Abstract
Sinorhizobium meliloti is a soil bacterium that elicits the formation of root organs called nodules on its host plant, Medicago sativa. Inside these structures, the bacteria are able to convert atmospheric nitrogen into ammonia, which is then used by the plant as a nitrogen source. The synthesis by S. meliloti of at least one exopolysaccharide, succinoglycan or EPS II, is essential for a successful symbiosis. While exopolysaccharide-deficient mutants induce the formation of nodules, they fail to invade them, and as a result, no nitrogen fixation occurs. Interestingly, the low-molecular-weight fractions of these exopolysaccharides are the symbiotically active forms, and it has been suggested that they act as signals to the host plant to initiate infection thread formation. In this work, we explored the role of these rhizobial exopolysaccharides in biofilm formation and their importance in the symbiotic relationship with the host. We showed that the ExpR/Sin quorum-sensing system controls biofilm formation in S. meliloti through the production of EPS II, which provides the matrix for the development of structured and highly organized biofilms. Moreover, the presence of the low-molecular-weight fraction of EPS II is vital for biofilm formation, both in vitro and in vivo. This is the first report where the symbiotically active fraction of EPS II is shown to be a critical factor for biofilm formation and root colonization. Thus, the ability of S. meliloti to properly attach to root surfaces and form biofilms conferred by the synthesis of exopolysaccharides may embody the main function of these symbiotically essential molecules.
The gram-negative soil bacterium Sinorhizobium meliloti fixes atmospheric nitrogen in association with its host plant, Medicago sativa (alfalfa). This symbiotic relationship involves a series of intricate signaling events between the two partners (23, 28). Initially, alfalfa releases flavonoids that attract bacteria from the surrounding environment to the roots and induce the production of bacterial lipochitooligosaccharide signal molecules referred to as Nod factors (29). The Nod factors elicit root hair deformation and trigger the plant meristematic cells to divide and differentiate, leading to the formation of plant nodules. Root nodule invasion requires the action of additional factors such as the exopolysaccharides produced by S. meliloti. Once inside the plant, bacteria differentiate into morphologically distinct forms called bacteroids that actively fix nitrogen (30).
Rhizobial exopolysaccharides are crucial for the establishment of a successful symbiosis with legumes. S. meliloti is capable of producing two exopolysaccharides, succinoglycan and EPS II. A 25-kb cluster of exo genes located in the second megaplasmid (pSymB) of S. meliloti is required for the production of succinoglycan (45), which is composed of repeating octasaccharide units of galactose and glucose residues (in a 1:7 molar ratio) decorated by acetyl, pyruvyl, and succinyl groups (44). This exopolysaccharide is secreted in two major fractions reflecting different degrees of subunit polymerization: high molecular weight (HMW), consisting of hundreds to thousands of octasaccharide subunits, and low molecular weight (LMW), represented by monomers, dimers, and trimers of the subunit (55). The presence of LMW succinoglycan is essential for nodule invasion to occur. Addition of the LMW fraction of succinoglycan in trans restores the ability of exo mutants to invade the host (2, 40, 54).
S. meliloti also has the ability to produce a second symbiotically active exopolysaccharide designated EPS II. Its synthesis is mediated by a 32-kb cluster of exp genes on pSymB, separate from the exo genes (18). EPS II is a polymer of repeating disaccharide units composed of an acetylated glucose and one pyruvylated galactose (22). As with succinoglycan, it is secreted in two major fractions, HMW and LMW (20, 37). This exopolysaccharide is produced by most isolates of S. meliloti but not by the well-studied Rm1021 strain under most conditions (18, 41). The explanation for this discrepancy lies in the fact that Rm1021 carries a spontaneous disruption in expR, a gene encoding a transcriptional regulator that is part of the ExpR/Sin quorum-sensing system (17, 31, 41). Restoration of the expR open reading frame leads to the synthesis of EPS II (31, 41). Interestingly, EPS II-producing strains frequently lose their ability to make this polymer when grown under laboratory conditions, suggesting that Rm1021 is a mutant derivative that arose during its use in research; therefore, we propose that a strain with an intact ExpR/Sin quorum-sensing system (capable of producing both succinoglycan and EPS II) should be regarded as the wild type (21, 24).
The ExpR/Sin quorum-sensing system in S. meliloti consists of (i) SinI, the autoinducer synthase, (ii) SinR, its regulator, and (iii) ExpR (32). SinI is responsible for the production of a series of long-chain N-acyl homoserine lactones (AHLs) (17, 33) which, in conjunction with ExpR, allows for the production of both the HMW and LMW fractions of EPS II (31, 41). When the ability to synthesize succinoglycan is abolished, strains with an intact quorum-sensing system continue to be nodulation competent since they produce LMW EPS II (20). In the absence of a functional quorum-sensing system, EPS II production is possible if a second transcriptional regulator, MucR, is disrupted; however, this only allows for symbiotically inactive HMW EPS II biosynthesis (20, 26, 59). As a result, these strains are incapable of invading M. sativa and establishing a symbiosis (20, 42). Therefore, the quorum-sensing activation of EPS II production and, more specifically, the resultant synthesis of the LMW fraction are critical for the ability of a succinoglycan-deficient strain to form a successful symbiosis with its host plant (31, 41). It has been suggested that these LMW fractions of the exopolysaccharides act as symbiotic signals during host infection (2, 20, 40); however, their precise role has, so far, been elusive.
In addition to facilitating nodule invasion, exopolysaccharides, together with other surface components, such as pili and lipopolysaccharides, may also play important roles in bacterial biofilm formation. Biofilms are formed when one or multiple bacterial species are enclosed in a self-produced polymeric matrix attached to a surface and to each other (8). These polysaccharide-encased microcolonies can eventually develop into complex three-dimensional microbial communities of variable depth and architecture and are often permeated by channels through which nutrients and water flow (52). The composition and structure of bacterially produced polysaccharides play an important role in primary biofilm conformation (53). Factors such as the effective polysaccharide concentration, degree of polymerization, type of carbohydrate linkages, ionic status, and the presence of other macromolecules determine the stability of the gel-like matrix (35).
