Abstract
Infection with high-risk (HR) human papillomavirus (HPV) is the major cause of cervical cancer. However, relatively few infections progress to malignant disease. Progression to malignancy requires the overexpression of the E6 and E7 genes in the integrated HPV genome. It follows that the E6 and E7 transcripts could be useful markers of disease progression. The study presented here tests this possibility, using data from colposcopy and from cytological and histological tests to compare RNA assays for the E6 and E7 genes with DNA testing. A total of 180 women underwent colposcopy, cytology, and biopsy of suspected lesions (143 cases). Cervical brush specimens were analyzed for HPV DNA and for E6 and E7 mRNA. DNA from HR HPV was found in 57.8% of the specimens; E6 and E7 transcripts were found in 45%. The rates of detection of HPV DNA and of E6 and E7 transcripts were 33.3% and 25%, respectively, for specimens with normal findings; 51.4% and 31.9%, respectively, for specimens with cervical intraepithelial neoplasia grade 1 (CIN1); and 61.1% and 44.2% for specimens with CIN2, respectively. All specimens with CIN3 and 95.5% of specimens from patients with squamous cell carcinoma were positive by both assays. Thirty-seven patients with normal colposcopy findings did not undergo biopsy. HPV DNA and mRNA transcripts were found in 32.4% and 18.9% of these cases, respectively. Comparisons with cytological tests produced similar results. Overall, the mRNA tests showed a higher specificity than the DNA tests for high-grade lesions (72.7% and 56.2%, respectively) and a higher positive predictive value (59.3% and 49.0%, respectively). These findings suggest that mRNA assays could be more powerful than DNA testing for predicting the risk of progression and offer a strong potential as a tool for triage and patient follow-up.
Carcinomas of the anogenital tract, particularly cancer of the cervix, represent the second most frequent type of neoplasm worldwide (43, 57). The major cause of these cancers is infection with high-risk (HR) human papillomavirus (HPV). DNA from HPV has been detected in more than 99% of cervical squamous cell carcinomas (SCCs) and a smaller proportion of adenocarcinomas (3, 4, 31, 33). However, most HPV infections regress spontaneously or progress only after a long period of latency. As a result, the number of infections is far higher than the number of women who develop cancer.
The most common types of HPV found in cancer patients are types 16, 18, 31, 33, and 45 (5, 10, 40, 53). Persistent infection with these types is regarded as a significant risk factor (40). The role of HPVs in the etiology of cervical cancer is tightly correlated with the overexpression of two oncogenes (E6 and E7) due to a specific opening in the E2 open reading frame in the integrated viral genome (23, 28). Studies of cervical cancer cell lines and cancer biopsy specimens have shown that the continuous expression of the genes is a necessary condition for the transformation and maintenance of neoplastic and dysplastic cells (46, 56, 57).
Cervical cancer is characterized by a well-defined premalignant phase that can be detected by cytological examination of exfoliated cervical cells and confirmed by histological examination of cervical material. Premalignant changes are reflected in a spectrum of histological abnormalities ranging from cervical intraepithelial neoplasia grade 1 (CIN1) or mild dysplasia, to moderate dysplasia (CIN2) and severe dysplasia or carcinoma in situ (CIN3 or CIS). Screening for these conditions has reduced the incidence of cervical cancer, especially in industrialized countries with effective screening programs. However, cervical cytology has limited sensitivity, specificity, and accuracy, especially in cases of low-grade or borderline lesions. As a result, cytological and histological examinations on their own are unable to distinguish the small number of women who will progress to invasive cancer from the vast majority of women whose abnormalities will spontaneously regress (8, 54).
In recent years, many studies have shown that testing for HPV DNA can improve the detection of high-grade squamous intraepithelial lesions (HSILs) and SCCs (33). This suggests that DNA testing can make a useful contribution to the triage of women with an equivocal cytology finding and to follow-up after the treatment of precursor lesions (9, 42, 45). However, the high prevalence of transient and asymptomatic HPV infections means that DNA tests have low specificities (38). Identification of the persistent infections likely to produce high-grade lesions currently requires repeated monitoring of the HPV DNA types (5, 19, 25). Commercial nucleic acid sequence-based amplification in a real-time format allows the reliable type-specific detection of E6 and E7 mRNA from HPV types 16, 18, 31, 33, and 45. Several authors have thus suggested that RNA-based assays could be more effective than DNA testing in risk assessment (16, 21, 22, 35, 36, 38, 48, 55). In the current study, we test this hypothesis using cytological and histological findings to compare the sensitivities, specificities, positive predictive values (PPVs), and negative predictive values (NPVs) of RNA and DNA testing.
MATERIALS AND METHODS
Study subjects and collection of specimens.
Specimens were collected from September 2007 to October 2008 from patients admitted for secondary screening to the Colposcopy Outpatient Service and the Gynecological Oncology Unit (Università Cattolica del Sacro Cuore, Rome, Italy) and the Department of Oncology (Università Cattolica del Sacro Cuore, Campobasso, Italy). The study group consisted of 180 women between 20 and 77 years of age (median age, 35 years; interquartile range [IQR], 13). Forty percent were aged between 30 and 39 years, 27% were under 30 years of age, and 32% were over 40 years of age. The age at first intercourse lay in the range of 13 to 39 years (median, 18 years; IQR, 3), with 49.7% of the patients having their first intercourse between 16 and 18 years of age. The number of partners was two to four for 45.6% of the women, and 38.3% of the women were nonsmokers. The study protocol was approved by the Ethical Committee of the Università Cattolica del Sacro Cuore, Rome, Italy. Written informed consent was obtained from all participants. All participants received a self-administered questionnaire requesting personal data, a gynecologic history, and information on exposure to risk factors. Pregnant women and women under treatment for invasive cervical cancer were excluded. All patients underwent cytology, colposcopy, and sampling for subsequent testing for HPV. In cases in which colposcopy suggested the presence of suspicious lesions, biopsy specimens were taken.
