Abstract
RNA viruses within a host exist as dynamic distributions of closely related mutants and recombinant genomes. These closely related mutants and recombinant genomes, which are subjected to a continuous process of genetic variation, competition, and selection, act as a unit of selection, termed viral quasispecies. Characterization of mutant spectra within hosts is essential for understanding viral evolution and pathogenesis resulting from the cooperative behavior of viral mutants within viral quasispecies. Furthermore, a detailed analysis of viral variability within hosts is needed to design control strategies, because viral quasispecies are reservoirs of viral variants that potentially can emerge with increased virulence or altered tropism. In this work, we report a detailed analysis of within-host viral populations in 13 field isolates of the bipartite Tomato chlorosis virus (ToCV) (genus Crinivirus, family Closteroviridae). The intraisolate genetic structure was analyzed based on sequencing data for 755 molecular clones distributed in four genomic regions within the RNA-dependent RNA polymerase (RNA1) and Hsp70h, CP, and CPm (RNA2) open reading frames. Our results showed that populations of ToCV within a host plant have a heterogeneous and complex genetic structure similar to that described for animal and plant RNA viral quasispecies. Moreover, the structures of these populations clearly differ depending on the RNA segment considered, being more complex for RNA1 (encoding replication-associated proteins) than for RNA2 (encoding encapsidation-, systemic-movement-, and insect transmission-relevant proteins). These results support the idea that, in multicomponent RNA viruses, function can generate profound differences in the genetic structures of the different genomic segments.
RNA viruses within a host exist as dynamic distributions of closely related mutants and recombinant genomes subjected to a continuous process of genetic variation, competition, and selection. These closely related mutants and recombinant genomes act as a unit of selection and are termed viral quasispecies (1, 12). Quasispecies complexity arises as a result of the genetic variation introduced by the low fidelity of RNA polymerases, and reverse transcriptases in the case of retroviruses, in combination with large progenies, small genome sizes, and short replicative cycles. Positive and negative selection and genetic drift and migration are the main evolutionary forces acting on the mutant spectrum. As a consequence, quasispecies evolve with a great capacity to adapt to changing environments and to survive even after strong population bottlenecks (15). Characterization of mutant spectra within hosts is essential for understanding viral evolution and pathogenesis resulting from the cooperative behavior of viral mutants within viral quasispecies (11). Furthermore, a detailed analysis of viral variability within hosts is needed to design appropriate control strategies because viral quasispecies are reservoirs of viral variants that potentially can emerge with increased virulence or altered tropism.
Most plant viruses have a genome constituted by RNA, and many of them cause very important diseases in agricultural crops (22). Analyses of quasispecies diversity in plant viruses are scarce. Studies have focused on experimental evolution, and little work has been done to analyze quasispecies in field isolates (reviewed in reference 44). Researchers have only recently begun to study the nature of plant RNA virus populations within individual host plants and the factors that affect the effective population sizes (the fraction of the population that passes its genes to the new generation), such as genetic bottlenecks and positive and negative selection (44).
Two main hypotheses have been proposed to explain the evolution of multicomponent RNA viruses (i.e., those with segmented genomes encapsidated in separate particles). Nee (36) suggested that evolution toward smaller RNA molecules could be favored by selection on RNAs within a host cell because smaller RNA molecules could be replicated and encapsidated more rapidly. In contrast, Chao (5) proposed that multicomponent reproduction evolved in RNA viruses as a form of sex. Sex can be advantageous to organisms that experience high mutation rates (like RNA viruses) because it can bring together genomes that have not been destroyed by deleterious mutations and thereby maintain or increase genetic population robustness (14).
The current paper concerns the within-host genetic structure of Tomato chlorosis virus (ToCV) (58), an economically important crinivirus within the family Closteroviridae, the most complex family of positive-strand RNA plant viruses. Criniviruses have two single-stranded RNA molecules (RNA1 and RNA2) of positive polarity, with the exception of the tripartite Potato yellow vein virus (28). Both RNA1 and RNA2 are needed for infectivity and are separately encapsidated in long and flexuous virions that are transmitted in nature by whiteflies (Hemiptera: Aleyrodidae) in a semipersistent manner.
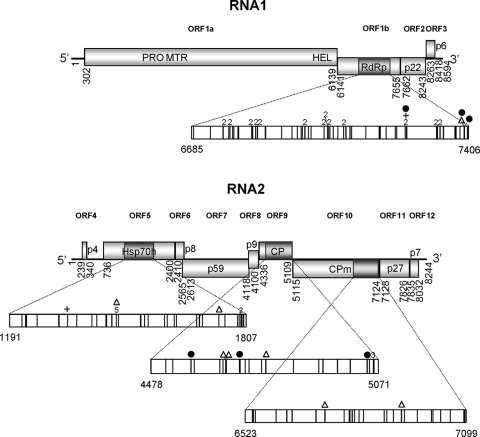
ToCV RNA1 encompasses four open reading frames (ORFs) (31, 57) (Fig. 1). The first two ORFs encode proteins involved in the replication of viral RNA. ORF 1a encodes a protein containing protease, methyltransferase, and helicase domains. ORF 1b encodes a protein containing the conserved motifs identified in the RNA-dependent RNA polymerases (RdRp) of positive-strand RNA viruses. ORF 2 encodes a putative protein of unknown function, and the predicted small ORF 3 encodes a putative protein of 6 kDa with a transmembrane domain similar to those of other 3′-end proteins of criniviruses. RNA2 includes nine ORFs that encompass the hallmark gene array of the family Closteroviridae; these ORFs are involved in encapsidation, movement, and host transmission (9) (Fig. 1). They encode a heat shock protein 70 family homologue (Hsp70h); a 59- to 60-kDa protein, the coat protein (CP); and a diverged CP (CPm) (30, 57).