Several possible biological functions for the S. meliloti exopolysaccharides have been postulated, including a role in protecting the bacteria against environmental stresses, determining host specificity, participating in the early steps of plant infection, such as adhesion of bacteria to the roots, inducing infection thread formation, modulating the plant defense responses, and acting as a plant developmental signal molecule (7, 9, 12, 27, 40, 50, 51). In order to determine if the S. meliloti ExpR/Sin quorum-sensing system and/or exopolysaccharide synthesis plays a role in biofilm formation, we used two different methods: a microtiter plate assay to quantify biofilm formation and a glass chamber assay to study the different stages of biofilm structure by confocal laser scanning microscopy (CLSM). In addition, we used fluorescence microscopy to evaluate biofilm formation directly on alfalfa roots. Here we show that the presence of EPS II, more specifically, the symbiotically active LMW fraction of this polymer, is essential for the proper and abundant formation of biofilms by S. meliloti in vitro and in vivo.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Starter cultures were grown in 5 ml of TYC broth (10 g of tryptone, 5 g of yeast extract, and 0.4 g of CaCl2/liter) for 48 h at 30°C. The strains were subcultured (1:100) in 2 ml of minimal glutamate mannitol (MGM) medium (50 mM morpholinepropanesulfonic acid [MOPS], 19 mM sodium glutamate, 55 mM mannitol, 1 mM MgSO4, 0.25 mM CaCl2, 0.004 mM biotin, pH 7) with a low or high phosphate concentration (0.1 or 10 mM K2HPO4-KH2PO4, respectively; the stock consists of an equimolar ratio of each compound) and grown at 30°C for an additional 48 h. Antibiotics were added, as appropriate, at the following final concentrations: streptomycin, 500 μg/ml; neomycin, 200 μg/ml; gentamicin, 50 μg/ml; tetracycline, 10 μg/ml; oxytetracycline, 0.75 μg/ml; chloramphenicol, 20 μg/ml. Strain Rm8530 exoY expA was constructed by general transduction using phage φM12 as previously described (10).
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| S. meliloti strains | ||
| Rm1021 | SU47 str-21 expR | 36 |
| Rm8530 | Rm1021 expR+ | 18 |
| Rm10002 | Rm8530 expA3::Tn5 | 37 |
| Rm11527 | Rm8530 sinI::Kmr | 31 |
| Rm11603 | Rm8530 exoY210::Tn5-233 | 18 |
| Rm11604 | Rm1021 mucR::Tn5-233 | 20 |
| Rm11605 | Rm8530 exoY210::Tn5-132 expA3::Tn5-233 | This work |
| Rm11606 | Rm1021 exoY210::Tn5-233 expA3::Tn5 | 37 |
| E. coli MT616 | pro-82 thi-1 hsdR17 supE44 recA56(pRK600) Cmr | 11 |
| Plasmids | ||
| pSWExpR | pSW213 containing an intact expR gene, Tcr | 19 |
| pTB93F | ptrp-GFP-S65Ta in pMB39, Spr Cmr | 16 |
| pDG77 | ptrp-DsRed, Tcr | 15 |
Kmr, kanamycin resistance; Cmr, chloramphenicol resistance; Tcr, tetracycline resistance; Spr, spectinomycin resistance.
Biofilm formation assay-microtiter plate method.
The biofilm formation assay used was based on the method of O'Toole and Kolter (39), with some modifications. Cultures were grown in low-phosphate MGM medium as described above, diluted to an optical density at 600 nm (OD600) of 0.8, and inoculated into the microplate wells in 100-μl aliquots. The plates were covered with a sterile microporous sealing film (AeraSeal catalog no. BS-25) to allow gas exchange and prevent evaporation and incubated with agitation (250 rpm) at 30°C for 48 h. Cells were gently resuspended by repeated pipetting of the contents in each well, and bacterial growth was quantified by measuring the OD600 at the end of the experiment. The contents of each well were then removed, and the wells were washed three times with 150 μl of sterile physiological saline solution in order to remove all nonadherent bacteria. The plates were emptied, air dried, stained for 15 min with 150 μl of 0.1% crystal violet per well, and then rinsed three times with water. Biofilm formation was quantified by the addition of 150 μl of 95% ethanol to each crystal violet-stained microtiter dish well, and the absorbance (at a wavelength of 560 nm) of the solubilized crystal violet was determined with a microplate reader (Infinite M200; Tecan Trading AG, Männedorf, Switzerland). Bacterial growth and adherence measurements were performed in duplicate, repeated at least four times, and averaged.
CLSM.
A confocal laser scanning microscope (Leica DM IRE2; Leica Microsystems, Wetzlar, Germany) was used to visualize the different events of biofilm formation in a 12-day time course experiment using chambered cover glass slides containing a 1-μm-thick borosilicate glass base (Lab-Tek no. 155411; Nunc) as described by Russo et al. (47). Confocal images were acquired from bacterial cultures carrying either plasmid pTB93F or pDG77, which constitutively produces the green fluorescent protein (GFP) (16) or DsRed (15), respectively. Fluorescence-labeled bacterial cultures were diluted to an OD600 of 0.001, and 500 μl of the diluted cultures was inoculated into each chamber and grown under static conditions at 30°C for up to 12 days. To prevent desiccation, the chambers were incubated in a sterile petri dish under humidified conditions. Three-dimensional images were reconstructed by using the Leica Confocal Software version 2.61 (Leica Microsystems, Wetzlar, Germany).
Microscopic observation of biofilm formation on roots.
Seeds of M. sativa cv. Iroquois (alfalfa) were surface sterilized in 50% bleach for 10 min, washed six times with sterile water, and germinated on 1.5% agar plates for 24 h. One-day-old seedlings were inoculated with twice-washed, GFP-labeled bacterial cultures (OD600 of 0.1) and maintained in a growth chamber at 22°C with an 18-h light/6-h dark photoperiod for 48 h. At least 10 roots per treatment (from two independent experiments) were extensively washed in sterile water, placed on a chambered cover glass slide (Lab-Tek no. 155361; Nunc), examined for biofilm formation with a Nikon Eclipse TE2000-U inverted microscope, and analyzed using the NIS-Elements AR Ver. 2.3 software, which allowed the overlapping of light and fluorescence images.
RESULTS
The presence of a functional ExpR/Sin quorum-sensing system in S. meliloti is necessary for biofilm formation.