Cytology was based on a conventional Pap smear. The cytological diagnosis was made by specialized cytopathologists by use of the Bethesda classification system. Colposcopy was performed by specialized gynecologists. The results were reported following guidelines issued by SICPCV (the Italian Society of Colposcopy and Cervico-Vaginal Pathology). Histology was performed with specimens collected by colposcopy-directed biopsy (traditional punch biopsy specimens) and/or cone specimens collected by the loop excision procedure. Histology results were obtained for 143 patients (79.5%). The pathologists involved in the cytological and histological assessments were not involved in testing for HPV. Cervical specimens for nucleic acid analyses were collected with a cervical brush by standard procedures. The material was preserved in PreservCyt/ThinPrep solution (Cytyc Corporation, Boxborough, MA). Analyses were performed by the Virology Laboratory at the Università Cattolica del Sacro Cuore, Rome, Italy.
Nucleic acid isolation.
Each ThinPrep sample was divided into two aliquots (4 ml and 10 ml), which were used for DNA and RNA detection, respectively. The aliquot for HPV DNA hybridization was prepared by using a sample conversion kit (Digene, Milan, Italy), according to the manufacturer's instructions. The aliquot used for analysis for HPV mRNA was centrifuged. Total nucleic acid was extracted from the concentrated cell pellet by the off-board protocol with the NucliSens easyMAG platform (bioMérieux, Rome, Italy), according to the manufacturer's instructions. The nucleic acids were eluted in 55 μl of elution buffer. Aliquots were appropriately stored for further processing.
HPV DNA detection and genotyping.
HPV DNA detection was performed with the Hybrid Capture II system (HC2; Digene), according to the manufacturer's directions. HC2 is a nonradioactive signal amplification method based on the hybridization of the target HPV DNA to a labeled RNA probe in solution. The assay, which is in routine use in our laboratory, differentiates between low-risk (LR) HPV types (types 6, 11, 42, 43, and 44) and HR HPV types (types 16, 18, 31, 33, 35, 39, 45, 51, 52, 58, 59, and 68) but does not allow the identification of specific genotypes. The results were expressed in relative light units (RLUs), computed as the ratio of light emission to the mean value for three concurrently tested controls, each of which contained 1 pg/ml HPV DNA. One RLU can be considered a proxy for a viral load of 1 pg/ml. Samples with RLUs of less than 5 were retested by multiplex PCR to confirm their positivity for HR HPV DNA.
HPV DNA was genotyped for HPV types 16, 18, 31, 33, and 45 by multiplex PCR. The PCR was based on modified versions of the type-specific primer sequences described by van den Brule and colleagues (34, 52). Each assay used 5 μl of eluted nucleic acids and 20 pmol of each of the four primer sets. The reaction mixture contained 25 μl of HotStart Taq master mixture (Qiagen S.p.a., Milan, Italy) and 10 μl of RNase-free water in a final volume of 50 μl. The amplification profile consisted of 15 min at 95°C to activate the HotStar Taq DNA polymerase (Qiagen S.p.a.), followed by 45 cycles of denaturation (94°C for 30 s), annealing (56°C for 40 s), and extension (72°C for 40 s). Assays were performed on an iCycler thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA). Amplicons were detected by electrophoresis of 20 μl of the amplification products in a 2% agarose gel and were visualized by ethidium bromide staining under a UV light transilluminator (Fluor-S). The molecular sizes of the amplicons (for HPV type 16 [HPV-16], 152 bp; for HPV-18, 216 bp; for HPV-31, 513 bp; for HPV-33, 455 bp; and for HPV-45, 124 bp) were determined by matching them against commercial DNA molecular size markers (molecular weight markers V and VIII; Roche Diagnostics, GmbH, Mannheim, Germany).
To assess sensitivity, plasmids containing genomic DNA of HPV-16 and HPV-18, kindly provided by E. M. de Villiers (Deutsches Krebsforschungszentrum, Heidelberg, Germany), and the specific PCR products of HPV-31, HPV-33, and HPV-45 cloned into the pCR2.1 vector (Invitrogen, San Diego, CA) were quantified by measurement of the optical density. The constructs were serially diluted and subsequently amplified with type-specific primers. The multiplex PCR was shown to detect HPV at concentrations as low as 5 copies per sample.
HPV mRNA detection.
Samples were analyzed for HPV E6 and E7 mRNA by real-time multiplex nucleic acid sequence-based amplification. Transcripts of HR HPV types 16, 18, 31, 33, and 45 were detected by the NucliSens EasyQ HPV assay (bioMerieux, Rome, Italy), according to the manufacturer's instructions.
Statistical analysis.
All analyses were performed with Stata (version 10.1) statistical software. Summary results are presented as counts and percentages, with 95% binomial exact confidence intervals being used for categorical data and medians and IQR values being used for continuous data. The concordance among the DNA and RNA test results was evaluated by using Cohen's kappa statistic, as described by Fleiss (18). The sensitivities, specificities, and PPVs for the HPV DNA and RNA assays were estimated by comparison with the cytological and histological findings. Cytological results were grouped as normal findings-low-grade squamous intraepithelial lesions (LSILs) versus HSILs-SCC (target condition [17], HSILs-SCC); histological results were grouped as normal-CIN1 versus CIN2-SCC (target condition, CIN2-SCC). The expected values and 95% confidence intervals for sensitivity, specificity, PPV, and PNV were calculated as described by Seed and Tobias (47).
RESULTS
Cytological and histological findings.
As mentioned earlier, all patients underwent a conventional Pap smear test. All except two Pap smears (1.1%) were of satisfactory quality. The results for 44.4% (n = 80) of the smears were normal; 54.5% showed various forms of cytological abnormality. A total of 11.1% of cases (n = 20) showed atypical squamous cells of uncertain significance (ASCUS); LSILs were found in 25.5% (n = 46) of the women, and HSILs were detected in 13.9% (n = 25) of the women. SCCs were detected in 3.9% (n = 7) of the women (Table 1).
TABLE 1.
