Abstract
Model fecal deposits from cattle fed or not fed antimicrobial growth promoters were examined over 175 days in the field for growth and persistence of total Escherichia coli and numbers and proportions of ampicillin-resistant (Ampr) and tetracycline-resistant (Tetr) E. coli. In addition, genotypic diversity and the frequency of genetic determinants encoded by Ampr and Tetr E. coli were investigated. Cattle were fed diets containing chlortetracycline (44 ppm; A44 treatment group), chlortetracycline plus sulfamethazine (both at 44 ppm; AS700 treatment group), or no antibiotics (control). Fecal deposits were sampled 12 times over 175 days. Numbers of Tetr E. coli in A44 and AS700 deposits were higher (P < 0.001) than those of controls and represented up to 35.6% and 20.2% of total E. coli, respectively. A time-by-treatment interaction (P < 0.001) was observed for the numbers of Tetr and Ampr E. coli. Except for Ampr E. coli in control deposits, all E. coli numbers increased (P < 0.001) in deposits up to day 56. Even after 175 days, high Tetr E. coli numbers were detected in A44 and AS700 deposits [5.9 log10 CFU (g dry matter)−1 and 5.4 log10 CFU (g dry matter)−1, respectively]. E. coli genotypes, as determined by pulsed-field gel electrophoresis, were diverse and were influenced by the antimicrobial growth promoter and the sampling time. Of the determinants screened, blaTEM1, tetA, tetB, tetC, sul1, and sul2 were frequently detected. Occurrence of determinants was influenced by the feeding of antimicrobials. Fecal deposits remain a source of resistant E. coli even after a considerable period of environmental exposure.
In North America antibiotics are widely used in beef cattle production as prophylactics or antimicrobial growth promoters (AGP). Used in this manner, antibiotics are generally administered in the diet either at times of high disease risk or on a continuous basis to improve feed efficiency. Employment of AGP in this manner may increase the prevalence of commensal antimicrobial-resistant (AR) bacteria (1, 41).
There is evidence that resistant bacteria can be transferred from livestock to humans (5, 39, 55), and consequently the use of AGP has raised public health concerns. While the modes of transmission of AR bacteria and gene determinants are complex (33), survivability in agriculture-related matrices may be a critical factor in their dissemination (54). Most research on the persistence of AR bacteria in livestock waste has focused on large-scale management systems, such as land-applied manure (20, 44) or storage lagoons and pits (28, 49). In some instances, these systems have been shown to decrease the survival of select bacteria and the presence of AR genes (35, 51). Nevertheless, there is limited knowledge of the fate of AR bacteria or resistance determinants in feces shed from individuals, such as the fecal deposits that are formed in feedlot pens on intensively managed farms. Previous work has shown that fecal deposits provide an environment that is conducive to the growth and survival of pathogenic and commensal Escherichia coli (4, 48, 57). Because cattle fed AGP excrete antimicrobial residues in their feces (3), the residues may exert a selection pressure on bacteria after deposition of feces and potentially confer a growth advantage to resistant bacteria.
The objective of the current study was to investigate both the longitudinal phenotypic and genotypic ecologies of AR E. coli in fecal deposits arising from groups of cattle administered either no AGP or one of two tetracycline-based AGP that are commonly used in the North American industry. We focused on ampicillin-resistant (Ampr) and tetracycline-resistant (Tetr) E. coli because both AGP treatments contained tetracycline, and we have previously found a link between the use of tetracycline-based AGP and Ampr E. coli (1). We hypothesized that resistant E. coli would be more numerous in fecal deposits from animals previously fed AGP.
MATERIALS AND METHODS
Animals and treatments.
A complete history of antimicrobial administration to the steers generating the fecal deposits was known and controlled. Ninety crossbred steers (6 to 8 months old, weighing 150 ± 20 kg) were randomly assigned to nine pens at the Lethbridge Research Centre feedlot (Lethbridge, Alberta). The steers were brought directly to the feedlot from a common range (Deseret Ranches, Raymond, Alberta, Canada) and received no antibiotics prior to the initiation of the experiment. Three pens (10 steers per pen) were assigned to each of three treatments: (i) control, no antibiotics; (ii) chlortetracycline (44 ppm; fed as Aureomycin-100 G; Alpharma; treatment denoted A44); (iii) chlortetracycline and sulfamethazine (each at 44 ppm; fed as Aureo S-700 G; Alpharma, Inc., Bridgewater, NJ; treatment denoted AS700). These antibiotics and concentrations were selected on the basis of conventional practices in the Canadian beef industry. Adjacent pens in the feedlot were supplied with a common watering bowl, but assignment of treatments to pens was arranged so that only cattle in the same treatment group drank from the same bowl.
Steers were administered antimicrobials for 197 days, starting on the day of arrival to the feedlot, up to the point of fecal collection for construction of deposits. At the time of fecal deposit setup, steers had been fed a concentrate-based diet for the previous 96 days that consisted of the following (dry-matter [DM] basis): 85% barley, 10% barley silage, and 5% supplement which contained calcium carbonate (34.8%), zinc sulfate (28.4%), manganous sulfate (14.6%), copper sulfate (10.3%), selenium (5.0%), vitamin E (4.7%), vitamin A (1.7%), vitamin D (0.2%), ethylenediamine dihydroiodide (0.1%), and cobalt sulfate (0.1%) (wt/wt basis). The steers were fed once daily in a manner that ensured that all feed was consumed daily. To avoid cross-contamination of feed, antibiotics were mixed with 5 kg of supplement and manually spread over the surface of the feed in each trough immediately after feed delivery. Steers assigned to the control treatment had no access to medicated feed at any time during the experiment. All cattle were cared for according to the guidelines of the Canadian Council on Animal Care (12).
Fecal deposit preparation and sampling.
For each pen, individual fecal samples (n = 10) were collected from each animal and mixed uniformly into a single composite (approximately 24 kg). The fecal material was collected in a manner that avoided feces that had contacted the ground and was added to the composite mixture within 1 min after defecation. Each composite mixture was then divided into duplicate artificial fecal deposits contained in metal pans (50 by 50 by 5 cm) to prevent possible contamination between treatments. The depth of the fecal deposits was ∼5 cm. The bottoms of the pans were perforated to allow water to drain to the subsoil in the event of rainfall. In total, 18 fecal deposits (2 replicates per pen) were prepared. The deposits were randomly placed outside on 1 March in two adjacent rows with 50-cm spacing between deposits. Deposit volume was predicated on the need to provide periodic samples over 175 days.
Fecal deposits were sampled after 0, 7, 14, 28, 42, 56, 70, 84, 98, 112, 126, and 175 days of environmental exposure. Deposits were sampled using sterile 60-ml syringes modified by cutting the end of the barrel of the syringe. At each sampling, two subsamples (∼15 g) 3 cm apart were collected and pooled. The two samples were collected by inserting the syringe into the bottom of the deposit and expelling the sample into a sterile cup using the plunger. Samples were immediately taken to the lab for processing. Average daily temperature and precipitation data for the Lethbridge Research Centre were obtained from the Agriculture and Agri-Food Canada weather observation station that was located 100 m from the site.
Microbial analysis.
Fecal material from each deposit was thoroughly mixed, and a subsample (5 g) was weighed into a pan and placed in a 105°C oven for 24 h to determine DM. A second sample (1 g) was placed in a stomacher bag (10.5 by 15 cm) and mixed with 9 ml of 0.05 M sodium phosphate buffer (pH 7.0). Bags were stomached for 2 min (setting 8; Interscience Mini-Mix), and the contents were used in a 1:10 dilution series with sodium phosphate buffer. Aliquots (100 μl) from selected dilutions were plated in duplicate on MacConkey agar (Difco/Becton Dickinson, Sparks, MD) containing no antibiotics (MAC) and on MAC amended with ampicillin (32 μg ml−1) (MAC+AMP) or tetracycline hydrochloride (16 μg ml−1) (MAC+TET). Plates were incubated for 18 to 24 h at 37°C before lactose-positive colonies were counted. Concentrations of antibiotics used in the MAC+AMP and MAC+TET plates were set at the breakpoint standards for defining resistance as described by the Clinical and Laboratory Standards Institute (15), and isolates that grew on these plates were considered resistant. Colony counts on each medium were reported as log10 CFU (g DM)−1. The percentage of E. coli isolates exhibiting resistance to the antibiotic concentrations in selective plates was calculated as follows: (number of colonies on the antibiotic-supplemented MAC plates/total colonies counted on nonselective MAC) × 100.
