Abstract
Phylogenomics reveals extreme gene loss in typhus group (TG) rickettsiae relative to the levels for other rickettsial lineages. We report here a curious protease-encoding gene (ppcE) that is conserved only in TG rickettsiae. As a possible determinant of host pathogenicity, ppcE warrants consideration in the development of therapeutics against epidemic and murine typhus.
Alphaproteobacteria of the genus Rickettsia are obligate intracellular residents of a wide range of eukaryotes (6, 27, 33). The biology and diversity of Rickettsia species are poorly understood, with much knowledge stemming from decades of research on the species that cause mild to severe human disease. Understandably, the first dozen sequenced rickettsial genomes comprised definitive or tentative human pathogens. However, recently sequenced genomes of the attenuated strain Iowa of Rickettsia rickettsii, along with two groups with no known host pathogenicity, Rickettsia peacockii and rickettsial endosymbiont of Ixodes scapularis (REIS), now provide a clearer framework for the elucidation of lineage specific virulence determinants.
Like other obligate intracellular bacteria, Rickettsia species have small genomes that reflect the reductive evolutionary processes of a lifestyle dependent on many eukaryotic host resources (3). Previously, we identified genes that define traits such as arthropod host species and disease phenotypes (13, 16). The smallest rickettsial genomes, belonging to the typhus group (TG) rickettsiae (Rickettsia prowazekii and Rickettsia typhi), lacked 53 genes present in all other sequenced rickettsial genomes. Additionally, TG genomes harbored very few pseudogenes, repetitive sequences, and genes typical of the bacterial mobile gene pool, relative to what was found for other species of Rickettsia. Thus, we concluded that some defining virulence factors associated with epidemic and murine typhus may comprise gene loss and strict reliance on host resources, a hypothesis previously put forward explaining the evolution of pathogenicity in all virulent rickettsiae (10).
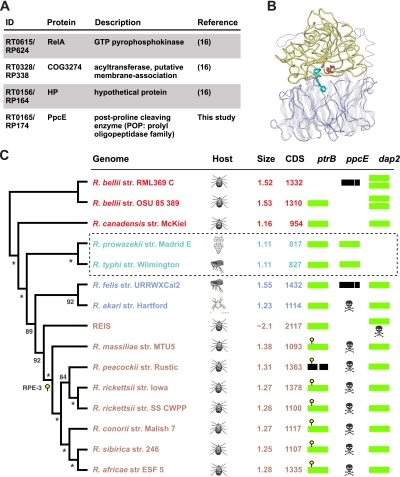
Our prior analyses identified only three TG rickettsia-specific genes; we report here another TG-specific gene that encodes a putative prolyl oligopeptidase (POP) protein (Fig. 1A). POPs belong to the S9 serine proteases and are characterized by the presence of an N-terminal seven-bladed β propeller and a C-terminal α/β hydrolase fold domain (Fig. 1B). The unusual β propeller is considered a gating filter for substrate entry into the catalytic triad of the peptidase domain (12). The detection of a conserved TG-specific POP gene, named the postproline cleaving enzyme (ppcE) gene, was previously hampered by the clustering of highly fragmented ppcE remnants into this ortholog group (16). Thus, ppcE has undergone substantial pseudogenization in all non-TG Rickettsia species (Fig. 1C) and is undetectable even as a pseudogene in all Rickettsiales genomes basal to the three derived rickettsial groups (see Fig. S1 in the supplemental material). Furthermore, it is completely absent from the REIS genome and split in the Rickettsia felis genome. Despite a stop codon separating the catalytic triad from the remaining open reading frame, R. felis ppcE is highly conserved with TG rickettsial ppcE; this prompted us to confirm the split gene with DNA sequencing (see Fig. S2 in the supplemental material). We also failed to detect a suppressor mutant tRNA in the R. felis genome that could incorporate Glu for UAA. Thus, while TG rickettsiae and R. felis are the only species in our analysis with definitive insect hosts, ppcE is apparently not essential for life inside insects.
FIG. 1.