S. meliloti strain Rm1021 (expR) does not have a functional ExpR/Sin quorum-sensing system due to an insertion element within the expR gene (31, 41). The Rm8530 (wild-type) strain contains a functional copy of this gene and thus has the capacity to regulate several important bacterial phenotypes based on population density, including the production of exopolysaccharides, motility, and others (21, 24, 25).
Biofilm development by wild-type S. meliloti and quorum-sensing mutants was analyzed in low-phosphate MGM medium, which attempts to replicate soil conditions, where nutrients and phosphate are scarce. S. meliloti grown in this medium produces large amounts of EPS II (18, 19, 24, 31, 37) and is highly motile at a low population density (21, 25). Moreover, we have demonstrated that quorum-sensing-controlled genes are maximally expressed in this medium (21, 24).
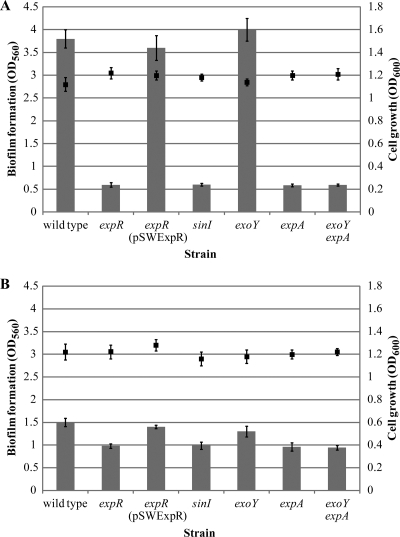
The wild-type strain produced a significantly larger mass of biofilm than the expR mutant, as measured by microtiter plate assay, suggesting that the ExpR quorum-sensing regulator plays a role in biofilm formation (Fig. 1A). The expR mutant formed only a slight biofilm ring at the liquid-air interface, compared with the full covering of the well walls by the wild type (data not shown).
FIG. 1.
Quantification of biofilm formation (bars) and bacterial growth (squares) of S. meliloti cultures under shaking conditions for 48 h in (A) low-phosphate (0.1 mM) and (B) high-phosphate (10 mM) MGM medium. Mean values and standard deviations are shown.
We performed a genetic complementation study to determine if biofilm formation in the expR strain could be restored by adding a functional copy of the expR gene in trans. When the pSWExpR plasmid was transferred into the mutant, biofilm development was restored to wild-type levels (Fig. 1A), again confirming that expR must be present in order for biofilm formation to occur.
The S. meliloti ExpR quorum-sensing regulator requires activation by AHLs to control a variety of cell functions (21). In order to determine if, in addition to ExpR, biofilm formation requires the presence of AHLs, we disrupted the sinI AHL synthase gene. Similar to the expR mutant strain, a sinI mutant showed a reduction in biofilm formation (Fig. 1A), indicating that an intact quorum-sensing system is required for maximum biofilm formation.
EPS II provides the matrix for biofilm formation.
The biosynthesis of both succinoglycan and EPS II is affected by quorum sensing (19, 31). In a quorum-sensing mutant, the average molecular weight of succinoglycan is increased (19) and production of EPS II is abolished (31). To examine the role of succinoglycan or EPS II in biofilm formation, independent of quorum sensing, we analyzed strain derivatives that were unable to produce one or both of these polymers. Assembly of the repeating unit of succinoglycan is initiated by the ExoY galactosyltransferase (45); therefore, a mutant lacking the gene for this enzyme is unable to produce this exopolysaccharide. The introduction of an exoY mutation into the wild-type strain led to no reduction in biofilm formation, suggesting that succinoglycan does not play a major role in this process (Fig. 1A). On the other hand, a marked decrease in biofilm development occurred when we introduced a mutation in the expA gene, which blocks EPS II production (18) (Fig. 1A). Elimination of succinoglycan production in an expA strain by the introduction of an exoY mutation resulted in no further reduction in biofilm formation (Fig. 1A). These results suggest that EPS II (but not succinoglycan) is crucial in S. meliloti for the formation of biofilms under these growth conditions.
Phosphate concentration affects biofilm formation.
Phosphorus is an important macronutrient which is found at limited concentrations (1 to 10 μM) in soil (6) since it is mostly present in immobilized and nonutilizable forms. Phosphate concentration constitutes an environmental signal which has been shown to affect the biosynthesis of EPS II (37, 46, 58). Low-phosphate conditions (0.1 mM) are required for maximal expression of the exp genes, while high phosphate concentrations (≥2 mM) dramatically reduce its production (4, 37). Interestingly, all EPS II-producing strains showed a dramatic decrease in biofilm formation when the microtiter assay was performed in high-phosphate (10 mM) MGM medium (Fig. 1B). This strongly correlates with previous observations that the amount of EPS II produced under those conditions is limited (4, 37). However, high-phosphate conditions slightly increased biofilm formation in the expR, sinI, expA, and exoY expA mutants (Fig. 1B) compared to that seen under the phosphate-limiting conditions provided by low-phosphate (0.1 mM) MGM medium (Fig. 1A). Since a mutant unable to produce any exopolysaccharides (exoY expA) shows a level of biofilm formation similar to that of the quorum-sensing (expR, sinI) and the EPS II-producing (expA) mutants, this effect cannot be attributed to the stimulation of succinoglycan production by high phosphate. The mechanisms used by non-EPS producing strains of S. meliloti to attach to abiotic surfaces and develop biofilms, as well as the role of phosphate in mediating this process, remain to be determined.
EPS II-producing strains develop structured biofilms.
