Abstract
The Escherichia coli chromosome encodes seven demonstrated type 2 toxin-antitoxin (TA) systems: cassettes of two or three cotranscribed genes, one encoding a stable toxin protein that can cause cell stasis or death, another encoding a labile antitoxin protein, and sometimes a third regulatory protein. We demonstrate that the yafNO genes constitute an additional chromosomal type 2 TA system that is upregulated during the SOS DNA damage response. The yafNOP genes are part of the dinB operon, of which dinB underlies stress-induced mutagenesis mechanisms. yafN was identified as a putative antitoxin by homology to known antitoxins, implicating yafO (and/or yafP) as a putative toxin. Using phage-mediated cotransduction assays for linkage disruption, we show first that yafN is an essential gene and second that it is essential only when yafO is present. Third, yafP is not a necessary part of either the toxin or the antitoxin. Fourth, although DinB is required, the yafNOP genes are not required for stress-induced mutagenesis in the Escherichia coli Lac assay. These results imply that yafN encodes an antitoxin that protects cells against a yafO-encoded toxin and show a protein-based TA system upregulated by the SOS response.
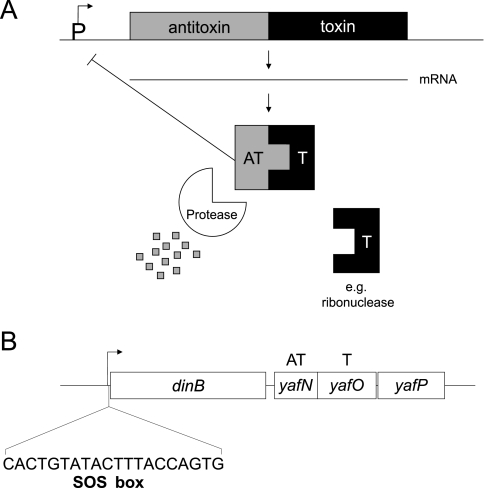
Toxin-antitoxin (TA) systems are modules in bacterial genomes that can cause growth arrest and/or programmed cell death in cells harboring them (1, 18, 19, 54). Type 1 TA systems consist of an RNA antitoxin and a protein toxin, in which the RNA antitoxin inhibits translation of the toxin mRNA. Type 2 TA systems typically consist of two genes in an operon, transcriptionally and translationally coupled, in which, usually, the upstream gene encodes a labile antitoxin protein and the downstream gene encodes a stable toxin protein (Fig. 1A). Continuous transcription of the operon, and thus continuous transcription of the antitoxin, ensures protection from the effects of the toxin. Interruption in transcription of the operon tips the balance in favor of the toxin because antitoxin is no longer made and is rapidly degraded.
FIG. 1.
Type 2 TA system organization and the dinB operon. (A) Type 2 TA system organization. Type 2 TA systems are usually two gene operons in which the antitoxin (upstream) and toxin (downstream) genes are cotranscribed/translated. The antitoxin protein is degraded more rapidly than the toxin, such that the cell requires continuous transcription/translation to avoid stasis/death induced by the toxin. (B) The E. coli dinB operon. The yafNOP genes, encoding a putative TA system, lie downstream of dinB in the operon. The operon is upregulated by the SOS DNA damage response (SOS box shown). yafN, encoding the putative antitoxin, is 297 bp; yafO, encoding the putative toxin, is 399 bp; and yafP, of unknown function, is 453 bp in length. Some strains used in this study (derivatives of SMR4562 or FC40) contain an additional copy of the dinB operon via a duplication of the operon in the F′ episome. T, toxin; AT, antitoxin.
TA systems were originally identified on plasmids as “plasmid-addiction modules” (34) that maintain the plasmid in the host cell. Failure of cells to inherit the plasmid results in rapid loss of the labile antitoxin, unmasking of the stable preexisting toxin, and death of the cell in a process known as postsegregational killing (13, 38).
More recently, TA systems have been discovered in the Escherichia coli chromosome, raising the question of their function there (1, 19). Proposed functions for chromosomal TA systems include roles in nutritional stress response (8), protection from phages (27), formation of “persister” cells that resist antibiotics (32), selfish genetic elements (38, 54), and antiaddiction modules that allow bacteria to resist plasmid addiction (54). Several type 2 chromosomal TA systems have been demonstrated in E. coli, including relBE, mazEF, dinJ-yafQ, prlF-yhaV, yefM-yoeB, chpBI-chpBK, and hipAB (1, 3, 19, 20, 39, 45, 50). Of these, relBE and mazEF exert their toxic effects during controlled responses such as during amino acid starvation (8, 28). Additionally, mazF kills cells in response to DNA-damaging and oxidative stresses (28). One type 2 (dinJ-yafQ) (45) and three characterized type 1 (hok, symER, and tisAB-istr1) (31, 46, 55) TA systems were previously reported to be controlled by the SOS response to DNA damage. However, dinJ-yafQ appears not to be SOS regulated. Although a consensus LexA binding site was identified upstream of the dinJ-yafQ operon (35) and LexA bound to this site in one study (56) but not in another (14), LexA does not regulate this operon in vivo upon exposure to mitomycin C (14) or UV light (11). The role of stress responses in regulating TA systems is an interesting problem. Here we provide evidence for the first E. coli chromosomal type 2 TA system under SOS response control, yafNO.
yafNOP was originally identified as a putative TA system by homology of yafN to known antitoxins, implicating yafO (and/or yafP) as a putative toxin(s) (19). yafNOP lies downstream of dinB in the dinB operon (43) (Fig. 1B), which is upregulated midway through the SOS DNA damage response (11). DinB is a central player in stress-induced mutagenesis mechanisms in E. coli (9, 17, 42), Salmonella enterica (48), Pseudomonas putida (53), and Bacillus subtilis (52). The proximity of yafNOP to dinB piqued our interest in these genes. Previously, Brown and Shaw demonstrated in classical TA system dissection that when yafO was overexpressed growth inhibition occurred (5). No additional work regarding either yafN or yafP was conducted (5). In contrast to this and to the notion that yafNOP was a TA system was the published yafN deletion strain of the Keio E. coli knockout collection, which should have been inviable if yafN were the antitoxin (2). Given these conflicting results, we reinvestigated yafNOP. Here, we show that yafN and yafO (not yafP) constitute a TA system and explore its possible function in the cell.