Cytological and histological findings for the 180 women enrolled in the study
| Cytology or histology result | No. (%) of women | Age (yr)a |
|---|---|---|
| Cytology result | ||
| Unsatisfactory | 2 (1.1) | |
| Normal | 80 (44.4) | 20-55 (33.5) |
| ASCUS | 20 (11.1) | 22-77 (33.5) |
| LSIL | 46 (25.6) | 22-59 (36.5) |
| HSIL | 25 (13.9) | 21-63 (35.0) |
| SCC | 7 (3.9) | 38-56 (43.0) |
| Total | 180 | |
| Histology result | ||
| Normal colposcopy findings, no biopsy | 37 (20.5) | 23-53 (37.0) |
| Normal-benign | 12 (6.7) | 21-53 (30.0) |
| CIN1 | 72 (40.0) | 20-58 (33.0) |
| CIN2 | 18 (10.0) | 22-59 (35.0) |
| CIN3-CIS | 19 (10.5) | 21-49 (34.0) |
| SCC | 22 (12.2) | 27-77 (44.5) |
| Total | 180 |
The data represent the range (median) ages.
Colposcopy was performed for all 180 cases. In 37 patients (20.5%), no suspect lesions were detected. These patients were subjected only to cytological surveillance. For the remaining 143 (79.5%) of the women, a specimen was taken by colposcopically directed biopsy. The majority of these patients had cytological findings within the normal limits and/or low-grade disease (Table 1). Among the 80 patients with normal cytology findings, biopsies were performed in 55 cases (68.7%), revealing 6 cases with normal histology findings, 34 with CIN1, 9 with CIN2, 3 with CIN3, and 3 with SCCs. High-grade lesions were found in 4 cases, and SCCs were found in 3 of 20 patients with ASCUS.
By consideration of the 143 patients, 12 biopsy samples (8.4%) were classified as normal/benign, 72 (50.3%) as CIN1, 18 (12.6%) as CIN2, and 19 (13.3%) as CIN3 or CIS. Twenty-two cases (15.4%) were classified as SCC (Table 1).
HPV DNA and mRNA tests.
All cervical specimens were tested for DNA and RNA from HR HPV. The tests were performed by investigators blinded to the cytology and histology results. The HC2 DNA assay detected HPV infection in 57.8% of the cases (104/180 patients). HR HPV DNA was found in 51.1% of the cases (92/180 patients). Mixed infections with LR types were detected in 11 cases (6.1%). Only one case was positive for LR HPV DNA (0.6%). The remaining 76 women (42.2%) tested HPV DNA negative.
The NucliSens EasyQ HPV assay, which detects E6 and E7 mRNA from HR HPV types 16, 18, 31, 33, and 45, identified specific transcripts in 81 of 180 (45%) samples.
To avoid false-negative results due to RNA degradation, all samples were tested with an RNA control (U1A) included in the HPV E6 and E7 mRNA test. All samples were positive.
In nine cases in which the HC2 test detected no DNA from HPV, the mRNA assay yielded positive results. These samples were retested by multiplex PCR. In each case, the assay confirmed the presence of the specific HPV genotype previously revealed by the RNA-based method.
The commonest HPV genotype revealed by RNA testing was HPV-16 (50/180 cases [27.8%]), followed by HPV-45 (16/180 cases [8.9%]), HPV-31 (11/180 cases [6.1%]), HPV-18 (9/180 cases [5.0%]), and HPV-33 (6 cases [3.3%]).
In 70/180 cases (38.9%), the test detected infections with single genotypes; 11/180 cases (6.1%) involved infections with multiple genotypes (Table 2). The most common were mixed infections with HPV-16 and HPV-45.
TABLE 2.
Prevalence of E6 and E7 transcripts from HPV types 16, 18, 31, 33, and 45
| HPV genotype | No. of patients infected with the indicated HPV type(s) as part of: |
% of patients | ||||||
|---|---|---|---|---|---|---|---|---|
| Single infection (n = 70) | Multiple infection (n = 11) |
Total | ||||||
| 16 | 18 | 31 | 33 | 45 | ||||
| 16 | 42 | 1 | 3 | 0 | 4 | 50/81 | 61.7 | |
| 18 | 6 | 1 | 0 | 0 | 2 | 9/81 | 11.1 | |
| 31 | 8 | 3 | 0 | 0 | 0 | 11/81 | 13.6 | |
| 33 | 5 | 0 | 0 | 0 | 1 | 6/81 | 7.4 | |
| 45 | 9 | 4 | 2 | 0 | 1 | 16/81 | 19.8 | |
Multiple infections were detected in 4 of 74 (5.4%) patients with CIN1, 1 of 18 with CIN2 (5.5%), 1 of 19 (5.2%) with CIN3-CIS, and 2 of 22 (9.1%) with SCC. Among 49 women with negative colposcopy findings who did not undergo biopsy (n = 37) or with normal histology findings (n = 12), there were 3 cases (6.1%) of double infection.
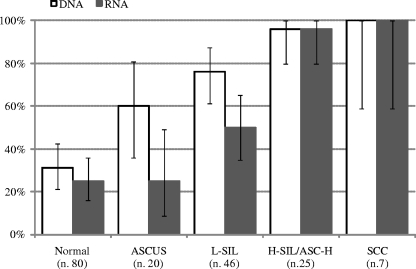
Comparing the data from our molecular assays with the cytological data, we found that the lowest prevalence rates for HR HPV DNA were in patients with normal cytology findings or ASCUS (31.2% and 60%, respectively) (Fig. 1). HPV DNA was also detected in one of the two smear samples unsuitable for cytological examination. A total of 76.1% (35/46) of the patients with LSILs and 96% (24/25) of the patients with HSILs or atypical squamous cells-cannot exclude HSILs (ASC-H) displayed simple or mixed infections with HR HPV. HPV DNA was found in all cases of carcinoma (n = 7).
FIG. 1.
Prevalence (95% confidence intervals) of positive results for HPV DNA and for E6 and E7 by cytological status of specimen.
E6 and E7 transcripts were detected in 20 of 80 patients with normal cytology findings (25%) by the mRNA test. The proportion of patients with detectable transcripts increased progressively with the grade of the lesions observed, rising from 25% for patients with ASCUS (5/20 patients) to 50% for those with LSILs (23/46 patients) and 96% for those with HSILs or ASC-H (24/25 patients). All cases Pap smear positive for SCC were positive for HPV RNA (Fig. 1).
The concordance between the DNA and RNA tests was fairly good for patients with normal cytological findings (81.3%; kappa = 0.53) and LSILs (69.6%; kappa = 0.39) but was slightly lower for patients with ASCUS (55%; kappa = 0.18); in cases of HSILs (92%) and SCCs (100%), the concordance was so high that the kappa statistics were no longer useful. Detailed results are shown in Table 3.