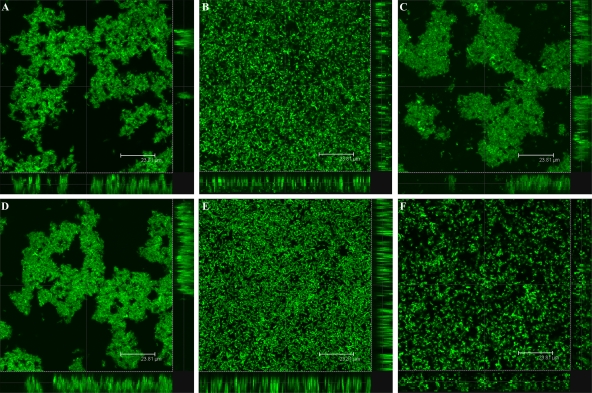
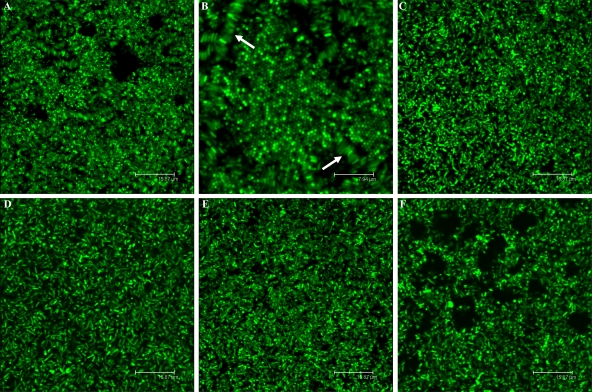
We used CLSM to analyze the structural characteristics of the S. meliloti biofilm formed in vitro over a 12-day time course experiment in a chambered cover glass. Inverted microscopy with GFP-labeled strains of S. meliloti allowed observation of the different stages of biofilm growth on the bottom of the chamber. As early as 2 days postinoculation (dpi), clear stages of structured biofilm development were observed in the wild-type strain (Fig. 2A), the pSWExpR-complemented expR mutant (Fig. 2C), and the exoY mutant (Fig. 2D). On the other hand, a flat, unorganized biofilm of uniform thickness composed of a confluent layer of cells covering the entire surface of the slide was observed in static cultures of the expR mutant over the course of the experiment (Fig. 2B). In order to investigate if EPS II is involved in the development of structured biofilms, we performed the same assay with an expA mutant strain carrying an intact quorum-sensing system. The biofilm formed by this mutant did not show any kind of structure or organization (Fig. 2E) and was similar to the one observed in the expR mutant strain, confirming that EPS II plays a major role in biofilm formation in S. meliloti. Formation of structured biofilms was abolished when EPS II-producing strains were grown in high-phosphate MGM medium, as flat, unstructured biofilms were formed by the wild-type strain, as well as by the expR and exoY mutants (data not shown).
FIG. 2.
CLSM observation of biofilm formation in (A) wild-type S. meliloti and the (B) expR, (C) expR (pSWExpR), (D) exoY, (E) expA, and (F) expR mucR mutants at 2 dpi. Bars, 23.8 μm.
Restoration of biofilm formation via extracellular complementation of quorum sensing.
To confirm that a functional quorum-sensing system and its corollary production of EPS II are essential for biofilm formation, we conducted a series of mixed-strain experiments. First, we combined the exoY sinI and exoY expA mutants. Both of these strains are unable to produce succinoglycan (which, as we demonstrated above, does not play a major role in biofilm formation) and possess an intact copy of the ExpR quorum-sensing regulator. In the exoY sinI mutant strain, while all of the exp genes are functional, no EPS II is synthesized due to a lack of ExpR activation in the absence of the Sin AHLs. In the second strain, the expA mutation blocks EPS II synthesis; therefore, this strain makes AHLs but does not produce any exopolysaccharides.
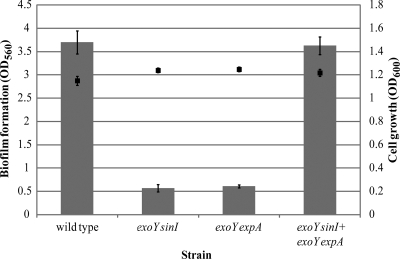
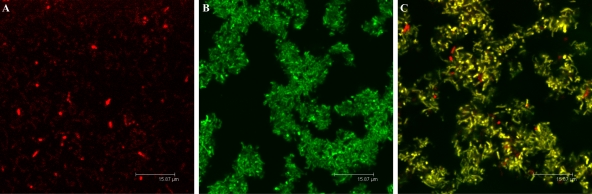
When biofilm formation was evaluated in both of these mutants individually, we observed a reduction in the biofilm biomass compared to that of the wild type (Fig. 3). However, biofilm formation was restored to wild-type levels when the two mutants were cocultured (Fig. 3). AHL complementation by the exoY expA mutant strain led to activation of the ExpR/Sin quorum-sensing system in the exoY sinI mutant strain and the eventual formation of biofilms. A similar effect on the formation of structured biofilms was observed by CLSM (Fig. 4). Both an exoY sinI mutant strain producing GFP and an exoY expA mutant expressing DsRed individually formed flat, unstructured biofilms when evaluated by CLSM (Fig. 4A and B, respectively). Again, coinoculation of these two strains led to the development of a mixed-structure biofilm (Fig. 4C) similar to the one shown by the wild-type strain (Fig. 2A). These findings confirm that the presence of a functional quorum-sensing system is essential for effective biofilm formation.
FIG. 3.
In vivo quorum-sensing complementation assay. Quantification of biofilm formation (bars) and bacterial growth (squares) of wild-type S. meliloti and exoY sinI, exoY expA, and mixed exoY sinI plus exoY expA mutant cultures under shaking conditions for 48 h in low-phosphate MGM medium. Mean values and standard deviations are shown.
FIG. 4.
Quorum-sensing complementation analysis. Shown are CLSM single-scan images from S. meliloti biofilms (3 dpi). (A) exoY sinI-GFP, (B) exoY expA-DsRed, and (C) restoration of biofilm formation by coculturing both of the strains mentioned above. Bars, 23.8 μm.
Restoration of biofilm formation via extracellular complementation of EPS II production.
