Abstract
Summary: Members of the ATP-binding cassette (ABC) transporter superfamily exist in bacteria, fungi, plants, and animals and play key roles in the efflux of xenobiotic compounds, physiological substrates, and toxic intracellular metabolites. Based on sequence relatedness, mammalian ABC proteins have been divided into seven subfamilies, ABC subfamily A (ABCA) to ABCG. This review focuses on recent advances in our understanding of ABC transporters in the model organism Saccharomyces cerevisiae. We propose a revised unified nomenclature for the six yeast ABC subfamilies to reflect the current mammalian designations ABCA to ABCG. In addition, we specifically review the well-studied yeast ABCC subfamily (formerly designated the MRP/CFTR subfamily), which includes six members (Ycf1p, Bpt1p, Ybt1p/Bat1p, Nft1p, Vmr1p, and Yor1p). We focus on Ycf1p, the best-characterized yeast ABCC transporter. Ycf1p is located in the vacuolar membrane in yeast and functions in a manner analogous to that of the human multidrug resistance-related protein (MRP1, also called ABCC1), mediating the transport of glutathione-conjugated toxic compounds. We review what is known about Ycf1p substrates, trafficking, processing, posttranslational modifications, regulation, and interactors. Finally, we discuss a powerful new yeast two-hybrid technology called integrated membrane yeast two-hybrid (iMYTH) technology, which was designed to identify interactors of membrane proteins. iMYTH technology has successfully identified novel interactors of Ycf1p and promises to be an invaluable tool in future efforts to comprehensively define the yeast ABC interactome.
INTRODUCTION
Members of the ATP-binding cassette (ABC) superfamily catalyze the ATP-dependent transport of chemically diverse compounds across cellular membranes, including the plasma membrane or intracellular organellar membranes (27, 29, 51, 61). In humans, ABC transporters are clinically important for maintaining the blood-brain barrier, which excludes cytotoxic drugs from the brain, and for mediating cellular resistance to chemotherapeutic drugs (52, 58, 153). Loss-of-function mutations in ABC transporter genes are implicated in a diverse and ever-increasing number of inherited human diseases, including the lung disease cystic fibrosis, the cholesterol transport disorder Tangier's disease, the retinal syndrome Stargardt's disease, the elastic tissue disorder pseudoxanthoma elasticum, and many others. A comprehensive list of the diseases that map to ABC genes is available (25-27; see also http://nutrigene.4t.com/humanabc.htm). In contrast to diseases that are associated with a loss of transporter function, the overexpression of ABC proteins such as the human MDR1 or MRP1 protein can enhance multidrug resistance in mammalian cells (3, 13, 58). Thus, an understanding of the mechanistic principles and substrate selectivity determinants of ABC transporters has important biological and medical implications. To gain insight into the workings of ABC transporters, investigators are currently employing methodologies ranging from X-ray crystallography to genetic analysis in model organisms. The latter approach is exemplified by the yeast studies discussed in this review.
Nucleotide-Binding Domains Are the Diagnostic Features of ABC Proteins
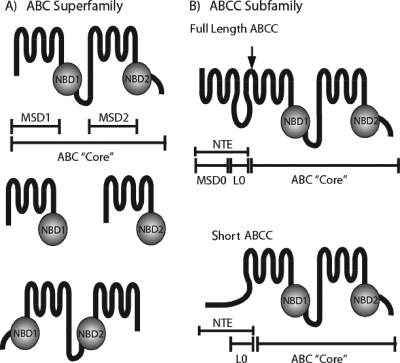
Members of the ABC superfamily share a conserved overall architecture (Fig. 1A). The ABC “core domain” consists of two homologous halves, each containing a membrane-spanning domain (MSD) with multiple transmembrane spans (generally, but not always, six) and a nucleotide-binding domain (NBD), which couples nucleotide hydrolysis to substrate transport (26, 61, 62, 97, 98). In eukaryotes, these homologous halves are encoded as a single polypeptide or as half-molecules that form homo- or heterodimers. The NBDs can be located N or C terminally to the MSDs (as shown in Fig. 1A) (13, 26, 61).
FIG. 1.
(A) Overall architecture of the ABC superfamily. ABC transporters (top) have an “ABC core” region consisting of two MSDs (MSD1 and MSD2) containing six transmembrane spans and two cytosolic NBDs connected by a linker region (not labeled). ABC transporters can also be expressed as half-molecules (middle), with each half containing a single MSD and NBD; the halves can homo- or heterodimerize to form a functioning transporter. Some ABC transporters are encoded in “reverse” (bottom), where the NBDs precede the MSDs. (B) Overall architecture of the ABCC subfamily. Members of the ABCC subfamily of ABC transporters contain a characteristic NTE in addition to the “ABC core.” In full-length ABCCs (top), the NTE contains an MSD (MSD0) with five transmembrane spans and a cytosolic loop (L0). In short ABCCs (bottom), an L0 domain, but no MSD0, is present. The arrow over the full-length ABCC indicates the site of Ycf1p posttranslational processing in L6 (discussed in the text).
Nucleotide hydrolysis is critical for ABC protein function. Each NBD is ∼200 residues in length with several conserved regions, including the Walker A and B motifs (separated by ∼90 to 120 residues) and a “signature” motif with the consensus sequence LSGGQ (also called the C motif and located just upstream from the Walker B region) (28, 61, 97, 98). X-ray crystallographic analysis of model ABC transporters from bacteria indicates that the two NBDs of a single transporter interact in a head-to-tail fashion, with the Walker A and B motifs of one NBD interacting with the C motif of the other (24, 64, 97, 98). It is notable that a subset of ABC proteins are comprised solely of NBDs and lack membrane spans entirely (ABCE and ABCF subfamilies) (Fig. 2 and see Fig. 4); these ABC proteins do not function as transporters but instead are likely to couple ATP hydrolysis to other processes such as DNA repair and protein translation (78, 174, 175).
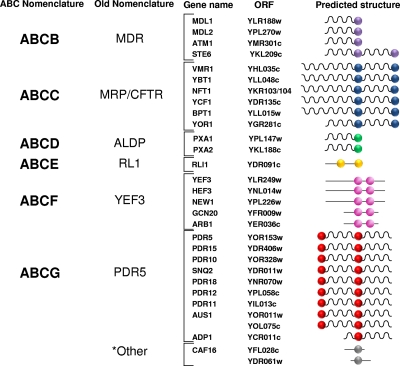
FIG. 2.
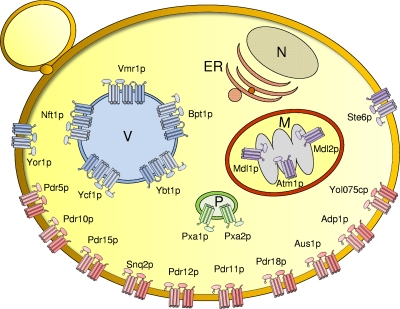
Assignment of yeast ABC proteins into subfamilies ABCB to ABCG using the mammalian nomenclature. Yeast ABC proteins are divided into subfamilies according to the sequence similarity within their NBDs. The ABCB to ABCG (left) (http://nutrigene.4t.com/humanabc.htm) (25-27) and traditional (right) (107, 155) yeast subfamilies are shown. We propose here to rename the yeast subfamilies using the standard ABC nomenclature employed for human ABC transporters. Each subfamily is separately colored: purple, ABCB; blue, ABCC; green, ABCD; yellow, ABCE; magenta, ABCF; red, ABCG. Note that the mammalian ABCA subfamily is absent in yeast. Two of the yeast ABC proteins, Caf16p and Ydr061w, are not closely homologous to any of the mammalian ABC transporter subfamilies and are labeled “other” (gray). Balls indicate NBDs, wavy lines indicate MSDs, and straight lines signify nonmembrane sequences. Only four of the subfamilies contain members with membrane spans (ABCB, ABCC, ABCD, and ABCG) and are thus likely to function as transporters.
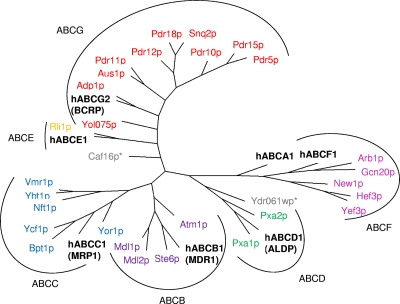
FIG. 4.
Yeast ABC phylogenetic tree. The protein sequences of the yeast ABC transporters have been subjected to a multiple-sequence alignment using CLUSTALW and phylogenetic analysis, and the resulting data are depicted in a radial-tree format (PHYLO). Subfamilies have been highlighted and grouped by black lines and arcs. As in Fig. 2, the nomenclature ABCB to ABCG is used to assign the yeast ABC proteins to their homologous subfamilies. Colors are as defined in the legend of Fig. 2. For each subfamily, a mammalian member (boldface type) was included in the analysis as a point of reference.
ABCA to ABCG Subfamilies
Through the efforts of ABC researchers and the Human Genome Organization (HUGO), the mammalian ABC superfamily has been divided into seven subfamilies (designated ABCA to ABCG) based on the relatedness of sequences within their NBDs (http://nutrigene.4t.com/humanabc.htm) (25-27). Members of each subfamily tend to have partially overlapping physiological and biochemical functions. This principle is well exemplified by studies of ABCC subfamily members in yeast, plants, and mammals (Fig. 3). ABCC transporters generally transport xenobiotic compounds or toxic metabolites that have first been conjugated to glutathione (GSH), glucuronide, or sulfur (3, 15, 33, 58, 66, 82, 125, 160).
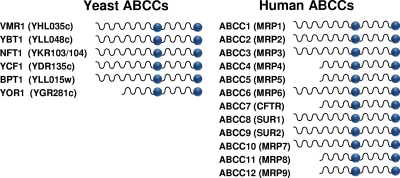
FIG. 3.
Yeast and human ABCC subfamily members. Yeast ABCC proteins are shown at left with their common names and ORF names (in parentheses). Human proteins are shown at right along with their ABCC names and common names (in parentheses) (http://nutrigene.4t.com/humanabc.htm). The diagram indicates whether the particular subfamily member is full length or short.
Below, we discuss the complete inventory of yeast ABC proteins and then focus on members of the ABCC subfamily in yeast, particularly Ycf1p, which has been proven an excellent model for improving our understanding of the shared features of ABCC subfamily members throughout the eukaryotes.
INVENTORY OF YEAST ABC PROTEINS
The genetic, biochemical, and cell biological tractability of yeast makes it a particularly well-suited model system for investigating protein function and protein interaction networks and for defining cellular pathways (12, 45, 67, 87, 140, 152). This has proven to be the case for many aspects of ABC transporter biology, particularly because upon the completion of the yeast genome sequence project in 1996, Saccharomyces cerevisiae became the first organism for which a glimpse of the complete inventory of ABC transporters was available (30, 48, 107, 155). Because ABCC transporters exhibit significant functional redundancy, the ability to easily knock out multiple genes in yeast has greatly facilitated our understanding of ABCC transporter function.
The yeast genome contains 30 ABC proteins (Fig. 2 and 4 and Table 1) originally identified based on BLAST searches for homologs of NBD1 of STE6 (30, 107, 155). Of these proteins, 22 are predicted to contain multiple membrane spans and are thus considered to be true ABC transporters, while the remaining 8 have no predicted spans and presumably carry out nontransport functions in the cell (Fig. 2). The localization and the function of the 22 yeast ABC transporters are shown in Table 1 and Fig. 5. Previous phylogenetic analyses established the existence of six ABC subfamilies in yeast, and each was assigned a subfamily name, with the name being based on a prominent mammalian or yeast representative of that subfamily (147, 155). Here, we propose to redesignate the old subfamily names in accordance with the now commonly accepted HUGO nomenclature for ABC proteins (ABCA to ABCG) (http://nutrigene.4t.com/humanabc.htm) (Fig. 2 and 4 and Table 1). Thus, we propose that the former yeast subfamily names MDR, MRP/CFTR, ALDP, RLI, YEF3, and PDR5 be redesignated ABCB to ABCG, respectively (Fig. 2 and 4 and Table 1). It should be noted that the mammalian ABCA subfamily is entirely absent in yeast. In addition, two yeast ABC open reading frames (ORFs) (CAF16 and YDR061w) cannot be classified into any of the HUGO subfamilies and are categorized here as “other” (Fig. 2 and 4). The redesignation of yeast subfamilies that we propose here should facilitate a better correlation of experimental findings between yeast and mammalian ABC transporters.
TABLE 1.
Classification, localization, and function of Saccharomyces cerevisiae ABC transportersa
| Family | Transporter | ORF | Localization | Description | Reference(s) |
|---|---|---|---|---|---|
| ABCB | MDL1 | YLR188w | Mitochondrial inner membrane | Involved in export of peptides from mitochondria, plays a role regulating oxidative stress; function may overlap with that of ATM1; homodimerizes | 20, 172 |
| MDL2 | YPL279w | Mitochondrial inner membrane | Unknown; homodimerizes | 96 | |
| ATM1 | YMR301c | Mitochondrial inner membrane | Essential for cytosolic iron-sulfur clusters and iron homeostasis; homodimerizes | 80 | |
| STE6 | YKL209c | Plasma membrane | Exporter of a-factor pheromone in MATa cells | 79, 106 | |
| ABCC | VMR1 | YHL035c | Vacuole | Unknown | 30 |
| YBT1 | YLL048c | Vacuole | Transports bile acid in vitro; transports Ade2 pigment in vivo | 113, 144 | |
| NFT1 | YKR103/104 | Vacuole | Unknown | 102 | |
| YCF1 | YDR135c | Vacuole | Glutathione S-conjugate transporter involved in cellular detoxification; transports heavy metals such as cadmium, mercury, lead, and arsenite; transports Ade2 pigment; bile pigments and MRI contrast agents are in vitro substrates and unconjugated bilirubin | 94, 120, 154 | |
| BPT1 | YLL015w | Vacuole | Glutathione S-conjugate transporter involved in cellular detoxification; transports cadmium and Ade2 pigment in vivo; function overlaps with that of Ycf1p | 83, 120 | |
| YOR1 | YGR281c | Plasma membrane | Multidrug transporter; exports oligomycin, organic anions, and many other compounds | 23, 76 | |
| ABCD | PXA1 | YPL147w | Peroxisome | Implicated in transport of acyl coenzyme A esters across the peroxisomal membrane; heterodimerizes with Pxa2p | 60, 142, 143 |
| PXA2 | YKL188c | Peroxisome | Implicated in transport of acyl coenzyme A esters across the peroxisomal membrane; heterodimerizes with Pxa1p | 60, 142, 143 | |
| ABCG | PDR5 | YOR153w | Plasma membrane | Multidrug transporter involved in resistance to xenobiotic compounds (mutagens and anticancer drugs) and cations and in steroid transport | 7, 100, 109 |
| PDR15 | YDR406w | Plasma membrane | Multidrug transporter involved in general stress response for cellular detoxification | 168, 169 | |
| PDR10 | YOR328w | Plasma membrane | Multidrug transporter involved in the pleiotropic drug resistance network | 6, 168 | |
| SNQ2 | YDR011w | Plasma membrane | Multidrug transporter involved in multidrug resistance and resistance to singlet-oxygen species | 141, 161 | |
| PDR18 | YNR070w | Plasma membrane | Putative transporter implicated in pleiotropic drug resistance | 9, 132 | |
| PDR12 | YPL058c | Plasma membrane | Multidrug transporter involved in weak organic acid resistance | 65, 121 | |
| PDR11 | YIL013c | Plasma membrane | Multidrug transporter involved in multidrug resistance and mediating sterol uptake | 134, 167 | |
| AUS1 | YOR011w | Plasma membrane | Involved in sterol uptake | 167 | |
| YOL075c | YOL075c | Plasma membrane | Unknown | 30 | |
| ADP1 | YCR011c | Plasma membrane | Unknown | 30 |
FIG. 5.