TABLE 3.
Concordance between HPV DNA and HPV E6 and E7 mRNA tests by cytological status of specimen
| Cytology result | No. of specimens | No. of specimens with the indicated result by: |
Concordancea | % | Kappa value | P | |||
|---|---|---|---|---|---|---|---|---|---|
| HPV DNA test |
HPV mRNA test |
||||||||
| + | − | + | − | ||||||
| Normal | 80 | 25 | 55 | 20 | 60 | 65/80 | 81.3 | 0.539 | <0.0001 |
| ASCUS | 20 | 12 | 8 | 5 | 15 | 11/20 | 55.0 | 0.182 | 0.1459 |
| LSIL | 46 | 35 | 11 | 23 | 23 | 32/46 | 69.6 | 0.391 | 0.0009 |
| Normal-LSIL | 146 | 72 | 74 | 48 | 98 | 108/146 | 74.0 | 0.477 | <0.0001 |
| HSIL | 25 | 24 | 1 | 24 | 1 | 23/25 | 92.0 | −0.042 | 0.5825 |
| SCC | 7 | 7 | 0 | 7 | 0 | 7/7 | 100.0 | ||
| HSIL-SCC | 32 | 31 | 1 | 31 | 1 | 30/32 | 93.8 | −0.032 | 0.5724 |
The data represent the number of specimens for which the test results were concordant/total number of specimens tested.
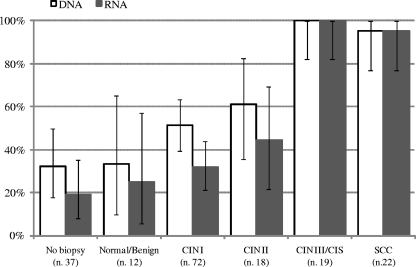
In terms of the histology findings, HPV DNA was detected in 33.3% of the 12 patients with normal-benign specimens and 32.4% of the 37 women with negative colposcopy findings who did not undergo biopsy. Infection with HR HPV was found in 51.4% (37/72) of cases of CIN1, 61.1% (11/18) of CIN2, 100% (19/19) of CIN3, and 95.5% (21/22) of SCC (Fig. 2).
FIG. 2.
Prevalence (95% confidence intervals) of positive results for HPV DNA and for E6 and E7 mRNA by histological status of specimens.
RNA tests showed a higher prevalence of E6 and E7 transcripts in patients with higher-grade lesions. Transcripts were detected in 25% (3/12) of the specimens with normal-benign findings, 31.9% (23/72) of those with CIN1, 44.4% (8/18) of those with CIN2, 100% (19/19) of those with CIN3, and 95.5% (21/22) of those with SCC. Among the 37 patients with normal colposcopy findings who did not undergo biopsy, HPV mRNA was present in 18.9% (Fig. 2).
The concordance between the DNA and the mRNA test results was fair for specimens from patients with normal colposcopy findings who did not undergo biopsy (64.9%; kappa = 0.10), very high for those with a normal-benign histology (91.7%; kappa = 0.80), and good for specimens classified as having CIN1 (72.2%; kappa = 0.45) or CIN2 (72.2%; kappa = 0.46). In the case of specimens classified as having CIN3-CIS or SCC, the concordance was so high (100% and 90.5%, respectively) that kappa statistics were no longer useful. Detailed results are shown in Table 4.
TABLE 4.
Concordance between HPV DNA and HPV E6 and E7 mRNA tests by histological status of specimen
| Histology result | No. of specimens | No. of specimens with the indicated result by: |
Concordancea | % | Kappa value | P | |||
|---|---|---|---|---|---|---|---|---|---|
| HPV DNA test |
HPV mRNA test |
||||||||
| + | − | + | − | ||||||
| No biopsy | 37 | 12 | 25 | 7 | 30 | 24/37 | 64.9 | 0.101 | 0.2565 |
| Normal | 12 | 4 | 8 | 3 | 9 | 11/12 | 91.7 | 0.800 | 0.0023 |
| CIN1 | 72 | 37 | 35 | 23 | 49 | 52/72 | 72.2 | 0.450 | <0.0001 |
| No biopsy CIN1 | 121 | 53 | 68 | 33 | 88 | 87/121 | 71.9 | 0.405 | <0.0001 |
| CIN2 | 18 | 11 | 7 | 8 | 10 | 13/18 | 72.2 | 0.458 | 0.0200 |
| CIN3-CIS | 19 | 19 | 0 | 19 | 0 | 19/19 | 100.0 | ||
| SCC | 22 | 21 | 1 | 21 | 1 | 20/22 | 90.9 | −0.048 | 0.5884 |
| CIN2-SCC | 59 | 51 | 8 | 48 | 11 | 52/59 | 79.4 | 0.563 | <0.0001 |
The data represent the number of specimens for which the test results were concordant/total number of specimens tested.
Sensitivity, specificity, and predictive values.
On the basis of the results described above, we used the cytology and histology findings to estimate the sensitivities, specificities, PPVs, and NPVs of positive DNA and RNA test results. In the cytology-based analysis, the target conditions defining disease were diagnoses of HSILs or SCC; in the histology-based analysis, the target conditions were diagnoses of CIN2, CIN3, CIS, or SCC. The results of the DNA and mRNA assays for patients who did not undergo biopsy were very similar. In view of this similarity, patients with normal findings for the biopsy specimens and patients who did not undergo biopsy were considered a single group. The results, presented in Table 5, show that in terms of the cytology target condition—currently considered the “gold standard”—the RNA and DNA assays had the same sensitivities. However the RNA assay had a higher specificity (67.1%) than the DNA-based test (50.7%). The histology results were similar. Although the DNA test was slightly more sensitive than the RNA assay (86.4% and 81.4%, respectively), the confidence intervals overlapped. The RNA assay had a significantly higher specificity than the DNA assay (72.7% and 56.2%, respectively). In this case, the overlap of the confidence intervals was minimal.
TABLE 5.