Four isolates from MAC and when possible up to six isolates from MAC+AMP and MAC+TET plates were selected, streaked onto Trypticase soy agar (Difco), and incubated at 37°C for 24 h. Colonies on the Trypticase soy agar plates were transferred and stored at −80°C in 20% (vol/vol) glycerol in brain heart infusion medium (Difco). AR isolates selected for pulsed-field gel electrophoresis (PFGE) (see below) were again tested for growth on plates containing the antimicrobial to which they were resistant (i.e., MAC+AMP or MAC+TET) and were confirmed to be E. coli on the basis of lactose utilization, methyl red reaction, and β-galactosidase and β-glucuronidase activities. Additionally, all Ampr E. coli from MAC+AMP plates that were analyzed by PFGE were also tested for resistance to tetracycline by plating on MAC+TET medium.
PFGE.
Ampr and Tetr E. coli from MAC+AMP and MAC+TET plates associated with each dietary treatment (A44, AS700, or control) were chosen for analysis by PFGE. Isolates were randomly selected from fecal deposits across six time points representing short (days 0 and 7), moderate (days 56 and 70), or long (days 126 and 175) periods of environmental exposure and included isolates from each pen. A total of 161 Ampr (A44, n = 56; AS700, n = 56; control, n = 49) and 237 Tetr (A44, n = 79; AS700, n = 83; control, n = 75) E. coli isolates were selected for PFGE.
Isolates were subtyped via XbaI-digested genomic DNA according to the Centers for Disease Control and Prevention (13) 1-day standardized laboratory protocol for molecular subtyping of E. coli. Electrophoresis was performed with a Chef DR II electrophoresis unit (Bio-Rad Laboratories, Mississauga, Ontario, Canada). Gels were documented using an AlphaImager imaging system (Alpha Innotech, San Leandro, CA). E. coli E318N (kindly made available by A. Borczyk, Enteric Reference Laboratory, Ministry of Health, Toronto, Ontario, Canada) was included in each PFGE gel as a control. Each time an E. coli isolate was cultured for PFGE, one to three plated colonies were suspended in 40 μl of 1× TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.0) and stored at −80°C for PCR analysis of resistance gene determinants.
Resistance determinants encoded by E. coli.
E. coli isolates analyzed by PFGE were tested by PCR for select genes encoding tetracycline (tetA, tetB, tetC, tetL, tetM, tetO, and tetW), ampicillin (blaOXA1, blaPSE1, and blaTEM1), and sulfonamide (sul1 and sul2) resistance. Primers and annealing temperatures were previously described for tetA, tetC, tetL, tetM, and tetO (37), tetB and tetW (38), blaOXA1 and blaPSE1 (22), blaTEM1 (6), and sul1 and sul2 (8). PCR for the detection of each gene was carried out individually. Each PCR mixture (20 μl) contained (final concentrations) 1× HotStarTaq Plus master mix (Qiagen Inc., Mississauga, Ontario, Canada) and 0.4 μM of each primer.
Colonies of E. coli were heat lysed (98°C for 4 min) and centrifuged (16,000 × g; 4 min), and the supernatant (1 μl) was added to the PCR mixture as a DNA template. The PCR conditions were as follows: 95°C for 5 min; 35 cycles of 95°C for 20 s, respective annealing temperatures for 30 s, and 72°C for 1 min; and 72°C for 10 min. The PCRs were carried out using an Eppendorf MasterCycler thermal cycler (Eppendorf, Mississauga, Ontario, Canada). Twenty microliters of product was visualized on a 1.5% (wt/vol) agarose gel following electrophoresis and staining with ethidium bromide. All PCRs were run with positive controls (plasmids with sequence-verified genes) for respective gene determinants and negative controls containing water in place of a DNA template.
Statistical analysis.
PFGE patterns of E. coli isolates were separated into two databases based on the selective medium used for isolation (MAC+AMP or MAC+TET). Within each database, PFGE patterns were either classified as unique or grouped into clusters based on ≥90% homology using Dice similarity coefficients with unweighted pair group methods with arithmetic average algorithms built into Bionumerics software (Bionumerics 5.1; Applied Maths BVBA, Sim-Martens-Latem, Belgium). The number of grouped clusters was used to describe the genetic diversity of E. coli from each treatment (A44, AS700, or control) and exposure period (short, moderate, or long), with larger numbers of clusters representing greater diversity. The position tolerance and optimization were set at 1% and 0.5%, respectively.
The Dimensioning Techniques package of the Bionumerics software program was used for analysis of within- and between-group similarities of PFGE groups that were categorized by treatment, duration of exposure within each treatment, and gene profiles of the presence or absence of resistance determinants. For this, binary character profiles using the band-matching option were created after clustering using the Dice coefficient evaluated by the unweighted pair group method. The significance of each group's average within-group similarity was tested using bootstrapping analysis (n = 1,000). The null hypothesis was that PFGE profiles within a single group were no more similar than those of a comparison group. The P value for genotyping analysis was the percentage of bootstrap resamplings in which the actual within-group similarity was less than that of a randomized group.
E. coli counts were analyzed using the “Proc Mixed” procedure of the SAS software program (42). The pen was considered the experimental unit. The model included the fixed effects of treatment (A44, AS700, or control), time (day of sampling), and the interaction between treatment and time. The repeated statement was applied to the day of sampling, using the pen nested within the treatment as the subject. Various error structures were tested, and the one giving the lowest Akaike information criterion was chosen for analysis. In all instances, P values of ≤0.05 were considered significant.
RESULTS
Environmental conditions.
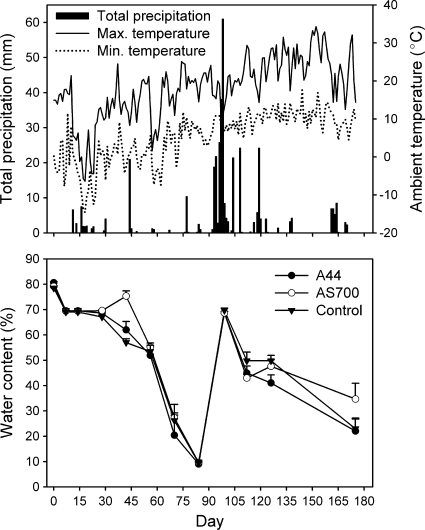
The minimum daily air temperature was −14.6°C on day 19, and the maximum was 34.3°C on day 153 (Fig. 1). Daily precipitation was greatest during the month of June (days 93 to 123). The water content of the fecal deposits appeared to correlate with precipitation trends (Fig. 1). By day 84, the water content of all deposits had declined to less than 10%, compared to an initial content of ca. 80% at the start of the experiment. On day 98, the moisture content of deposits increased as a result of the June rainfall.
FIG. 1.
Daily total precipitation, ambient minimum and maximum air temperatures (top), and mean (n = 3; plus standard error) water content of fecal deposits from each treatment group (bottom) over the 175-day experimental period. The treatments were as follows: control, no antimicrobial agents added to the diets of steers from which fecal deposits originated; A44, chlortetracycline (44 ppm); AS700, chlortetracycline and sulfamethazine (each at 44 ppm).
Escherichia coli persistence in artificial fecal deposits.
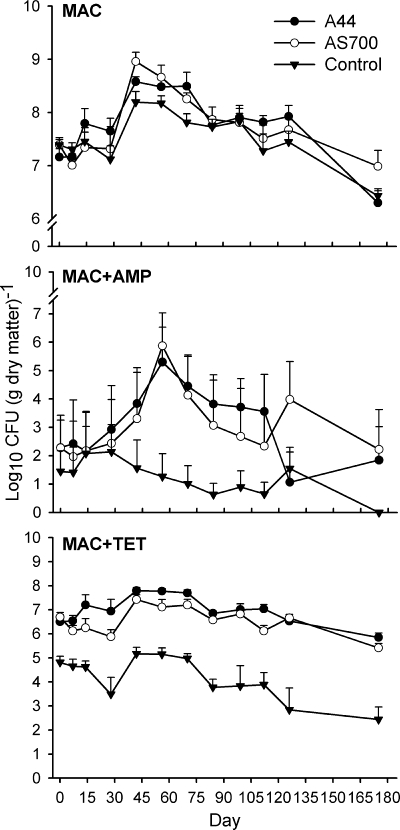
Generic E. coli counts were similar among treatments (Fig. 2, MAC). Only time affected total counts of E. coli (P < 0.001), with a 1- to 2-log increase in numbers between day 0 and day 42. Numbers of total E. coli isolates remained greater than day 0 levels up to day 98 (P < 0.05) and decreased (P < 0.001) below starting counts only on day 175.
FIG. 2.
Mean counts (n = 3; plus standard error) of total and ampicillin- and tetracycline-resistant E. coli isolated from fecal deposits by using MacConkey agar (MAC) or MAC amended with ampicillin (MAC+AMP, 32 μg ml−1) or tetracycline (MAC+TET, 16 μg ml−1), respectively. The treatments were as follows: control, no antimicrobial agents added to the diets of steers from which fecal deposits originated; A44, chlortetracycline (44 ppm); AS700, chlortetracycline and sulfamethazine (each at 44 ppm).