Subcellular localization of S. cerevisiae ABC transporters. The 22 yeast ABC proteins containing membrane spans are colored by their subfamily, ABCB (purple), ABCC (blue), ABCD (green), and ABCG (red), and are localized to the indicated intracellular organelles (P, peroxisomes; V, vacuole; M, mitochondria) and the plasma membrane (not labeled). No ABC proteins localize to the nucleus (N) or ER. The three mitochondrial ABC transporters are localized to the inner mitochondrial membrane. Because they are sequestered from the cytosol by the outer mitochondrial membrane, iMYTH studies cannot be performed with these three ABC transporters. Cylinders indicate MSDs. Full-length transporters and half-transporters are depicted, as are the full-length and short MRPs. NBDs are represented by ellipses.
Over the years, studies of yeast ABC transporters have made important contributions to our understanding of many areas of cell biology and drug resistance. Examples include the identification of physiological ABC transporter substrates (notably, the lipopeptide mating pheromone a-factor for Ste6p) (11, 88, 104, 106), the elucidation of ubiquitin as a trafficking signal for the endocytosis of certain membrane proteins (Ste6p being one of the first examples) (10, 77, 86), the analysis of endoplasmic reticulum (ER) quality control machinery and the identification of ER-associated compartments (34, 68, 69, 112), the discovery of new roles of ABC half-transporters in organellar function (96, 142), and the elucidation of pleiotropic drug resistance networks that are transcriptionally coregulated and that may contribute to the growing problem of drug resistance of fungal pathogens such as Candida albicans or Candida glabrata (37, 57, 85). The reader is referred to several comprehensive reviews that have catalogued the contributions of yeast ABC transporter studies in discovering basic cell biological principles and in providing clues to the roles of ABC transporters in human health and disease (9, 74, 107, 147, 155).
The remainder of this review will focus on one of the best-studied subfamilies in yeast, the ABCC subfamily. We highlight the many advances that have been made in our understanding of the role of ABCC transporters in a variety of cellular processes and discuss the role of these transporters in heavy metal detoxification, fungal multidrug resistance, and the development of integrated membrane yeast two-hybrid (iMYTH) technology to identify membrane protein interactors. Because of the functional and structural similarities between yeast and mammalian ABC transporters, advances derived from the yeast studies have impacted our understanding of all ABCC transporters, as discussed in further detail below.
THE ABCC (MRP) SUBFAMILY
ABCC (MRP) Transporters Are Glutathione Conjugate (GS-X) Pumps and Can Contain an N-Terminal Extension
Members of the ABCC (also called MRP) subfamily play key roles in the efflux of xenobiotic compounds in eukaryotic cells. Human MRP1 (ABCC1) is the founding member of the ABCC subfamily and was discovered based on its ability, when overexpressed in a human cancer-derived cell line, to confer multidrug resistance to a variety of chemotherapeutic cancer drugs (21). The discovery of other mammalian, yeast, and plant MRP1 homologues soon followed (3, 13, 33, 74, 76, 124, 125, 154, 155). Generally, ABCC subfamily members are distinguished from other ABC transporters by two striking hallmarks, one structural, the other functional: (i) in addition to the “ABC core” domain, they contain a characteristic “N-terminal extension” (NTE), which comprises an MSD (MSD0) with five transmembrane spans and a short cytosolic loop (L0) (Fig. 1) (although it should be noted that a subset of ABCCs have only an L0 and no MSD0), and (ii) ABCC proteins transport most substrates in the form of GSH conjugates and, in some cases, glucuronide or sulfate conjugates, rather than transporting the unmodified substrates themselves (14, 22, 58, 125). The connection between the NTE and GSH remains unclear at present. However, as discussed below, the discovery that human MRP1 transported GSH conjugates (53, 63, 89, 90) helped answer a long-standing question in the field of pharmacology, namely, which class of cellular pumps is responsible for the efflux of GSH-conjugated toxins out of the cell (33, 70).
MRP1 (ABCC1) Is the Long-Sought-After GS-X Pump
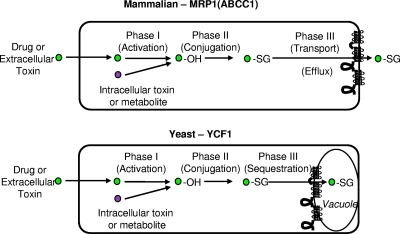
Pharmacologists had appreciated for many decades, even before the identification of the corresponding machinery, that cells possess an effective cellular detoxification system involving three phases that act to detoxify and ultimately remove exogenous toxins that have gained entry into mammalian cells (Fig. 6) (72, 160, 173). In phase I, the chemical “activation” of a toxin, for instance, by the addition of an epoxide mediated by cytochrome P-450, results in its increased chemical reactivity. Phase II involves the enzymatic conjugation of GSH, glucuronide, or sulfate to the activated toxin, which further increases its solubility. Finally, in phase III, the ATP-dependent efflux of activated toxins out of the cell occurs via a transporter (170). While the biochemical activity of the phase III GSH conjugate transporters, also referred to as the GS-X pumps, was well established for decades, their molecular identity remained elusive until MRP1 was discovered in 1994 and shown to be a major GS-X pump in human cells (21, 90, 123). Later, other ABCC subfamily members were also shown to be GS-X pumps (3, 13, 33, 66).
FIG. 6.
Cellular detoxification, phases I, II, and III. The cellular detoxification of intracellular and extracellular toxins in yeast and mammalian cells generally utilizes the phases of detoxification shown here and described in the text. Toxins or metabolites acted upon by this system are indicated by a circle. For the phase I “activation” step, the addition of OH is indicated. For phase II, the conjugation of GSH (−SG) to the toxin is shown. Phase III is mediated by an ABC transporter to move the conjugated toxin across a cellular membrane, either the plasma membrane for MRP1 or the vacuole membrane for yeast Ycf1p, as discussed in the text.
In addition to xenobiotic toxins, ABCC transporters also efflux endogenous physiological substrates, including the signaling molecule leukotriene C4 (LTC4) (mediated by MRP1) and metabolites destined for bile, including bilirubin, glucuronide conjugates, and sulfated bile salts (mediated by a variety of ABCC transporters) (Fig. 6) (3, 13, 33, 66). Taken together, it has become clear that the ABCC subfamily plays an important protective role in the cell, in processes ranging from cellular assault by externally derived chemicals to the physiological transport of signaling molecules or endogenously generated toxic products.
The Mammalian ABCC Subfamily Contains Full-Length and Short Members
Human ABCC subfamily members are shown in Fig. 3 (25, 26). Although most of these proteins are likely to function as transporters, three are not: CFTR (ABCC7) is an ion channel, and SUR1 (ABCC8) and SUR2 (ABCC9) are ion channel regulators. However, the possibility that these ABCCs also have undiscovered transport functions should be considered.
The ABCC subfamily of ABC transporters consists of two structurally distinct classes of proteins. The first class of human ABCC proteins (7 of 12) possesses a complete NTE (containing both MSD0 and L0) (Fig. 1B and 3) and are designated “full-length” ABCC proteins. The second class of proteins (5 of 12) contains only a partial NTE (L0 is present, but an MSD0 region is not). The latter proteins are designated “short” ABCC proteins (Fig. 1B and 3). The MSD0 portion within the NTE of “full-length” ABCCs is implicated in the proper trafficking of several ABCC proteins, while L0 is required for substrate transport (but not localization) in all cases where it has been tested, but whether the NTE also contributes to substrate specificity is not known (8, 41, 103, 165, 166).
A complete understanding of the role of the NTE in “full-length” transporter function remains to be established and is a major subject of current research. An issue under study is whether the NTE, so distinctive for the ABCC subfamily, also plays a special role in the unique ability of this subfamily to specifically recognize GSH-conjugated substrates (123). Because the weight of current evidence suggests that GSH-binding sites of ABCC transporters lie in the core domain and not the NTE, the role of the NTE in the recognition of GSH conjugates remains questionable (22).
MEMBERS OF THE YEAST ABCC SUBFAMILY
The ABCC subfamily in yeast is shown in Fig. 2 to 4 and Table 1. Five members are “full length” (Ycf1p, Bpt1p, Ybt1p, Nft1p, and Vmr1p), and one is “short” (Yor1p) (107, 155). Interestingly, while Yor1p localizes to the plasma membrane, the others all appear to be localized to the vacuolar membrane (Table 1 and Fig. 5) (75, 83, 94, 105, 113, 145). However, it should be noted that the localization data for Nft1p and Vmr1p are preliminary, and a punctate pattern in addition to a vacuolar membrane pattern have been observed for these transporters (105). Both the efflux of harmful compounds out of the cell and their transport into the vacuole have the same benefit of removing dangerous toxic molecules from the cytosol and, thus, away from intracellular targets. In many cases, yeast ABCCs exhibit overlapping substrate specificity (144, 145). The fact that both Ycf1p, a “full-length” ABCC transporter, and Yor1p, the plasma membrane-localized “short” transporter, can confer cadmium resistance (discussed in detail below) indicates that there is not a critical functional distinction between full-length and short ABCCs and that their distinct cellular locations may not necessarily confer unique properties (111). The yeast system is ideal for further dissecting these issues.
Below, we discuss what is currently known about each of the yeast ABCC transporters, discussing “full-length” ABCC transporters first, with a focus on Ycf1p, the best-studied member of the family. In the last section, we provide a comprehensive discussion of the newly developed split-ubiquitin MYTH system, which is specifically designed to identify membrane protein interactors and has been used with great success to identify functional interactors of Ycf1p.
Ycf1p
Ycf1p is the prototypical yeast ABCC transporter with a broad range of xenobiotic substrates. (i) Discovery of Ycf1p as a GSH-cadmium transporter and complementation of yeast ycf1Δ by human and plant MRPs.
Ycf1p is the first member of the ABCC subfamily identified in yeast and remains the best-characterized member to date. The YCF1 gene was discovered in 1994 based upon its ability to confer cadmium resistance when overexpressed (hence the name yeast cadmium factor) (154). Conversely, the deletion of YCF1 results in cadmium hypersensitivity. A surprising early finding was that Ycf1p is not localized to the plasma membrane, where efflux pumps had previously been found. Instead, Ycf1p was shown to reside on the vacuolar membrane (the vacuole in yeast being equivalent to the mammalian lysosome) (94).
Motivated by the structural similarity between human MRP1 and yeast Ycf1p, investigators proposed that like MRP1, Ycf1p might be a GSH conjugate transporter. Using an in vitro transport assay, this was shown to be the case. A radiolabeled substrate, GS-conjugated dinitrophenyl (3H-DNP-GS), was efficiently transported into vesiculated vacuoles prepared from a wild-type (WT) (YCF1) strain but not from a ycf1Δ strain (94). Notably, it was shown that cadmium is transported by Ycf1p as a cadmium-bis-GSH complex (Cd-GS2), providing the first evidence for the involvement of GSH in heavy metal transport (93). Those early studies indicated that Ycf1p, like MRP1 (ABCC1) in mammalian cells, functions as a phase III export pump. However, rather than excreting GSH conjugates into the extracellular space, Ycf1p transports conjugated substrates across the vacuolar membrane, sequestering them within the vacuolar lumen (Fig. 6) (1, 93, 145, 154).
Strikingly, human MRP1, heterologously expressed in a yeast ycf1Δ mutant, was shown to complement its cadmium sensitivity in vivo and to restore GSH S-conjugate transport activity into yeast vacuoles in vitro (157). Furthermore, the expression of a plant MRP from Arabidopsis thaliana, AtMrp1, also complements a ycf1Δ mutant (158). This remarkable functional interchangeability of yeast, human, and plant MRPs suggests that ABCC activities are likely to be similar in all organisms, reflecting an impressive level of cross-organismal conservation in function. As discussed below, attempts are currently being made by researchers to harness this property for the purpose of phytoremediation, in which plants are used to remove harmful toxins from the soil and water.
(ii) Transcriptional regulation of the YCF1 gene by Yap1p.
Shortly after its discovery, YCF1 was shown to be regulated by Yap1p, a transcription factor which belongs to the b-Zip class of DNA-binding proteins and is related to mammalian Ap-1 and yeast GCN4 (164). The overexpression of Yap1p confers resistance to cadmium in yeast that is dependent on Ycf1p (50, 114, 164). Yap1p binds to a Yap1p response element located upstream of the YCF1 ORF and promotes increased levels of expression of YCF1. A number of studies have implicated Yap1p as being the single most critical transcriptional regulator in defending cells against oxidative stress (57, 133).
(iii) Ycf1p transports a broad range of xenobiotic substrates: in vivo studies.