MATERIALS AND METHODS
Media, antibiotics, and growth conditions.
The media used included Luria-Bertani-Herskowitz (LBH; 1% tryptone, 0.5% NaCl, 0.5% yeast extract, 2 μg/ml thymine), tryptone broth (TB; 1% tryptone, 0.5% NaCl), and BBL plates (0.5% NaCl, 1% BBL Trypticase peptone solidified with 1.5% agar). K-glucose consists of 2× M9 (44) with the following additions: 7.5% Casamino Acids, 5 mM MgSO4, 0.05% NaCl, 0.2% glucose, 10 μg/ml vitamin B1, 3 μM FeCl3, and 50 μM CaCl2. Plating culture broth is TB with the following additions: 0.2% maltose, 5 mM MgSO4, 10 μg/ml thymine, and 10 μg/ml vitamin B1. Carbon sources glucose, maltose, glycerol, and lactose were all used at 0.1% unless otherwise stated. The antibiotics used included tetracycline (Tet; 10 μg/ml, 3.33 μg/ml when sodium citrate [CIT] is present), kanamycin (Kan; 30 μg/ml), chloramphenicol (25 μg/ml), bleomycin (various concentrations, specified in the text), mitomycin C (various concentrations, specified in the text), and rifampin (100 μg/ml). 5-Bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal; 40 μg/ml) was also used. The buffers used included TM (Tris buffer + MgSO4, pH 7.5) and M9 (44). Bacteria were grown at 32°C or 37°C, as stated for each experiment.
Strains and new alleles.
E. coli K-12 strains and plasmids used in this study are listed in Table 1. New deletion alleles created for this study are shown in Table 2, and the PCR primers used to construct the new deletions by short-homology recombination methods (12) are shown in Table 3. Phage P1-mediated transductions were performed as described previously (44). To construct λ lysogens, stationary-phase TB cultures of nonlysogens were diluted 200× into plating culture broth, shaken for 6 hours at 32°C, pelleted, and resuspended in TM at a 1:4 dilution. Phage were added at a multiplicity of infection of 10 per cell, allowed to sit at room temperature for 10 minutes, diluted 10× with TB, shaken for 2 hours at 32°C, and then plated onto MacConkey-maltose plates seeded with 108 to 109 λcI and 108 to 109 λcIh80 phage to kill nonlysogens, and red (maltose-positive) colonies were picked, purified, and verified. λcIh80 adsorbs via an alternate receptor and therefore kills nonlysogens with mutated maltose uptake receptors, which are resistant to wild-type λ adsorption.
TABLE 1.
E. coli strains, plasmids, and lambda phage used in this studya
| Strain, phage, or plasmid | Relevant genotype | Reference(s) or source |
|---|---|---|
| Strains | ||
| BT340 | DH5α(pCP20) | 7 (via CGSC) |
| BW25113(pKD46) | pKD46 | 12 (via CGSC) |
| BW25141(pKD3) | pKD3 | 12 (via CGSC) |
| BW25141(pKD13) | pKD13 | 12 (via CGSC) |
| CAG18436 | MG1655 zae-502::Tn10 | 51 |
| DH5α | endA1 hsdR17 (rK− mK+) supE44 thi-1 λ−recA1 gyrA96 relA1 deoR Δ(lacZYA-argF)U169 φ80dlacZΔM15 | 21 (via W. Bridger [Edmonton, Alberta, Canada]) |
| FC36 | Δ(lac-proAB)XIIIthi ara Rifr | 6 |
| FC40 | Δ(lac-proAB)XIIIthi ara Rifr [F′ proAB+lacI33ΩlacZ] | 6 |
| JW0222 | ΔyafN::FRTKanFRT | 2 |
| MG1655 | Wild type | 4 |
| N2731 | recG258::Tn10 mini-Kan | 36 (via R. Kolodner [San Diego, CA]) |
| SMR4562 | Genotype same as that of FC40, independent construction | 41 |
| SMR5833 | SMR4562(pKD46) | 43 |
| SMR5889 | SMR4562 ΔdinB50::FRT [F′ ΔdinB50::FRT] | 43 |
| SMR6068 | SMR4562 ΔyafN11::FRTKanFRT [F′ yaf+] | SMR5833 × DNA of ΔyafN::FRTKanFRT amplified from pKD13 [from BW25141(pKD13)], location screened by mating |
| SMR6074 | SMR4562 [F′ ΔyafO14::FRTKanFRT] | SMR5833 × DNA of ΔyafO::FRTKanFRT amplified from pKD13 [from BW25141(pKD13)], location screened by mating |
| SMR6076 | FC36 ΔyafO14::FRTKanFRT | FC36 × P1(SMR6074) |
| SMR6080 | SMR4562 ΔyafP18::FRTKanFRT [F′ yaf+] | SMR5833 × DNA of ΔyafP::FRTKanFRT amplified from pKD13 [from BW25141(pKD13)], location screened by mating |
| SMR6082 | FC36 ΔyafP18::FRTKanFRT | FC36 × P1(SMR6080) |
| SMR6221 | FC36 ΔyafP20::FRT | SMR6082 × pCP20 (from BT340) |
| SMR6233 | MG1655(pKD46) | MG1655 × pKD46 [from BW25113(pKD46)] |
| SMR6353 | MG1655 Δ(yafN-yafP)776::FRTKanFRT | SMR6233 × DNA of Δ(yafN-yafP)::FRTKanFRT from pKD13 [from BW25141(pKD13)] |
| SMR6669 | MG1655 Δattλ::PsulAΩgfp-mut2 | 26, 40 |
| SMR7491 | SMR4562 Δ(yafN-yafP)602 [F′ Δ(yafN-yafP)602] | 43 |