An interaction between time and treatment (P < 0.001) was observed for the numbers and persistence of Ampr E. coli in deposits, with numbers being higher and persistence longer for deposits from A44 and AS700 treatments than for the control deposits (Fig. 2, MAC+AMP). Throughout the study, Ampr E. coli accounted for less than 1% of the total generic E. coli population. On day 56, Ampr E. coli populations peaked in A44 and AS700 treatment deposits and accounted for 0.07% and 0.16%, respectively, of the E. coli population. In contrast, Ampr E. coli numbers in the control treatment exhibited a gradual decline over the experiment, and Ampr E. coli was not detected in control fecal deposits on day 175.
Tetr E. coli in control fecal deposits did not exceed more than 0.3% of the total population but represented from 4.1% (day 126) to 35.6% (day 175) of total E. coli in A44 fecal deposits. Similarly, 2.8% (day 56) to 20.2% (day 0) of the E. coli in AS700 fecal deposits were Tetr. An interaction between time and treatment (P < 0.005) was also observed for Tetr E. coli (Fig. 2, MAC+TET). Generally, the numbers of Tetr E. coli bacteria in fecal deposits from the controls were less (P < 0.05) than those from the A44 and AS700 deposits. In contrast, counts of Tetr E. coli were similar between A44 and AS700 fecal deposits across time. In control and AS700 fecal deposits, levels of Tetr E. coli declined between days 0 and 28 and then increased and remained greater than day 28 levels for the next three samplings (days 42, 56, and 70). At day 175, the number of Tetr E. coli in control and AS700 fecal deposits declined (P < 0.05) to levels below those on day 0. Tetr E. coli levels from A44 fecal deposits initially increased and were greater (P < 0.05) on days 42 and 56 than on day 0. Thereafter, levels of Tetr E. coli in the A44 fecal deposits declined, but after 175 days the counts were still not different (P > 0.05) from those on day 0 [6.6 versus 5.9 log10 CFU (g DM)−1].
Genetic diversity of Ampr and Tetr Escherichia coli.
PFGE profiles were analyzed by using the Dice similarity index to assess the genetic diversity among Ampr and Tetr E. coli isolates in fecal deposits across treatments and durations of environmental exposure. A total of 13 clusters of Ampr isolates having ≥90% similarity were identified, while eight isolates had PFGE patterns considered unique (Table 1). In general, Ampr isolates tended to cluster by treatment. Thirty-five isolates (71.4%) from the control deposits grouped into C8A. Isolates falling into this cluster were present only at the short and moderate exposure periods, accounting for more than 80% of the genetic background, and were exclusive to control deposits. Clusters C2A, C5A, and C7A were isolated only after long exposure, with C2A and C7A being exclusive to control deposits. Most Ampr isolates from the A44 and AS700 fecal deposits grouped into one of two major clusters, C3A and C10A, which persisted as the dominant genotypes from early to long exposure intervals. Clusters C5A, C9A, and C12A were exclusive to A44 deposits, whereas C6A, C11A, and C13A were exclusive to AS700 deposits. In A44 and AS700 treatments, Ampr isolates from some clusters were collected solely during a single exposure period.
TABLE 1.
Temporal PFGE clusters (≥90% relatedness) of ampicillin-resistant E. coli isolated from fecal deposits of steers fed antimicrobial agentsa
| PFGE cluster group | No. (%) of isolates within antimicrobial treatment and time intervalb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A44 |
AS700 |
Control |
||||||||||
| Short | Moderate | Long | Overall | Short | Moderate | Long | Overall | Short | Moderate | Long | Overall | |
| C1A | 1 (5.0) | 1 (1.8) | 1 (5.3) | 1 (2.0) | ||||||||
| C2A | 2 (25.0) | 2 (4.1) | ||||||||||
| C3A | 18 (90.0) | 6 (33.3) | 12 (66.7) | 36 (64.3) | 3 (14.3) | 3 (5.4) | 4 (18.2) | 4 (8.2) | ||||
| C4A | 1 (5.6) | 1 (4.8) | 1 (1.8) | |||||||||
| C5A | 2 (11.1) | 2 (3.6) | 4 (50.0) | 4 (8.2) | ||||||||
| C6A | 2 (13.3) | 2 (3.6) | ||||||||||
| C7A | 2 (25.0) | 2 (4.1) | ||||||||||
| C8A | 17 (89.5) | 18 (81.8) | 35 (71.4) | |||||||||
| C9A | 4 (22.2) | 4 (7.1) | ||||||||||
| C10A | 1 (5.0) | 8 (44.4) | 9 (16.1) | 18 (90.0) | 8 (38.1) | 10 (66.7) | 36 (64.3) | |||||
| C11A | 6 (28.6) | 6 (10.7) | ||||||||||
| C12A | 2 (11.1) | 2 (3.6) | ||||||||||
| C13A | 2 (10.0) | 2 (3.6) | ||||||||||
| Uniquec | 1 (5.6) | 1 (1.8) | 3 (14.3) | 3 (20.0) | 6 (10.7) | 1 (5.3) | 1 (2.0) | |||||
| Total | 20 | 18 | 18 | 56 | 20 | 21 | 15 | 56 | 19 | 22 | 8 | 49 |
The degree of relatedness was calculated by using the Dice similarity index of PFGE profiles from isolates across all treatments and time points. E. coli was isolated from MacConkey agar amended with ampicillin (32 μg ml−1).
The treatments were as follows: control, no antimicrobial agents added to the diets of steers from which fecal pats originated; A44, chlortetracycline (44 ppm); AS700, chlortetracycline and sulfamethazine (each at 44 ppm). Each time interval represents the duration of environmental exposure of fecal deposits prior to sampling: short, days 0 and 7; moderate, days 56 and 70; long, days 126 and 175. “Overall” represents the genotype across all time intervals within each treatment.
E. coli PFGE patterns categorized as unique did not show ≥90% homology to any other isolate patterns.
Compared to the Ampr isolates, there was greater strain diversity among the Tetr isolates (Table 2). In total, 22 clusters of Tetr isolates having ≥90% similarity and 8 unique PFGE patterns were identified. As observed with Ampr isolates, Tetr isolates also tended to cluster by treatment. Tetr E. coli from control deposits grouped into nine different clusters, and two isolates were unique. Most control Tetr isolates (n = 40 [53.3%]) grouped into C7T, while no Tetr isolates from A44 or AS700 were associated with this cluster. Cluster C7T persisted throughout the experiment, representing 57.1%, 70.6%, and 34.8% of the control genotypes at short, moderate, and long exposures, respectively. Tetr isolates from A44 deposits clustered into nine groups, and six isolates had unique genetic profiles. Clusters C8T, C13T, and C20T accounted for most of the A44 Tetr isolates and were obtained at all exposure periods. Clusters C6T and C8T accounted for the majority (67.4% total) of Tetr E. coli from AS700 deposits and were also present at all exposure periods.
TABLE 2.
Temporal PFGE clusters (≥90% relatedness) of tetracycline-resistant E. coli isolated from fecal deposits of steers fed antimicrobial agentsa
| PFGE cluster group | No. (%) of isolates within antimicrobial treatment and time intervalb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A44 |
AS700 |
Control |
||||||||||
| Short | Moderate | Long | Overall | Short | Moderate | Long | Overall | Short | Moderate | Long | Overall | |
| C1T | 6 (17.1) | 2 (11.8) | 4 (17.4) | 12 (16.0) | ||||||||
| C2T | 2 (5.7) | 2 (8.7) | 4 (5.3) | |||||||||
| C3T | 3 (8.6) | 3 (4.0) | ||||||||||
| C4T | 1 (3.2) | 3 (17.6) | 2 (5.7) | 6 (7.2) | ||||||||
| C5T | 2 (11.8) | 2 (8.7) | 4 (5.3) | |||||||||
| C6T | 11 (35.5) | 6 (35.3) | 11 (31.4) | 28 (33.7) | 3 (13.0) | 3 (4.0) | ||||||
| C7T | 20 (57.1) | 12 (70.6) | 8 (34.8) | 40 (53.3) | ||||||||
| C8T | 6 (20.7) | 5 (27.8) | 8 (25.0) | 19 (24.1) | 4 (12.9) | 6 (35.3) | 18 (51.4) | 28 (33.7) | ||||
| C9T | 2 (6.5) | 2 (2.4) | ||||||||||
| C10T | 4 (12.9) | 1 (2.9) | 5 (6.0) | |||||||||
| C11T | 8 (25.8) | 8 (9.6) | 1 (5.9) | 1 (1.3) | ||||||||
| C12T | 1 (3.2) | 1 (1.2) | 3 (13.0) | 3 (4.0) | ||||||||
| C13T | 5 (17.2) | 4 (22.2) | 9 (28.1) | 18 (22.8) | 2 (11.8) | 2 (2.4) | ||||||
| C14T | 2 (6.9) | 2 (2.5) | ||||||||||
| C15T | 2 (6.9) | 1 (5.6) | 1 (3.1) | 4 (5.1) | ||||||||
| C16T | 2 (6.9) | 2 (2.5) | ||||||||||
| C17T | 3 (8.6) | 3 (3.6) | ||||||||||
| C18T | 5 (17.2) | 5 (6.3) | ||||||||||
| C19T | 2 (11.1) | 2 (2.5) | ||||||||||
| C20T | 5 (17.2) | 1 (5.6) | 12 (37.5) | 18 (22.8) | ||||||||
| C21T | 3 (16.7) | 3 (3.8) | ||||||||||
| C22T | 3 (8.6) | 3 (4.0) | ||||||||||
| Uniquec | 2 (6.9) | 2 (11.1) | 2 (6.3) | 6 (7.6) | 1 (2.9) | 1 (4.3) | 2 (2.7) | |||||
| Total | 29 | 18 | 32 | 79 | 31 | 17 | 35 | 83 | 35 | 17 | 23 | 75 |
The degree of relatedness was calculated by using the Dice similarity index of PFGE profiles from isolates across all treatments and time points. E. coli was isolated on MacConkey agar amended with tetracycline (16 μg ml−1).