A series of reports have shown that Ycf1p can transport many different heavy metals in addition to cadmium, as evidenced by the inability of a ycf1Δ strain to grow in media containing these metals. These metals include lead (Pb), antimony (Sb), mercury (Hg), and arsenite (As) (46, 56, 122, 149). Gueldry and coworkers pointed out that care must be taken in carrying out such growth tests, as the composition of the medium can influence the apparent efficacy of heavy metals or other compounds to inhibit yeast growth (56). In particular, certain supplements, such as the amino acid histidine, can chelate heavy metals, reducing their effective concentration. It is likely that all heavy metal substrates of Ycf1p are transported in the form of a GS-metal complex, as shown for Cd-GS2 and Hg-GS2 (56, 93). However, this has yet to be experimentally confirmed. Results of a number of studies examining As3+ resistance in yeast, bacteria, and mammalian cells have shown that As3+ forms multiple metabolites, including both GSH and methyl conjugates (135, 136).
In addition to heavy metals, a number of other cytotoxic compounds have been shown to require Ycf1p for their detoxification. Examples include the antioxidant/oxidizing agents diamide and 1-chloro-2,4-dinitrobenzene (94, 163). A potentially useful Ycf1p substrate that has yet to be fully exploited is monochlorobimane, which can be added to living yeast cells and visualized by fluorescence microscopy. In a WT YCF1 strain, monochlorobimane is concentrated in the vacuole, whereas in a ycf1Δ mutant, it is excluded (94). Thus, this fluorescent compound may serve as a sensitive in vivo indicator of Ycf1p function, or the lack thereof, facilitating the analysis of mutant forms of Ycf1p and Ycf1p-protein interactions.
Clearly, Ycf1p can transport a range of exogenous compounds, many of which are cytotoxic. However, a potential role of Ycf1p in sequestering endogenous cellular toxins, possibly arising from normal metabolic processes, has remained unclear. With the increasing ability of researchers to carry out large-scale metabolic systems biology studies of yeast, one tantalizing avenue of research is to identify candidate endogenous substrates for Ycf1p and other yeast MRPs by high-throughput metabolomics, comparing ycf1Δ deletion or YCF1-overexpressing strains to a WT strain.
(iv) Ycf1p transports many different xenobiotic compounds in vitro.
Ycf1p can transport GSH conjugates (e.g., DNP-GS), glucuronate conjugates (e.g., E217βG), and reduced GSH, all three of which are human MRP1 substrates (94, 126-128, 145). The transport of these substrates by Ycf1p indicates that Ycf1p is a phase III transporter in yeast, similar to its human counterpart, MRP1 (Fig. 6). Transport assays are performed utilizing right-side-out-vesiculated yeast vacuoles prepared from WT (YCF1) and mutant (ycf1Δ) yeast strains (93, 94, 119). In general, these assays are designed to measure the transport of radioactively labeled substrates into the vesiculated vacuoles. Ycf1p has also been expressed in Sf21 insect cells, and membrane vesicles from these cells show Ycf1p-dependent transport properties similar to those of yeast vacuole preparations (131).
In addition to the “standard” MRP substrates noted above, additional substrates for Ycf1p have been demonstrated by assays of in vitro transport into yeast vacuoles. These substrates include unconjugated bile pigment and several magnetic resonance imaging (MRI) contrast agents (117, 120). As discussed below, for nearly all substrates tested to date, Ycf1p exhibits overlapping substrate specificity with its closest relative, Bpt1p (Fig. 2 and Table 1), and, in some cases, other yeast ABCCs as well (117, 120, 145). One exciting potential use for yeast Ycf1p transport assays is for the identification of inhibitors that ultimately might also be useful for inhibiting human MRP activity.
In addition to examining particular substrates, transport assays in vitro have also been important for examining the energy requirements for ABC transporter activity. We have demonstrated that Ycf1p can use either ATP or GTP to power substrate transport in vitro, as has also been reported for several other ABC transporters (49, 119, 162). The significance or universality of alternative nucleotides is not clear, although it has been proposed that Pdr5p increases its multidrug transport specificity by using more than one nucleotide as an energy source (49).
(v) Potential of harnessing Ycf1p for phytoremediation.
There has been significant interest in harnessing Ycf1p's ability to transport heavy metals for phytoremediation, which refers to the use of plants to reduce the levels of environmental contaminants (54, 149, 159). Heavy metals such as Pb, Cd, Hg, and As represent some of the most significant toxic soil and dust contaminants in industrialized countries. Phytoremediation has been proposed to be a low-cost, effective way to remove heavy metal contaminants from soil. Notably, Ycf1p expressed in the model plant Arabidopsis promotes the improved tolerance of these plants to Pb(II) and Cd(II) and results in a high intracellular concentration of the metals, presumably due to their sequestration in the vacuole (149, 159). An extension of this work involving the expression of yeast Ycf1p in poplar, a tree that grows rapidly and to a high mass, making it suitable for bioremediation, was recently suggested (146). Although not yet accomplished, it has been proposed that ultimately, full-grown transgenic poplars, replete with vacuolar heavy metals resulting from Ycf1p-mediated transport, could be cut and removed from a contamination site, proving an effective mechanism by which soil can be decontaminated relatively inexpensively and in an environmentally friendly way.
Ycf1p transports endogenous metabolites and could be involved in metabolic “quality control” in yeast.
Ycf1p plays an important role in the striking pink/red pigmentation of yeast colonies that arise from adenine biosynthetic mutants in yeast (144, 145). Specifically, strains carrying ade1 or ade2 mutations accumulate an intense red pigment in their vacuoles when they are grown under conditions of adenine limitation. The formation of this pigment involves a series of steps, including the conjugation of the adenine biosynthetic intermediate phosphoribosylaminoimidazole (AIR) to GSH, the Ycf1p-dependent transport of AIR-GS into the vacuole, and further metabolism within the vacuole, ultimately giving rise to the familiar red pigment associated with an ade2 yeast strain (18, 144, 145). In a ycf1Δ mutant, the pigmentation of an ade2 mutant colony is greatly diminished due to the significant lack of transport of AIR-GS into the vacuole. The residual pigmentation in a ycf1Δ mutant is due to a low level of transport activity mediated by two other ABCC transporters, Bpt1p and Ybt1p (144, 145). The “red ade2 assay” has been used experimentally as a sensitive in vivo assay for the function of Ycf1p and/or its functional interactors (119).
Importantly, the cytosolic elimination of the ade2 red intermediate may reflect an important cellular role for Ycf1p and other ABCC transporters in a “metabolic quality control” system that prevents the aberrant cytosolic accumulation of potentially toxic biosynthetic intermediates. A role for Ycf1p in metabolic quality control is being experimentally sought but is complicated by the redundancy of other yeast ABCCs. To most effectively determine the role of Ycf1p or any ABCCs, human or yeast alike, in metabolic quality control, the optimal model would be cells that are missing multiple ABCC proteins. This type of analysis could be carried out using strains with a deletion of any number and combination of yeast ABCCs (F. Roth, Harvard University, personal communication).
Ycf1p proteolytic processing and trafficking.
Studies of Ycf1p trafficking and processing have relied on antibodies that recognize Ycf1p or a C-terminally green fluorescent protein-tagged version of Ycf1p that retains normal function and trafficking properties (102, 103). An unusual aspect of Ycf1p biology is that this transporter is posttranslationally processed to yield N- and C-terminal cleavage products that are stable and remain tightly associated with one another (102, 163). Cleavage occurs within luminal loop 6 (L6) in a region representing an insertion that is specific to Ycf1p (Fig. 1). The “L6 insertion” appears to be functionally transplantable in that it can promote processing when moved to new luminal locations in Ycf1p or another transporter, Bpt1p (102). Cleavage occurs only after Ycf1p reaches its final vacuolar destination and is dependent on the vacuolar proteases Pep4p and Prb1p. Pep4p, the master vacuolar protease, and Prb1p have reciprocal roles in activating each other. Surprisingly, the processing of Ycf1p is not required for its activity (103). However, certain mutations within the Ycf1p “L6 insertion” appear to alter substrate specificity, suggesting that both cytosolic and luminal domains can influence substrate binding or, possibly, substrate release (102). The recent publication of the crystal structure for the mammalian ABC transporter MDR1 (ABCB1; also called P-glycoprotein) and the bacterial ABC transporter Sav1866 suggests the possibility that luminal L6 of Ycf1p may be in very close proximity to the predicted pore from which substrates might exit; however, it should be noted that such a model has yet to be tested (2, 24, 64).
Mutant forms of Ycf1p also appear to hold promise for gaining insight into the ER quality control processes that handle misfolded membrane proteins, such as the mutant form of CFTR (CFTR-ΔF508) that results in cystic fibrosis. Such misfolded membrane proteins are not permitted to exit the ER and undergo degradation by the ubiquitin-proteasome system. Most ABCC proteins, including Ycf1p, have a phenylalanine analogous to CFTR-F508. When mutated in Ycf1p (Ycf1-Δ713), the mutant form of Ycf1p is retained in the ER, in structures called ER-associated compartments, and degraded by the proteasome, providing an excellent model for studying ER quality control components (101, 163). Current studies in our laboratory are focused on using Ycf1p and the ABCB transporter Ste6p as models to study protein misfolding and ER-associated degradation (68, 101, 105).
Mutational structure-function analysis of Ycf1p: partial-molecule studies and ABC mutants.
Ycf1p can be subdivided into two “partial molecules” containing the NTE (MSD0 plus L0) and the “core” domain that upon coexpression can reconstitute functional Ycf1p in the vacuole membrane. Studies with mutant forms of these Ycf1p partial molecules have provided evidence that the L0 portion of the NTE is absolutely required for Ycf1p transport function (103). Furthermore, the deletion of a highly conserved 17-amino-acid amphipathic helical region within L0 is sufficient to abolish Ycf1p function (102). In contrast, data from partial-molecule studies indicate that MSD0 is required for the proper localization of Ycf1p (103). A functional role for MSD0 could not be assessed, because when MSD0 is compromised, Ycf1p mislocalizes. Similar to Ycf1p, the L0 region in mammalian ABCC transporters is critical for the function of human MRP1 and MRP2, and likewise, MSD0 is required for proper MRP1 and MRP2 localization to the plasma membrane (4, 41, 165, 166). Because analysis of the yeast Ycf1p NTE has provided generalizable insights into this intriguing module that sets ABCC transporters apart from other ABC subfamilies, its further analysis in Ycf1p is warranted.
To perform mutational structure-function analyses of Ycf1p, investigators have carefully analyzed an extensive series of site-directed mutants within NBD1, NBD2, a region within the linker called the R domain, and the so-called intracellular loop 4 (CL4) positioned between membrane spans 15 and 16 of Ycf1p (which has a total of 17 spans). Residues chosen for mutagenesis are conserved in MRP1 and CFTR, and some of the mutations were analogous to cystic fibrosis-associated mutations (39, 40, 163). Mutants were assessed for Cd(II) resistance in vivo and vacuolar transport activity in vitro (transport of radiolabeled LTC4 into vacuolar vesicles). Most of the mutations were found to be deleterious, due either to a loss of function or to biogenesis defects (the latter class manifests as a lack of protein due to degradation). However, a few of the mutants showed differential effects on the ability of yeast to grow on Cd(II) and Ycf1p-dependent LTC4 transport, potentially suggesting alterations in substrate binding (39, 40, 163). Follow-up studies using these mutants may be useful for elucidating the basis of Ycf1p substrate specificity.
In addition to the primary mutational analysis described above, intragenic suppressor studies were carried out, starting with mutations that inactivate Ycf1p function. One of these was D777N in the Walker B motif of NBD1, which inactivates Ycf1p function (39). Gain-of-function suppressor mutations that restored Ycf1p function to ycf1-D777N were identified in multiple regions of Ycf1p, including NBD1 itself, NBD2, MSD1, and MSD2 (39). These results suggest that NBD1 may functionally interact with NBD2, MSD1, and MSD2 for proper Ycf1p function. Ultimately, an analysis of the ATPase catalytic cycle of Ycf1p will be an important area for further research and would be invaluable for assessing the mutants discussed above. However, in light of the low ATPase activity of most ABC transporters in vitro and the lack of substrate stimulation of ATPase activity in most cases, analysis of the ATPase catalytic cycle of Ycf1p promises to be challenging (79, 137).
Posttranslational modulation of Ycf1p function by phosphorylation and interaction partners.
Ycf1p appears to be strongly positively regulated by phosphorylation. Two residues (S908 and T911) within the linker of the core domain of Ycf1p are important for its positive regulation by phosphorylation (36, 118). The alteration of either residue to alanine (S908A and T911A) lowers cadmium resistance in vivo and LTC4 transport in vitro. In the double mutant (S908A T908A), cells show an even more severe phenotype in both assays, and this mutant form of Ycf1p exhibits a marked gel mobility shift, reflecting diminished phosphorylation (36). Furthermore, the mutation of these residues to aspartic acid (S908D T911D), which mimics phosphorylation, restores WT Ycf1p-dependent function. To date, the kinases responsible for phosphorylating these residues are not known (36).
Recently a number of large-scale proteomic studies aimed at identifying the yeast “phosphoproteome” have been reported. In several of those studies, phosphopeptides derived from Ycf1p were observed among a vast repertoire of total yeast phosphopeptides (19, 92, 148). The phosphorylated residues in the Ycf1p core domain include S908 and T911 (discussed above) as well as S869, S870, S872, S873, S903, and S914 (19, 92, 148). One additional phosphorylated residue, S251, lies within the L0 domain of Ycf1p. Recent studies from our group suggest that phosphorylation at S251 may negatively regulate Ycf1p function, since an S251A mutant exhibits increased resistance to cadmium in vivo and increased Ycf1p transport activity in vitro compared to WT YCF1 (118). Both activities are restored to the WT level in an S251E mutant. Notably, through interactor studies (discussed below), a kinase gene, CKA1, that encodes a subunit of the protein kinase, CKII, was identified. Interestingly, a cka1Δ mutant is cadmium sensitive, and additional genetic evidence points to the possibility that Cka1p may act by phosphorylating S251 of Ycf1p (118).
In general, the role of phosphorylation in regulating ABC protein function remains unclear. The studies discussed above suggest an important mechanistic role of phosphorylation in both positively and negatively regulating Ycf1p transporter function. Interestingly, many putative Ycf1p phosphorylated residues are highly conserved in human MRPs (118). Thus, findings for Ycf1p phosphorylation at particular sites could pave the way for examining whether mutations that ablate or mimic phosphorylation have a predictable pattern of positive or negative impact on function in mammalian ABCC transporters.
Regulation of Ycf1p function by Tus1p, a guanine nucleotide exchange factor for Rho1p.