Sensitivities, specificities, PPVs, and NPVs of DNA and mRNA tests for prediction of cytological and histological findingsa
| Finding and test | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|
| Cytology finding of HSIL-SCC (sample prevalence, 18.0%) | ||||
| DNA test | 96.9 (83.8-99.9) | 50.7 (42.3-59.0) | 30.1 (21.5-39.9) | 98.7 (92.8-100.0) |
| mRNA test | 96.9 (83.8-99.9) | 67.1 (58.9-74.7) | 39.2 (28.4-50.9) | 99.0 (94.5-100.0) |
| Histology finding of CIN2, CIN3, CIS, and SCC (sample prevalence, 33.0%) | ||||
| DNA test | 86.4 (75.0-94.0) | 56.2 (46.9-65.2) | 49.0 (39.1-59.0) | 89.5 (80.3-95.3) |
| mRNA test | 81.4 (69.1-90.3) | 72.7 (63.9-80.4) | 59.3 (47.8-70.1) | 88.9 (81.0-94.3) |
The data represent point estimates (95% confidence intervals).
DISCUSSION
Cervical cancer is strongly associated with HPV infection (2, 5, 53). Progression to cervical carcinoma often extends over decades and is partly driven by the overexpression of HPV oncogenes. Although little is known regarding the possible transient nature of such expression, it is certain that the E6 and E7 proteins are consistently expressed in neoplastic tissue and play a significant role in malignant transformation.
Against this background, the goal of the study reported in this paper was to assess whether tests for these transcripts could be a better predictor of disease progression than screening for HR HPV DNA.
Few of the specimens investigated in our study came from a primary screening program; rather, they came from the secondary screening of patients referred for the evaluation of preneoplastic lesions. The histological findings from our study confirm earlier suggestions (8, 11, 44) that the conventional Pap smear test commonly used for both primary and secondary screening has a relatively poor sensitivity for high-grade cervical lesions.
Compared to the findings of earlier studies (24, 36), the results presented here are for a relatively large number of samples evaluated histologically. To the knowledge of the authors, the only study with a larger sample (n = 383) is that of Lie et al. (29).
In women displaying cervical abnormalities of any grade, the RNA assay produced fewer positive results than the DNA test. Given that not all HR HPV infections express E6 and E7, this result was expected. Although the HC2 test used for DNA detection detects 13 types of HR HPV while the NucliSens EasyQ HPV assay detects only 5 types (HPV types 6, 18, 31, 33, and 45), the HR genotypes included in the RNA assay are far more common than the rarer types in cancer specimens (10, 11, 26, 27, 30). It is thus very unlikely that differences in type coverage explain the gap in detection rates.
The results from the DNA and RNA assays associated well with the grade of lesion. The lowest rates of concordance were for patients with normal findings and low-grade lesions. In these cases, DNA from HPV was detected more frequently than the E6 and E7 transcripts. This result probably reflects the transient nature of most HPV infections, only a few of which produce precancerous lesions. However, the detection of E6 and E7 mRNA in a number of women with cytologically normal findings or low-grade lesions indicates that HR HPV may be oncogenically active before it produces detectable changes in cells and that the E6 and E7 transcripts could provide a sensitive, early predictor of persistent infection and subsequent severe dysplasia. Moreover, we cannot exclude the possibility that the condition in patients positive for HPV DNA but negative for mRNA will never progress to invasive cancer. Anyway, for this subset of patients, the timing of follow-up examinations can be less intensive. The value of repeated RNA testing as part of the follow-up examination remains to be assessed. We are currently studying these possibilities in a longitudinal follow-up study.
For patients with high-grade dysplasia and cancer (Fig. 1 and 2), the concordance between the RNA and DNA test results was high. This suggests that the presence of the E6 and E7 proteins is a specific marker for high-grade lesions and that positive RNA test results have a greater prognostic value than positive results from the DNA-based assays. In a single patient diagnosed with SCC, a DNA-positive specimen tested negative for RNA. It is possible that this negative result was due to a very low level of viral transcriptional activity. Alternatively, the DNA may have come from an HPV genotype not covered by the RNA test (27).
In 9 of the 81 HPV E6 and E7 RNA-positive cases, the HC2 test detected no HPV DNA. As reported earlier, subsequent retesting by multiplex PCR consistently confirmed the presence of DNA from the E6 region of the viral genome. This suggests that the virus was present only at very low copy numbers and/or that only a specific region of viral DNA was integrated into the host genome. The loss of the L1 gene and its impact on viral replication after integration could lead to relatively low viral loads (7, 15, 24, 32). Given that malignant phenotypes require continuous expression of the E6 and E7 oncogenes (13, 27, 39, 57) and that they produce transcripts throughout the epithelium and the surface layers (12, 15), it is not surprising that RNA assays detect more clinically significant infections than DNA testing, especially when samples come from the surface layers of the cervical epithelium, as in the present study.
In our series, the most frequent single type that caused infection (42/81 samples) was HPV-16, followed by HPV-45 (9/81), HPV-31 (8/81), HPV-18 (6/81), and HPV-33 (5/81). These findings are consistent with the HPV DNA prevalence rates reported by other Italian groups (1, 6, 14, 51) and match the rates of prevalence from combined DNA- and RNA-based studies (2, 7, 10, 13, 24).
As reported earlier, the study detected a number of mixed infections (Table 4). The most common were mixed infections with HPV-16 and HPV-45 (6.1% of cases). Although previous studies have suggested that disease progression may depend on the overexpression of E6 and E7 RNA by a single dominant type (7, 13), the precise role of these infections in carcinogenesis remains unclear.
For patients with histological diagnoses of CIN2, we found E6 and E7 transcripts in only 44.4% of the cases. It is likely that many of these lesions will regress. This prediction is supported by the results of earlier studies (20, 41) that showed that 32% of lesions resolve, even in the presence of high-grade dysplasia. For reasons of safety, these lesions are usually surgically resected. Our findings support previous suggestions that some women who undergo surgical resection may be overtreated (12, 26, 27, 29, 32).
As reported earlier, comparison of the results of the RNA assay with the results of cytology—the current gold standard—confirms the superior sensitivity and higher specificity of the RNA assay. This finding is supported by the results of earlier studies (2, 24, 36, 37, 49, 50). Keegan et al. (24) actually found a stronger difference (75.8% and 43.7% for the RNA assay and cytology, respectively), with the discrepancy possibly being due to differences in the sensitivity and the specificity of cytological diagnoses.