Prolyl oligopeptidase (POP) genes of rickettsiae. (A) TG-specific genes identified previously (16) or in this study. Annotation is from PATRIC (32). (B) Tertiary structure of Myxococcus xanthus prolyl endopeptidase (31), illustrating the major features of POP proteins: blue, β-propeller domain; yellow, α/β hydrolase fold domain; red, catalytic triad (Ser-Asp-His); cyan, substrate. (C) Distribution of POP genes across 15 rickettsial genomes. Phylogeny was estimated from 18 genes encoding the rvh type IV secretion system as previously described (14). Branch support is from 1 million bootstrap replications; asterisks indicate 100% bootstrap support. The rickettsial classification scheme (15, 16) was as follows: red, ancestral group; turquoise, TG; blue, transitional group; and brown, SFG. Ticks are listed as the principal arthropod vector for all species except R. prowazekii (louse), R. typhi (flea), R. felis (flea), and R. akari (mite). Genome statistics were compiled from the PATRIC database (32). ptrB, oligopeptidase B gene (EC 3.4.21.83); ppcE, postproline cleaving enzyme gene (EC 3.4.21.26); dap2, dipeptidyl aminopeptidase 2 gene (EC 3.4.19.1). Full-length POP genes are colored light green, with truncated and split open reading frames colored black. Skulls and crossbones symbols depict gene fragments (three or more) detected with a TBLASTN search against the NCBI Rickettsiales database (taxid:766). Rickettsial palindromic element 3 (RPE-3) is depicted with a yellow ball-and-stick at its approximated insertion point in all derived SFG rickettsiae.
The POP family contains at least four subfamilies: the type prolyl oligopeptidase, dipeptidyl peptidase, oligopeptidase B, and acylaminoacyl peptidase (28, 29). PpcE is a member of the type prolyl oligopeptidase subfamily. Rickettsia species also harbor a ubiquitous POP-encoding gene, typically annotated as ptrB or the protease II gene, that is split in the genomes of both the RML369 C strain of Rickettsia bellii and R. peacockii. Furthermore, all spotted fever group (SFG) rickettsiae derived from REIS contain an insertion sequence (Rickettsia palindromic element 3) in the 5′ region of the gene. This nearly ubiquitous POP gene, herein named ptrB, is best described as a member of the oligopeptidase B subfamily. ptrB is also present in the sequenced genomes of Orientia and Pelagibacter species (see Fig. S1 in the supplemental material), suggesting that the gene has been lost from all Anaplasmataceae genomes and possesses typical characteristics of many other genes within the Rickettsiales that are undergoing pseudogenization.
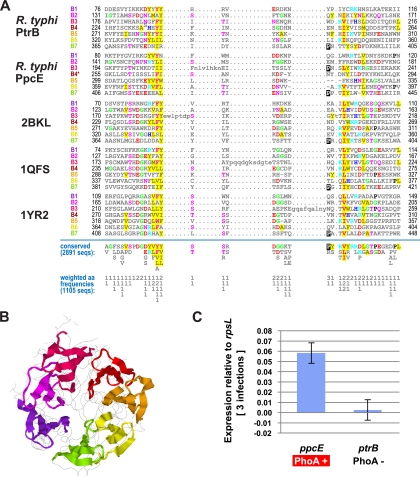
Comparison of R. typhi PtrB and PpcE with five diverse POP sequences illustrates the limited conservation in the β-propeller domain relative to the α/β hydrolase fold domain (see Fig. S3 in the supplemental material). Structural analysis of β-propeller domains from a large number of divergent POPs suggests that both PtrB and PpcE contain at least seven blades (Fig. 2A). Despite minimal sequence similarity, refined alignment with reference to several crystal structures (e.g., Fig. 2B) illustrates a hypervariable β-propeller domain. This is not surprising, as catalytic divergences have been demonstrated for POPs, ranging from hydrolysis of small peptides to cleavage of larger polypeptides, including triple-helical collagen fibers, fibronectin, and histones (7, 8, 17, 19, 20, 26, 30). Of note, a third POP-encoding rickettsial gene, named here dap2, was uncovered in our structural analysis despite having a β-propeller domain that is undetectable in BLASTP searches using PpcE or PtrB as a query (data not shown). Interestingly, dap2 is present at least once in all rickettsial genomes except TG rickettsiae (Fig. 1C) and, like ppcE, is undetectable in other Rickettsiales genomes (see Fig. S1 in the supplemental material). The converse distribution of these two highly divergent genes, relative to what was found for ptrB, further defines TG rickettsiae and accentuates another novel correlate to murine and epidemic typhus.
FIG. 2.