Multispanning membrane proteins are underrepresented in protein interaction studies because many of the screens employed in those studies are not amenable for use with an integral membrane protein such as Ycf1p. However, interaction partners are just as likely to regulate the activity and function of membrane proteins as they do for soluble proteins. By using iMYTH technology, which is discussed in detail below, we identified a number of potential Ycf1p interactors. Strains bearing deletions of several of these interactors exhibit a decreased level of cadmium resistance compared to the level exhibited by a WT strain (119). One of these is the TUS1 gene, which encodes a well-characterized cytosolic guanine nucleotide exchange factor for the small GTPase Rho1p (139, 171). We showed that Tus1p is a Rho1p-dependent positive regulator of Ycf1p function (119). Tus1p interacts with Ycf1p via the Tus1p C-terminal region that contains a citron domain, whose function remains uncharacterized.
Previous to our study, cytosolic modulators of the ABC transporters had not been generally considered, because transporter-containing vesicles, nucleotides, and substrate are sufficient to promote ABC transport activity in vitro. However, we showed that the addition of cytosol from a WT TUS1 strain increases Ycf1p-dependent transport activity by about twofold and requires the presence of GTP. Importantly, this activation fails to occur with cytosol prepared from a tus1Δ strain, indicating that Tus1p is required for the observed stimulation of Ycf1p activity (119). In addition, Ycf1p-dependent transport activity was diminished in vacuoles prepared from a rho1ts mutant strain and in this case could not be further stimulated by the addition of Tus1p. Taken together, these results suggest that Rho1p stimulates Ycf1p function above the basal level when it is recruited to Ycf1p by Tus1p (119). Rho1p is indeed known to stimulate the activity of two other enzymes in yeast [protein kinase C and β(1-3)-glucan synthase] (16, 17, 91, 110). These studies suggest diverse roles for yeast Rho1p in regulating numerous cellular functions, including Ycf1p activity.
Bpt1p and Ybt1p Functionally Overlap with Ycf1p
Bpt1p and Ybt1p have not been extensively characterized. However, to the extent to which they have been characterized, both appear to have some overlapping function with Ycf1p. As discussed below, both are localized to the vacuole membrane (Fig. 5 and Table 1) and play a role in the cellular detoxification of both endogenous and exogenous compounds (105, 113, 117, 120, 145). However, neither Bpt1p nor Ybt1p undergoes Pep4p-dependent cleavage like Ycf1p (102), and both have dissimilar expression patterns from one another and from that of Ycf1p (145) (see the Saccharomyces Genome Database [SGD] [www.yeastgenome.org/] for expression profiles).
Bpt1p is the closest yeast homolog to Ycf1p and is named for its ability to function as a bile pigment transporter in vitro, an activity which it shares with Ycf1p (120). Bile pigments are degradation products of heme-containing proteins that are excreted by hepatocytes in humans. In vitro transport assays showed that one of these bile pigments, unconjugated bilirubin, can be transported into yeast vacuolar vesicles. Bpt1p and Ycf1p make roughly equal contributions to unconjugated bilirubin transport (120). In addition, gadolinium (Gd)-based MRI contrast reagents, which are also excreted by hepatocytes, can be transported into yeast vacuoles. Both Bpt1 and Ycf1p can mediate their transport, but Bpt1p plays the major role for some contrast agents, while Ycf1p does so for others (117). Finally, Bpt1p also shares the ability to mediate the in vitro transport of free GSH, GSH conjugates (DNP-GS and [14C]S-2,4-dinitrobenzeneglutathione {DNB-GS}), and a glucuronide conjugate (E217βG) with Ycf1p, although in all of these cases, the activity of Ycf1p predominates (5, 83, 145). In vivo, Bpt1p contributes to a small extent to cadmium resistance and the formation of the red Ade2 pigment that is associated with Ycf1p (144, 145). A similar pattern of overlapping but nonidentical substrate specificity as seen for yeast Ycf1p and Bpt1p has also been documented for mammalian MRPs, reflecting a recurring theme within the ABCC subfamily in different organisms (5, 13, 33, 66, 125).
Bpt1p does not appear to be regulated by the oxidative stress regulator Yap1p, as is the case for Ycf1p (145). However, the Bpt1p expression level was shown to be strongly elevated upon the entry of yeast into stationary phase (83). Since yeast faces a number of metabolic challenges during high-density growth, it was suggested that the pattern of expression of Bpt1p is consistent with a direct role for Bpt1p in the detoxification of cellular metabolites (83). However, Bpt1p, like Ycf1p, may also protect cells from the products of environmental challenges such as oxidative stress.
Considerably less is known about Ybt1p than Bpt1p. Ybt1p was first named Bat1p (for bile acid transporter) based on the ability of WT yeast vacuoles, but not vacuoles prepared from a bat1Δ mutant, to transport bile acids such as taurocholate (113). However, due to a nomenclature conflict, the gene name was changed to YBT1 (for yeast bile acid transporter) (see the SGD [www.yeastgenome.org/]). In vivo, Ybt1p has been shown to provide a minor (less than 5%) contribution to the formation of red Ade2 pigment, along with Ycf1p and Bpt1p, indicating that it is capable of transporting GSH conjugates (144, 145). However, other Ybt1p substrates have not been reported.
Vmr1p and Nft1p
Vmr1p and Nft1p are the least characterized of the yeast ABCC transporters. No substrates are known for either one. Preliminary data suggest that both proteins appear to be localized to the vacuolar membrane (Fig. 5 and Table 1) (105), yet both proteins show an additional “punctate” pattern, which may reflect Golgi apparatus localization, endosomal localization, or some other organellar site of localization. The name Vmr1 stands for vacuolar multidrug resistance transporter (D. Wawrzycka, SGD [www.yeastgenome.org/], personal communication); however, no Vmr1p substrates are known to date (56). However, interestingly, recent high-throughput proteomic studies of yeast have found Vmr1p to be associated with both ribosomal complexes and the mitochondria (43, 129, 130).
Nft1p originally presented a confusing picture because in the major laboratory strain of Saccharomyces cerevisiae, S288c, it is present as two contiguous ORFs, YKR103w and YKR104w, separated by a nonsense codon. S288c is the strain that was used for the sequencing of the yeast genome. However, in other laboratory strains of Saccharomyces cerevisiae and in most other fungal species, YKR103w and YKR104w comprise a single continuous full-length ORF, which we named NFT1, for new full-length transporter (102). One potential explanation for the apparently mutated form of NFT1 in S288c is that the manner in which this strain was cultivated in the laboratory led to unintentional selection against full-length NFT1 (102). We considered the possibility that the mutation of NFT1 might provide lithium resistance, which is potentially advantageous for cells subjected to LiCl during transformation experiments. However, no evidence that Nft1p plays a role in LiCl resistance was found. Indeed, no function has yet been ascribed to Nft1p. One way to learn about the activities of the NFT1 and VMR1 gene products would be to challenge strains overexpressing these genes with an arsenal of toxic compounds and determine if resistance occurs in any case.
Yor1p, a Short ABCC at the Plasma Membrane That Mediates Pleiotropic Drug Resistance
In vivo studies of Yor1p activity.
Yor1p is the second-most-studied yeast ABCC. Yor1p is atypical among the yeast ABCCs in that it is the only “short” ABCC and the only one thus far that localizes to the plasma membrane (Fig. 2 and 3 and Table 1) (75, 76). Whether these two characteristics are related is unclear. The name Yor1p (for yeast oligomycin resistance) reflects that fact that the YOR1 gene was discovered based on its ability to confer resistance to the mitochondrial poison oligomycin when overexpressed and to confer sensitivity when deleted (76). Early on, Yor1p overexpression was also shown to confer resistance to the anionic antifungal drug reveromycin A and the anionic fluorescent compound rhodamine B, leading to the suggestion that Yor1p was a multispecific organic anion transporter (23). However, a large-scale study of several hundred compounds identified many more Yor1p substrates and indicated that Yor1p could transport an impressive variety of compounds with quite distinct chemical features (85, 134). Thus, Yor1p is now more appropriately considered to be a pleiotropic drug transporter.
Indeed, Yor1p is now viewed as being one of the three major pleiotropic drug transporters in yeast (Yor1p, Pdr5p, and Snq2p) that comprise the pleiotropic drug resistance network (57, 84, 85, 147). Yor1p is coregulated with Pdr5 and Snq2 by the positively acting transcription factors Pdr1p and Pdr3p, and it exhibits overlapping substrate specificity with Pdr5p and Snq2p but also has distinct substrate preferences (134). It has become quite common to delete these three transporters, Yor1p, Pdr5p, and Snq2p, yielding a strain called AD1-3 (also called AD123), to sensitize strains to various drugs for the purpose of conducting high-throughput drug screens (95, 134). One such example involved using a strain with deletions of these three genes to screen the effects of estrogenic compounds on signaling through the estrogen receptor (59). In other cases, it has been advantageous to use a strain with deletions of eight of the major ABC transporters, called AD1-8 (or AD12345678) (31, 111, 134). Both AD1-3 and AD1-8 have been used to examine the cellular uptake of the yeast vital stain diS-C3(3), a substrate for Yor1p, Pdr5p, and Snq2p. The cellular uptake and efflux of this dye permit the analysis of pump function in vivo by simple measurements of fluorescence intensity. In this assay, strains AD1-3 and AD1-8 show a high level of accumulation of dye in comparison to their WT counterparts due to a diminished efflux capacity (44).
An interesting issue is whether Yor1p, like Ycf1p, is a GS-X transporter. Data from a recent report showed that Yor1p confers resistance to Cd2+ but only at 23°C and not at higher temperatures (111) (although it should be noted that an early study implicated Yor1p in Cd2+ tolerance even at 30°C [23]; however, this finding could not be repeated by others). In contrast, Ycf1p confers Cd2+ resistance at all temperatures. Interestingly, the overexpression of Yor1p can suppress a ycf1Δ Cd2+ sensitivity phenotype at 23°C (111). Furthermore, the Yor1p-mediated efflux of cadmium is accompanied by the efflux of GSH, suggesting that Yor1p transports both together as a Cd-(GS)2 complex, just as Ycf1p does. Thus, because Yor1p lacks MSD0, it is tantalizing to suggest that the MSD0 of ABCC transporters is not required for GSH conjugate recognition.
Biochemical analysis of Yor1p.
For biochemical studies, a strain called SUPERYOR has been useful, in which Yor1p comprises roughly 10% of the total protein in the plasma membrane due to its overexpression (in strain AD1-8), so that Yor1p is the major ABC transporter present (31, 55). In membrane preparations from this strain, Yor1p could be photolabeled by azido-ATP and exhibits ATPase activity that is inhibited by vanadate, as is the case for other ABC transporters. Furthermore, mutations in NBD2 of Yor1p reduced the level of activity to that measured for substrates derived from a yor1Δ mutant (55). However, unlike some other ABC transporters, Yor1p ATPase activity was inhibited, rather than stimulated, upon the addition of substrates, an observation also made for yeast Pdr5p (31, 38, 55, 83). It was speculated that an unknown Yor1p substrate might be present in membrane preparations and that an added substrate could act as a competitor, leading to a reduction in the rate of ATP hydrolysis (55). Alternatively, some of the Yor1p substrates tested may have dual binding sites within the pore and the NBDs, and therefore, the binding of the substrate to the transporter would also inhibit the ATPase activity of the NBDs (55). An interesting yet unclear finding is that Yor1p can be phosphorylated in vitro, as observed upon the incubation of Yor1p-containing membranes with [γ-32P]ATP (32). Whether phosphorylation occurs in vivo, and, if so, whether it is stimulatory or inhibitory, has not been examined.
Trafficking, folding, and assembly of Yor1p.
Yor1p has proven to be an excellent model protein for analyzing the processes of ER exit and ER-associated degradation (75, 116). Yor1p has two diacidic (DxE) ER exit motifs, one within the NTE and the second at the C terminus (35). The N-terminal DxE is the dominant one of the two motifs. When this DxE is deleted, Yor1p is retained in the ER in vivo and cannot be packaged into CopII-coated ER-to-Golgi membrane transport vesicles in vitro (116). The DxE motif of Yor1p is needed to interact with the “B”-site cargo recognition domain of the CopII subunit Sec24p (116).
The Yor1p-ΔF670 mutant (equivalent to the cystic fibrosis mutation CFTR-ΔF508) results in ER retention followed by ER-associated degradation (75). The ER retention of Yor1p-ΔF670 is due to its misfolding, as assayed by limited-proteolysis, cross-linking, and gel migration assays (116). Ultimately, ER-retained Yor1p-ΔF670 is degraded by the ubiquitin-proteasome system. Interestingly, when Yor1p-ΔF670 is stabilized, by preventing its degradation in a mutant (ubc7Δ) defective for its ubiquitination, it still cannot exit the ER and is precluded from entering CopII vesicles in an in vitro packaging assay. It is possible that chaperones bound to misfolded Yor1p-ΔF670 may block the access of its DxE ER exit motif to the B site of Sec24p and, hence, inhibit its ER exit (116). An understanding of precisely how Yor1p-ΔF670 engages the ER quality control checkpoint may eventually shed light onto mechanisms for releasing ER-trapped CFTR-ΔF508 into the plasma membrane, which in turn could potentially provide an important clue to treatments that might reduce the symptoms of cystic fibrosis.
In addition to providing insights into protein trafficking and quality control, an understanding of how Yor1p is folded and assembled is likely to provide principles that are generalizable for all ABC proteins. To this end, Pagant and coworkers (115) have been attempting to genetically and biochemically define the intramolecular interactions that contribute to the tertiary assembly of Yor1p. Using chemical cross-linking, they defined interactions between the NBDs of Yor1p and between the NBDs and specific intracellular loops (CLs) that are consistent with current structural models of bacterial ABC transporters. Furthermore, starting with a yor1 mutation that alters an intracellular loop (CL2) and causes oligomycin sensitivity due to the misassembly of Yor1p, intragenic suppressors that restored oligomycin resistance were found. These suppressing mutations were shown to restore multiple interdomain interfaces (115). In addition to providing a greater understanding of ABC transporter architecture, the continued application of this approach is also expected to reveal the molecular components and mechanisms that govern ER retention and quality control for ABC proteins, which, at this point, are largely unknown.
iMYTH TECHNOLOGY, A POWERFUL TOOL FOR IDENTIFYING ABC INTERACTORS IN YEAST AND POSSIBLY HUMANS: YCF1 AS A TESTING GROUND FOR THE VALIDITY OF iMYTH TECHNOLOGY
Considering the importance of the ABC transporter family in diverse diseases and multidrug resistance, obtaining an understanding of the function and mechanism of action of these proteins is of crucial importance. One of the major steps toward obtaining this understanding is through the elucidation of the complete ABC transporter interactome. The identification of the interaction partners of various ABC transporters will provide valuable information about the specific pathways in which these proteins are involved as well as the elements involved in the regulation and mediation of their function. Constructing such an interactome, however, is particularly challenging since the hydrophobic nature of these multispanning membrane proteins makes them recalcitrant to analysis using conventional interaction assays, particularly those which are amenable to use in a high-throughput format.