| SMR10285 | SMR4562 Δ(yafN-yafO)779::FRTKanFRT [F′ yaf+] | SMR5833 × DNA of Δ(yafN-yafO)::FRTKanFRT amplified from pKD13 [from BW25141(pKD13)] |
| SMR10287 | FC36 Δ(yafN-yafO)779::FRTKanFRT | FC36 × P1(SMR10285) |
| SMR10291 | SMR4562 ΔyafN11::FRTKanFRT zae-502::Tn10 [F′ yaf+] | SMR6068 × P1(CAG18436) |
| SMR10483 | FC36 Δ(yafN-yafO)779::FRTKanFRT zae-502::Tn10 | SMR10287 × P1(CAG18436) |
| SMR10943 | FC36(pBAD24) | FC36 × pBAD24 (from ATCC 87399) |
| SMR10946 | DH5α(pLS1) | DH5α × pLS1 (initial plasmid construction) |
| SMR10948 | FC36(pLS1) | FC36 × pLS1 (from SMR10946) |
| SMR10953 | SMR4562 ΔyafP18::FRTKanFRT zae-502::Tn10 [F′ yaf+] | SMR6080 × P1(CAG18436) |
| SMR10988 | MG1655 Δattλ::PsulAΩgfp-mut2 Δ(yafN-yafP)776::FRTKanFRT | SMR6669 × P1(SMR6353) |
| SMR10990 | MG1655 recG258::Tn10 mini-Kan | MG1655 × P1(N2731) |
| SMR10995 | SMR4562 ΔyafN11::FRTKanFRT [F′ yaf+] [pKD46] | SMR6068 × pKD46 [from BW25113(pKD46)] |
| SMR11008 | SMR4562 ΔyafN11::FRTKanFRT ΔyafP19::FRTcatFRT zae-502::Tn10 [F′ yaf+] | SMR11168 × P1(CAG18436) |
| SMR11168 | SMR4562 ΔyafN11::FRTKanFRT ΔyafP19::FRTcatFRT [F′ yaf+] | SMR10995 × DNA of ΔyafP::FRTcatFRT amplified from pKD3 [from BW25141(pKD3)] |
| SMR11228 | MG1655(λ) | MG1655 × λSR108 |
| SMR11229 | MG1655(λ) Δ(yafN-yafP)776::FRTKanFRT | SMR11228 × P1(SMR6353) |
| SMR11231 | MG1655(λ) recG258::Tn10 mini-Kan | SMR11228 × P1(N2731) |
| Phage lambda | ||
| λSR108 | λ wild type | F. Stahl (Oregon) |
| Plasmids | ||
| pBAD24 | Plasmid containing arabinose-inducible promoter PBAD, Ampr | 23 (via ATCC 87399) |
| pLS1 | Plasmid containing yafN under the control of PBAD | This work |
| pKD3 | cat-containing plasmid, template for PCR | 12 |
| pKD13 | kan-containing plasmid, template for PCR | 12 |
| pCP20 | FLP-containing plasmid, used to excise drug markers flanked by FRT sequences | 12 |
| pKD46 | Red recombinase expression plasmid | 12 |
FRT, FLP recombination target; CGSC, E. coli Genetic Stock Center (Yale University); ATCC, American Type Culture Collection.
TABLE 2.
Description of new deletion alleles
| Strain | New allele | Wild-type gene length (bp) | Deletion coordinates (distance from gene translation start) | Replacement cassettea |
|---|---|---|---|---|
| SMR6068 | ΔyafN11::FRTKanFRT | 294 | +13 to +279 | FLPable Kan |
| SMR6074 | ΔyafO14::FRTKanFRT | 399 | +13 to +384 | FLPable Kan |
| SMR6080 | ΔyafP18::FRTKanFRT | 453 | +16 to +438 | FLPable Kan |
| SMR6221 | ΔyafP20::FRT | 453 | +16 to +438 | FRT scar |
| SMR10285 | Δ(yafN-yafO)779::FRTKanFRT | See individual genes above | +13 (yafN) to +384 (yafO) | FLPable Kan |
| SMR11168 | ΔyafP19::FRTcatFRT | 453 | +40 to +416 | FLPable cat |
For each replacement cassette, the sequence of the inserted DNA begins with ATCC (Kan cassette) or TCATA (cat cassette) and ends with CTACA (Kan cassette) or TACAC (cat cassette). Sequences are listed 5′ to 3′. FRT, FLP recombination target.
TABLE 3.
Linear replacement gene deletion primers
| Strain | Linear replacement primer name | Sequencea | Template |
|---|---|---|---|
| SMR6068 | yafNwL | tgtatattctggtgtgcattattatgagggtatcactgtatgcatcgaattATTCCGGGGATCCGTCGACC | pKD13 |
| yafNwR | gctgtaagttgcaggcgaataagttttgttttgaatacccgcatccttattccttaaagtcTGTAGGCTGGAGCTGCTTC | ||
| SMR6074 | yafOwL | tatgacggatgatgatttcaatgactttaaggaataaggatgcgggtattcATTCCGGGGATCCGTCGACC | pKD13 |
| yafOwR | atagtttcttatttgtatgttattcataatataaattcaaaaacgcatgcgTGTAGGCTGGAGCTGCTTC | ||
| SMR6080/6221 | yafPwL | gcagaagcgtttcgcatgcgtttttgaatttatattatgaataacatacaaATTCCGGGGATCCGTCGACC | pKD13 |
| yafPwR | ataccaggcgggcgttattttcattgcaagctggatttaatgttgcggtttTGTAGGCTGGAGCTGCTTC | ||
| SMR6353 | yafNwL | tgtatattctggtgtgcattattatgagggtatcactgtatgcatcgaattATTCCGGGGATCCGTCGACC | pKD13 |
| yafPwR | ataccaggcgggcgttattttcattgcaagctggatttaatgttgcggtttTGTAGGCTGGAGCTGCTTC | ||
| SMR10285 | yafNwL | tgtatattctggtgtgcattattatgagggtatcactgtatgcatcgaattATTCCGGGGATCCGTCGACC | pKD13 |
| yafOwR | atagtttcttatttgtatgttattcataatataaattcaaaaacgcatgcgTGTAGGCTGGAGCTGCTTC | ||
| SMR11168 | yafP::CAT-L | atgaataacatacaaataagaaactatcagcctggcgatTCATATGAATATCCTCCTTAG | pKD3 |
| yafP::CAT-R | atgttgcggtttatatcgcatataaaaattagtaaacGTGTAGGCTGGAGCTGCTTC |
Lowercase letters refer to chromosomal sequence; uppercase letters refer to template sequence. Sequences are listed 5′ to 3′.