Predicted sequence locations of β-propeller blades within R. typhi PpcE and PtrB. Using as queries those sequence regions corresponding to the β-propeller domains of bacterial prolyl endopeptidases of known structure, PSI-BLAST (1) iterative searches of the NCBI NR database were preformed with a very conservative E value cutoff of 10−40. Those database sequence regions homologous to the queries were pooled, and both closely related sequences (those sharing ≥80% identity) and sequences less than 315 amino acids in length were removed. The remaining 380 sequence regions were used as input to a Bayesian Markov chain Monte Carlo sampling procedure for detecting subtly conserved internal repeats (22-24). This resulted in the detection and alignment of 2,891 putative β-propeller blades, including those shown in the alignment in panel A, which corresponds to regions within the R. typhi PpcE and PtrB sequences and to β-propeller blades of known structure within prolyl endopeptidases from Myxococcus xanthus (31) (as indicated in panel B), Sus scrofa (11), and Novosphingobium capsulatum (31) (Protein Data Bank accession numbers 2BKL, 1QFS, and 1YR2, respectively). The seven propeller blades are labeled in the N- to C-terminal direction as B1 to B7 in panel A and are colored purple, magenta, scarlet, red, orange, yellow, and green, respectively, in panel B. Note that the Bayesian sampler failed to detect the putative blade B4 within R. typhi PpcE (as indicated by an asterisk); instead, an optimal alignment of this region against a profile of the detected repeats was obtained independent of the sampler. The most commonly occurring residues at each position (within all 2,891 blades) are shown directly below the alignment, and directly below these, the corresponding (weighted) residue frequencies are given in integer tenths; for example, “2” indicates that the corresponding pattern residue occurs in 20% to 30% of the aligned sequences at that position. The most commonly occurring residue types are highlighted in the alignment, with chemically similar residues colored similarly, as previously described (21). The β-propeller domains in panel B are oriented perpendicularly to the structure shown in Fig. 1B. (C) Gene expression of R. typhi PpcE and PtrB in HeLa cells was as previously described (2). On the basis of Student's t test, the P value (for RT0165 versus RT0272) was 0.022503 (P < 0.05). The ability of the proximal N-terminal sequence of PpcE to translocate E. coli alkaline phosphatase (PhoA) to the periplasm is illustrated.
In addition to the strong bioinformatics-based evidence that PpcE represents a unique and functional protein specific to the TG rickettsiae, we found that ppcE is transcribed by R. typhi during infections in mammalian cells (Fig. 2C). HeLa cells were infected with R. typhi strain Wilmington with a multiplicity of infection of approximately 10; at 4 days postinfection, the rickettsiae were isolated for RNA extraction, and gene expression was analyzed using two-step, real-time quantitative reverse transcription-PCR, as previously described (2). In this model system, ppcE was significantly more highly expressed than ptrB, indicating that PpcE is likely important for the R. typhi intracellular lifestyle. Both of the TG PpcE homologs are predicted to encode an N-terminal signal peptide (18), suggesting possible extracytoplasmic secretion. We analyzed the function of the R. typhi PpcE putative signal peptide in an Escherichia coli-based system by fusing the predicted PpcE signal peptide to the E. coli alkaline phosphatase (PhoA) moiety lacking its native signal peptide (2). In E. coli, PhoA is a Sec-dependent protein that is enzymatically active only after its translocation into the bacterial periplasm, and its phosphatase activity can be visualized by the addition of a colorimetric substrate to the culture medium. We found that the putative PpcE signal peptide was able to direct the translocation of PhoA into the periplasmic space of E. coli, indicating that its cognate protein (PpcE) is likely extracytoplasmic. Taken together, these data indicate that PpcE has a biologically relevant role within the TG rickettsiae, with its probable extracytoplasmic localization suggesting it could be involved in host cell interactions. In support of this, it was recently found that R. prowazekii ppcE (RP174) is expressed in L929 cells and significantly upregulated following heat shock (4).
Recently sequenced rickettsial genomes have revealed the prevalence of plasmids and a high number of laterally acquired genes (32), exemplifying the streamlined nature of TG rickettsial genomes. A TG-specific ppcE gene warrants attention, as POPs have been implicated in virulence in other systems (5, 20, 25) and are present in the secretome of Bacillus anthracis (9). Interestingly, PpcE of Trypanosoma cruzi is secreted extracellularly by trypomastigotes and involved in nonphagocytic mammalian cell invasion (17, 30). TG rickettsial ppcE presents a new target for elucidating the mechanisms of pathogenicity underlying murine and epidemic typhus.
Supplementary Material
Acknowledgments
The project described here was supported by awards R01AI017828 and R01AI59118 from the National Institute of Allergy and Infectious Diseases (NIAID) to A.F.A., by NIH Division of General Medicine grant GM078541 to A.F.N., and through NIAID contract HHSN266200400035C to B.W.S.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the National Institutes of Health.