A powerful tool traditionally used for performing large-scale in vivo analysis of protein-protein interactions is the YTH system (42, 152). The YTH system exploits the modular nature of eukaryotic transcription factors. A “bait” protein is fused to the DNA-binding domain and a “prey” protein is fused to the activation domain of a transcription factor. Neither portion is functional on its own, but a functional transcription factor can be reconstituted upon physical interactions between the bait and prey, resulting in the activation of a reporter system and typically yielding the expression of β-galactosidase or His3p. Major advantages of this classical YTH system include the ability to rapidly screen for interactions within the natural environment of a cell as well as facile determinations of the DNA encoding any detected interaction partners. Because the assay relies on the reconstitution of a protein-protein interaction inside the nucleus of the cell, however, it requires that the proteins be both soluble and targeted to the nucleus. As such, it is poorly suited for the screening of membrane proteins, which, due to their hydrophobic nature, tend to form insoluble aggregates outside of the membrane environment (150). Thus, a different approach is required for the meaningful analysis of membrane protein interactions.
One such approach is the MYTH system, which has recently been developed into a powerful alternative to the traditional YTH system, retaining all of the latter method's advantages yet being suitable for the analysis of proteins localized to a membrane environment (47, 71, 81, 119, 138, 151, 156) (Fig. 7). The MYTH system is based upon the concept of “split ubiquitin.” Ubiquitin is a highly conserved, 76-amino-acid protein that is covalently linked to the lysine residues of substrate proteins, thereby regulating their function and/or targeting them for degradation (99). Research has shown that ubiquitin can be expressed as two separate N- and C-terminal fragments (termed Nub and Cub, respectively) capable of interacting and reconstituting a quasinative molecule (73). This “pseudoubiquitin” is similar enough to native ubiquitin that it can be recognized by cytosolic deubiquitinating enzymes (DUBs), a class of proteases responsible for deconjugating ubiquitin from modified proteins via the hydrolysis of the amide bond formed between the protein and the C-terminal residue of ubiquitin (73, 99). The spontaneous reconstitution of Nub and Cub can be disrupted by a mutation of the isoleucine 13 residue in the Nub fragment to glycine, forming a fragment referred to as NubG. An interaction between proteins covalently attached to the NubG and Cub fragments, however, is sufficient to allow the reconstitution of the quasinative ubiquitin, thus allowing the system to be adapted for use as a “sensor” of protein-protein interactions (73).
FIG. 7.
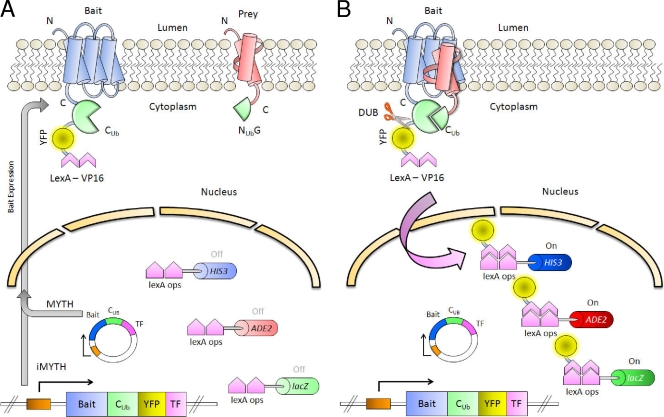
Outline of the iMYTH system. (A) The bait, a membrane protein of interest (in blue), is fused to the C-terminal moiety of ubiquitin (CUb), yellow fluorescent protein (YFP), and transcription factor LexA-VP16. The prey protein (in red) is fused to the N-terminal moiety of ubiquitin (NUbG). Both traditional MYTH (cassette on an exogenous plasmid) and iMYTH (genomically integrated cassette) systems are illustrated. The promoter is shown as an orange box. (B) If the bait and prey interact, the half-ubiquitin moieties reconstitute into a “pseudoubiquitin.” Cytosolic DUBs recognize this “pseudoubiquitin” and cleave its C-terminal end, releasing the transcription factor into the nucleus. The transcription factor binds to the LexA operator sites (lexA ops) and activates the reporter genes HIS3, ADE2, and lacZ.
In the MYTH system, the Cub moiety is fused to a reporter molecule, “TF,” consisting of the Escherichia coli DNA-binding protein LexA fused to the transcriptional activation domain of VP16 from herpes simplex virus. This TF molecule is capable of activating the transcription of reporter genes (typically HIS3, lacZ, and ADE2) under the control of promoters containing LexA-binding sites (Fig. 7) (71, 81, 151). “Baits” are generated by the fusion of Cub-TF to the C terminus of a membrane-anchored protein of interest, which prevents TF from diffusing into the nucleus and activating the transcription of the reporter genes (151). “Preys,” in the form of either specifically selected proteins or entire libraries, are generated by the fusion of the NubG moiety to their N or C termini (71, 81). Yeast cells expressing the bait are transformed with prey constructs and plated onto selective medium. Cells in which particular bait and prey proteins interact reconstitute the “pseudoubiquitin,” leading to DUB recognition and the proteolytic release of TF, which can then enter the nucleus and activate reporter gene transcription, thereby allowing growth on selective medium and subsequent verification using an X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) screen (71, 81, 151). There are currently two major forms of the MYTH system, which are distinguished primarily by whether the bait is expressed ectopically from a plasmid (traditional MYTH system) or endogenously under the control of its native promoter (iMYTH screening). The latter is suitable only for the screening of yeast proteins but provides the advantage of maintaining WT expression levels, which helps reduce the number of false positives, a problem inherent in any YTH assay (71, 81, 119). The iMYTH system is also currently configured with the option of using a Cub-yellow fluorescent protein-TF (CYT) tag instead the traditional Cub-TF tag, allowing easier verification of proper bait localization within the cell (81, 119). A detailed overview of iMYTH and traditional MYTH systems is provided in Fig. 7. A recent study demonstrated the utility of using the iMYTH approach to detect interacting proteins of ABC transporters. In that work, the yeast ABC transporter Ycf1p was screened using an iMYTH approach with endogenous CYT tagging. We found that the use of iMYTH was necessary in order to prevent the strong, nonspecific self-activation of the Ycf1p bait (presumably a result of its mislocalization and misfolding) previously observed when this transporter was overexpressed from a plasmid in a traditional MYTH screen (108, 119). We showed that when expressed from its endogenous promoter, this transporter was present at WT levels and was properly localized to the vacuolar membrane. Additionally, the ability of Ycf1p-CYT to confer cadmium resistance to yeast cells was also measured and was shown to be unaffected by the presence of the C-terminal tag.
A full iMYTH screen using Ycf1p-CYT as a bait and a genomic DNA prey library consisting of genomic DNA fragments (500 to 2,000 bp in length) inserted upstream of an NubG sequence detected a total of six candidate proteins, which interacted specifically with Ycf1p and not with an unrelated vacuolar membrane bait (Vps55-CYT). These proteins included Tus1p (a cytosolic guanine nucleotide exchange factor for Rho1p), Fab1p (a vacuolar membrane kinase involved in vacuolar sorting), Psa1p (a cytosolic GDP-mannose pyrophosphorylase needed for normal cell wall structure), Num1p (a cytosolic protein involved in nuclear migration and mediating microtubular interactions), Kin4p (a cytosolic protein kinase), and YDR115w (unknown function). We then attempted to establish the biological relevance of the observed interactions. Intriguingly, deletion mutants of Tus1p, Num1p, and Fab1p displayed reduced resistance to cadmium and arsenite, providing strong evidence that they work in concert with, or positively regulate, Ycf1p function (119). Additional experiments with Tus1p also showed that it appeared to be involved in the formation of the red color in ade2Δ cells, a phenotype known to be associated with the Ycf1p-mediated uptake of an adenine biosynthetic intermediate into the vacuole (18, 118, 119, 145), as discussed above. Vacuolar uptake experiments using a radiolabeled substrate also revealed that the addition of cytosolic extracts from cells expressing Tus1p stimulated Ycf1p-mediated uptake in a GTP-dependent manner. No such stimulation was observed when cytosolic extracts from tus1Δ cells were used. Purified Tus1p was also observed to enhance Ycf1p-mediated uptake into vacuoles (119). This enhancement was dependent upon the presence of functional Rho1p, a membrane-bound GTPase known to be activated by Tus1p, in cells from which the vacuoles were isolated, indicating that Tus1p exerts its effect on Ycf1p via Rho1p GTPase (119, 139). A physical interaction between Tus1p and Ycf1p was also demonstrated by using immunoprecipitation experiments with the unrelated vacuolar ABC transporter Ybt1p as a noninteracting control. The iMYTH approach was also used with a variety of Tus1p truncation mutants as baits, mapping the Ycf1p interaction site to the C-terminal citron-like domain (119).
It is therefore readily apparent that the iMYTH system represents a powerful tool for the detection of membrane protein interactions and appears to be particularly well suited for the study of ABC transporters. Work using Ycf1p as a bait protein clearly illustrates the robustness of the iMYTH system in detecting interactions of biological relevance as well as its adaptability for use in other in vivo interaction experiments, such as for the determination of specific interaction regions between proteins. Our laboratory is currently in the process of using the iMYTH approach to determine the ABC transporter interactome in the yeast Saccharomyces cerevisiae. Of the 22 membrane-bound ABC proteins in yeast, 19 are predicted to be suitable for iMYTH analysis (the remaining 3 proteins are reportedly localized to the mitochondrial inner membrane, where they are not accessible to cystolic DUBs) (Table 1 and Fig. 5). Currently, we have constructed endogenous, C-terminally-tagged Cub-TF and Cub-CYT baits from all 19 of these transporters in a total of three different strain backgrounds. Localization studies and screening of these baits, using both genomic and cDNA NubG libraries, have recently been completed in our laboratory. Preliminary results have already detected a number of intriguing interactions, and the final, completely constructed interactome promises to provide a host of valuable insights into the function and regulation of ABC transporter proteins as a whole.
YEAST ABCs, AN IMPORTANT ROLE IN ABC TRANSPORTER RESEARCH
The study of the yeast ABC transporters has played a integral part in expanding our current understanding of the cellular function of ABC transporters and the mechanisms by which their function is regulated (expression, posttranslational modification, trafficking, and/or localization). Methodological advances in yeast genetics and biochemistry, such as iMYTH, will keep yeast, as a model organism, at the forefront of all aspects of ABC transporter research, from cellular biology and biochemistry to pharmacology and metabolism. To this end, yeast will continue to be an important tool in aiding us in our understanding of the role of ABC transporters in human health and disease.
In this review we have discussed the current state of ABCC research in Saccharomyces cerevisiae and have described our current understanding of the biochemistry and cellular biology of each member of the ABCC subfamily. We have also described how a new YTH system called iMYTH, which is designed specifically to identify protein interactors for membrane-bound proteins, has resulted in the identification of new functional regulators of the yeast ABCC transporter, Ycf1p. The application of this technology to other members of the ABC transporter family will potentially prove extremely useful in identifying protein interactors that may regulate protein function and/or protein localization. We believe that our work and the work of others using yeast as a tool to determine the role of ABC transporters in cellular metabolism and human health and disease are only in their infancy. It is reasonable to assume that the research carried out on the yeast ABC transporters will continue to make important contributions and will play a major role in directing current and future studies.
Acknowledgments
We thank James Mullally for critical comments on the manuscript. We also thank the many researchers in our laboratories over the years that have contributed to our studies.
This work was supported by grants from the National Institutes of Health to S.M. (grant R01 GM51508) and to C.P. (grant P20 RR020171 COBRE). The Stagljar group is supported by grants from the Canadian Foundation for Innovation, the Canadian Institute for Health Research, the Canadian Cancer Society, the Heart and Stroke Foundation of Canada, and Novartis.