Cloning of yafN into pBAD24.
yafN was amplified from strain FC36 using primers yafN-F-KpnI (AAAAAAGGTACCATTCTGGTGTGCATTATTATG) and yafN-R-HindIII (AAAAAAAAGTTCTTATTCCTTAAAGTCATTG) with Platinum Pfx polymerase (Invitrogen) and cloned into pCR-BluntII-Topo-Kanr using Invitrogen's Zero Blunt Topo PCR cloning kit. yafN was excised from the cloning plasmid with KpnI and XhoI and ligated into pBAD24 using T4 DNA ligase (Invitrogen), creating the plasmid pLS1. Insertion of yafN into pBAD24 was confirmed by sequencing (SeqWright, Houston, TX).
Quantitative phage P1-mediated cotransduction assays.
Phage P1-mediated cotransductions were performed based on the Miller protocol (44) with the following modifications. Transductants selected on LBH-Tet with 20 mM CIT were then patched in 100-colony grids onto LBH-Kan-CIT (and LBH-chloramphenicol-CIT for ΔyafN ΔyafP cotransduction) to screen for recovery of markers of interest. Plates were monitored over several days of incubation for possible slowly growing colonies.
Sequencing of JW0222 yafO.
The yafO gene of strain JW0222 was amplified using PCR primers yafN-F-KpnI and yafPExtR (AGTTTGTGGAATCAGAAAACG). PCR product was purified with Qiagen's QIAquick PCR purification kit and sequenced using the above-mentioned primers (SeqWright, Houston, TX).
Mutagenesis and DNA damage sensitivity assays.
Stress-induced mutagenesis assays were performed according to the method of Harris et al. (25). For UV light kill curves, cultures were grown to early/mid-log phase (optical density at 600 nm, 0.1 to 0.3) in LBH and then spread onto LBH agar plates, exposed to the indicated UV light dose, incubated in the dark at 37°C for 16 h, and scored for colonies. For bleomycin and mitomycin sensitivity assays, saturated LBH cultures were diluted and grown to mid-log phase (optical density at 600 nm, 0.3 to 0.6) and spotted onto fresh drug-containing plates at indicated dilutions. Bleomycin was dissolved in 0.9% NaCl. Mitomycin C was dissolved in sterile H2O (final pH, 6.0), per the manufacturer's recommendation. Plates were incubated for 16 h at 32°C and scored for CFU.
Lambda burst size assay from UV-induced lysogens.
Saturated LBH cultures of E. coli λ lysogens were diluted in K-glucose and grown to 1.5 × 108 cells/ml. An aliquot of cells was removed, lysed with lysozyme and chloroform, and submitted to plaque assay to quantify phage that was spontaneously induced from the prophage state. Cells were UV irradiated (50 J/m2) to induce the lambda prophage to lytic growth and shaken at ≥300 rpm for 1.5 h at 37°C in darkness. Lysozyme (50 mg/ml) and chloroform were added to lyse cells and release intracellular phage, debris were pelleted, and the supernatants were assayed for PFU. Phage titers were calculated and used to determine burst size (PFU/cell, corrected for preexisting phage).
Flow cytometry and cell sorting.
Flow cytometry analyses and fluorescence-activated cell sorting (FACS) were performed per the method in reference 47 with the following modifications. Log-phase cells were analyzed using the BD FACSAria cell sorter (BD Biosciences). Data shown are the means of five experiments (one culture/experiment for experiments 1 to 4 and two cultures/experiment for experiment 5). At least 106 nongreen and 8 × 103 (typically >104) green cells were sorted per culture. Sorting purity controls were performed before each experiment (per the method in reference 47). Propidium iodide (PI) staining was performed per the method in reference 47.
RESULTS
yafN is essential in the presence of yafO.
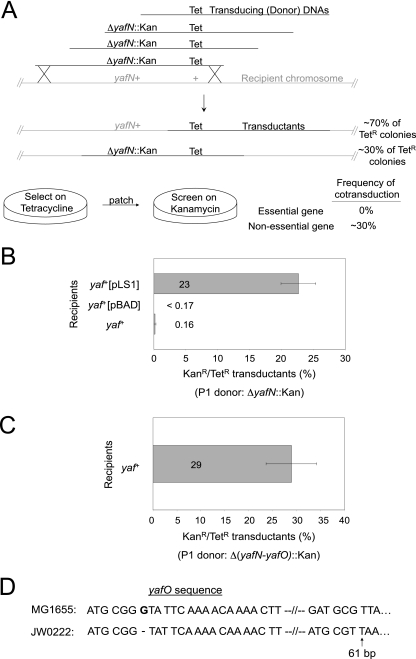
To test whether yafNOP is a functional TA system, we examined whether the putative antitoxin gene, yafN, is an essential gene but only in the presence of a functional putative toxin gene, yafO. As diagrammed in Fig. 2A, using a cotransduction assay, we transduced E. coli cells with phage P1 that had been grown on cells carrying the zae-502::Tn10 Tet resistance (Tetr) gene partially linked with a chromosomal ΔyafN::Kan deletion, conferring Kan resistance (Kanr), or other mutation of interest. The P1 donor cells carried a yafN+-expressing plasmid to allow their viability (if essential) and prevent the accumulation/selection of extragenic suppressor mutations that would allow a cell with a mutated essential gene to grow. We initially selected for the neutral Tetr marker and then screened colonies for the Kan-replaced gene of interest (either yafN, yafNO, yafP, or yafN and yafP simultaneously). The expected frequency of cotransduction of these two markers (Kanr and Tetr) is approximately 30% based on their physical distance. Cotransduction frequencies of ∼30% imply that the gene of interest is not essential, whereas cotransduction frequencies that are significantly lower imply reduced viability of transductants that received the deletion, that is, that the gene deleted is essential.
FIG. 2.
Cotransduction assay for quantitative determination of inviability of various mutant strains. (A) The assay design. Phage P1 grown on strains containing gene knockouts of interest (Kanr) with known linkage to a nonlethal Tetr marker, zae-502::Tn10, were transduced into recipient cells and selected on Tet. Colonies were patched to Kan plates to determine the frequency of cotransduction of the markers, to determine whether the gene of interest is essential. If a gene is essential, the frequency of cotransduction of the Kanr gene deletion among Tetr transductants will be lower than that predicted by the distance between the markers. (B) yafN is essential. The ΔyafN::Kan deletion is cotransduced efficiently with zae-502::Tn10 into cells harboring plasmid pLS1, which expresses yafN, but not into cells carrying no plasmid or the vector only. P1 donor: ΔyafN::Kan zae-502::Tn10 (SMR10291). Recipient cells: yaf+ (FC36), yaf+[pBAD] (SMR10943), or yaf+[pLS1] (SMR10948). Average ± 1 SEM of three experiments. (C) yafN is essential only in the presence of yafO. Δ(yafN-yafO)::Kan zae-502::Tn10 cotransduction (P1 donor: SMR10483) into yaf+ strain FC36. Average ± 1 SEM of three experiments. (D) The yafO gene is mutated in yafN deletion strain JW0222 of the E. coli gene knockout Keio collection. Sequencing of the JW0222 yafO gene revealed a −1-bp deletion at bp 7 that results in a premature stop codon at bp 61 of the 399-bp gene.