The treatments were as follows: control, no antimicrobial agents added to the diets of steers from which fecal deposits originated; A44, chlortetracycline (44 ppm); AS700, chlortetracycline and sulfamethazine (each at 44 ppm). Each time interval represents the duration of environmental exposure of fecal deposits prior to sampling: short, days 0 and 7; moderate, days 56 and 70; long, days 126 and 175. “Overall” represents the genotype across all time intervals, within each treatment group.
E. coli PFGE patterns categorized as unique did not show ≥90% homology to any other isolate patterns.
Impact of treatment and exposure time on genetic relatedness of Ampr and Tetr E. coli.
PFGE patterns of Ampr and Tetr E. coli isolates from each exposure period were compared within treatments to assess the genetic similarity within and between groups (Table 3). Ampr E. coli isolated at short and moderate exposure times from control deposits showed a high (P < 0.001) level of within-group similarity (81.2 and 89.1%, respectively). The high degree of similarity between isolates with short and moderate exposures (89.9%) was supported by the fact that the majority of these isolates fell into C8A (Table 1). This indicated that the Ampr E. coli isolates were closely related when first deposited in control feces and that this population persisted for moderate periods of exposure but diversified with prolonged periods of exposure (long exposure, 52.6% similarity to short exposure). The within-group diversity of Ampr isolates (56.5%) also increased (P < 0.001) with the long exposure interval. As with the control, Ampr E. coli isolates from AS700 deposits were closely (P < 0.001) related after a short exposure time (within-group similarity of 82.5%), with this population becoming more diverse after a moderate exposure period (within-group similarity of 59.1%) and remaining so even after long exposure (within-group similarity, 66.1%). The degree of similarity of Ampr isolates collected from AS700 deposits after moderate and long exposure periods remained relatively constant at 70.6 and 70.2%, respectively. As with AS700, Ampr E. coli isolates from A44 deposits were highly (P < 0.001) related after short exposure periods (within-group similarity of 87.9%) but diversified after moderate exposure periods (59.4%). After prolonged exposure, the within group similarity of Ampr isolates from A44 increased (74.3%; P < 0.05). This pattern was also reflected in Ampr isolates collected after long exposure, in that they also exhibited a high degree of relatedness (82.1% similarity) to those that were collected after a short exposure period.
TABLE 3.
Degrees of similarity of PFGE group patterns of resistant E. coli isolated from fecal deposits of steers as affected by duration of environmental exposure
| Resistance and exposure interval groupb | Value for antimicrobial treatment groupa |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A44 |
AS700 |
Control |
||||||||||
| No. of isolates | Similarity to “short” group | Within- group similarity | P valuec | No. of isolates | Similarity to “short” group | Within- group similarity | P value | No. of isolates | Similarity to “short” group | Within- group similarity | P value | |
| Ampicillin | ||||||||||||
| Short | 20 | 87.9 | <0.001 | 20 | 82.5 | <0.001 | 19 | 81.2 | <0.001 | |||
| Moderate | 18 | 60.5 | 58.4 | 0.999 | 21 | 70.6 | 59.1 | 0.759 | 22 | 89.9 | 89.1 | <0.001 |
| Long | 18 | 82.1 | 74.3 | 0.050 | 15 | 70.2 | 66.1 | 0.994 | 8 | 52.6 | 56.5 | <0.001 |
| Tetracycline | ||||||||||||
| Short | 29 | 61.1 | 0.979 | 31 | 59.5 | <0.001 | 35 | 66.1 | 0.101 | |||
| Moderate | 18 | 54.6 | 57.2 | 0.998 | 17 | 56.3 | 60.8 | 0.19 | 17 | 67.8 | 73.7 | <0.001 |
| Long | 32 | 61.0 | 70.4 | <0.001 | 35 | 53.7 | 59.1 | 0.002 | 23 | 63.6 | 61.1 | 0.868 |
Control, no antimicrobial agents added to the diets of steers from which fecal deposits originated; A44, chlortetracycline (44 ppm); AS700, chlortetracycline and sulfamethazine (each at 44 ppm).
Each interval represents the duration of environmental exposure of fecal deposits prior to sampling: short, days 0 and 7; moderate, days 56 and 70; long, days 126 and 175. Ampicillin- or tetracycline-resistant E. coli was isolated from MacConkey agar amended with ampicillin (32 μg ml−1) or tetracycline (16 μg ml−1), respectively. The groups of resistant E. coli were analyzed separately.
The effect of the exposure interval on the significance of within-group similarity (P value) was analyzed within each antimicrobial treatment group. PFGE profiles were grouped by short, moderate, and long exposure intervals and analyzed by bootstrapping (n = 1,000).
In general, Tetr isolates exhibited a higher diversity than Ampr isolates (Table 3), a result that reflects the greater number of clusters identified within this group (Table 2). At early exposure periods, only Tetr isolates from the AS700 deposits were a distinct population (P < 0.001) (within-group similarity of 59.5%). Within-group similarity of Tetr isolates from AS700 fecal deposits remained relatively constant over the exposure period and was still significant (P < 0.01) after long exposure (59.1%). Within-group similarity of Tetr isolates from A44 deposits increased (P < 0.001) after prolonged periods of exposure (70.4%). Similarity of Tetr isolates collected after moderate and long exposure periods to those collected after short exposure decreased in control deposits. In contrast, Tetr isolates collected from A44 deposits after long exposure became more similar to those collected during the period of short exposure.
Analysis of isolates across all exposure periods demonstrated that all Ampr and Tetr isolates within treatment groups exhibited a high (P < 0.001) degree of relatedness. Analysis of exposure periods across all treatments indicated that Ampr isolates within short and moderate exposure periods were closely (P < 0.001) related, whereas this characteristic was observed only in Tetr isolates collected after long exposure.
Encoded resistance determinants.
The gene determinants tetL, tetM, tetO, tetW, blaOXA1, and blaPSE1 were not detected in any of the isolates. Most of the Ampr isolates (Table 4) had gene profile G8A (34.8%; positive for tetC), followed by profile 2A (25.6%; positive for tetA, sul1, and blaTEM1). Ninety-eight (60.9%) of the Ampr isolates did not test positive for blaTEM1, indicating that most of the Ampr E. coli isolates in the study were resistant by a mechanism other than that associated with the Ampr genes screened. Tetr genes were detected in 79.1% of Ampr isolates. tetC (37.3%) was most commonly detected, followed by tetA (34.3%) and tetB (6.9%). In every instance in which an isolate was positive for tetA, the same isolate was also positive for tem1. Because of the high frequency of Tetr genes detected in Ampr E. coli, these bacteria were also tested for phenotypic resistance to tetracycline. Each isolate from the A44 and AS700 fecal deposits that encoded a Tetr gene was resistant to tetracycline, excepting the following: from A44, two isolates and one isolate within the gene profiles G2A and G8A, respectively; from AS700, two isolates within gene profile G8A. Every isolate from control deposits that encoded a Tetr gene was resistant to tetracycline. Additionally, three and five control isolates within G12A and G13A, respectively, were resistant to tetracycline despite testing negative for any Tetr genes (data not shown).
TABLE 4.