REFERENCES
- 1.Adamis, P. D. B., A. D. Panek, and E. C. A. Eleutherio. 2007. Vacuolar compartmentation of the cadmium-glutathione complex protects Saccharomyces cerevisiae from mutagenesis. Toxicol. Lett. 173:1-7. [DOI] [PubMed] [Google Scholar]
- 2.Aller, S. G., J. Yu, A. Ward, Y. Weng, S. Chittaboina, R. Zhuo, P. M. Harrell, Y. T. Trinh, Q. Zhang, I. L. Urbatsch, and G. Chang. 2009. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323:1718-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambudkar, S. V., S. Dey, C. A. Hrycyna, M. Ramachandra, I. Pastan, and M. M. Gottesman. 1999. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu. Rev. Pharmacol. Toxicol. 39:361-398. [DOI] [PubMed] [Google Scholar]
- 4.Bakos, E., R. Evers, G. Calenda, G. E. Tusnady, G. Szakacs, A. Varadi, and B. Sarkadi. 2000. Characterization of the amino-terminal regions in the human multidrug resistance protein (MRP1). J. Cell Sci. 113(Pt. 24):4451-4461. [DOI] [PubMed] [Google Scholar]
- 5.Ballatori, N., C. L. Hammond, J. B. Cunningham, S. M. Krance, and R. Marchan. 2005. Molecular mechanisms of reduced glutathione transport: role of the MRP/CFTR/ABCC and OATP/SLC21A families of membrane proteins. Toxicol. Appl. Pharmacol. 204:238-255. [DOI] [PubMed] [Google Scholar]
- 6.Balzi, E., and A. Goffeau. 1995. Yeast multidrug resistance: the PDR network. J. Bioenerg. Biomembr. 27:71-76. [DOI] [PubMed] [Google Scholar]
- 7.Balzi, E., M. Wang, S. Leterme, L. Van Dyck, and A. Goffeau. 1994. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 269:2206-2214. [PubMed] [Google Scholar]
- 8.Bandler, P. E., C. J. Westlake, C. E. Grant, S. P. Cole, and R. G. Deeley. 2008. Identification of regions required for apical membrane localization of human multidrug resistance protein 2. Mol. Pharmacol. 74:9-19. [DOI] [PubMed] [Google Scholar]
- 9.Bauer, B. E., H. Wolfger, and K. Kuchler. 1999. Inventory and function of yeast ABC proteins: about sex, stress, pleiotropic drug and heavy metal resistance. Biochim. Biophys. Acta 1461:217-236. [DOI] [PubMed] [Google Scholar]
- 10.Berkower, C., D. Loayza, and S. Michaelis. 1994. Metabolic instability and constitutive endocytosis of STE6, the a-factor transporter of Saccharomyces cerevisiae. Mol. Biol. Cell 5:1185-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berkower, C., and S. Michaelis. 1991. Mutational analysis of the yeast a-factor transporter STE6, a member of the ATP binding cassette (ABC) protein superfamily. EMBO J. 10:3777-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boone, C., H. Bussey, and B. J. Andrews. 2007. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 8:437-449. [DOI] [PubMed] [Google Scholar]
- 13.Borst, P., and R. O. Elferink. 2002. Mammalian ABC transporters in health and disease. Annu. Rev. Biochem. 71:537-592. [DOI] [PubMed] [Google Scholar]
- 14.Borst, P., R. Evers, M. Kool, and J. Wijnholds. 2000. A family of drug transporters: the multidrug resistance-associated proteins. J. Natl. Cancer Inst. 92:1295-1302. [DOI] [PubMed] [Google Scholar]
- 15.Borst, P., R. Evers, M. Kool, and J. Wijnholds. 1999. The multidrug resistance protein family. Biochim. Biophys. Acta 1461:347-357. [DOI] [PubMed] [Google Scholar]
- 16.Cabib, E., T. Drgon, J. Drgonova, R. A. Ford, and R. Kollar. 1997. The yeast cell wall, a dynamic structure engaged in growth and morphogenesis. Biochem. Soc. Trans. 25:200-204. [DOI] [PubMed] [Google Scholar]
- 17.Cabib, E., J. Drgonova, and T. Drgon. 1998. Role of small G proteins in yeast cell polarization and wall biosynthesis. Annu. Rev. Biochem. 67:307-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaudhuri, B., S. Ingavale, and A. K. Bachhawat. 1997. apd1+, a gene required for red pigment formation in ade6 mutants of Schizosaccharomyces pombe, encodes an enzyme required for glutathione biosynthesis: a role for glutathione and a glutathione-conjugate pump. Genetics 145:75-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chi, A., C. Huttenhower, L. Y. Geer, J. J. Coon, J. E. Syka, D. L. Bai, J. Shabanowitz, D. J. Burke, O. G. Troyanskaya, and D. F. Hunt. 2007. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. USA 104:2193-2198. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chloupkova, M., L. S. LeBard, and D. M. Koeller. 2003. MDL1 is a high copy suppressor of ATM1: evidence for a role in resistance to oxidative stress. J. Mol. Biol. 331:155-165. [DOI] [PubMed] [Google Scholar]
- 21.Cole, S. P., G. Bhardwaj, J. H. Gerlach, J. E. Mackie, C. E. Grant, K. C. Almquist, A. J. Stewart, E. U. Kurz, A. M. Duncan, and R. G. Deeley. 1992. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science 258:1650-1654. [DOI] [PubMed] [Google Scholar]
- 22.Cole, S. P., and R. G. Deeley. 2006. Transport of glutathione and glutathione conjugates by MRP1. Trends Pharmacol. Sci. 27:438-446. [DOI] [PubMed] [Google Scholar]
- 23.Cui, Z., D. Hirata, E. Tsuchiya, H. Osada, and T. Miyakawa. 1996. The multidrug resistance-associated protein (MRP) subfamily (Yrs1/Yor1) of Saccharomyces cerevisiae is important for the tolerance to a broad range of organic anions. J. Biol. Chem. 271:14712-14716. [DOI] [PubMed] [Google Scholar]
- 24.Dawson, R. J., and K. P. Locher. 2006. Structure of a bacterial multidrug ABC transporter. Nature 443:180-185. [DOI] [PubMed] [Google Scholar]
- 25.Dean, M. 2005. The genetics of ATP-binding cassette transporters. Methods Enzymol. 400:409-429. [DOI] [PubMed] [Google Scholar]
- 26.Dean, M., A. Rzhetsky, and R. Allikmets. 2001. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 11:1156-1166. [DOI] [PubMed] [Google Scholar]
- 27.Dean, M., and R. Allikmets. 2001. Complete characterization of the human ABC gene family. J. Bioenerg. Biomembr. 33:475-479. [DOI] [PubMed] [Google Scholar]
- 28.Dean, M., and R. Allikmets. 1995. Evolution of ATP-binding cassette transporter genes. Curr. Opin. Genet. Dev. 5:779-785. [DOI] [PubMed] [Google Scholar]
- 29.Dean, M., Y. Hamon, and G. Chimini. 2001. The human ATP-binding cassette (ABC) transporter superfamily. J. Lipid Res. 42:1007-1017. [PubMed] [Google Scholar]
- 30.Decottignies, A., and A. Goffeau. 1997. Complete inventory of the yeast ABC proteins. Nat. Genet. 15:137-145. [DOI] [PubMed] [Google Scholar]
- 31.Decottignies, A., A. M. Grant, J. W. Nichols, H. de Wet, D. B. McIntosh, and A. Goffeau. 1998. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 273:12612-12622. [DOI] [PubMed] [Google Scholar]
- 32.Decottignies, A., G. Owsianik, and M. Ghislain. 1999. Casein kinase I-dependent phosphorylation and stability of the yeast multidrug transporter Pdr5p. J. Biol. Chem. 274:37139-37146. [DOI] [PubMed] [Google Scholar]
- 33.Deeley, R. G., C. Westlake, and S. P. Cole. 2006. Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol. Rev. 86:849-899. [DOI] [PubMed] [Google Scholar]
- 34.Egner, R., and K. Kuchler. 1996. The yeast multidrug transporter Pdr5 of the plasma membrane is ubiquitinated prior to endocytosis and degradation in the vacuole. FEBS Lett. 378:177-181. [DOI] [PubMed] [Google Scholar]
- 35.Epping, E. A., and W. S. Moye-Rowley. 2002. Identification of interdependent signals required for anterograde traffic of the ATP-binding cassette transporter protein Yor1p. J. Biol. Chem. 277:34860-34869. [DOI] [PubMed] [Google Scholar]
- 36.Eraso, P., M. Martinez-Burgos, J. M. Falcon-Perez, F. Portillo, and M. J. Mazon. 2004. Ycf1-dependent cadmium detoxification by yeast requires phosphorylation of residues Ser908 and Thr911. FEBS Lett. 577:322-326. [DOI] [PubMed] [Google Scholar]
- 37.Ernst, R., R. Klemm, L. Schmitt, and K. Kuchler. 2005. Yeast ATP-binding cassette transporters: cellular cleaning pumps. Methods Enzymol. 400:460-484. [DOI] [PubMed] [Google Scholar]
- 38.Ernst, R., P. Kueppers, C. M. Klein, T. Schwarzmueller, K. Kuchler, and L. Schmitt. 2008. A mutation of the H-loop selectively affects rhodamine transport by the yeast multidrug ABC transporter Pdr5. Proc. Natl. Acad. Sci. USA 105:5069-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falcon-Perez, J. M., M. Martinez-Burgos, J. Molano, M. J. Mazon, and P. Eraso. 2001. Domain interactions in the yeast ATP binding cassette transporter Ycf1p: intragenic suppressor analysis of mutations in the nucleotide binding domains. J. Bacteriol. 183:4761-4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falcon-Perez, J. M., M. J. Mazon, J. Molano, and P. Eraso. 1999. Functional domain analysis of the yeast ABC transporter Ycf1p by site-directed mutagenesis. J. Biol. Chem. 274:23584-23590. [DOI] [PubMed] [Google Scholar]
- 41.Fernandez, S. B., Z. Hollo, A. Kern, E. Bakos, P. A. Fischer, P. Borst, and R. Evers. 2002. Role of the N-terminal transmembrane region of the multidrug resistance protein MRP2 in routing to the apical membrane in MDCKII cells. J. Biol. Chem. 277:31048-31055. [DOI] [PubMed] [Google Scholar]
- 42.Fields, S. 2005. High-throughput two-hybrid analysis. The promise and the peril. FEBS J. 272:5391-5399. [DOI] [PubMed] [Google Scholar]
- 43.Fleischer, T. C., C. M. Weaver, K. J. McAfee, J. L. Jennings, and A. J. Link. 2006. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 20:1294-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaskova, D., R. Cadek, R. Chaloupka, V. Vacata, J. Gebel, and K. Sigler. 2002. Monitoring the kinetics and performance of yeast membrane ABC transporters by diS-C3(3) fluorescence. Int. J. Biochem. Cell Biol. 34:931-937. [DOI] [PubMed] [Google Scholar]
- 45.Gavin, A. C., P. Aloy, P. Grandi, R. Krause, M. Boesche, M. Marzioch, C. Rau, L. J. Jensen, S. Bastuck, B. Dumpelfeld, A. Edelmann, M. A. Heurtier, V. Hoffman, C. Hoefert, K. Klein, M. Hudak, A. M. Michon, M. Schelder, M. Schirle, M. Remor, T. Rudi, S. Hooper, A. Bauer, T. Bouwmeester, G. Casari, G. Drewes, G. Neubauer, J. M. Rick, B. Kuster, P. Bork, R. B. Russell, and G. Superti-Furga. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440:631-636. [DOI] [PubMed] [Google Scholar]
- 46.Ghosh, M., J. Shen, and B. P. Rosen. 1999. Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:5001-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gisler, S. M., S. Kittanakom, D. Fuster, V. Wong, M. Bertic, T. Radanovic, R. A. Hall, H. Murer, J. Biber, D. Markovich, O. W. Moe, and I. Stagljar. 2008. Monitoring protein-protein interactions between the mammalian integral membrane transporters and PDZ-interacting partners using a modified split-ubiquitin membrane yeast two-hybrid system. Mol. Cell. Proteomics 7:1362-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274:546, 563-567. [DOI] [PubMed] [Google Scholar]
- 49.Golin, J., Z. N. Kon, C. P. Wu, J. Martello, L. Hanson, S. Supernavage, S. V. Ambudkar, and Z. E. Sauna. 2007. Complete inhibition of the Pdr5p multidrug efflux pump ATPase activity by its transport substrate clotrimazole suggests that GTP as well as ATP may be used as an energy source. Biochemistry 46:13109-13119. [DOI] [PubMed] [Google Scholar]
- 50.Gomes, D. S., L. C. Fragoso, C. J. Riger, A. D. Panek, and E. C. A. Eleutherio. 2002. Regulation of cadmium uptake by Saccharomyces cerevisiae. Biochim. Biophys. Acta 1573:21-25. [DOI] [PubMed] [Google Scholar]
- 51.Gottesman, M. M., and S. V. Ambudkar. 2001. ABC transporters and human disease. J. Bioenerg. Biomembr. 33:453-458. [DOI] [PubMed] [Google Scholar]
- 52.Gottesman, M. M., and V. Ling. 2006. The molecular basis of multidrug resistance in cancer: the early years of P-glycoprotein research. FEBS Lett. 580:998-1009. [DOI] [PubMed] [Google Scholar]
- 53.Grant, C. E., G. Valdimarsson, D. R. Hipfner, K. C. Almquist, S. P. Cole, and R. G. Deeley. 1994. Overexpression of multidrug resistance-associated protein (MRP) increases resistance to natural product drugs. Cancer Res. 54:357-361. [PubMed] [Google Scholar]