We found that ΔyafN::Kan zae-502::Tn10 cells were cotransduced in only 0.16% ± 0.16% of Tetr transductants (Fig. 2B). These data imply that yafN is an essential gene. Supporting this interpretation, we found that loss of the chromosomal copy of yafN by transduction could be achieved efficiently (23% ± 2.7% ΔyafN::Kan zae-502::Tn10 cotransductants) in recipient cells carrying a yafN-containing plasmid (pLS1) (Fig. 2B) but not in cells carrying only the empty plasmid vector (Fig. 2B). We conclude that yafN is an essential gene.
If the yafNO(P) genes were a TA system, then yafN would be expected to be essential only when the putative toxin, YafO, was present. We found that when Δ(yafN-yafO)::Kan zae-502::Tn10 cells were transduced, the frequency of cotransduction of the two markers with Tetr was 29% ± 5% (Fig. 2C). These data show that yafN is essential only if YafO is functional and so support the interpretation that yafN and yafO are a TA system.
In contrast with our results, there is a published yafN deletion strain, JW0222, of the E. coli Keio deletion collection (2). We hypothesized that to be viable, this strain might carry a spontaneous mutation inactivating yafO or alternatively a duplication of the operon in which only one of two copies of yafN was deleted. Such mutants are likely to have been responsible for the 0.16% frequency of cotransduction seen in the ΔyafN::Kan zae-502::Tn10 cotransduction (Fig. 2B). We sequenced the yafO gene of strain JW0222 and report that the yafO gene contains a single base pair deletion of a guanine at bp 7, which, because of the frameshift, creates an early stop codon (TAA) at bp 61 of the 399-bp yafO gene (Fig. 2D). The resultant YafO protein is prematurely truncated and likely to be nonfunctional.
YafP is neither a toxin nor an antitoxin component.
We sought to understand whether yafP was integral to the function of either the antitoxin or the toxin. If yafP was an integral part of the antitoxin, it, like yafN, would be essential for viability in the presence of yafO. We found that ΔyafP::Kan zae-502::Tn10 is cotransduced efficiently at 20% ± 1.8% into a strain without an additional plasmid-borne yafP gene and at 26% ± 2% into strains harboring an extra yafP gene (Fig. 3A). Thus, yafP is not essential in the presence of yafO and therefore YafP is not an integral part of the antitoxin.
FIG. 3.
YafP is not a necessary component of either toxin or antitoxin. (A) yafP is not essential for viability and thus not part of the antitoxin. ΔyafP::Kan is cotransduced efficiently with zae-502::Tn10 (P1 donor: SMR10953) into recipient cells: yaf+ (FC36) or yaf+[F′ yaf+] (SMR4562). Average ± SEM of three experiments. (B) Loss of yafP does not allow recovery of ΔyafN cotransductants as would be expected if yafP were required for toxin function. Cotransduction with P1 donor ΔyafN::Kan ΔyafP::cat zae-502::Tn10 (SMR11008) into recipient cells: yaf+ (FC36), ΔyafP (SMR6221), yaf+[pBAD] (SMR10943), or yaf+[pLS1] (SMR10948). Average ± SEM of three experiments.
If yafP was an integral part of the toxin, then the loss of yafP would be expected to allow efficient cotransduction of ΔyafN::Kan with the linked Tetr marker because yafN would no longer be required for viability. We found that the frequency of cotransduction of the double deletion ΔyafN::Kan ΔyafP::cat with the linked Tetr marker was <0.33% in cells with no extra copy of the yafN gene but was efficient in cells carrying the yafN expression plasmid pLS1 (Fig. 3B), indicating that yafN remains essential even when yafP is deleted. To address the possible concern that lingering toxin in the recipient cell (present prior to transduction of ΔyafP) might cause inviability after cotransduction, an additional cotransduction was performed into a ΔyafP recipient strain. The frequency of cotransductants remained <0.33% (Fig. 3B). We conclude that yafN remains essential despite the absence of yafP. Therefore, YafP is not a necessary part of the toxin.
No role in stress-induced mutagenesis.
Because dinB, the first gene in the dinB-yafNOP operon, is a key player in stress-induced point mutagenesis in the E. coli Lac system (17, 42) and one could imagine roles for a TA system in the process, we tested for possible involvement of yafNOP in stress-induced lac reversion. We observed that the yafNOP strain showed slightly but not significantly higher stress-induced mutagenesis than did the yaf+ strain, unlike dinB, which is required (Fig. 4A and B). Thus, the yafNOP genes are not required for stress-induced mutagenesis. The yafNOP genes were shown previously to contribute only slightly to generation-dependent mutagenesis (43).
FIG. 4.
The yafNOP genes are not required for stress-induced mutagenesis. Lac assay strains were starved on lactose for several days and monitored each day for Lac+ reversion mutant colonies according to the method in reference 25. (A) Representative experiment. Lac+ revertants per 108 cells plotted over time. Average ± SEM of five cultures. (B) Summary of mutation rates from multiple experiments. Mutation rates, calculated according to the method in reference 37, are Lac+ revertants per 108 cells per day on days 3 to 5. Mean ± SEM from five experiments. The yaf+ strain is SMR4562, the ΔyafNOP strain is SMR7491, and the ΔdinB strain is SMR5889.
The SOS DNA damage response.