Patterns of encoded genetic determinants in ampicillin-resistanta E. coli strains isolated from fecal deposits of steers fed antimicrobial agents
| Gene profile | Detection of gene |
No. (%) of isolates within treatment groupb |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| tetA | tetB | tetC | sul1 | sul2 | blaTEM1 | A44 | AS700 | Control | Overall | |
| G1A | + | − | − | − | − | + | 3 (5.4) | 0 (0) | 5 (10.4) | 8 (5.0) |
| G2A | + | − | − | + | − | + | 34 (60.7) | 7 (12.5) | 0 (0) | 41 (25.6) |
| G3A | + | − | − | − | + | + | 0 (0) | 0 (0) | 1 (2.1) | 1 (0.6) |
| G4A | + | − | + | + | − | + | 0 (0) | 1 (1.8) | 0 (0) | 1 (0.6) |
| G5A | + | − | − | + | + | + | 5 (8.9) | 0 (0) | 0 (0) | 5 (3.1) |
| G6A | − | + | − | − | + | − | 0 (0) | 3 (5.4) | 7 (14.6) | 10 (6.3) |
| G7A | − | + | − | + | + | − | 0 (0) | 1 (1.8) | 0 (0) | 1 (0.6) |
| G8A | − | − | + | − | − | − | 9 (16.1) | 38 (67.9) | 9 (18.4) | 56 (34.8) |
| G9A | − | − | + | − | + | − | 1 (1.8) | 3 (5.4) | 0 (0) | 4 (2.5) |
| G10A | − | − | − | + | − | − | 0 (0) | 0 (0) | 1 (2.1) | 1 (0.6) |
| G11A | − | − | − | − | + | − | 1 (1.8) | 1 (1.8) | 0 (0) | 2 (1.3) |
| G12A | − | − | − | − | − | + | 2 (3.6) | 0 (0) | 5 (10.4) | 7 (4.4) |
| G13A | − | − | − | − | − | − | 1 (1.8) | 2 (3.6) | 21 (43.8) | 24 (15.0) |
| Total | 56 | 56 | 49 | 161 | ||||||
E. coli was isolated using MacConkey agar amended with ampicillin (32 μg ml−1), and fecal material was sampled from deposits on six occasions over 175 days, representing short, moderate, and long periods of environmental exposure.
Control, no antimicrobial agents added to the diet; the A44 and AS700 antimicrobial feeding treatments are defined in Materials and Methods.
Sulfonamide-resistant genes occurred in 41.2% of Ampr E. coli isolates. sul1 and sul2 were detected separately in 26.8% and 10.7% of the Ampr isolates, respectively, and together in only 3.7% of isolates. Of the Ampr isolates from A44 deposits, 41 (73.2%) possessed one or both sul genes, which was more frequent than the occurrence among Ampr isolates collected from AS700 fecal deposits (26.8%).
Overall, 12 patterns of encoded resistance genes were detected among the Tetr isolates (Table 5). The majority of isolates from all treatments possessed profile G6T (overall, 62.9%; positive for tetB and sul2). G3T was the second most common (13.3%; positive for tetA, sul1, and tem1), and almost all G3T isolates originated from the A44 deposits. Profiles G9T and G10T were detected only in Tetr E. coli from AS700 deposits. Six other profiles (G1T, G2T, G4T, G7T, G8T, and G11T) were also specific to a single treatment. Of the Tetr isolates examined, 96.3% possessed one or more of the tetA, tetB, and tetC determinants. The most common Tetr gene was tetB (68.5%), followed by tetA (13.7%), tetC (12.0%), and the combination of tetA and tetB (1.7%) or tetB and tetC (0.4%). Most of the Tetr E. coli isolates with TetC were from AS700 deposits.
TABLE 5.
Patterns of encoded genetic determinants in tetracycline-resistanta E. coli isolated from fecal deposits of steers fed antimicrobial agents
| Gene profile | Detection of gene |
No. (%) of isolates within treatment groupb |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| tetA | tetB | tetC | sul1 | sul2 | blaTEM1 | A44 | AS700 | Control | Overall | |
| G1T | + | − | − | − | − | + | 1 (1.3) | 0 (0) | 0 (0) | 1 (0.4) |
| G2T | + | + | − | − | + | − | 0 (0) | 0 (0) | 1 (1.4) | 1 (0.4) |
| G3T | + | − | − | + | − | + | 30 (38.0) | 1 (1.2) | 0 (0) | 31 (13.3) |
| G4T | + | + | − | + | + | + | 3 (3.8) | 0 (0) | 0 (0) | 3 (1.3) |
| G5T | − | + | − | − | − | − | 0 (0) | 2 (2.4) | 10 (14.1) | 12 (5.2) |
| G6T | − | + | − | − | + | − | 41 (51.9) | 48 (57.8) | 60 (80.0) | 149 (62.9) |
| G7T | − | + | + | − | + | − | 0 (0) | 0 (0) | 1 (1.4) | 1 (0.4) |
| G8T | − | + | − | + | + | − | 0 (0) | 0 (0) | 1 (1.4) | 1 (0.4) |
| G9T | − | − | + | − | − | − | 0 (0) | 10 (12.0) | 0 (0) | 10 (4.3) |
| G10T | − | − | + | − | + | − | 0 (0) | 18 (21.7) | 0 (0) | 18 (7.7) |
| G11T | − | − | − | − | + | − | 2 (2.5) | 0 (0) | 0 (0) | 2 (0.9) |
| G12T | − | − | − | − | − | − | 2 (2.5) | 4 (4.8) | 1 (1.4) | 7 (3.0) |
| Total | 79 | 83 | 75 | 237 | ||||||
E. coli was isolated using MacConkey agar amended with tetracycline (16 μg ml−1), and fecal material was sampled from deposits on six occasions over 175 days, representing short, moderate, and long periods of environmental exposure.
Control, no antimicrobial agents added to the diet; the A44 and AS700 antimicrobial feeding treatments are defined in Materials and Methods.
Sulfonamide resistance genes were detected in 87.1% of Tetr isolates, with sul2 (72.3%) being the most prevalent, followed by sul1 (13.3%) and the combination of sul1 and sul2 (1.7%). sul determinants were detected in 96.2%, 80.7%, and 84.2% of Tetr E. coli isolates from A44, AS700, and control deposits, respectively. Approximately 15% of Tetr isolates possessed blaTEM1, and all, except one isolate, were from A44 deposits. Only in one instance was tetA detected in Tetr isolates without being associated with blaTEM1.
DISCUSSION
Persistence of E. coli in fecal deposits.
The present study examined the persistence and diversity of AR E. coli in fecal deposits from cattle fed one of three AGP treatments. E. coli was isolated using selective plates, a method that has been described previously (24, 43).
Bacteria from fecal material can be transferred in runoff water, potentially contaminating water sources for humans (45). Our work suggests that feces could pose a risk of contaminating watersheds with AR E. coli for months after defecation. The fecal deposits presented a matrix that enabled growth and prolonged survival of AR E. coli beyond 175 days. Except for Ampr E. coli in control deposits, E. coli numbers increased after defecation. This suggests that the initial load of total and resistant E. coli deposited in feces may underestimate populations that develop after feces enter the environment. Other studies have also reported a growth phase of E. coli in fecal deposits, although most have shown that growth occurred within 21 days of incubation (36, 48). Growth of E. coli in fecal material is related to temperature, with a proposed optimal range between 20 and 35°C (48, 56). The delayed growth in our study may have resulted from the low environmental temperatures at the beginning of the experiment, given that growth of E. coli occurred when the maximum daily temperature ranged between 14°C and 21°C. It is also possible that E. coli growth was partially attributable to reductions in competing bacteria as oxygen penetrated the feces with time (34). This may have reduced numbers of obligate anaerobes, perhaps changing the niche to one more favorable for E. coli.
Declines the in E. coli population corresponded with a loss of moisture from fecal deposits. However, generic E. coli numbers still exceeded 107 (g dry matter)−1 even when the moisture content of the deposits declined below 10% after 84 days of exposure. Sinton et al. (48) showed that rehydration of fecal deposits promoted regrowth of enterococci and reduced the rate of decline of viable E. coli, although growth was more prolific in fecal deposits that contained more than 80% water. In our study, heavy precipitation occurred in June (days 93 to 123). Although this event did not dramatically stimulate the growth of E. coli, it did appear to reduce the rate of decline in viable E. coli and may have contributed to persistence. It is also possible that by this time, nutrients as opposed to moisture limited growth.
Although cattle from the control treatment were not fed antimicrobials, their fecal deposits had relatively high numbers of Tetr E. coli on day 0. Similar results have been described for fecal samples that have been collected directly from the rectums of feedlot cattle (1, 46) and dairy calves (25) that have not been fed AGP. Given the widespread prevalence of tetracycline resistance (40), presumably these cattle were colonized by Tetr bacteria from other environmental sources, most likely prior to their arrival at the feedlot. The fact that Tetr E. coli can survive for up to 175 days in fecal deposits suggests that fecal matter on pasture, surface water sources, herd cohorts, or possibly even wildlife could have served as acquisition sources of Tetr E. coli for cattle prior to their arrival at the feedlot.