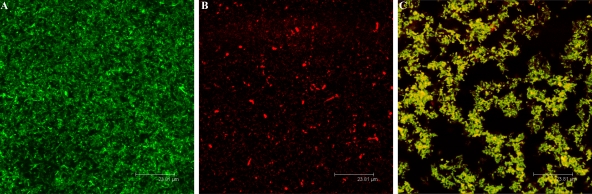
The biofilm formation shown above could be due to the restoration of EPS II production in the complemented strains or to the reestablishment of another quorum-sensing-dependent factor. To determine the specific basis of biofilm formation, we performed a different mixed-strain complementation experiment. In an expR exoY expA triple mutant, the synthesis of both EPS II and succinoglycan is genetically blocked. In addition, the ExpR/Sin quorum-sensing system is nonfunctional due to the absence of an intact expR gene. This constitutively DsRed-producing strain develops flat biofilms in which no structure or organization is observed by CLSM (Fig. 5A), a phenotype analogous to that of the expR mutant strain (Fig. 2B). The DsRed-producing expR exoY expA mutant strain was cocultured with a GFP-expressing exoY mutant, a strain previously shown to form structured and robust biofilms (Fig. 2D and 5B). This allowed us to evaluate if an EPS II-producing strain is able to complement the mutant. When the strains were combined, the DsRed-expressing expR exoY expA mutant was incorporated into the microcolonies developed by the exoY-GFP strain, leading to the formation of a mixed biofilm (Fig. 5C). This, together with the results shown above, suggests that the presence of EPS II is crucial for the proper formation of biofilms.
FIG. 5.
EPS II complementation analysis. Shown are CLSM single-scan images of S. meliloti biofilms (3 dpi). (A) DsRed-producing expR exoY expA mutant (a quorum-sensing-deficient mutant capable of producing neither succinoglycan nor EPS II and therefore non-biofilm forming), (B) GFP-producing exoY mutant (a biofilm-forming succinoglycan mutant), and (C) a mixed biofilm showing how the expR exoY expA mutant integrates into a typical wild-type biofilm when cocultured with the exoY EPS II-producing strain. Bars, 15.8 μm.
EPS II-producing strains develop highly organized biofilms.
Microbial species have been shown to form complex biofilm structures, including honeycombs and veils (47, 48). It has been speculated that they may provide mechanical stability and protection against physical stress and may maximize reactive surface area, lowering the energy costs of individual cells (48). Moreover, the tertiary structure of honeycombs may provide a larger surface area for the absorption of nutrients. After our initial observations, we continued to monitor the ability of S. meliloti to form biofilms. Following a period of cell dispersal at 4 dpi, the wild type, as well as the exoY mutant, formed interconnected complex cell clusters or honeycomb-like biofilms starting at 10 dpi (Fig. 6A). Bacteria were predominantly attached to each other through lateral interactions, forming rows of cells identically oriented (Fig. 6B), although pole-to-pole interactions were also observed. These honeycomb-like arrangements were apparent until the end of the experiment at 12 dpi. On the other hand, no honeycomb structures were observed in the expR, sinI, or expA mutant strain (Fig. 6C to E, respectively).
FIG. 6.
CLSM images of S. meliloti biofilms in low-phosphate MGM medium (12 dpi). The (A and B) exoY, (C) expR, (D) sinI (E) expA, and (F) expR mucR mutant strains are shown. Bars (except for panel B), 15.8 μm. Panel B shows honeycomb-like structures and lateral interactions indicated by arrows (bar, 7.9 μm).
The LMW fraction of EPS II is required for biofilm formation.
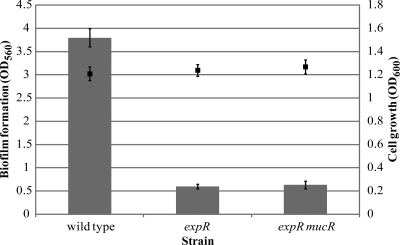
MucR is a repressor of selected EPS II biosynthetic genes (26). In an expR mutant strain, this repression leads to no EPS II production. A mucR mutation results in EPS II production, but this polymer lacks the symbiotically critical LMW fraction (20). In an expR mucR mutant, biofilm formation was dramatically decreased compared to that of the wild-type strain (Fig. 7). In glass chambers, the expR mucR mutant formed small channels but failed to develop microcolonies (Fig. 2F) or honeycomb-like structures (Fig. 6F), analogous to the phenotype observed in the non-EPS II-producing expR mutant strain (Fig. 2D). This suggests that not only is the production of EPS II important, but the presence of the specific LMW fraction of EPS II is apparently essential for biofilm formation by S. meliloti.
FIG. 7.
Quantification of biofilm formation (bars) and bacterial growth (squares) of wild-type and expR and expR mucR mutant S. meliloti cultures under shaking conditions for 48 h in low-phosphate MGM medium. Mean values and standard deviations are shown.
EPS II-producing strains colonize root surfaces more efficiently.
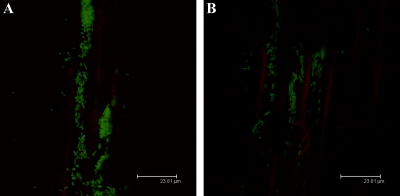
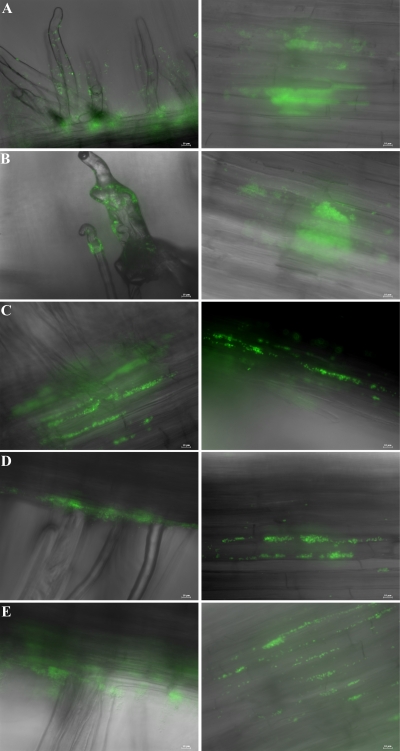
Plant-associated bacteria interact with host surfaces during pathogenesis and symbiosis (13, 38, 49). The effect of EPS II or succinoglycan on alfalfa root colonization was examined for the different S. meliloti derivatives. At 2 dpi, clusters of rhizobia were observed attached to epidermal cells, especially along epidermal fissures. The wild type, as well as an exoY strain, developed dense, confluent biofilms (Fig. 8A) that covered the entire surface of the root, including the root hairs, where the initial symbiotic interaction takes place (Fig. 9A and B, respectively). Conversely, expR and expA mutants (lacking EPS II) formed smaller clusters of cells (Fig. 8B), attached predominantly to the principal root (Fig. 9C and D, respectively). This strongly suggests that, in vivo, EPS II-producing strains are more efficient root hair colonizers and that non-EPS II-producing strains at this early stage do not develop the extensive networks elaborated by the wild type or the exoY mutant strain. On the other hand, the expR mucR mutant strain, which only produces HMW EPS II, showed a phenotype intermediate between those of the wild type and the expR mutant (Fig. 9E). Similar to the expR mutant, this strain colonized mostly the principal root, forming patchy colonies, but on occasion could be seen associated with root hairs, suggesting that LMW EPS II is necessary for maximal host colonization.