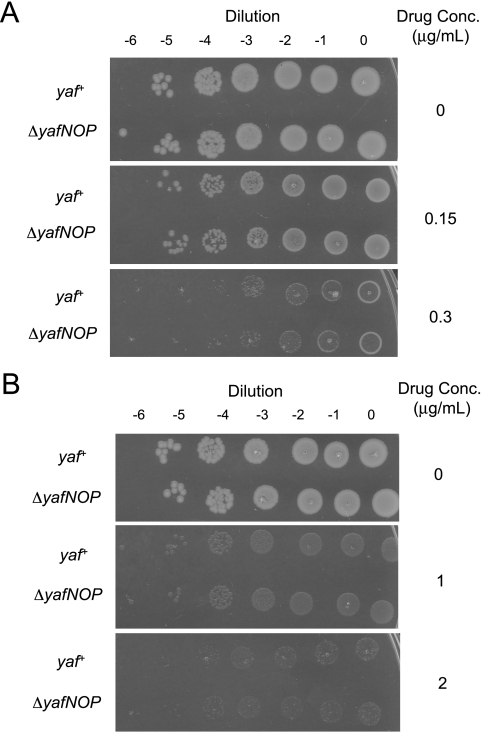
In hopes of discovering a possible role for yafNOP during the SOS response, we explored several assays in which DNA damage affects cell survival to test whether yafNO might contribute to survival or loss-of-survival phenotypes. We used several different DNA damage-inducing agents including UV light, bleomycin, and mitomycin C to induce the SOS response. We saw no significant difference between ΔyafNOP and yaf+ cells in survival of UV light (Fig. 5A); bleomycin, a double-strand break-inducing agent (29) (Fig. 5B); or DNA cross-linking agent mitomycin C (30) (Fig. 5C).
FIG. 5.
Sensitivity of nonlysogens to DNA-damaging agents. (A) UV light kill curve. Log-phase cells were UV irradiated and monitored for colony formation. Average ± 1 SEM of three experiments shown. (B) Nonlysogen bleomycin kill plates. Log-phase cells were plated onto bleomycin-containing plates and monitored for viability. Bleomycin creates both single- and double-strand breaks in DNA (29). A ΔrecG strain monitored concurrently showed sensitivity (not shown). Results shown in panels B and C are representative of two experiments. (C) Nonlysogen mitomycin C kill plates. Log-phase cells were plated onto mitomycin C-containing plates (various concentrations) and monitored for viability. Mitomycin C is a powerful DNA interstrand cross-linker (30). A ΔrecG strain monitored concurrently showed sensitivity (not shown). For all panels, the yaf+ strain is MG1655, the ΔyafNOP strain is SMR6353, and the ΔrecG strain is SMR10990. Log dilutions are indicated.
A previous report suggested that the MazEF TA system protects cells against phage P1 infection (27). We found no evidence that yafNOP might serve as a protective unit against E. coli phage λ. First, the burst size of a λ lytic infection was not different when yaf+ cells were infected than when ΔyafNOP cells were infected; the ratio of yaf+ to ΔyafNOP was 1.4 ± 0.6 (mean ± standard error of the mean [SEM] of three experiments). Next, to test whether the yafNOP genes either promote or prevent the progression of λ during induction from lysogenic state, a transition controlled by the SOS response (49), we spotted various dilutions of λ lysogens of E. coli yafNOP+ or ΔyafNOP cells onto different concentrations of either bleomycin- or mitomycin C-containing plates. By inducing the SOS response via DNA-damaging drugs, we simultaneously induced the λ prophage to become lytic, leading to far more severe killing in the lysogens (Fig. 6A and B) than the nonlysogens (Fig. 5B and C), indicating that the major mode of killing in lysogens was by prophage induction. However, there was no difference between the survival of ΔyafNOP lysogens and that of yaf+ lysogens (Fig. 6A and B).
FIG. 6.
YafNOP does not affect killing by prophage induction by DNA-damaging agents. DNA damage induces the SOS response and activates the λ lytic cycle, causing killing by prophage induction, as can be seen by the greater sensitivity to DNA-damaging agents of lysogens (this figure) than of nonlysogens (Fig. 5). YafNOP does not affect this killing by prophage induction. (A) Lambda lysogen bleomycin kill plates. Log-phase E. coli (λ) lysogens were plated onto bleomycin-containing plates (various concentrations) and monitored for viability. A ΔrecG strain monitored concurrently showed sensitivity (not shown). (B) Lambda lysogen mitomycin C kill plates. Log-phase E. coli λ lysogens were plated onto mitomycin C-containing plates (various concentrations) and monitored for viability. A ΔrecG strain monitored concurrently showed sensitivity (not shown). For both panels, the yaf+ strain is SMR11228, the ΔyafNOP strain is SMR11229, and the ΔrecG strain is SMR11231. Results shown are representative of two experiments.
SOS-induced senescence is not caused by yafNO.
Previously, Pennington and Rosenberg found that ∼65% of cells that undergo spontaneous SOS induction were unable to form colonies, despite the fact that nearly all of the SOS-induced cells were viable, as determined by their ability to exclude the dye PI (47), suggesting that they were in a senescence-like state (47). To test whether this senescence-like state might result from the toxic (and possibly bacteriostatic) yafO, we repeated the experiment using FACS to sort yaf+ and ΔyafNOP reporter strains carrying the chromosomal gfp gene controlled by the SOS-inducible sulA promoter. These were sorted into spontaneously SOS-induced green and SOS-uninduced nongreen subpopulations, which compose ∼1% and ∼99% of the cell population, respectively (reference 47 and this study). We found that there was little difference in colony-forming abilities of the spontaneously SOS-induced green populations. We observed that 38% ± 12% of yafNOP+ SOS-induced green cells formed colonies (35% ± 9% of green cells forming colonies normalized by 94% ± 10% of nongreen cells forming colonies to control for FACS-induced effects on colony formation) (Fig. 7). Similarly, 26% ± 5% of ΔyafNOP green cells formed colonies (25% ± 5% of green cells forming colonies normalized by 95% ± 2% of nongreen cells forming colonies) (Fig. 7). Also, both strains' cells were nearly all viable, with 3.6% ± 1.4% of yafNOP+ green cells being PI+ (dead) versus 4.5% ± 1.4% of ΔyafNOP green cells being PI+. Thus, the numbers of cells in a senescence-like state of being alive (PI−) but unable to form colonies were not different between yafNOP+ and ΔyafNOP cells (Fig. 7).
FIG. 7.