As expected, the numbers of Tetr E. coli in fecal deposits from cattle fed AGP containing chlortetracycline were greater than those of Tetr E. coli in fecal deposits from cattle fed no AGP. The AGP affected the concentrations of Tetr E. coli detected in A44 and AS700 fecal deposits to the same extent. This was likely because A44 and AS700 contained the same levels of chlortetracycline (44 ppm), although the AS700 mixture also contained sulfamethazine (44 ppm). Generally, Tetr E. coli from all treatments displayed similar trends, with an initial growth followed by a death phase. Approximately 75% of chlortetracycline administered in the diets of cattle is excreted in feces and urine (29). Elmund et al. (18) reported that levels of chlortetracycline decreased from 14 ppm in fresh bovine feces to 0.34 ppm in aged feces. However, degradation of chlortetracycline is temperature dependent (19). Over 30 days, Gavalchin and Katz (19) showed that soil amended with manure at 4°C, 20°C, or 30°C resulted in 0%, 12%, or 56% degradation of chlortetracycline, respectively. These data would suggest that chlortetracycline concentrations would have been greater in fecal deposits early in our experiment when temperatures were low, subsequently degrading more rapidly when temperatures increased at later time points. Throughout the entire course of our study, the percentage of Tetr E. coli in the total population remained relatively constant in fecal deposits regardless of treatment. Consequently, it appears that the selective pressure of subtherapeutic tetracycline was more potent for Tetr E. coli in the digestive tracts of cattle than in deposited feces. This would also imply that in the absence of selection pressure in control fecal deposits, the fitness of Tetr E. coli was not inhibited by retaining encoded resistance to chlortetracycline. This is in agreement with an earlier study that reported selection and maintenance of E. coli with a streptomycin-sulfadiazine-tetracycline-resistant phenotype in dairy cattle that was independent of antimicrobial administration (26). The same authors suggested that resistance elements were maintained because they were linked to additional genetic traits conferring a selective advantage (27). There may also be advantages to E. coli maintaining resistance elements in secondary environments.
Although initial Ampr E. coli counts in control deposits were similar to those of A44 and AS700 deposits, numbers of Ampr E. coli in control deposits did not increase significantly after defecation. In contrast, numbers of Ampr E. coli in A44 and AS700 deposits increased dramatically from initial levels. Despite a large standard error of the means, counts of Ampr E. coli from treatment fecal deposits were statistically different and, on day 56, were approximately four log units greater than those for control Ampr E. coli. The reasons for these differences are unclear but may be related to the AGP fed to cattle. The genetic backgrounds of Ampr E. coli in A44 and AS700 fecal deposits were significantly different from those in control deposits. It may be possible that while within the ruminant digestive tract, the AGP selected for Ampr E. coli populations that in some unknown way were better able to persist in secondary environments than were Ampr E. coli populations from cattle that were not administered AGP.
Genetic diversity of Escherichia coli.
Despite the genetic analysis being limited to only Ampr and Tetr E. coli isolates, considerable genetic diversity among isolates was observed. Others have also shown high diversity in the E. coli populations from cattle (2, 43). Survival of E. coli outside the primary habitat of the host's intestinal tract has been hypothesized to be limited and transient as a result of fluctuations in nutrient availability and temperature (58). However, there is evidence that certain strains of E. coli are adapted to survive in secondary habitats, such as soil and water (11) and manure storage systems (17). It has been hypothesized that once the bacteria are shed from the digestive tract, a multitude of factors select for fit subpopulations of E. coli, leading to an expansion in genetic diversity as strains persist in a variety of environments (21, 53). Our results show that the diversity of Ampr E. coli within treatment groups increased after environmental exposure, even within the relatively restricted habitat of the fecal deposit. The increase in diversity of Ampr E. coli isolated from A44 and AS700 deposits was apparent after only moderate exposure periods. The fact that E. coli in feces with a short duration of exposure would have recently emerged from the comparatively steady-state conditions of the digestive tract may have led to a higher degree of relatedness among isolates obtained at this point in the study.
None of the eight Ampr clusters associated with long-exposed control deposits had been identified previously. Because the fecal deposits were exposed to environmental conditions, it cannot be ruled out that contaminating sources, including cross-contamination from adjacent samples, contributed to the Ampr E. coli populations and the detection of new PFGE clusters after long exposure. Regardless, there was a clear contrast between Ampr E. coli isolates in A44 and AS700 deposits and those from control deposits in that among Ampr E. coli isolates from the treatment deposits, main PFGE cluster groups survived throughout the experiment. This led us to speculate that the Ampr E. coli present initially in control fecal deposits was less fit than that in the A44 or AS700 fecal deposit, although it is difficult to explain why. This speculation may also be relevant to why Ampr E. coli numbers in control fecal deposits did not increase; they may have been outcompeted by other bacteria in the fecal deposits.
Tetr E. coli isolates were genotypically more diverse than Ampr E. coli, a result that is perhaps not surprising given the far higher numbers of Tetr than Ampr E. coli isolates in deposits. There were numerous instances where distinct PFGE clusters were detected in fecal deposits only after short exposure periods, a result suggesting that there was differential Tetr E. coli survival in deposits after defecation. However, as with Ampr E. coli in A44 and AS700 fecal deposits, the Tetr E. coli PFGE cluster groups that encompassed the majority of E. coli within each treatment at the beginning of the experiment were stable across time.
Overall, there was a clear relationship between treatment and the genetic diversity of resistant E. coli in fecal deposits. It was interesting that the within-group similarities were highest for Ampr and Tetr E. coli isolates from control deposits compared to E. coli isolates from A44 and AS700 deposits. We hypothesized that the selective pressure of including AGP in the diet would reduce the within-group diversity of E. coli by selecting for resistant strains, but in the present experiment this was clearly not the case. The time period of environmental exposure also influenced the genotypic nature of E. coli in fecal deposits, perhaps indicating that some genotypes survive better in secondary environments. These findings have implications for microbial source tracking since the nature of E. coli released from the point source may change with both diet and time of environmental exposure (21). Knowledge of genetic markers and of the external factors that influence their frequency is important for accurate bacterial source tracking (31).
Resistance determinants encoded by Escherichia coli.
The only β-lactamase gene detected in E. coli in this study was blaTEM1. This determinant is generally plasmid mediated (9) and is one of the more prevalent β-lactamases (32). Similar to our results, blaTEM1 has been reported to be the most common β-lactamase gene detected among Ampr E. coli isolates from livestock (9). The genes tetL, tetM, and tetW were not detected in any of the E. coli in our study. Generally, these genes are associated with gram-positive bacteria (40), but a recent investigation detected tetM and tetW in E. coli isolated from farm soils (50). These results would suggest that E. coli may acquire these more-obscure tet determinants through interaction with microbes in the environment as opposed to those in the digestive tract.
Gene profiles were clearly related to treatment. This outcome indicates that the type of AGP administered to cattle had an impact on the nature of Ampr E. coli in deposits. More than 90% of Ampr E. coli from A44 and AS700 fecal deposits possessed Tetr genes, although these populations were still clearly distinct. Almost all of the Ampr E. coli isolates encoding tetA were from A44 deposits, whereas most encoding tetC were from AS700 deposits. Presumably, this difference arises from the presence of a mixture of tetracycline and sulfamethazine in AS700 as opposed to tetracycline alone in A44. Sulfamethazine is relatively stable in manure (3, 16) and would likely have been prevalent in AS700 fecal deposits throughout our study. Even though AS700 contained sulfamethazine, a large number of the isolates from these deposits lacked sul1 or sul2. In fact, the majority of Ampr isolates that possessed sul1 and sul2 were associated with A44 deposits. It is not clear why this occurred.
Except for five isolates, all Ampr E. coli isolates that possessed tetA, tetB, or tetC were resistant to tetracycline. Tetracycline-susceptible E. coli strains that encode resistance genes have been reported previously (14). Additionally, some E. coli isolates from control deposits tested negative for tetracycline resistance genes yet were resistant to tetracycline. Likely these E. coli isolates encoded a resistance gene not tested in our investigation. The high level of both phenotypic resistance (80.7%) and tetracycline resistance genes (79%) in Ampr E. coli supports previous research showing a link between tetracycline and additional resistance determinants. Our laboratory and others have previously reported that the feeding of tetracycline-based AGP to cattle can promote multiresistance to ampicillin and tetracycline in E. coli (1, 23). In addition, resistance determinants for ampicillin and tetracycline have been identified on a common plasmid (30, 47). These data further highlight the selection of multiresistant E. coli when subtherapeutic levels of tetracycline are administered to cattle. Our results show that multiresistant E. coli can survive for extended periods in the environment.
