanii: development of biocide resistance. J. Eukaryot. Microbiol. 48:11-16. [DOI] [PubMed] [Google Scholar]
- 31.Lundman, T. 2005. Watch out for tularemia also in Southern Sweden! Lakartidningen 102:1986-1987. [PubMed] [Google Scholar]
- 32.McCoy, G. W., and C. W. Chapin. 1912. Further observations of a plague-like disease of rodents with a preliminary note of the causative agent, Bacterium tularensis. J. Infect. Dis. 10:61-72. [Google Scholar]
- 33.Moon, E. K., D. I. Chung, Y. C. Hong, T. I. Ahn, and H. H. Kong. 2008. Acanthamoeba castellanii: gene profile of encystation by ESTs analysis and KOG assignment. Exp. Parasitol. 119:111-116. [DOI] [PubMed] [Google Scholar]
- 34.Moon, E. K., D. I. Chung, Y. C. Hong, and H. H. Kong. 2008. Characterization of a serine proteinase mediating encystation of Acanthamoeba. Eukaryot. Cell 7:1513-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morner, T. 1992. The ecology of tularaemia. Rev. Sci. Tech. 11:1123-1130. [PubMed] [Google Scholar]
- 36.Oyston, P. C. 2008. Francisella tularensis: unravelling the secrets of an intracellular pathogen. J. Med. Microbiol. 57:921-930. [DOI] [PubMed] [Google Scholar]
- 37.Oyston, P. C., A. Sjostedt, and R. W. Titball. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2:967-978. [DOI] [PubMed] [Google Scholar]
- 38.Pagano, M. 1997. Cell cycle regulation by the ubiquitin pathway. FASEB J. 11:1067-1075. [DOI] [PubMed] [Google Scholar]
- 39.Paoletti, M., M. Castroviejo, J. Begueret, and C. Clave. 2001. Identification and characterization of a gene encoding a subtilisin-like serine protease induced during the vegetative incompatibility reaction in Podospora anserina. Curr. Genet. 39:244-252. [DOI] [PubMed] [Google Scholar]
- 40.Parker, R. R., E. A. Steinhaus, G. M. Kohls, and W. L. Jellison. 1951. Contamination of natural waters and mud with Pasteurella tularensis and tularemia in beavers and muskrats in the northwestern United States. Bull. Natl. Inst. Health. 193:1-161. [PubMed] [Google Scholar]
- 41.Penn, R. L. 2005. Francisella tularensis (tularemia). vol. 1. Churchill Livingstone, New York, NY.
- 42.Petersen, J. M., and M. E. Schriefer. 2005. Tularemia: emergence/re-emergence. Vet. Res. 36:455-467. [DOI] [PubMed] [Google Scholar]
- 43.Santic, M., M. Molmeret, K. E. Klose, S. Jones, and Y. A. Kwaik. 2005. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell Microbiol. 7:969-979. [DOI] [PubMed] [Google Scholar]
- 44.Saslaw, S., and S. Carhart. 1961. Studies with tularemia vaccines in volunteers. III. Serologic aspects following intracutaneous or respiratory challenge in both vaccinated and nonvaccinated volunteers. Am. J. Med. Sci. 241:689-699. [PubMed] [Google Scholar]
- 45.Saslaw, S., H. T. Eigelsbach, J. A. Prior, H. E. Wilson, and S. Carhart. 1961. Tularemia vaccine study. II. Respiratory challenge. Arch. Intern. Med. 107:702-714. [DOI] [PubMed] [Google Scholar]
- 46.Saslaw, S., H. T. Eigelsbach, H. E. Wilson, J. A. Prior, and S. Carhart. 1961. Tularemia vaccine study. I. Intracutaneous challenge. Arch. Intern. Med. 107:689-701. [DOI] [PubMed] [Google Scholar]
- 47.Sata, T. 2005. Bioterrorism. Nihon Hoigaku Zasshi 59:119-125. [PubMed] [Google Scholar]
- 48.Schuster, F. L. 2002. Cultivation of pathogenic and opportunistic free-living amebas. Clin. Microbiol. Rev. 15:342-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sjostedt, A. 2007. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann. N. Y. Acad. Sci. 1105:1-29. [DOI] [PubMed] [Google Scholar]
- 50.Takeshige, K., M. Baba, S. Tsuboi, T. Noda, and Y. Ohsumi. 1992. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119:301-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vandekerckhove, J. S., D. A. Kaiser, and T. D. Pollard. 1989. Acanthamoeba actin and profilin can be cross-linked between glutamic acid 364 of actin and lysine 115 of profilin. J. Cell Biol. 109:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, X., and D. G. Ahearn. 1997. Effect of bacteria on survival and growth of Acanthamoeba castellanii. Curr. Microbiol. 34:212-215. [DOI] [PubMed] [Google Scholar]
- 53.Weber, A. 2004. Current epidemiology of selected bacterial zoonoses. Gesundheitswesen 66(Suppl. 1):S26-S30. [DOI] [PubMed] [Google Scholar]
- 54.Yan, L., R. L. Cerny, and J. D. Cirillo. 2004. Evidence that hsp90 is involved in the altered interactions of Acanthamoeba castellanii variants with bacteria. Eukaryot. Cell 3:567-578. [DOI] [PMC free article] [PubMed] [Google Scholar]