The yafNOP genes were not responsible for the senescence-like state of spontaneously SOS-induced cells. Strains with a chromosomal gfp gene controlled by the SOS-inducible sulA promoter were sorted into SOS-induced green and SOS-uninduced nongreen populations by FACS and plated for CFU. The decreased ability of SOS-induced green cells to form colonies was not reversed by the ΔyafNOP mutation. Percentage of live/dead cells in each population was determined by PI staining and flow cytometry to quantify dead cells, which take up the dye. The percentage of cells in a senescence-like state, excluding PI but unable to form colonies, was not different for yaf+ (SMR6669) and ΔyafNOP (SMR10988) cells. Average ± SEM of five experiments.
DISCUSSION
We have shown that the yafN gene of the dinB-yafN-yafO-yafP operon (43) is essential for viability only in the presence of a functional yafO gene (Fig. 2B and C), providing strong evidence that these two genes constitute the antitoxin and toxin genes, respectively, of a type 2 TA pair. Whereas previous in vivo demonstrations of type 2 TA systems have used the method of separate cloning of each gene into differentially inducible plasmids and showing that the toxin induces cell stasis when solely expressed, but not when the antitoxin is also expressed (1, 50), we used an alternative genetic approach that allows us to rule out possible effects specific to overexpression of either the toxin or the antitoxin. We can therefore conclude that YafO exerts its toxic effect, and YafN can quell that effect, when each gene is expressed at normal levels from its native promoter, in single copy in its normal chromosomal position.
Typically, chromosomal TA systems consist of two genes in an operon, the toxin and the antitoxin. Although there is precedent for a TA system with a third gene element, mazEFG (22), we found that yafP, the third yaf gene in the operon, is not an integral component of either the toxin or the antitoxin (Fig. 3A and B). This conclusion does not preclude the possibility that YafP might play a regulatory role in the YafNO TA system which we have not detected, as mazG does for mazEF (22).
The possible function of a TA system controlled by the SOS response is an interesting problem. We found that unlike dinB (42), the first gene in the operon (43), the yafNOP genes do not contribute to stress-induced mutagenesis significantly (Fig. 4), a process that requires SOS-induced levels of dinB, but not SOS-induced levels of any other SOS-controlled component (17). Other SOS-controlled genes such as recA (6, 24) and ruvA and ruvB (15, 25) are required for stress-induced Lac mutagenesis, we now appreciate, at their constitutive levels of expression, not at induced levels (17). The results presented here rule out a requirement for YafNOP in stress-induced mutagenesis even at their constitutive expression levels. Previously, YafNOP had little effect on spontaneous generation-dependent mutagenesis in nonstressed growing cells (43).
We also did not detect effects of yafNOP on survival of cells following various SOS-inducing treatments, for nonlysogens (Fig. 5A to C) and also for cells harboring a wild-type lambda prophage, which is induced leading to cell lysis when the SOS response is activated (Fig. 6A and B). Finally, although we could recapitulate the previous results of Pennington and Rosenberg, showing that many spontaneously SOS-induced green fluorescent cells (bearing an SOS-controlled chromosomal gfp reporter gene) are apparently viable but unable to form colonies when recovered by FACS (47), we found that yafNOP is not responsible for their senescence-like state (Fig. 7).
What might be the function of an SOS-controlled TA system? For TA systems, the toxic effects ensue when the operon's expression is decreased, so we would expect possible effects of YafO to be manifested as cells recover from an SOS response and return to normal after DNA repair, even though these genes are expressed mid-range in the SOS response (11). Perhaps YafO induces a transient cell stasis upon recovery. This might function to extend the cell cycle checkpoint caused by SOS-induced expression of the SulA inhibitor of cell division (16). Although we did not find effects of YafNO on cells after SOS induction, we cannot rule out the possibility of an important role in SOS recovery that our assays might not have detected. Alternatively or in addition, dinB, and presumably the rest of its operon, is also upregulated slightly by the RpoS general stress response (33). Perhaps YafO plays a role in promoting cell stasis upon recovery from the RpoS response. Additionally, although the yafNO genes are transcribed from the upstream SOS-controlled (and RpoS-controlled) dinB promoter (43), there is in vitro evidence that an additional promoter may exist immediately upstream of yafN, possibly creating an SOS (or RpoS)-independent yafNOP operon (57). If so, yafNOP might act outside the contexts of the SOS (or RpoS) response.
A recent report shows that DinB and also another SOS-inducible DNA polymerase, Pol V, interact directly with the NusA transcription and antitermination factor (10). The authors suggest that NusA might direct the translesion DNA synthesis activity of DinB to sites of active transcription. Similarly, we can imagine that DinB might affect NusA-dependent transcription termination, which might then provide another level of SOS control of gene expression (negative or positive). Perhaps YafNO or YafP functions in such a process, and perhaps the toxic effect of YafO is related to a transcription-termination-specific effect.
While the manuscript was being prepared, another group reported that the YafO protein is an RNase (58). Their results support our conclusions and provide a mechanism for the toxic action of the YafO toxin. Important next steps toward understanding the yafNO TA system include defining when as well as on what targets the toxin acts.
Acknowledgments
This work was supported by National Institutes of Health grant R01-GM53158.
We thank N. Fonville, R. S. Galhardo, P. J. Hastings, C. Herman, A. Al Mamun, and C. Shee for valuable input throughout the course of this work.
Footnotes
Published ahead of print on 16 October 2009.
REFERENCES
- 1.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, D. S., A. J. Kelly, M. J. Mardis, and H. S. Moyed. 1991. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 173:5732-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Brown, J. M., and K. J. Shaw. 2003. A novel family of Escherichia coli toxin-antitoxin gene pairs. J. Bacteriol. 185:6600-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cairns, J., and P. L. Foster. 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 98:14328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirz, R. T., and F. E. Romesberg. 2006. Induction and inhibition of ciprofloxacin resistance-conferring mutations in hypermutator bacteria. Antimicrob. Agents Chemother. 50:220-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen, S. E., V. G. Godoy, and G. C. Walker. 2009. Transcriptional modulator NusA interacts with translesion DNA polymerases in Escherichia coli. J. Bacteriol. 191:665-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courcelle, J., A. Khodursky, B. Peter, P. O. Brown, and P. C. Hanawalt. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez De Henestrosa, A. R., T. Ogi, S. Aoyagi, D. Chafin, J. J. Hayes, H. Ohmori, and R. Woodgate. 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35:1560-1572. [DOI] [PubMed] [Google Scholar]
- 15.Foster, P. L., J. M. Trimarchi, and R. A. Maurer. 1996. Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation in Escherichia coli. Genetics 142:25-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedberg, E. C., W. Siede, and G. C. Walker. 2006. DNA repair and mutagenesis. ASM Press, Washington, DC.