- 54.Gravot, A., A. Lieutaud, F. Verret, P. Auroy, A. Vavasseur, and P. Richaud. 2004. AtHMA3, a plant P1B-ATPase, functions as a Cd/Pb transporter in yeast. FEBS Lett. 561:22-28. [DOI] [PubMed] [Google Scholar]
- 55.Grigoras, I., M. Lazard, P. Plateau, and S. Blanquet. 2008. Functional characterization of the Saccharomyces cerevisiae ABC-transporter Yor1p overexpressed in plasma membranes. Biochim. Biophys. Acta 1778:68-78. [DOI] [PubMed] [Google Scholar]
- 56.Gueldry, O., M. Lazard, F. Delort, M. Dauplais, I. Grigoras, S. Blanquet, and P. Plateau. 2003. Ycf1p-dependent Hg(II) detoxification in Saccharomyces cerevisiae. Eur. J. Biochem. 270:2486-2496. [DOI] [PubMed] [Google Scholar]
- 57.Gulshan, K., and W. S. Moye-Rowley. 2007. Multidrug resistance in fungi. Eukaryot. Cell 6:1933-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haimeur, A., G. Conseil, R. G. Deeley, and S. P. Cole. 2004. The MRP-related and BCRP/ABCG2 multidrug resistance proteins: biology, substrate specificity and regulation. Curr. Drug Metab. 5:21-53. [DOI] [PubMed] [Google Scholar]
- 59.Hasenbrink, G., A. Sievernich, L. Wildt, J. Ludwig, and H. Lichtenberg-Frate. 2006. Estrogenic effects of natural and synthetic compounds including tibolone assessed in Saccharomyces cerevisiae expressing the human estrogen alpha and beta receptors. FASEB J. 20:1552-1554. [DOI] [PubMed] [Google Scholar]
- 60.Hettema, E. H., C. W. van Roermund, B. Distel, M. van den Berg, C. Vilela, C. Rodrigues-Pousada, R. J. Wanders, and H. F. Tabak. 1996. The ABC transporter proteins Pat1 and Pat2 are required for import of long-chain fatty acids into peroxisomes of Saccharomyces cerevisiae. EMBO J. 15:3813-3822. [PMC free article] [PubMed] [Google Scholar]
- 61.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 62.Higgins, C. F., and K. J. Linton. 2004. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 11:918-926. [DOI] [PubMed] [Google Scholar]
- 63.Hipfner, D. R., S. D. Gauldie, R. G. Deeley, and S. P. Cole. 1994. Detection of the M(r) 190,000 multidrug resistance protein, MRP, with monoclonal antibodies. Cancer Res. 54:5788-5792. [PubMed] [Google Scholar]
- 64.Hollenstein, K., D. C. Frei, and K. P. Locher. 2007. Structure of an ABC transporter in complex with its binding protein. Nature 446:213-216. [DOI] [PubMed] [Google Scholar]
- 65.Holyoak, C. D., D. Bracey, P. W. Piper, K. Kuchler, and P. J. Coote. 1999. The Saccharomyces cerevisiae weak-acid-inducible ABC transporter Pdr12 transports fluorescein and preservative anions from the cytosol by an energy-dependent mechanism. J. Bacteriol. 181:4644-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Homolya, L., A. Varadi, and B. Sarkadi. 2003. Multidrug resistance-associated proteins: export pumps for conjugates with glutathione, glucuronate or sulfate. Biofactors 17:103-114. [DOI] [PubMed] [Google Scholar]
- 67.Hughes, T. R., M. D. Robinson, N. Mitsakakis, and M. Johnston. 2004. The promise of functional genomics: completing the encyclopedia of a cell. Curr. Opin. Microbiol. 7:546-554. [DOI] [PubMed] [Google Scholar]
- 68.Huyer, G., G. L. Longsworth, D. L. Mason, M. P. Mallampalli, J. M. McCaffery, R. L. Wright, and S. Michaelis. 2004. A striking quality control subcompartment in Saccharomyces cerevisiae: the endoplasmic reticulum-associated compartment. Mol. Biol. Cell 15:908-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huyer, G., W. F. Piluek, Z. Fansler, S. G. Kreft, M. Hochstrasser, J. L. Brodsky, and S. Michaelis. 2004. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 279:38369-38378. [DOI] [PubMed] [Google Scholar]
- 70.Ishikawa, T., Z. S. Li, Y. P. Lu, and P. A. Rea. 1997. The GS-X pump in plant, yeast, and animal cells: structure, function, and gene expression. Biosci. Rep. 17:189-207. [DOI] [PubMed] [Google Scholar]
- 71.Iyer, K., L. Burkle, D. Auerbach, S. Thaminy, M. Dinkel, K. Engels, and I. Stagljar. 2005. Utilizing the split-ubiquitin membrane yeast two-hybrid system to identify protein-protein interactions of integral membrane proteins. Sci. STKE 2005:pl3. [DOI] [PubMed] [Google Scholar]
- 72.Jeffery, E. H. 2007. Detoxification basics. Altern. Ther. Health Med. 13:S96-S97. [PubMed] [Google Scholar]
- 73.Johnsson, N., and A. Varshavsky. 1994. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. USA 91:10340-10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jungwirth, H., and K. Kuchler. 2006. Yeast ABC transporters—a tale of sex, stress, drugs and aging. FEBS Lett. 580:1131-1138. [DOI] [PubMed] [Google Scholar]
- 75.Katzmann, D. J., E. A. Epping, and W. S. Moye-Rowley. 1999. Mutational disruption of plasma membrane trafficking of Saccharomyces cerevisiae Yor1p, a homologue of mammalian multidrug resistance protein. Mol. Cell. Biol. 19:2998-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Katzmann, D. J., T. C. Hallstrom, M. Voet, W. Wysock, J. Golin, G. Volckaert, and W. S. Moye-Rowley. 1995. Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6875-6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kelm, K. B., G. Huyer, J. C. Huang, and S. Michaelis. 2004. The internalization of yeast Ste6p follows an ordered series of events involving phosphorylation, ubiquitination, recognition and endocytosis. Traffic 5:165-180. [DOI] [PubMed] [Google Scholar]
- 78.Kerr, I. D. 2004. Sequence analysis of twin ATP binding cassette proteins involved in translational control, antibiotic resistance, and ribonuclease L inhibition. Biochem. Biophys. Res. Commun. 315:166-173. [DOI] [PubMed] [Google Scholar]
- 79.Ketchum, C. J., W. K. Schmidt, G. V. Rajendrakumar, S. Michaelis, and P. C. Maloney. 2001. The yeast a-factor transporter Ste6p, a member of the ABC superfamily, couples ATP hydrolysis to pheromone export. J. Biol. Chem. 276:29007-29011. [DOI] [PubMed] [Google Scholar]
- 80.Kispal, G., P. Csere, C. Prohl, and R. Lill. 1999. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18:3981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kittanakom, S., M. Chuk, V. Wong, J. Snyder, D. Edmonds, A. Lydakis, Z. Zhang, D. Auerbach, and I. Stagljar. 2009. Analysis of membrane protein complexes using the split-ubiquitin membrane yeast two-hybrid (MYTH) system. Methods Mol. Biol. 548:247-271. [DOI] [PubMed] [Google Scholar]
- 82.Klein, M., B. Burla, and E. Martinoia. 2006. The multidrug resistance-associated protein (MRP/ABCC) subfamily of ATP-binding cassette transporters in plants. FEBS Lett. 580:1112-1122. [DOI] [PubMed] [Google Scholar]
- 83.Klein, M., Y. M. Mamnun, T. Eggmann, C. Schuller, H. Wolfger, E. Martinoia, and K. Kuchler. 2002. The ATP-binding cassette (ABC) transporter Bpt1p mediates vacuolar sequestration of glutathione conjugates in yeast. FEBS Lett. 520:63-67. [DOI] [PubMed] [Google Scholar]
- 84.Kolaczkowska, A., M. Kolaczkowski, A. Goffeau, and W. S. Moye-Rowley. 2008. Compensatory activation of the multidrug transporters Pdr5p, Snq2p, and Yor1p by Pdr1p in Saccharomyces cerevisiae. FEBS Lett. 582:977-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kolaczkowski, M., A. Kolaczowska, J. Luczynski, S. Witek, and A. Goffeau. 1998. In vivo characterization of the drug resistance profile of the major ABC transporters and other components of the yeast pleiotropic drug resistance network. Microb. Drug Resist. 4:143-158. [DOI] [PubMed] [Google Scholar]
- 86.Kolling, R., and C. P. Hollenberg. 1994. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 13:3261-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo, A. Ignatchenko, J. Li, S. Pu, N. Datta, A. P. Tikuisis, T. Punna, J. M. Peregrin-Alvarez, M. Shales, X. Zhang, M. Davey, M. D. Robinson, A. Paccanaro, J. E. Bray, A. Sheung, B. Beattie, D. P. Richards, V. Canadien, A. Lalev, F. Mena, P. Wong, A. Starostine, M. M. Canete, J. Vlasblom, S. Wu, C. Orsi, S. R. Collins, S. Chandran, R. Haw, J. J. Rilstone, K. Gandi, N. J. Thompson, G. Musso, P. St. Onge, S. Ghanny, M. H. Lam, G. Butland, A. M. Altaf-Ul, S. Kanaya, A. Shilatifard, E. O'Shea, J. S. Weissman, C. J. Ingles, T. R. Hughes, J. Parkinson, M. Gerstein, S. J. Wodak, A. Emili, and J. F. Greenblatt. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637-643. [DOI] [PubMed] [Google Scholar]
- 88.Kuchler, K., R. E. Sterne, and J. Thorner. 1989. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 8:3973-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leier, I., G. Jedlitschky, U. Buchholz, M. Center, S. P. Cole, R. G. Deeley, and D. Keppler. 1996. ATP-dependent glutathione disulphide transport mediated by the MRP gene-encoded conjugate export pump. Biochem. J. 314(Pt. 2):433-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leier, I., G. Jedlitschky, U. Buchholz, S. P. Cole, R. G. Deeley, and D. Keppler. 1994. The MRP gene encodes an ATP-dependent export pump for leukotriene C4 and structurally related conjugates. J. Biol. Chem. 269:27807-27810. [PubMed] [Google Scholar]
- 91.Levin, D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li, X., S. A. Gerber, A. D. Rudner, S. A. Beausoleil, W. Haas, J. Villen, J. E. Elias, and S. P. Gygi. 2007. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 6:1190-1197. [DOI] [PubMed] [Google Scholar]
- 93.Li, Z. S., Y. P. Lu, R. G. Zhen, M. Szczypka, D. J. Thiele, and P. A. Rea. 1997. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc. Natl. Acad. Sci. USA 94:42-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li, Z. S., M. Szczypka, Y. P. Lu, D. J. Thiele, and P. A. Rea. 1996. The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. J. Biol. Chem. 271:6509-6517. [DOI] [PubMed] [Google Scholar]
- 95.Lichtenberg-Frate, H., M. Schmitt, G. Gellert, and J. Ludwig. 2003. A yeast-based method for the detection of cyto and genotoxicity. Toxicol. In Vitro 17:709-716. [DOI] [PubMed] [Google Scholar]
- 96.Lill, R., and G. Kispal. 2001. Mitochondrial ABC transporters. Res. Microbiol. 152:331-340. [DOI] [PubMed] [Google Scholar]
- 97.Linton, K. J. 2007. Structure and function of ABC transporters. Physiology (Bethesda) 22:122-130. [DOI] [PubMed] [Google Scholar]
- 98.Locher, K. P. 2009. Structure and mechanism of ATP-binding cassette transporters. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:239-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Love, K. R., A. Catic, C. Schlieker, and H. L. Ploegh. 2007. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat. Chem. Biol. 3:697-705. [DOI] [PubMed] [Google Scholar]
- 100.Mahe, Y., Y. Lemoine, and K. Kuchler. 1996. The ATP binding cassette transporters Pdr5 and Snq2 of Saccharomyces cerevisiae can mediate transport of steroids in vivo. J. Biol. Chem. 271:25167-25172. [DOI] [PubMed] [Google Scholar]
- 101.Mason, D. L. 2002. Functional analysis of MRP transporters in Saccharomyces cerevisiae. Ph.D. thesis. Johns Hopkins University, Baltimore, MD.
- 102.Mason, D. L., M. P. Mallampalli, G. Huyer, and S. Michaelis. 2003. A region within a lumenal loop of Saccharomyces cerevisiae Ycf1p directs proteolytic processing and substrate specificity. Eukaryot. Cell 2:588-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mason, D. L., and S. Michaelis. 2002. Requirement of the N-terminal extension for vacuolar trafficking and transport activity of yeast Ycf1p, an ATP-binding cassette transporter. Mol. Biol. Cell 13:4443-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McGrath, J. P., and A. Varshavsky. 1989. The yeast STE6 gene encodes a homologue of the mammalian multidrug resistance P-glycoprotein. Nature 340:400-404. [DOI] [PubMed] [Google Scholar]
- 105.Metzger, M. B. 2009. Protein quality control in the endoplasmic reticulum and cytosol. Ph.D. thesis. Johns Hopkins University, Baltimore, MD.