FIG. 8.
CLSM images of biofilms established on alfalfa roots by S. meliloti (A) wild-type and (B) expR mutant strains at 1 dpi. Bars, 23.8 μm.
FIG. 9.
Images of S. meliloti (A) wild-type and (B) exoY, (C) expR, (D) expA, and (E) expR mucR mutant strains on the root hairs and main root of alfalfa at 2 dpi. Bars, 10 μm.
DISCUSSION
Bacteria in biofilms are better able to resist antimicrobial agents, adapt to environmental changes, defend themselves against host attack, and exchange genetic information more efficiently (14). Biofilms are clearly formed by rhizobial species, but it is not apparent what role they play in a successful symbiotic association (12, 13, 27, 47). The capacity that biofilms have demonstrated in other settings to protect bacteria from desiccation, starvation, and host responses and to mediate attachment (3) could potentially impact the overall fitness of rhizobia in the soil and in the rhizosphere microenvironments and therefore contribute to the symbiotic process.
Previous work has shown that the LMW fractions of succinoglycan and EPS II produced by S. meliloti independently mediate root nodule invasion in alfalfa (2, 20, 40, 54). Here we show that the LMW fraction of EPS II is also essential for biofilm formation. The ExpR/Sin quorum-sensing-proficient strain synthesizes both the HMW and LMW forms of EPS II and develops complex and robust biofilms, both in vitro and in vivo. A strain which lacks EPS II or produces only the HMW fraction of this polymer forms very low levels of biofilm. Therefore, the ExpR/Sin quorum-sensing system controls biofilm formation through LMW EPS II production in S. meliloti. Interestingly, a succinoglycan-deficient expR mucR mutant, despite its ability to make HMW EPS II, is also nodule invasion deficient, indicating that LMW EPS II is vital for invasion as well (20).
We did not detect any differences in biofilm formation between the wild-type strain and an exoY mutant. Therefore, our results suggest that a normal amount of succinoglycan (LMW or HMW) does not play a major role in biofilm formation.
Recently, Wells et al. showed that overproducers of succinoglycan can form biofilms (56). Strains with a mutation in exoR, a negative regulator of the exo genes, produce at least five times more succinoglycan. This increase in succinoglycan synthesis leads to extensive biofilm formation and attachment to abiotic surfaces (56). Interestingly, exoR mutants are also unable to effectively invade alfalfa nodules (56). Perhaps the composition, quantity, and specific structure of the polymer generated by the symbiont are critical for proper colonization of and effective interaction with the host plant.
Unorganized biofilms (no prevalence of lateral or polar interactions between the cells) permeated by water channels were observed in the wild-type and exoY mutant strains producing EPS II during the first days postinoculation. The non-EPS II-producing strains, as well as the expR mucR mutant, formed a flat, uniform layer of cells attached to the glass surface but with no apparent order. After a period of dispersal, the wild-type and exoY mutant strains developed a new kind of biofilm where the bacteria formed very organized honeycomb-like structures. Again, non-EPS II-producing strains failed to show any reorganization of the biofilm. These data indicate that the presence of EPS II, specifically, the LMW fraction of this polymer, is responsible not only for biofilm formation by S. meliloti but also for its eventual highly ordered structure. The formation of honeycomb-like biofilms has also been described in Rhizobium leguminosarum (47, 57) and shown to be dependent on the acidic EPS.
Bacteria associated with plants frequently form biofilms on leaves, on root surfaces, and within the intercellular spaces of plant tissues (1, 3, 5, 13, 34, 38, 43). Microscopic studies show that the rhizobial cells migrate down the infection threads as biofilm-like filaments toward the root interior (15, 43). Here we show that EPS II-producing strains are able to colonize efficiently the principal roots, as well as the root hairs, of alfalfa, while the expR and expA mutant strains develop patchy microcolonies primarily on the principal root. Moreover, the expR mucR mutant strain, which produces only the HMW fraction of EPS II, has an intermediate phenotype. HMW EPS II could be mediating a basal level of root colonization, since the expR mucR mutant shows a transitional phenotype both in vitro and in vivo. However, the efficiency of colonization seems to be dramatically impaired by the absence of LMW EPS II. EPS II biosynthesis, specifically, production of the LMW fraction of this polymer, plays a crucial role in biofilm formation, root colonization, and nodule invasion. Our recent findings that quorum sensing not only controls the production of EPS II in S. meliloti but also strongly represses flagellar synthesis at high population densities bring together two critical requirements for proper biofilm formation, matrix production and motility suppression (21, 25). It has been proposed that S. meliloti exopolysaccharides may play a signaling role in the process of nodule invasion, perhaps through the induction of infection thread formation (20, 40). In light of the data presented here, it is tempting to speculate that the main role of LMW EPS II is in positioning the bacteria appropriately on the host root so that invasion can occur. The biofilm matrix could be providing S. meliloti with the suitable microenvironment necessary for colonization and eventual invasion of the root hairs, while the absence of exopolysaccharides might lead to poor root association and host invasion failure. From that point of view, LMW EPS II acts as an essential structural component of the colonization matrix for correct interaction with the host. Our observations set the stage for the study of the specific mechanisms through which biofilm formation mediates attachment and/or invasion. Elucidating potential plant surface interactions with the bacterial matrix, possible host plant responses to biofilm-forming and non-biofilm-forming bacteria, and any other host-symbiont interactions will further advance our understanding of the particular S. meliloti-alfalfa symbiosis and bacterium-host interactions in general.
Acknowledgments
We thank the members of our laboratory for helpful discussions and critical reading of the manuscript. We also thank Audry Almengor and Daniel Gage for providing strains.