The most common gene profile detected in Tetr E. coli isolates was G6T, which included tetB and sul2, a result that confirms the possible link between these two determinants as observed for Ampr isolates. Similarly, Boerlin et al. (8) found an association between tetB and sul2. In that same study, an association was also noted between tetA and sul1. Our results support their conclusion, in that G3T, which included tetA, sul1, and blaTEM1, was the second most prevalent profile in Tetr E. coli. Only in a single isolate was tetA not detected along with blaTEM1.
There were clear differences in the resistance genes found in Tetr E. coli compared to those in Ampr E. coli. Notably, tetB was the most prevalent determinant in Tetr E. coli. The prevalence of tetB in generic E. coli from cattle has previously been shown to be higher than that of tetA (10, 43). Although tetB may be the most common determinant in generic E. coli, there clearly are subpopulations of E. coli (e.g., Ampr E. coli) in which other tet determinants dominate. Blake and colleagues (7) reported differences in Tetr genes in E. coli isolated from pigs fed or not fed antimicrobials. We also report differences in Tetr genes carried by E. coli in feces from cattle fed diets with or without AGP. An interesting finding from our study was that chlortetracycline administered to animals at similar concentrations can select equally for phenotypic resistance (viz., Tetr E. coli counts) through different genes encoding the resistance. As noted previously, differential selection of resistance genes may alter multiresistance through carriage of linked resistance determinants (7). A genetic link between tetA and class 1 integrons on the same conjugative plasmid has recently been reported (52), which may suggest that selection for tetA (largely seen in Ampr E. coli isolates from A44 deposits) could also select for class 1 integrons. Characterization of the selective pressures exerted by AGP that are responsible for a differential prevalence of resistance determinants could provide additional insight into the ecology of antimicrobial resistance.
In conclusion, bovine fecal deposits present a matrix for the growth and persistence of Tetr, Ampr, and generic E. coli beyond 175 days. Resistant E. coli populations in fecal material can increase after defecation, increasing the true microbial load introduced into the environment. Inclusion of AGP in the diet promotes the presence and preferential persistence of AR E. coli in fecal deposits. However, Tetr E. coli persisted for 175 days even in fecal deposits that arose from cattle that had not been fed AGP. This suggests that encoding of Tet resistance had minimal effects on environmental fitness, even in the absence of antimicrobials. Genotypic diversity of Ampr E. coli isolates tended to increase with longer periods of environmental exposure, whereas this trend was less apparent for Tetr E. coli, a result that may reflect the greater initial diversity of Tetr isolates. Fecal deposits can serve as a reservoir of AR E. coli for months after defecation and should be considered a potential point-source contaminant in the development of watershed management plans.
Acknowledgments
Financial support for this research project was provided by the GAPS funding program of Agriculture and Agri-Food Canada.
We thank Kim Stanford for her assistance with the PFGE analysis and Toby Entz for his statistical guidance. We gratefully acknowledge Lorna Selinger, Ruth Barbieri, Wendi Smart, Zexun Lu, Yu-Hung Hsu, and Cassidy Klima for their technical assistance. We appreciate the excellent animal care skills of the staff at the Lethbridge Research Centre Research Feedlot.
Footnotes
Published ahead of print on 2 October 2009.
REFERENCES
- 1.Alexander, T. W., L. J. Yanke, E. Topp, M. E. Olson, R. R. Read, D. W. Morck, and T. A. McAllister. 2008. Effect of subtherapeutic administration of antibiotics on the prevalence of antibiotic-resistant Escherichia coli bacteria in feedlot cattle. Appl. Environ. Microbiol. 74:4405-4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, M. A., J. E. Whitlock, and V. J. Harwood. 2006. Diversity and distribution of Escherichia coli genotypes and antibiotic resistance phenotypes in feces of humans, cattle, and horses. Appl. Environ. Microbiol. 72:6914-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aust, M. O., F. Godlinski, G. R. Travis, X. Hao, T. A. McAllister, P. Leinweber, and S. Thiele-Bruhn. 2008. Distribution of sulfamethazine, chlortetracycline and tylosin in manure and soil of Canadian feedlots after subtherapeutic use in cattle. Environ. Pollut. 156:1243-1251. [DOI] [PubMed] [Google Scholar]
- 4.Bach, S. J., K. Stanford, and T. A. McAllister. 2005. Survival of Escherichia coli O157:H7 in feces from corn- and barley-fed steers. FEMS Microb. Lett. 252:25-33. [DOI] [PubMed] [Google Scholar]
- 5.Bezanson, G. S., R. Khakhria, and E. Bollegraaf. 1983. Nosocomial outbreak caused by antibiotic-resistant strain of Salmonella typhimurium acquired from dairy cattle. Can. Med. Assoc. J. 128:426-427. [PMC free article] [PubMed] [Google Scholar]
- 6.Bibbal, D., V. Dupouy, J. P. Ferré, P. L. Toutain, O. Fayet, M. F. Prère, and A. Bousquet-Mélou. 2007. Impact of three ampicillin dosage regimens on selection of ampicillin resistance in Enterobacteriaceae and excretion of blaTEM genes in swine feces. Appl. Environ. Microbiol. 73:4785-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blake, D. P., R. W. Humphry, K. P. Scott, K. Hillman, D. R. Fenlon, and J. C. Low. 2003. Influence of tetracycline exposure on tetracycline resistance and the carriage of tetracycline resistance genes within commensal Escherichia coli populations. J. Appl. Microbiol. 94:1087-1097. [DOI] [PubMed] [Google Scholar]
- 8.Boerlin, P., R. Travis, C. L. Gyles, R. Reid-Smith, N. Janecko, H. Lim, V. Nicholson, S. A. McEwen, R. Friendship, and M. Archambault. 2005. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl. Environ. Microbiol. 71:6753-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinas, L., M. Zarazaga, Y. Saenz, F. Ruiz-Larrea, and C. Torres. 2002. β-Lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans and healthy animals. Antimicrob. Agents Chemother. 46:3156-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryan, A., N. Shapir, and M. J. Sadowsky. 2004. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 70:2503-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byappanahalli, M. N., R. L. Whitman, D. A. Shively, M. J. Sadowsky, and S. Ishii. 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8:504-513. [DOI] [PubMed] [Google Scholar]
- 12.Canadian Council on Animal Care. 1993. Guide to the care and use of experimental animals, vol. 1. Canadian Council on Animal Care, Ottawa, Ontario, Canada.
- 13.Centers for Disease Control and Prevention. 1996. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis: a manual. National Center for Infectious Diseases, Atlanta, GA.