- 17.Galhardo, R. S., R. Do, M. Yamada, E. C. Friedberg, P. J. Hastings, T. Nohmi, and S. M. Rosenberg. 2009. DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics 182:55-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 19.Gotfredsen, M., and K. Gerdes. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29:1065-1076. [DOI] [PubMed] [Google Scholar]
- 20.Grady, R., and F. Hayes. 2003. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 47:1419-1432. [DOI] [PubMed] [Google Scholar]
- 21.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross, M., I. Marianovsky, and G. Glaser. 2006. MazG—a regulator of programmed cell death in Escherichia coli. Mol. Microbiol. 59:590-601. [DOI] [PubMed] [Google Scholar]
- 23.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris, R. S., S. Longerich, and S. M. Rosenberg. 1994. Recombination in adaptive mutation. Science 264:258-260. [DOI] [PubMed] [Google Scholar]
- 25.Harris, R. S., K. J. Ross, and S. M. Rosenberg. 1996. Opposing roles of the Holliday junction processing systems of Escherichia coli in recombination-dependent adaptive mutation. Genetics 142:681-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hastings, P. J., A. Slack, J. F. Petrosino, and S. M. Rosenberg. 2004. Adaptive amplification and point mutation are independent mechanisms: evidence for various stress-inducible mutation mechanisms. PLoS Biol. 2:e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazan, R., and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics 272:227-234. [DOI] [PubMed] [Google Scholar]
- 28.Hazan, R., B. Sat, and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186:3663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hecht, S. M. 2000. Bleomycin: new perspectives on the mechanism of action. J. Nat. Prod. 63:158-168. [DOI] [PubMed] [Google Scholar]
- 30.Iyer, V. N., and W. Szybalski. 1963. A molecular mechanism of mitomycin action: linking of complementary DNA strands. Proc. Natl. Acad. Sci. USA 50:355-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawano, M., L. Aravind, and G. Storz. 2007. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 64:738-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korch, S. B., and T. M. Hill. 2006. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J. Bacteriol. 188:3826-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Layton, J. C., and P. L. Foster. 2003. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 50:549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehnherr, H., E. Maguin, S. Jafri, and M. B. Yarmolinsky. 1993. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 233:414-428. [DOI] [PubMed] [Google Scholar]
- 35.Lewis, L. K., G. R. Harlow, L. A. Gregg-Jolly, and D. W. Mount. 1994. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J. Mol. Biol. 241:507-523. [DOI] [PubMed] [Google Scholar]
- 36.Lloyd, R. G., and C. Buckman. 1991. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J. Bacteriol. 173:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lombardo, M. J., I. Aponyi, and S. M. Rosenberg. 2004. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics 166:669-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magnuson, R. D. 2007. Hypothetical functions of toxin-antitoxin systems. J. Bacteriol. 189:6089-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masuda, Y., K. Miyakawa, Y. Nishimura, and E. Ohtsubo. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 175:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCool, J. D., E. Long, J. F. Petrosino, H. A. Sandler, S. M. Rosenberg, and S. J. Sandler. 2004. Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol. Microbiol. 53:1343-1357. [DOI] [PubMed] [Google Scholar]
- 41.McKenzie, G. J., R. S. Harris, P. L. Lee, and S. M. Rosenberg. 2000. The SOS response regulates adaptive mutation. Proc. Natl. Acad. Sci. USA 97:6646-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenzie, G. J., P. L. Lee, M. J. Lombardo, P. J. Hastings, and S. M. Rosenberg. 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell 7:571-579. [DOI] [PubMed] [Google Scholar]
- 43.McKenzie, G. J., D. B. Magner, P. L. Lee, and S. M. Rosenberg. 2003. The dinB operon and spontaneous mutation in Escherichia coli. J. Bacteriol. 185:3972-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 45.Motiejunaite, R., J. Armalyte, A. Markuckas, and E. Suziedeliene. 2007. Escherichia coli dinJ-yafQ genes act as a toxin-antitoxin module. FEMS Microbiol. Lett. 268:112-119. [DOI] [PubMed] [Google Scholar]
- 46.Pedersen, K., and K. Gerdes. 1999. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 32:1090-1102. [DOI] [PubMed] [Google Scholar]
- 47.Pennington, J. M., and S. M. Rosenberg. 2007. Spontaneous DNA breakage in single living Escherichia coli cells. Nat. Genet. 39:797-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prieto, A. I., F. Ramos-Morales, and J. Casadesus. 2006. Repair of DNA damage induced by bile salts in Salmonella enterica. Genetics 174:575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts, J. W., and R. Devoret. 1983. Lysogenic induction, p. 123-144. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 50.Schmidt, O., V. J. Schuenemann, N. J. Hand, T. J. Silhavy, J. Martin, A. N. Lupas, and S. Djuranovic. 2007. prlF and yhaV encode a new toxin-antitoxin system in Escherichia coli. J. Mol. Biol. 372:894-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sung, H. M., G. Yeamans, C. A. Ross, and R. E. Yasbin. 2003. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J. Bacteriol. 185:2153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tegova, R., A. Tover, K. Tarassova, M. Tark, and M. Kivisaar. 2004. Involvement of error-prone DNA polymerase IV in stationary-phase mutagenesis in Pseudomonas putida. J. Bacteriol. 186:2735-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Melderen, L., and M. Saavedra De Bast. 2009. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 5:e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vogel, J., L. Argaman, E. G. Wagner, and S. Altuvia. 2004. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 14:2271-2276. [DOI] [PubMed] [Google Scholar]
- 56.Wade, J. T., N. B. Reppas, G. M. Church, and K. Struhl. 2005. Genomic analysis of binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional targest sites. Genes 19:2619-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zaslaver, A., A. Bren, M. Ronen, S. Itzkovitz, I. Kikoin, S. Shavit, W. Liebermeister, M. G. Surette, and U. Alon. 2006. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3:623-628. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Y., Y. Yamaguchi, and M. Inouye. 2009. Characterization of YafO, an Escherichia coli toxin. J. Biol. Chem. 284:25522-25531. [DOI] [PMC free article] [PubMed] [Google Scholar]