Comparison with histology findings confirms the superior specificity of the RNA assay. Given the positive impact on PPVs, it is likely that in subjects with a high expected prevalence of disease (e.g., groups at risk, symptomatic patients, and patients with persistent cytological abnormalities after negative colposcopy results), RNA assays will provide better risk predictions than DNA tests.
Our results so far suggest that the RNA assay has approximately the same sensitivity as DNA assays and a higher specificity. The test can provide sensitive, early-stage detection of persistent infections at risk of progression and can also identify lesions that are likely to regress. Under these conditions, it is possible that a single test by use of an RNA assay could be more effective at detecting an HR HPV infection than repeated DNA testing (13). If this is true, RNA assays could take on a valuable role in the triage of patients with abnormal cytology findings and the follow-up of patients who have been treated for neoplastic lesions. Potential benefits include reductions in the number of cases referred for colposcopy, improved patient well-being, and significant reductions in costs.
Assessment of this possibility requires investigation of the prospective sensitivities and specificities of RNA-based tests. To date, there have been few studies in this direction. The most significant, from Molden et al. (35), compared the value of HPV mRNA and DNA testing for the prediction of CIN2 or worse at 2 years after the detection of HPV DNA or RNA. The study, which involved 77 women from a set of 4,136 cases, showed that the RNA assay is more specific than DNA testing (84.8% and 50%, respectively). With the support of these preliminary findings, we have recently begun a longitudinal study of RNA-positive patients to better assess the role of RNA testing as a predictor of clinical outcomes.
Acknowledgments
This work was partly supported by grants 7021327 and 70200167 D.1 from the Università Cattolica del Sacro Cuore, Rome, Italy.
We thank Simona Galuppi for her expert technical assistance and Richard Walker for critical reading of the manuscript.
Footnotes
Published ahead of print on 14 October 2009.
REFERENCES
- 1.Ammatuna, P., L. Giovannelli, D. Matranga, S. Ciriminna, and A. Perino. 2008. Prevalence of genital human papilloma virus infection and genotypes among young women in Sicily, South Italy. Cancer Epidemiol. Biomarkers Prev. 17:2002-2006. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, S., B. Hansson, I. Norman, V. Gaberi, M. Mints, A. Hjerpe, F. Karlsen, and B. Johansson. 2006. Expression of E6/E7 mRNA from ‘high risk’ human papillomavirus in relation to CIN grade, viral load and p16INK4a. Int. J. Oncol. 29:705-711. [PubMed] [Google Scholar]
- 3.Bosch, F. X., M. M. Manos, N. Muñoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, K. V. Shah, et al. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl. Cancer Inst. 87:796-802. [DOI] [PubMed] [Google Scholar]
- 4.Bosch, F. X., and N. Munoz. 2002. The viral etiology of cervical cancer. Virus Res. 89:183-190. [DOI] [PubMed] [Google Scholar]
- 5.Bosch, F. X., A. Lorincz, N. Munoz, C. J. Meijer, and K. V. Shah. 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55:244-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broccolo, F., S. Chiari, A. Piana, P. Castiglia, T. Dell'Anna, R. Garcia-Parra, A. Maneo, A. Villa, E. B. Leone, P. Perego, A. Maida, C. Mangioni, and C. E. Cocuzza. 2009. Prevalence and viral load of oncogenic human papillomavirus types associated with cervical carcinoma in a population of north Italy. J. Med. Virol. 81:278-287. [DOI] [PubMed] [Google Scholar]
- 7.Cattani, P., A. Siddu, S. D'Onghia, S. Marchetti, R. Santangelo, V. G. Vellone, G. F. Zannoni, and G. Fadda. 2009. RNA (E6/E7) assays versus DNA (E6/E7) assays for risk evaluation for women infected with human papillomavirus. J. Clin. Microbiol. 47:2136-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clary, K. M., J. F. Silverman, Y. Liu, C. D. Sturgis, D. M. Grzybicki, L. K. Mahood, and S. S. Raab. 2002. Cytohistologic discrepancies: a means to improve pathology practice and patient outcomes. Am. J. Clin. Pathol. 117:567-573. [DOI] [PubMed] [Google Scholar]
- 9.Clavel, C., M. Masure, J. P. Bory, I. Putaud, C. Mangeonjean, M. Lorenzato, P. Nazeyrollas, R. Gabriel, C. Quereux, and P. Birembaut. 2001. Human papillomavirus testing in primary screening for the detection of high-grade cervical lesions: a study of 7932 women. Br. J. Cancer 84:1616-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clifford, G. M., J. S. Smith, M. Plummer, N. Munoz, and S. Franceschi. 2003. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br. J. Cancer 88:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox, T., and J. Cuzick. 2006. HPV DNA testing in cervical cancer screening: from evidence to policies. Gynecol. Oncol. 103:8-11. [DOI] [PubMed] [Google Scholar]
- 12.Cuschieri, K., and N. Wentzensen. 2008. Human papillomavirus mRNA and p16 detection as biomarkers for the improved diagnosis of cervical neoplasia. Cancer Epidemiol. Biomarkers Prev. 17:2536-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuschieri, K. S., M. J. Whitley, and H. A. Cubie. 2004. Human papillomavirus type specific DNA and RNA persistence—implications for cervical disease progression and monitoring. J. Med. Virol. 73:65-70. [DOI] [PubMed] [Google Scholar]
- 14.Del Prete, R., A. M. Di Taranto, M. R. Lipsi, V. Nirchio, R. Antonetti, and G. Miragliotta. 2008. Prevalence and genotypes identification of human papillomavirus infection in a population of south Italy. J. Clin. Virol. 42:211-214. [DOI] [PubMed] [Google Scholar]
- 15.Doorbar, J. 2006. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. (London) 110:525-541. [DOI] [PubMed] [Google Scholar]
- 16.Falcinelli, C., E. Claas, B. Kleter, and W. G. Quint. 1992. Detection of the human papillomavirus type 16 mRNA-transcripts in cytological abnormal scrapings. J. Med. Virol. 37:93-98. [DOI] [PubMed] [Google Scholar]
- 17.FDA. 13 March 2007, posting date. Statistical guidance on reporting results from studies evaluating diagnostic tests. http://www.fda.gov/cdrh/osb/guidance/1620.pdf.