L.V.R. has a doctoral fellowship from CONICET, and her work was supported in part by a UNESCO-ASM travel award. This work was supported by a National Science Foundation grant (MCB-9733532) and a National Institutes of Health grant (1R01GM069925) to J.E.G.
Footnotes
Published ahead of print on 25 September 2009.
REFERENCES
- 1.Assmus, B., P. Hutzler, G. Kirchhof, R. Amann, J. R. Lawrence, and A. Hartmann. 1995. In situ localization of Azospirillum brasilense in the rhizosphere of wheat with fluorescently labeled, rRNA-targeted oligonucleotide probes and scanning confocal laser microscopy. Appl. Environ. Microbiol. 61:1013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battisti, L., J. C. Lara, and J. A. Leigh. 1992. Specific oligosaccharide form of the Rhizobium meliloti exopolysaccharide promotes nodule invasion in alfalfa. Proc. Natl. Acad. Sci. USA 89:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beattie, G. A., and S. E. Lindow. 1999. Bacterial colonization of leaves: a spectrum of strategies. Phytopathology 89:353-359. [DOI] [PubMed] [Google Scholar]
- 4.Becker, A., S. Rüberg, B. Baumgarth, P. A. Bertram-Drogatz, I. Quester, and A. Pühler. 2002. Regulation of succinoglycan and galactoglucan biosynthesis in Sinorhizobium meliloti. J. Mol. Microbiol. Biotechnol. 4:187-190. [PubMed] [Google Scholar]
- 5.Bianciotto, V., S. Andreotti, R. Balestrini, P. Bonfante, and S. Perotto. 2001. Mucoid mutants of the biocontrol strain Pseudomonas fluorescens CHA0 show increased ability in biofilm formation on mycorrhizal and nonmycorrhizal carrot roots. Mol. Plant-Microbe Interact. 14:255-260. [DOI] [PubMed] [Google Scholar]
- 6.Bieleski, R. 1973. Phosphate pools, phosphate transport, and phosphate availability. Annu. Rev. Plant Physiol. 24:225-252. [Google Scholar]
- 7.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180:5183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 9.D'Haeze, W., and M. Holsters. 2004. Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol. 12:555-561. [DOI] [PubMed] [Google Scholar]
- 10.Finan, T. M., E. Hartweig, K. LeMieux, K. Bergman, G. C. Walker, and E. R. Signer. 1984. General transduction in Rhizobium meliloti. J. Bacteriol. 159:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraysse, N., F. Couderc, and V. Poinsot. 2003. Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. Eur. J. Biochem. 270:1365-1380. [DOI] [PubMed] [Google Scholar]
- 13.Fujishige, N. A., N. N. Kapadia, P. L. De Hoff, and A. M. Hirsch. 2006. Investigations of Rhizobium biofilm formation. FEMS Microbiol. Ecol. 56:195-206. [DOI] [PubMed] [Google Scholar]
- 14.Fux, C. A., J. W. Costerton, P. S. Stewart, and P. Stoodley. 2005. Survival strategies of infectious biofilms. Trends Microbiol. 13:34-40. [DOI] [PubMed] [Google Scholar]
- 15.Gage, D. J. 2002. Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J. Bacteriol. 184:7042-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gage, D. J., T. Bobo, and S. R. Long. 1996. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J. Bacteriol. 178:7159-7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, M., H. Chen, A. Eberhard, M. R. Gronquist, J. B. Robinson, B. G. Rolfe, and W. D. Bauer. 2005. sinI- and expR-dependent quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 187:7931-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glazebrook, J., and G. C. Walker. 1989. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661-672. [DOI] [PubMed] [Google Scholar]
- 19.Glenn, S. A., N. Gurich, M. A. Feeney, and J. E. González. 2007. The ExpR/Sin quorum-sensing system controls succinoglycan production in Sinorhizobium meliloti. J. Bacteriol. 189:7077-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González, J. E., B. L. Reuhs, and G. C. Walker. 1996. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc. Natl. Acad. Sci. USA 93:8636-8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurich, N., and J. E. González. 2009. Role of quorum sensing in Sinorhizobium meliloti-alfalfa symbiosis. J. Bacteriol. 191:4372-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Her, G. R., J. Glazebrook, G. C. Walker, and V. N. Reinhold. 1990. Structural studies of a novel exopolysaccharide produced by a mutant of Rhizobium meliloti strain Rm1021. Carbohydr. Res. 198:305-312. [DOI] [PubMed] [Google Scholar]
- 23.Hirsch, A. M., M. R. Lum, and J. A. Downie. 2001. What makes the rhizobia-legume symbiosis so special? Plant Physiol. 127:1484-1492. [PMC free article] [PubMed] [Google Scholar]
- 24.Hoang, H. H., A. Becker, and J. E. González. 2004. The LuxR homolog ExpR, in combination with the Sin quorum sensing system, plays a central role in Sinorhizobium meliloti gene expression. J. Bacteriol. 186:5460-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoang, H. H., N. Gurich, and J. E. González. 2008. Regulation of motility by the ExpR/Sin quorum-sensing system in Sinorhizobium meliloti. J. Bacteriol. 190:861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller, M., A. Roxlau, W. M. Weng, M. Schmidt, J. Quandt, N. Karsten, D. Jording, W. Arnold, and A. Pühler. 1995. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol. Plant-Microbe Interact. 8:267-277. [DOI] [PubMed] [Google Scholar]
- 27.Laus, M. C., A. A. van Brussel, and J. W. Kijne. 2005. Exopolysaccharide structure is not a determinant of host-plant specificity in nodulation of Vicia sativa roots. Mol. Plant-Microbe Interact. 18:1123-1129. [DOI] [PubMed] [Google Scholar]
- 28.Long, S. R. 2001. Genes and signals in the Rhizobium-legume symbiosis. Plant Physiol. 125:69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long, S. R. 1996. Rhizobium symbiosis: nod factors in perspective. Plant Cell 8:1885-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long, S. R., and B. J. Staskawicz. 1993. Prokaryotic plant parasites. Cell 73:921-935. [DOI] [PubMed] [Google Scholar]