We thank members of the Azad laboratory (UMB) and the Cyberinfrastructure Group (VBI) for critical advice during the completion of this work.
Footnotes
Published ahead of print on 9 October 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ammerman, N. C., M. S. Rahman, and A. F. Azad. 2008. Characterization of Sec-translocon-dependent extracytoplasmic proteins of Rickettsia typhi. J. Bacteriol. 190:6234-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, S. G., and C. G. Kurland. 1995. Genomic evolution drives the evolution of the translation system. Biochem. Cell Biol. 73:775-787. [DOI] [PubMed] [Google Scholar]
- 4.Audia, J. P., M. C. Patton, and H. H. Winkler. 2008. DNA microarray analysis of the heat shock transcriptome of the obligate intracytoplasmic pathogen Rickettsia prowazekii. Appl. Environ. Microbiol. 74:7809-7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banbula, A., P. Mak, M. Bugno, J. Silberring, A. Dubin, D. Nelson, J. Travis, and J. Potempa. 1999. Prolyl tripeptidyl peptidase from Porphyromonas gingivalis. A novel enzyme with possible pathological implications for the development of periodontitis. J. Biol. Chem. 274:9246-9252. [DOI] [PubMed] [Google Scholar]
- 6.Braig, H. R., B. D. Turner, and M. A. Perotti. 2008. Symbiotic rickettsia, p. 221-249. In K. Bourtzis and T. A. Miller (ed.), Insect symbiosis, vol. 3. CRC Press, Boca Raton, FL.
- 7.Cavasin, M. A., N. E. Rhaleb, X. P. Yang, and O. A. Carretero. 2004. Prolyl oligopeptidase is involved in release of the antifibrotic peptide Ac-SDKP. Hypertension 43:1140-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, H. C., M. Abdel-Ghany, R. C. Elble, and B. U. Pauli. 1998. Lung endothelial dipeptidyl peptidase IV promotes adhesion and metastasis of rat breast cancer cells via tumor cell surface-associated fibronectin. J. Biol. Chem. 273:24207-24215. [DOI] [PubMed] [Google Scholar]
- 9.Chitlaru, T., O. Gat, Y. Gozlan, N. Ariel, and A. Shafferman. 2006. Differential proteomic analysis of the Bacillus anthracis secretome: distinct plasmid and chromosome CO2-dependent cross talk mechanisms modulate extracellular proteolytic activities. J. Bacteriol. 188:3551-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darby, A. C., N. H. Cho, H. H. Fuxelius, J. Westberg, and S. G. Andersson. 2007. Intracellular pathogens go extreme: genome evolution in the Rickettsiales. Trends Genet. 23:511-520. [DOI] [PubMed] [Google Scholar]
- 11.Fulop, V., Z. Bocskei, and L. Polgar. 1998. Prolyl oligopeptidase: an unusual beta-propeller domain regulates proteolysis. Cell 94:161-170. [DOI] [PubMed] [Google Scholar]
- 12.Fulop, V., Z. Szeltner, and L. Polgar. 2000. Catalysis of serine oligopeptidases is controlled by a gating filter mechanism. EMBO Rep. 1:277-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillespie, J. J., N. C. Ammerman, M. Beier-Sexton, B. S. Sobral, and A. F. Azad. 2009. Louse- and flea-borne rickettsioses: biological and genomic analyses. Vet. Res. 40:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillespie, J. J., N. C. Ammerman, S. M. Dreher-Lesnick, M. S. Rahman, M. J. Worley, J. C. Setubal, B. S. Sobral, and A. F. Azad. 2009. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS ONE 4:e4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillespie, J. J., M. S. Beier, M. S. Rahman, N. C. Ammerman, J. M. Shallom, A. Purkayastha, B. S. Sobral, and A. F. Azad. 2007. Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS ONE 2:e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillespie, J. J., K. Williams, M. Shukla, E. E. Snyder, E. K. Nordberg, S. M. Ceraul, C. Dharmanolla, D. Rainey, J. Soneja, J. M. Shallom, N. D. Vishnubhat, R. Wattam, A. Purkayastha, M. Czar, O. Crasta, J. C. Setubal, A. F. Azad, and B. S. Sobral. 2008. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS ONE 3:e2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grellier, P., S. Vendeville, R. Joyeau, I. M. Bastos, H. Drobecq, F. Frappier, A. R. Teixeira, J. Schrevel, E. Davioud-Charvet, C. Sergheraert, and J. M. Santana. 2001. Trypanosoma cruzi prolyl oligopeptidase Tc80 is involved in nonphagocytic mammalian cell invasion by trypomastigotes. J. Biol. Chem. 276:47078-47086. [DOI] [PubMed] [Google Scholar]