- 18.Fleiss, J. L. 1981. Statistical methods for rates and proportions. John Wiley & Sons, Inc., New York, NY.
- 19.Ho, G. Y. F., R. Bierman, L. Beardsley, C. J. Chang, and R. D. Burk. 1998. Natural history of cervicovaginal papillomavirus infection in young women. N. Engl. J. Med. 338:423-428. [DOI] [PubMed] [Google Scholar]
- 20.Holowaty, P., A. B. Miller, T. Rohan, and T. To. 1999. Natural history of dysplasia of the uterine cervix. J. Natl. Cancer Inst. 91:252-258. [DOI] [PubMed] [Google Scholar]
- 21.Hsu, E. M., P. J. McNicol, F. B. Guijon, and M. Paraskevas. 1993. Quantification of HPV-16 E6-E7 transcription in cervical intraepithelial neoplasia by reverse transcriptase polymerase chain reaction. Int. J. Cancer 55:397-401. [DOI] [PubMed] [Google Scholar]
- 22.Jeon, S., B. L. Allen-Hoffmann, and P. F. Lambert. 1995. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 69:2989-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalantari, M., F. Karlsen, G. Kristensen, R. Holm, B. Hagmar, and B. Johansson. 1998. Disruption of the E1 and E2 reading frames of HPV 16 in cervical carcinoma is associated with poor prognosis. Int. J. Gynecol. Pathol. 17:146-153. [DOI] [PubMed] [Google Scholar]
- 24.Keegan, H., J. McInerney, L. Pilkington, P. Grønn, I. Silva, F. Karlsen, N. Bolger, C. Logan, L. Furuberg, J. O'Leary, and C. Martin. 2009. Comparison of HPV detection technologies: Hybrid Capture 2, PreTect HPV-Proofer and analysis of HPV DNA viral load in HPV16, HPV18 and HPV33 E6/E7 mRNA positive specimens. J. Virol. Methods 155:61-66. [DOI] [PubMed] [Google Scholar]
- 25.Kjaer, S. K., A. J. van den Brule, G. Paull, E. I. Svare, M. E. Sherman, B. L. Thomsen, M. Suntum, J. E. Bock, P. A. Poll, and C. J. Meijer. 2002. Type specific persistence of high risk human papillomavirus (HPV) as indicator of high grade cervical squamous intraepithelial lesions in young women: population based prospective follow up study. BMJ 325:572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraus, I., T. Molden, L. E. Ernø, H. Skomedal, F. Karlsen, and B. Hagmar. 2004. Human papillomavirus oncogenic expression in the dysplastic portio; an investigation of biopsies from 190 cervical cones. Br. J. Cancer 90:1407-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraus, I., T. Molden, R. Holm, A. K. Lie, F. Karlsen, G. B. Kristensen, and H. Skomedal. 2006. Presence of E6 and E7 mRNA from human papillomavirus types 16, 18, 31, 33, and 45 in the majority of cervical carcinomas. J. Clin. Microbiol. 44:1310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazo, P. A. 1997. Papillomavirus integration: prognostic marker in cervical cancer. Am. J. Obstet. Gynecol. 176:1121-1122. [DOI] [PubMed] [Google Scholar]
- 29.Lie, A. K., B. Risberg, B. Borge, B. Sandstad, J. Delabie, R. Rimala, M. Onsrud, and S. Thoresen. 2005. DNA-versus RNA-based methods for human papillomavirus detection in cervical neoplasia. Gynecol. Oncol. 97:908-915. [DOI] [PubMed] [Google Scholar]
- 30.Lie, A. K., and G. Kristensen. 2008. Human papillomavirus E6/E7 mRNA testing as a predictive marker for cervical carcinoma. Expert Rev. Mol. Diagn. 8:405-415. [DOI] [PubMed] [Google Scholar]
- 31.Lorincz, A. T., R. Reid, A. B. Jenson, M. D. Greenberg, W. Lancaster, and R. J. Kurman. 1992. Human papillomavirus infection of the cervix: relative risk associations of 15 common anogenital types. Obstet. Gynecol. 79:328-337. [DOI] [PubMed] [Google Scholar]
- 32.Manavi, M., G. Hudelist, A. Fink-Retter, D. Gschwantler-Kaulich, K. Pischinger, and K. Czerwenka. 2008. Human papillomavirus DNA integration and messenger RNA transcription in cervical low- and high-risk squamous intraepithelial lesions in Austrian women. Int. J. Gynecol. Cancer 18:285-294. [DOI] [PubMed] [Google Scholar]
- 33.Meijer, C. J., A. J. Van Den Brule, P. J. Snijders, T. Helmerhorst, P. Kenemans, and J. M. Walboomers. 1992. Detection of human papillomavirus in cervical scrapes by the polymerase chain reaction in relation to cytology: possible implications for cervical cancer screening. IARC Sci. Publ. 119:271-281. [PubMed] [Google Scholar]
- 34.Melchers, W., A. van den Brule, J. Walboomers, M. de Bruin, M. Burger, P. Herbrink, C. Meijer, J. Lindeman, and W. Quint. 1989. Increased detection rate of human papillomavirus in cervical scrapes by the polymerase chain reaction compared to modified FISH and Southern-blot analysis. J. Med. Virol. 27:329-335. [DOI] [PubMed] [Google Scholar]
- 35.Molden, T., I. Kraus, F. Karlsen, H. Skomedal, J. F. Nygård, and B. Hagmar. 2005. Comparison of human papillomavirus messenger RNA and DNA detection: a cross-sectional study of 4,136 women >30 years of age with a 2-year follow-up of high-grade squamous intraepithelial lesion. Cancer Epidemiol. Biomarkers Prev. 14:367-372. [DOI] [PubMed] [Google Scholar]
- 36.Molden, T., J. F. Nygård, I. Kraus, F. Karlsen, M. Nygård, G. B. Skare, H. Skomedal, S. O. Thoresen, and B. Hagmar. 2005. Predicting CIN2+ when detecting HPV mRNA and DNA by PreTect HPV-Proofer and consensus PCR: a 2-year follow-up of women with ASCUS or LSIL Pap smear. Int. J. Cancer 114:973-976. [DOI] [PubMed] [Google Scholar]