- 106.Michaelis, S. 1993. STE6, the yeast a-factor transporter. Semin. Cell Biol. 4:17-27. [DOI] [PubMed] [Google Scholar]
- 107.Michaelis, S., and C. Berkower. 1995. Sequence comparison of yeast ATP-binding cassette proteins. Cold Spring Harb. Symp. Quant. Biol. 60:291-307. [DOI] [PubMed] [Google Scholar]
- 108.Miller, J. P., R. S. Lo, A. Ben-Hur, C. Desmarais, I. Stagljar, W. S. Noble, and S. Fields. 2005. Large-scale identification of yeast integral membrane protein interactions. Proc. Natl. Acad. Sci. USA 102:12123-12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miyahara, K., M. Mizunuma, D. Hirata, E. Tsuchiya, and T. Miyakawa. 1996. The involvement of the Saccharomyces cerevisiae multidrug resistance transporters Pdr5p and Snq2p in cation resistance. FEBS Lett. 399:317-320. [DOI] [PubMed] [Google Scholar]
- 110.Momany, M. 2002. Polarity in filamentous fungi: establishment, maintenance and new axes. Curr. Opin. Microbiol. 5:580-585. [DOI] [PubMed] [Google Scholar]
- 111.Nagy, Z., C. Montigny, P. Leverrier, S. Yeh, A. Goffeau, M. Garrigos, and P. Falson. 2006. Role of the yeast ABC transporter Yor1p in cadmium detoxification. Biochimie 88:1665-1671. [DOI] [PubMed] [Google Scholar]
- 112.Nakatsukasa, K., G. Huyer, S. Michaelis, and J. L. Brodsky. 2008. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132:101-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ortiz, D. F., M. V. St. Pierre, A. Abdulmessih, and I. M. Arias. 1997. A yeast ATP-binding cassette-type protein mediating ATP-dependent bile acid transport. J. Biol. Chem. 272:15358-15365. [DOI] [PubMed] [Google Scholar]
- 114.Oskouian, B., and J. D. Saba. 1999. YAP1 confers resistance to the fatty acid synthase inhibitor cerulenin through the transporter Flr1p in Saccharomyces cerevisiae. Mol. Gen. Genet. 261:346-353. [DOI] [PubMed] [Google Scholar]
- 115.Pagant, S., E. Y. Brovman, J. J. Halliday, and E. A. Miller. 2008. Mapping of interdomain interfaces required for the functional architecture of Yor1p, a eukaryotic ATP-binding cassette (ABC) transporter. J. Biol. Chem. 283:26444-26451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pagant, S., L. Kung, M. Dorrington, M. C. Lee, and E. A. Miller. 2007. Inhibiting endoplasmic reticulum (ER)-associated degradation of misfolded Yor1p does not permit ER export despite the presence of a diacidic sorting signal. Mol. Biol. Cell 18:3398-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pascolo, L., S. Petrovic, F. Cupelli, C. V. Bruschi, P. L. Anelli, V. Lorusso, M. Visigalli, F. Uggeri, and C. Tiribelli. 2001. ABC protein transport of MRI contrast agents in canalicular rat liver plasma vesicles and yeast vacuoles. Biochem. Biophys. Res. Commun. 282:60-66. [DOI] [PubMed] [Google Scholar]
- 118.Paumi, C. M., M. Chuk, I. Chevelev, I. Stagljar, and S. Michaelis. 2008. Negative regulation of the yeast ABC transporter Ycf1p by phosphorylation within its N-terminal extension. J. Biol. Chem. 283:27079-27088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Paumi, C. M., J. Menendez, A. Arnoldo, K. Engels, K. R. Iyer, S. Thaminy, O. Georgiev, Y. Barral, S. Michaelis, and I. Stagljar. 2007. Mapping protein-protein interactions for the yeast ABC transporter Ycf1p by integrated split-ubiquitin membrane yeast two-hybrid analysis. Mol. Cell 26:15-25. [DOI] [PubMed] [Google Scholar]
- 120.Petrovic, S., L. Pascolo, R. Gallo, F. Cupelli, J. D. Ostrow, A. Goffeau, C. Tiribelli, and C. V. Bruschi. 2000. The products of YCF1 and YLL015w (BPT1) cooperate for the ATP-dependent vacuolar transport of unconjugated bilirubin in Saccharomyces cerevisiae. Yeast 16:561-571. [DOI] [PubMed] [Google Scholar]
- 121.Piper, P., Y. Mahe, S. Thompson, R. Pandjaitan, C. Holyoak, R. Egner, M. Muhlbauer, P. Coote, and K. Kuchler. 1998. The pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 17:4257-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Preveral, S., E. Ansoborlo, S. Mari, A. Vavasseur, and C. Forestier. 2006. Metal(loid)s and radionuclides cytotoxicity in Saccharomyces cerevisiae. Role of YCF1, glutathione and effect of buthionine sulfoximine. Biochimie 88:1651-1663. [DOI] [PubMed] [Google Scholar]
- 123.Qian, Y. M., W. Qiu, M. Gao, C. J. Westlake, S. P. Cole, and R. G. Deeley. 2001. Characterization of binding of leukotriene C4 by human multidrug resistance protein 1: evidence of differential interactions with NH2- and COOH-proximal halves of the protein. J. Biol. Chem. 276:38636-38644. [DOI] [PubMed] [Google Scholar]
- 124.Rea, P. A. 1999. MRP subfamily ABC transporters from plants and yeast. J. Exp. Bot. 50:895-913. [Google Scholar]
- 125.Rea, P. A. 2007. Plant ATP-binding cassette transporters. Annu. Rev. Plant Biol. 58:347-375. [DOI] [PubMed] [Google Scholar]
- 126.Rebbeor, J. F., G. C. Connolly, and N. Ballatori. 2002. Inhibition of Mrp2- and Ycf1p-mediated transport by reducing agents: evidence for GSH transport on rat Mrp2. Biochim. Biophys. Acta 1559:171-178. [DOI] [PubMed] [Google Scholar]
- 127.Rebbeor, J. F., G. C. Connolly, M. E. Dumont, and N. Ballatori. 1998. ATP-dependent transport of reduced glutathione in yeast secretory vesicles. Biochem. J. 334(Pt. 3):723-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rebbeor, J. F., G. C. Connolly, M. E. Dumont, and N. Ballatori. 1998. ATP-dependent transport of reduced glutathione on YCF1, the yeast orthologue of mammalian multidrug resistance associated proteins. J. Biol. Chem. 273:33449-33454. [DOI] [PubMed] [Google Scholar]
- 129.Reinders, J., and A. Sickmann. 2007. Proteomics of yeast mitochondria. Methods Mol. Biol. 372:543-557. [DOI] [PubMed] [Google Scholar]
- 130.Reinders, J., R. P. Zahedi, N. Pfanner, C. Meisinger, and A. Sickmann. 2006. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 5:1543-1554. [DOI] [PubMed] [Google Scholar]
- 131.Ren, X.-Q., T. Furukawa, Z.-S. Chen, H. Okumura, S. Aoki, T. Sumizawa, A. Tani, M. Komatsu, X.-D. Mei, and S.-I. Akiyama. 2000. Functional comparison between YCF1 and MRP1 expressed in Sf21 insect cells. Biochem. Biophys. Res. Commun. 270:608-615. [DOI] [PubMed] [Google Scholar]
- 132.Rockwell, N. C., H. Wolfger, K. Kuchler, and J. Thorner. 2009. ABC transporter Pdr10 regulates the membrane microenvironment of Pdr12 in Saccharomyces cerevisiae. J. Membr. Biol. 229:27-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rodrigues-Pousada, C. A., T. Nevitt, R. Menezes, D. Azevedo, J. Pereira, and C. Amaral. 2004. Yeast activator proteins and stress response: an overview. FEBS Lett. 567:80-85. [DOI] [PubMed] [Google Scholar]
- 134.Rogers, B., A. Decottignies, M. Kolaczkowski, E. Carvajal, E. Balzi, and A. Goffeau. 2001. The pleitropic drug ABC transporters from Saccharomyces cerevisiae. J. Mol. Microbiol. Biotechnol. 3:207-214. [PubMed] [Google Scholar]
- 135.Rosen, B. P. 1999. Families of arsenic transporters. Trends Microbiol. 7:207-212. [DOI] [PubMed] [Google Scholar]
- 136.Rosen, B. P. 2002. Transport and detoxification systems for transition metals, heavy metals and metalloids in eukaryotic and prokaryotic microbes. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 133:689-693. [DOI] [PubMed] [Google Scholar]
- 137.Sauna, Z. E., and S. V. Ambudkar. 2007. About a switch: how P-glycoprotein (ABCB1) harnesses the energy of ATP binding and hydrolysis to do mechanical work. Mol. Cancer Ther. 6:13-23. [DOI] [PubMed] [Google Scholar]
- 138.Scheper, W., S. Thaminy, S. Kais, I. Stagljar, and K. Romisch. 2003. Coordination of N-glycosylation and protein translocation across the endoplasmic reticulum membrane by Sss1 protein. J. Biol. Chem. 278:37998-38003. [DOI] [PubMed] [Google Scholar]
- 139.Schmelzle, T., S. B. Helliwell, and M. N. Hall. 2002. Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol. Cell. Biol. 22:1329-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schuldiner, M., S. R. Collins, N. J. Thompson, V. Denic, A. Bhamidipati, T. Punna, J. Ihmels, B. Andrews, C. Boone, J. F. Greenblatt, J. S. Weissman, and N. J. Krogan. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123:507-519. [DOI] [PubMed] [Google Scholar]
- 141.Servos, J., E. Haase, and M. Brendel. 1993. Gene SNQ2 of Saccharomyces cerevisiae, which confers resistance to 4-nitroquinoline-N-oxide and other chemicals, encodes a 169 kDa protein homologous to ATP-dependent permeases. Mol. Gen. Genet. 236:214-218. [DOI] [PubMed] [Google Scholar]
- 142.Shani, N., and D. Valle. 1996. A Saccharomyces cerevisiae homolog of the human adrenoleukodystrophy transporter is a heterodimer of two half ATP-binding cassette transporters. Proc. Natl. Acad. Sci. USA 93:11901-11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shani, N., P. A. Watkins, and D. Valle. 1995. PXA1, a possible Saccharomyces cerevisiae ortholog of the human adrenoleukodystrophy gene. Proc. Natl. Acad. Sci. USA 92:6012-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sharma, K. G., R. Kaur, and A. K. Bachhawat. 2003. The glutathione-mediated detoxification pathway in yeast: an analysis using the red pigment that accumulates in certain adenine biosynthetic mutants of yeasts reveals the involvement of novel genes. Arch. Microbiol. 180:108-117. [DOI] [PubMed] [Google Scholar]
- 145.Sharma, K. G., D. L. Mason, G. Liu, P. A. Rea, A. K. Bachhawat, and S. Michaelis. 2002. Localization, regulation, and substrate transport properties of Bpt1p, a Saccharomyces cerevisiae MRP-type ABC transporter. Eukaryot. Cell 1:391-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shim, D., D.-Y. Kim, J. Park, J.-Y. Jin, W.-Y. Song, Y.-I. Choi, E.-W. Noh, E. Martinoia, and Y. Lee. 2008. Heavy metal resistance of poplar plants transformed with an MRP-type ABC transporter of budding yeast, p. 154. Abstr. 2nd FEBS Meet. ATP-Binding Cassette Proteins, Innsbruck, Austria.
- 147.Sipos, G., and K. Kuchler. 2006. Fungal ATP-binding cassette (ABC) transporters in drug resistance & detoxification. Curr. Drug Targets 7:471-481. [DOI] [PubMed] [Google Scholar]
- 148.Smolka, M. B., C. P. Albuquerque, S. H. Chen, and H. Zhou. 2007. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. USA 104:10364-10369. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Song, W. Y., E. J. Sohn, E. Martinoia, Y. J. Lee, Y. Y. Yang, M. Jasinski, C. Forestier, I. Hwang, and Y. Lee. 2003. Engineering tolerance and accumulation of lead and cadmium in transgenic plants. Nat. Biotechnol. 21:914-919. [DOI] [PubMed] [Google Scholar]
- 150.Stagljar, I., and S. Fields. 2002. Analysis of membrane protein interactions using yeast-based technologies. Trends Biochem. Sci. 27:559-563. [DOI] [PubMed] [Google Scholar]
- 151.Stagljar, I., C. Korostensky, N. Johnsson, and S. te Heesen. 1998. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. USA 95:5187-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Suter, B., D. Auerbach, and I. Stagljar. 2006. Yeast-based functional genomics and proteomics technologies: the first 15 years and beyond. BioTechniques 40:625-644. [DOI] [PubMed] [Google Scholar]
- 153.Szakacs, G., J. K. Paterson, J. A. Ludwig, C. Booth-Genthe, and M. M. Gottesman. 2006. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 5:219-234. [DOI] [PubMed] [Google Scholar]
- 154.Szczypka, M. S., J. A. Wemmie, W. S. Moye-Rowley, and D. J. Thiele. 1994. A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J. Biol. Chem. 269:22853-22857. [PubMed] [Google Scholar]
- 155.Taglicht, D., and S. Michaelis. 1998. Saccharomyces cerevisiae ABC proteins and their relevance to human health and disease. Methods Enzymol. 292:130-162. [DOI] [PubMed] [Google Scholar]
- 156.Thaminy, S., D. Auerbach, A. Arnoldo, and I. Stagljar. 2003. Identification of novel ErbB3-interacting factors using the split-ubiquitin membrane yeast two-hybrid system. Genome Res. 13:1744-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tommasini, R., R. Evers, E. Vogt, C. Mornet, G. J. Zaman, A. H. Schinkel, P. Borst, and E. Martinoia. 1996. The human multidrug resistance-associated protein functionally complements the yeast cadmium resistance factor 1. Proc. Natl. Acad. Sci. USA 93:6743-6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tommasini, R., E. Vogt, M. Fromenteau, S. Hortensteiner, P. Matile, N. Amrhein, and E. Martinoia. 1998. An ABC-transporter of Arabidopsis thaliana has both glutathione-conjugate and chlorophyll catabolite transport activity. Plant J. 13:773-780. [DOI] [PubMed] [Google Scholar]
- 159.Tong, Y.-P., R. Kneer, and Y.-G. Zhu. 2004. Vacuolar compartmentalization: a second-generation approach to engineering plants for phytoremediation. Trends Plant Sci. 9:7-9. [DOI] [PubMed] [Google Scholar]
- 160.Toyoda, Y., Y. Hagiya, T. Adachi, K. Hoshijima, M. T. Kuo, and T. Ishikawa. 2008. MRP class of human ATP binding cassette (ABC) transporters: historical background and new research directions. Xenobiotica 38:833-862. [DOI] [PubMed] [Google Scholar]
- 161.Ververidis, P., F. Davrazou, G. Diallinas, D. Georgakopoulos, A. K. Kanellis, and N. Panopoulos. 2001. A novel putative reductase (Cpd1p) and the multidrug exporter Snq2p are involved in resistance to cercosporin and other singlet oxygen-generating photosensitizers in Saccharomyces cerevisiae. Curr. Genet. 39:127-136. [DOI] [PubMed] [Google Scholar]
- 162.Wang, J., F. Sun, D. W. Zhang, Y. Ma, F. Xu, J. D. Belani, J. C. Cohen, H. H. Hobbs, and X. S. Xie. 2006. Sterol transfer by ABCG5 and ABCG8: in vitro assay and reconstitution. J. Biol. Chem. 281:27894-27904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wemmie, J. A., and W. S. Moye-Rowley. 1997. Mutational analysis of the Saccharomyces cerevisiae ATP-binding cassette transporter protein Ycf1p. Mol. Microbiol. 25:683-694. [DOI] [PubMed] [Google Scholar]
- 164.Wemmie, J. A., M. S. Szczypka, D. J. Thiele, and W. S. Moye-Rowley. 1994. Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J. Biol. Chem. 269:32592-32597. [PubMed] [Google Scholar]
- 165.Westlake, C. J., S. P. Cole, and R. G. Deeley. 2005. Role of the NH2-terminal membrane spanning domain of multidrug resistance protein 1/ABCC1 in protein processing and trafficking. Mol. Biol. Cell 16:2483-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Westlake, C. J., Y. M. Qian, M. Gao, M. Vasa, S. P. Cole, and R. G. Deeley. 2003. Identification of the structural and functional boundaries of the multidrug resistance protein 1 cytoplasmic loop 3. Biochemistry 42:14099-14113. [DOI] [PubMed] [Google Scholar]
- 167.Wilcox, L. J., D. A. Balderes, B. Wharton, A. H. Tinkelenberg, G. Rao, and S. L. Sturley. 2002. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 277:32466-32472. [DOI] [PubMed] [Google Scholar]
- 168.Wolfger, H., Y. Mahe, A. Parle-McDermott, A. Delahodde, and K. Kuchler. 1997. The yeast ATP binding cassette (ABC) protein genes PDR10 and PDR15 are novel targets for the Pdr1 and Pdr3 transcriptional regulators. FEBS Lett. 418:269-274. [DOI] [PubMed] [Google Scholar]
- 169.Wolfger, H., Y. M. Mamnun, and K. Kuchler. 2004. The yeast Pdr15p ATP-binding cassette (ABC) protein is a general stress response factor implicated in cellular detoxification. J. Biol. Chem. 279:11593-11599. [DOI] [PubMed] [Google Scholar]
- 170.Xu, C., C. Y. Li, and A. N. Kong. 2005. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch. Pharm. Res. 28:249-268. [DOI] [PubMed] [Google Scholar]
- 171.Yoshida, S., K. Kono, D. M. Lowery, S. Bartolini, M. B. Yaffe, Y. Ohya, and D. Pellman. 2006. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science 313:108-111. [DOI] [PubMed] [Google Scholar]
- 172.Young, L., K. Leonhard, T. Tatsuta, J. Trowsdale, and T. Langer. 2001. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science 291:2135-2138. [DOI] [PubMed] [Google Scholar]
- 173.Zhang, J. Y., Y. Wang, and C. Prakash. 2006. Xenobiotic-metabolizing enzymes in human lung. Curr. Drug Metab. 7:939-948. [DOI] [PubMed] [Google Scholar]
- 174.Zhao, Z., L. L. Fang, R. Johnsen, and D. L. Baillie. 2004. ATP-binding cassette protein E is involved in gene transcription and translation in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 323:104-111. [DOI] [PubMed] [Google Scholar]
- 175.Zhao, Z., J. H. Thomas, N. Chen, J. A. Sheps, and D. L. Baillie. 2007. Comparative genomics and adaptive selection of the ATP-binding-cassette gene family in Caenorhabditis species. Genetics 175:1407-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]