- 31.Marketon, M. M., S. A. Glenn, A. Eberhard, and J. E. González. 2003. Quorum sensing controls exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 185:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marketon, M. M., and J. E. González. 2002. Identification of two quorum-sensing systems in Sinorhizobium meliloti. J. Bacteriol. 184:3466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marketon, M. M., M. R. Gronquist, A. Eberhard, and J. E. González. 2002. Characterization of the Sinorhizobium meliloti sinR/sinI locus and the production of novel N-acyl homoserine lactones. J. Bacteriol. 184:5686-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthysse, A. G., and S. McMahan. 2001. The effect of the Agrobacterium tumefaciens attR mutation on attachment and root colonization differs between legumes and other dicots. Appl. Environ. Microbiol. 67:1070-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayer, C., R. Moritz, C. Kirschner, W. Borchard, R. Maibaum, J. Wingender, and H. C. Flemming. 1999. The role of intermolecular interactions: studies on model systems for bacterial biofilms. Int. J. Biol. Macromol. 26:3-16. [DOI] [PubMed] [Google Scholar]
- 36.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mendrygal, K. E., and J. E. González. 2000. Environmental regulation of exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 182:599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris, C. E., and J. M. Monier. 2003. The ecological significance of biofilm formation by plant-associated bacteria. Annu. Rev. Phytopathol. 41:429-453. [DOI] [PubMed] [Google Scholar]
- 39.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 40.Pellock, B. J., H. P. Cheng, and G. C. Walker. 2000. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J. Bacteriol. 182:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pellock, B. J., M. Teplitski, R. P. Boinay, W. D. Bauer, and G. C. Walker. 2002. A LuxR homolog controls production of symbiotically active extracellular polysaccharide II by Sinorhizobium meliloti. J. Bacteriol. 184:5067-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pühler, A. M., W. Arnold, A. Becker, A. Roxlau, M. Keller, D. Kapp, A. Lagares, J. Lorenzen, and K. Niehaus. 1993. The role of Rhizobium meliloti surface polysaccharides in nodule development, p. 207-212. In R. Palacios, J. Mora, and W. E. Newton (ed.), New horizons in nitrogen fixation. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 43.Ramey, B. E., M. Koutsoudis, S. B. von Bodman, and C. Fuqua. 2004. Biofilm formation in plant-microbe associations. Curr. Opin. Microbiol. 7:602-609. [DOI] [PubMed] [Google Scholar]
- 44.Reinhold, B. B., S. Y. Chan, T. L. Reuber, A. Marra, G. C. Walker, and V. N. Reinhold. 1994. Detailed structural characterization of succinoglycan, the major symbiotically important exopolysaccharide of Rhizobium meliloti Rm1021. J. Bacteriol. 176:1997-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reuber, T. L., and G. C. Walker. 1993. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell 74:269-280. [DOI] [PubMed] [Google Scholar]
- 46.Rüberg, S., A. Pühler, and A. Becker. 1999. Biosynthesis of the exopolysaccharide galactoglucan in Sinorhizobium meliloti is subject to a complex control by the phosphate-dependent regulator PhoB and the proteins ExpG and MucR. Microbiology 145:603-611. [DOI] [PubMed] [Google Scholar]
- 47.Russo, D. M., A. Williams, A. Edwards, D. M. Posadas, C. Finnie, M. Dankert, J. A. Downie, and A. Zorreguieta. 2006. Proteins exported via the PrsD-PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum. J. Bacteriol. 188:4474-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaudinn, C., P. Stoodley, A. Kainovic, T. Okeeffe, B. Costerton, D. Robinson, M. Baum, G. Ehrlich, and P. Webster. 2007. Bacterial biofilms, other structures seen as mainstream concepts. Microbe 2:231-237. [Google Scholar]
- 49.Schloter, M., W. Wiehe, B. Assmus, H. Steindl, H. Becke, G. Höflich, and A. Hartmann. 1997. Root colonization of different plants by plant-growth-promoting Rhizobium leguminosarum bv. trifolii R39 studied with monospecific polyclonal antisera. Appl. Environ. Microbiol. 63:2038-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shepherd, R. W., and S. E. Lindow. 2009. Two dissimilar N-acyl-homoserine lactone acylases of Pseudomonas syringae influence colony and biofilm morphology. Appl. Environ. Microbiol. 75:45-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skorupska, A., M. Janczarek, M. Marczak, A. Mazur, and J. Król. 2006. Rhizobial exopolysaccharides: genetic control and symbiotic functions. Microb. Cell Fact. 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanley, N. R., and B. A. Lazazzera. 2004. Environmental signals and regulatory pathways that influence biofilm formation. Mol. Microbiol. 52:917-924. [DOI] [PubMed] [Google Scholar]
- 53.Sutherland, I. 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3-9. [DOI] [PubMed] [Google Scholar]
- 54.Urzainqui, A., and G. C. Walker. 1992. Exogenous suppression of the symbiotic deficiencies of Rhizobium meliloti exo mutants. J. Bacteriol. 174:3403-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, L. X., Y. Wang, B. Pellock, and G. C. Walker. 1999. Structural characterization of the symbiotically important low-molecular-weight succinoglycan of Sinorhizobium meliloti. J. Bacteriol. 181:6788-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wells, D. H., E. J. Chen, R. F. Fisher, and S. R. Long. 2007. ExoR is genetically coupled to the ExoS-ChvI two-component system and located in the periplasm of Sinorhizobium meliloti. Mol. Microbiol. 64:647-664. [DOI] [PubMed] [Google Scholar]
- 57.Williams, A., A. Wilkinson, M. Krehenbrink, D. M. Russo, A. Zorreguieta, and J. A. Downie. 2008. Glucomannan-mediated attachment of Rhizobium leguminosarum to pea root hairs is required for competitive nodule infection. J. Bacteriol. 190:4706-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhan, H. J., C. C. Lee, and J. A. Leigh. 1991. Induction of the second exopolysaccharide (EPSb) in Rhizobium meliloti SU47 by low phosphate concentrations. J. Bacteriol. 173:7391-7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhan, H. J., S. B. Levery, C. C. Lee, and J. A. Leigh. 1989. A second exopolysaccharide of Rhizobium meliloti strain SU47 that can function in root nodule invasion. Proc. Natl. Acad. Sci. USA 86:3055-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]