- 37.Molden, T., I. Kraus, F. Karlsen, H. Skomedal, and B. Hagmar. 2006. Human papillomavirus E6/E7 mRNA expression in women younger than 30 years of age. Gynecol. Oncol. 100:95-100. [DOI] [PubMed] [Google Scholar]
- 38.Molden, T., I. Kraus, H. Skomedal, T. Nordstrøm, and F. Karlsen. 2007. PreTect HPV-Proofer: real-time detection and typing of E6/E7 mRNA from carcinogenic human papillomaviruses. J. Virol. Methods 142:204-212. [DOI] [PubMed] [Google Scholar]
- 39.Münger, K., A. Baldwin, K. M. Edwards, H. Hayakawa, C. L. Nguyen, M. Owens, M. Grace, and K. Huh. 2004. Mechanisms of human papillomavirus-induced oncogenesis. J. Virol. 78:11451-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muñoz, N., F. X. Bosch, S. de Sanjosé, R. Herrero, X. Castellsagué, K. V. Shah, P. J. Snijders, C. J. Meijer, and the International Agency for Research on Cancer Multicenter Cervical Cancer Study Group. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518-527. [DOI] [PubMed] [Google Scholar]
- 41.Ostör, A. G. 1993. Natural history of cervical intraepithelial neoplasia: a critical review. Int. J. Gynecol. Pathol. 12:186-192. [PubMed] [Google Scholar]
- 42.Petry, K. U., S. Menton, M. Menton, F. van Loenen-Frosch, H. de Carvalho Gomes, B. Holz, B. Schopp, S. Garbrecht-Buettner, P. Davies, G. Boehmer, E. van den Akker, and T. Iftner. 2003. Inclusion of HPV testing in routine cervical cancer screening for women above 29 years in Germany: results for 8466 patients. Br. J. Cancer 88:1570-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pisani, P., F. Bray, and D. M. Parkin. 2002. Estimates of the worldwide prevalence of cancer for 25 sites in the adult population. Int. J. Cancer 97:72-81. [DOI] [PubMed] [Google Scholar]
- 44.Sasieni, P. D., J. Cuzick, and E. Lynch-Farmery. 1996. Estimating the efficacy of screening by auditing smear histories of women with and without cervical cancer. Br. J. Cancer 73:1001-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider, A., H. Hoyer, B. Lotz, S. Leistritza, R. Kühne-Heid, I. Nindl, B. Müller, J. Haerting, and M. Dürst. 2000. Screening for high-grade cervical intraepithelial neoplasia and cancer by testing for high-risk HPV, routine cytology or colposcopy. Int. J. Cancer 89:529-534. [PubMed] [Google Scholar]
- 46.Schwarz, E., U. K. Freese, L. Gissmann, W. Mayer, B. Roggenbuck, A. Stremlau, and H. zur Hausen. 1985. Structure and transcription of human papillomavirus type 18 and 16 sequences in cervical carcinoma cells. Nature 314:111-114. [DOI] [PubMed] [Google Scholar]
- 47.Seed, P. T., and A. Tobias. 2001. Summary statistics for diagnostic tests. Stata Tech. Bull., sbe36.1. 59:9-12. [Google Scholar]
- 48.Sotlar, K., H. C. Selinka, M. Menton, R. Kandolf, and B. Bultmann. 1998. Detection of human papillomavirus type 16 E6/E7 oncogene transcripts in dysplastic and non dysplastic cervical scrapes by nested RT-PCR. Gynecol. Oncol. 69:114-121. [DOI] [PubMed] [Google Scholar]
- 49.Sotlar, K., A. Stubner, D. Diemer, S. Menton, M. Menton, K. Dietz, D. Wallwiener, R. Kandolf, and B. Bültmann. 2004. Detection of high-risk human papillomavirus E6 and E7 oncogene transcripts in cervical scrapes by nested RT-polymerase chain reaction. J. Med. Virol. 74:107-116. [DOI] [PubMed] [Google Scholar]
- 50.Szarewski, A., L. Ambroisine, L. Cadman, J. Austin, L. Ho, G. Terry, S. Liddle, R. Dina, J. McCarthy, H. Buckley, C. Bergeron, P. Soutter, D. Lyons, and J. Cuzick. 2008. Comparison of predictors for high-grade cervical intraepithelial neoplasia in women with abnormal smears. Cancer Epidemiol. Biomarkers Prev. 17:3033-3042. [DOI] [PubMed] [Google Scholar]
- 51.Tornesello, M. L., M. L. Duraturo, G. Botti, S. Greggi, R. Piccoli, G. De Palo, M. Montella, L. Buonaguro, F. M. Buonaguro, and the Italian HPV Working Group. 2006. Prevalence of alpha-papillomavirus genotypes in cervical squamous intraepithelial lesions and invasive cervical carcinoma in the Italian population. J. Med. Virol. 78:1663-1672. [DOI] [PubMed] [Google Scholar]
- 52.van den Brule, A. J., C. J. Meijer, V. Bakels, P. Kenemans, and J. M. Walboomers. 1990. Rapid detection of human papillomavirus in cervical scrapes by combined general primer-mediated and type-specific polymerase chain reaction. J. Clin. Microbiol. 28:2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Muñoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 54.Woodman, C. B. J., S. I. Collins, and L. S. Young. 2007. The natural history of cervical HPV infection: unresolved issues. Nat. Rev. Cancer 7:11-22. [DOI] [PubMed] [Google Scholar]
- 55.zur Hausen, H., and E. M. de Villiers. 1994. Human papillomaviruses. Annu. Rev. Microbiol. 48:427-447. [DOI] [PubMed] [Google Scholar]
- 56.zur Hausen, H. 1996. Papillomavirus infections: a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]
- 57.zur Hausen, H. 2002. Papillomavirus and cancer: from basic studies to clinical application. Nat. Rev. 2:342-350. [DOI] [PubMed] [Google Scholar]