- 14.Chalmers, G., G. K. Kozak, E. Hillyer, R. J. Reid-Smith, and P. Boerlin. Low minimum inhibitory concentrations associated with the tetracycline-resistance gene tet(C) in Escherichia coli. Can. J. Vet. Res., in press. [PMC free article] [PubMed]
- 15.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing, 18th informational supplement, M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 16.Dolliver, H., S. Gupta, and S. Noll. 2008. Antibiotic degradation during manure composting. J. Environ. Qual. 37:1245-1253. [DOI] [PubMed] [Google Scholar]
- 17.Duriez, P., and E. Topp. 2007. Temporal dynamics and impact of manure storage on antibiotic resistance patterns and population structure of Escherichia coli isolates from a commercial swine farm. Appl. Environ. Microbiol. 73:5486-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elmund, G. K., S. M. Morrison, D. W. Grant, and M. P. Nevins. 1971. Role of excreted chlortetracycline in modifying decomposition process in feedlot waste. Bull. Environ. Contam. Toxicol. 6:129-132. [DOI] [PubMed] [Google Scholar]
- 19.Gavalchin, J. and S. E. Katz. 1994. The persistence of fecal-borne antibiotics in soil. J. AOAC Int. 77:481-485. [Google Scholar]
- 20.Ghosh, S., and T. M. LaPara. 2007. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 1:191-203. [DOI] [PubMed] [Google Scholar]
- 21.Gordon, D. N., S. Bauer, and J. R. Johnson. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513-1522. [DOI] [PubMed] [Google Scholar]
- 22.Guerra, B., E. Junker, A. Miko, R. Helmuth, and M. C. Mendoza. 2004. Characterization and localization of drug resistance determinants in multidrug-resistant, integron-carrying Salmonella enterica serotype Typhimurium strains. Microb. Drug Resist. 10:83-91. [DOI] [PubMed] [Google Scholar]
- 23.Hoyle, D. V., C. M. Yates, M. E. Chase-Topping, E. J. Turner, S. E. Davies, J. C. Low, G. J. Gunn, M. E. Woolhouse, and S. G. Amyes. 2005. Molecular epidemiology of antimicrobial-resistant commensal Escherichia coli strains in a cohort of newborn calves. Appl. Environ. Microbiol. 71:6680-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hummel, R., H. Tschape, and W. Witte. 1986. Spread of plasmid-mediated nourseothricin resistance due to antibiotic use in animal husbandry. J. Basic Microbiol. 8:461-466. [DOI] [PubMed] [Google Scholar]
- 25.Khachatryan, A. R., D. D. Hancock, T. E. Besser, and D. R. Call. 2004. Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in dairy calves. Appl. Environ. Microbiol. 70:752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khachatryan, A. R., T. E. Besser, D. D. Hancock, and D. R. Call. 2006. Use of a nonmedicated dietary supplement correlates with increased prevalence of streptomycin-sulfa-tetracycline-resistant Escherichia coli on a dairy farm. Appl. Environ. Microbiol. 72:4583-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khachatryan, A. R., T. E. Besser, and D. R. Call. 2008. The streptomycin-sulfadiazine-tetracycline antimicrobial resistance element of calf-adapted Escherichia coli is widely distributed among isolates from Washington State cattle. Appl. Environ. Microbiol. 74:391-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koike, S., I. G. Krapac, H. D. Oliver, A. C. Yannarell, J. C. Chee-Sanford, R. I. Aminov, and R. I. Mackie. 2007. Monitoring and source tracking of tetracycline resistance genes in lagoons and groundwater adjacent to swine production facilities over a 3-year period. Appl. Environ. Microbiol. 73:4813-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar, K., S. C. Gupta, Y. Chander, and A. K. Singh. 2005. Antibiotic use in agriculture and its impact on the terrestrial environment. Adv. Agron. 87:1-54. [Google Scholar]
- 30.Lavollay, M., K. Mamlouk, T. Frank, A. Akpabie, B. Burghoffer, S. Ben Redjeb, R. Bercion, V. Gautier, and G. Arlet. 2006. Clonal dissemination of a CTX-M-15 β-lactamase-producing Escherichia coli strain in the Paris area, Tunis, and Bangui. Antimicrob. Agents Chemother. 50:2433-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leach, M., S. Broschat, and D. R. Call. 2008. A discrete, stochastic model and correction method for bacterial source tracking. Envrion. Sci. Technol. 42:524-529. [DOI] [PubMed] [Google Scholar]
- 32.Li, X. Z., M. Mehrotra, S. Ghimire, and L. Adewoye. 2007. Beta-lactam resistance and beta-lactamases in bacteria of animal origin. Vet. Microbiol. 121:197-214. [DOI] [PubMed] [Google Scholar]
- 33.McEwen, S. A., and P. J. Feorka-Cray. 2002. Antimicrobial use and resistance in animals. Clin. Infect. Dis. 34(Suppl. 3):S93-S106. [DOI] [PubMed] [Google Scholar]
- 34.McGarvey, J. A., W. G. Miller, R. Zhang, Y. Ma, and F. Mitloehner. 2007. Bacterial population dynamics in dairy waste during aerobic and anaerobic treatment and subsequent storage. Appl. Environ. Microbiol. 73:193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meals, D. W., and D. C. Braun. 2006. Demonstration of methods to reduce E. coli runoff from dairy manure application sites. J. Environ. Qual. 35:1088-1100. [DOI] [PubMed] [Google Scholar]
- 36.Muirhead, R. W., R. P. Collins, and P. J. Bremer. 2005. Erosion and subsequent transport state of Escherichia coli from cow pats. Appl. Environ. Microbiol. 71:2875-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng, L.-K., I. Martin, M. Alfa, and M. Mulvey. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15:209-215. [DOI] [PubMed] [Google Scholar]
- 38.Peak, N., C. W. Knapp, R. K. Yang, M. M. Hanfelt, M. S. Smith, D. S. Aga, and D. W. Graham. 2007. Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies. Environ. Microbiol. 9:143-151. [DOI] [PubMed] [Google Scholar]
- 39.Price, L. B., J. P. Graham, L. G. Lackey, A. Roess, R. Vailes, and E. Silbergeld. 2007. Elevated risk of carrying gentamicin-resistant Escherichia coli among U.S. poultry workers. Environ. Health Perspect. 115:1738-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts, M. C. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245:195-203. [DOI] [PubMed] [Google Scholar]
- 41.Salyers, A. A., A. Gupta, and Y. Wang. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12:412-416. [DOI] [PubMed] [Google Scholar]
- 42.SAS Institute Inc. 2001. SAS/STAT user's guide. SAS Institute Inc., Cary, NC.
- 43.Sawant, A. A., N. V. Hegde, B. A. Straley, S. C. Donaldson, B. C. Love, S. J. Knabel, and B. M. Jayarao. 2007. Antimicrobial-resistant enteric bacteria from dairy cattle. Appl. Environ. Microbiol. 73:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitt, H., K. Stoob, G. Hamscher, E. Smit, and W. Seinen. 2006. Tetracyclines and tetracycline resistance in agricultural soils: microcosm and field studies. Microb. Ecol. 51:267-276. [DOI] [PubMed] [Google Scholar]
- 45.Schuster, C. J., A. G. Ellis, W. J. Robertson, D. E. Charron, J. J. Aramini, B. J. Marshall, and D. T. Medeiros. 2005. Infectious disease outbreaks related to drinking water in Canada, 1974-2001. Can. J. Public Health 96:254-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma, R., K. Munns, T. Alexander, T. Entz, P. Mirzaagha, L. J. Yanke, M. Mulvey, E. Topp, and T. McAllister. 2008. Diversity and distribution of commensal fecal Escherichia coli bacteria in beef cattle administered selected subtherapeutic antimicrobials in a feedlot setting. Appl. Environ. Microbiol. 74:6178-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shehabi, A. A., J. F. Odeh, and M. Fayyad. 2006. Characterizing of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from human stools and drinking water sources in Jordan. J. Chemother. 18:468-472. [DOI] [PubMed] [Google Scholar]
- 48.Sinton, L. W., R. R. Braithwaite, C. H. Hall, and M. L. Mackenzie. 2007. Survival of indicator and pathogenic bacteria in bovine feces on pasture. Appl. Environ. Microbiol. 73:7917-7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, M. S., R. K. Yang, C. W. Knapp, Y. Niu, N. Peak, M. M. Hanfelt, J. C. Galland, and D. W. Graham. 2004. Quantification of tetracycline resistance genes in feedlot lagoons by real-time PCR. Appl. Environ. Microbiol. 70:7372-7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srinivasan, V., H. M. Nam, A. A. Sawant, S. I. Headrick, L. T. Nguyen, and S. P. Oliver. 2008. Distribution of tetracycline and streptomycin resistance genes and class 1 integrons in Enterobacteriaceae isolated from dairy and nondairy farm soils. Microb. Ecol. 55:184-193. [DOI] [PubMed] [Google Scholar]
- 51.Storteboom, H. N., S. C. Kim, K. C. Doesken, K. H. Carlson, J. G. Davis, and A. Pruden. 2007. Response of antibiotics and resistance genes to high-intensity and low-intensity manure management. J. Environ. Qual. 36:1695-1703. [DOI] [PubMed] [Google Scholar]
- 52.Sunde, M., and M. Norstrom. 2006. The prevalence of associations between and conjugal transfer of antibiotic resistance genes in Escherichia coli isolated from Norwegian meat and meat products. J. Antimicrob. Chemother. 58:741-747. [DOI] [PubMed] [Google Scholar]
- 53.Topp, E., M. Welsh, Y.-C. Tien, A. Dang, G. Lazarovitis, K. Conn, and H. Zhu. 2003. Strain-dependent variability in growth and survival of Escherichia coli in agricultural soil. FEMS Microbiol. Ecol. 44:303-308. [DOI] [PubMed] [Google Scholar]
- 54.Unc, A., and M. J. Goss. 2004. Transport of bacteria from manure and protection of water resources. Appl. Soil Ecol. 25:1-18. [Google Scholar]
- 55.van den Bogaard, A. E., and E. E. Stobberingh. 2000. Epidemiology of resistance to antibiotics: links between animals and humans. Int. J. Antimicrob. Agents 14:327-335. [DOI] [PubMed] [Google Scholar]
- 56.Van Kessel, J. S., Y. A. Pachepsky, D. R. Shelton, and J. S. Karns. 2007. Survival of Escherichia coli in cow pats in pasture and in laboratory conditions. J. Appl. Microbiol. 103:1122-1127. [DOI] [PubMed] [Google Scholar]
- 57.Weaver, R. W., J. A. Entry, and A. Graves. 2005. Numbers of fecal streptococci and Escherichia coli in fresh and dry cattle, horse, and sheep manure. Can. J. Microbiol. 51:847-851. [DOI] [PubMed] [Google Scholar]
- 58.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]