- 18.Kall, L., A. Krogh, and E. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027-1036. [DOI] [PubMed] [Google Scholar]
- 19.Loster, K., K. Zeilinger, D. Schuppan, and W. Reutter. 1995. The cysteine-rich region of dipeptidyl peptidase IV (CD 26) is the collagen-binding site. Biochem. Biophys. Res. Commun. 217:341-348. [DOI] [PubMed] [Google Scholar]
- 20.Morty, R. E., V. Fulop, and N. W. Andrews. 2002. Substrate recognition properties of oligopeptidase B from Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3329-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neuwald, A. F., N. Kannan, A. Poleksic, N. Hata, and J. S. Liu. 2003. Ran's C-terminal, basic patch, and nucleotide exchange mechanisms in light of a canonical structure for Rab, Rho, Ras, and Ran GTPases. Genome Res. 13:673-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neuwald, A. F., and J. S. Liu. 2004. Gapped alignment of protein sequence motifs through Monte Carlo optimization of a hidden Markov model. BMC Bioinformatics 5:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuwald, A. F., J. S. Liu, and C. E. Lawrence. 1995. Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Sci. 4:1618-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuwald, A. F., and A. Poleksic. 2000. PSI-BLAST searches using hidden Markov models of structural repeats: prediction of an unusual sliding DNA clamp and of beta-propellers in UV-damaged DNA-binding protein. Nucleic Acids Res. 28:3570-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Overweg, K., A. Kerr, M. Sluijter, M. H. Jackson, T. J. Mitchell, A. P. de Jong, R. de Groot, and P. W. Hermans. 2000. The putative proteinase maturation protein A of Streptococcus pneumoniae is a conserved surface protein with potential to elicit protective immune responses. Infect. Immun. 68:4180-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, J. E., M. C. Lenter, R. N. Zimmermann, P. Garin-Chesa, L. J. Old, and W. J. Rettig. 1999. Fibroblast activation protein, a dual specificity serine protease expressed in reactive human tumor stromal fibroblasts. J. Biol. Chem. 274:36505-36512. [DOI] [PubMed] [Google Scholar]
- 27.Perlman, S. J., M. S. Hunter, and E. Zchori-Fein. 2006. The emerging diversity of Rickettsia. Proc. Biol. Sci. 273:2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polgar, L. 2002. The prolyl oligopeptidase family. Cell. Mol. Life Sci. 59:349-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rea, D., and V. Fulop. 2006. Structure-function properties of prolyl oligopeptidase family enzymes. Cell Biochem. Biophys. 44:349-365. [DOI] [PubMed] [Google Scholar]
- 30.Santana, J. M., P. Grellier, J. Schrevel, and A. R. Teixeira. 1997. A Trypanosoma cruzi-secreted 80 kDa proteinase with specificity for human collagen types I and IV. Biochem. J. 325:129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shan, L., I. I. Mathews, and C. Khosla. 2005. Structural and mechanistic analysis of two prolyl endopeptidases: role of interdomain dynamics in catalysis and specificity. Proc. Natl. Acad. Sci. USA 102:3599-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder, E. E., N. Kampanya, J. Lu, E. K. Nordberg, H. R. Karur, M. Shukla, J. Soneja, Y. Tian, T. Xue, H. Yoo, F. Zhang, C. Dharmanolla, N. V. Dongre, J. J. Gillespie, J. Hamelius, M. Hance, K. I. Huntington, D. Jukneliene, J. Koziski, L. Mackasmiel, S. P. Mane, V. Nguyen, A. Purkayastha, J. Shallom, G. Yu, Y. Guo, J. Gabbard, D. Hix, A. F. Azad, S. C. Baker, S. M. Boyle, Y. Khudyakov, X. J. Meng, C. Rupprecht, J. Vinje, O. R. Crasta, M. J. Czar, A. Dickerman, J. D. Eckart, R. Kenyon, R. Will, J. C. Setubal, and B. W. Sobral. 2007. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 35:D401-D406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinert, L. A., J. H. Werren, A. Aebi, G. N. Stone, and F. M. Jiggins. 2009. Evolution and diversity of Rickettsia bacteria. BMC Biol. 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.