FIG. 1.
Schematic representation of the ToCV genome in which analyzed regions are represented as dark shaded boxes. Mutations, identified by comparing each individual sequence with its corresponding consensus sequence are indicated with bars in the enlarged fragments. Mutations detected in more than one sequence are indicated by the number of times they have been identified. ▵, deletion; +, insertion; •, stop codon.
Intraisolate genetic diversity has been studied for a few members of the Closteroviridae family, namely the closterovirus Citrus tristeza virus (CTV) (2, 3, 25, 47, 56), the ampeloviruses Grapevine leafroll-associated virus 1 (GLRaV-1) and GLRaV-3 (24, 51), and the criniviruses Cucurbit yellow stunt disease virus (CYSDV) and Lettuce infectious yellows virus (LIYV) (45, 46).
This paper reports a detailed analysis of within-host viral populations in field isolates of ToCV from southeastern Spain. The intraisolate genetic structures of 13 infected tomato plants were analyzed based on sequencing data for 755 molecular clones distributed in four genomic regions within the RdRp, Hsp70h, CP, and CPm ORFs. Our results demonstrate that within-host populations of ToCV have a heterogeneous and complex genetic structure similar to that described for animal and plant RNA viral quasispecies. Moreover, the structures of these populations clearly differ depending on the RNA segment considered. Our results suggest that these differences are related to the specific function of each genomic segment.
MATERIALS AND METHODS
Virus isolates.
ToCV isolates were obtained from tomato plants sampled from commercial field-grown or greenhouse-grown crops in the provinces of Málaga, Almería, and Murcia in southeastern Spain from 1997 to 2004 (see Table 2 for a summary). The plants were tested for ToCV infection by petiole tissue printing hybridization with a digoxigenin-labeled probe recognizing the CP gene. Leaf samples from the ToCV-infected plants were stored at −80°C until RNA extraction. RNA was extracted with TRIzol reagent (Sigma) following the manufacturer's instructions plus an extra step of centrifugation at 12,000 × g at 4°C for 10 min after the addition of the reagent to remove insoluble glycopolysaccharides. RNA obtained from 0.1 g leaf tissue was resuspended in 50 μl of diethyl pyrocarbonate-treated water.
TABLE 2.
Intraisolate genetic complexity and heterogeneity of mutant spectra in 13 field isolates of ToCV
| Isolate | Origin | RNA1 |
RNA2 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| RdRpc |
Hsp70hd |
CPe |
CPmf |
||||||
| Mutation frequencya (10−4) | Shannon entropyb | Mutation frequency (10−4) | Shannon entropy | Mutation frequency (10−4) | Shannon entropy | Mutation frequency (10−4) | Shannon entropy | ||
| AT 1/97 | Málaga | 9.22 | 0.680 | 3.24 | 0.213 | 1.60 | 0.089 | 3.47 | 0.263 |
| AT 3/98 | Almería | 3.41 | 0.256 | <1.08 | 0.000 | 2.22 | 0.121 | 6.12 | 0.232 |
| AT 24/99 | Málaga | 3.42 | 0.265 | 4.32 | 0.395 | <1.16 | 0.000 | 1.02 | 0.077 |
| AT 80/99 | Málaga | 22.70 | 0.706 | NDg | ND | ND | ND | ND | ND |
| AT 198/00 | Málaga | 13.00 | 0.321 | 2.16 | 0.177 | 1.09 | 0.083 | 3.47 | 0.263 |
| AT 205/00 | Murcia | 4.55 | 0.404 | 3.60 | 0.33 | 1.84 | 0.069 | <1.08 | 0.000 |
| AT 230/00 | Almería | 1.80 | 0.209 | 5.71 | 0.443 | 3.50 | 0.275 | 2.17 | 0.165 |
| AT 36/02 | Murcia | 2.76 | 0.296 | 4.05 | 0.325 | 3.08 | 0.179 | 2.31 | 0.177 |
| AT 45/02 | Málaga | 0.90 | 0.089 | 4.05 | 0.246 | 5.82 | 0.429 | 5.78 | 0.347 |
| AT 31/03 | Almería | 2.95 | 0.177 | 2.16 | 0.299 | 2.05 | 0.154 | 1.15 | 0.089 |
| AT 138/03 | Murcia | 0.98 | 0.097 | 3.24 | 0.452 | 2.33 | 0.416 | 3.47 | 0.229 |
| AT 204/04 | Almería | 2.76 | 0.296 | 6.07 | 0.477 | 3.50 | 0.263 | 1.15 | 0.089 |
| AT 379/04 | Murcia | 6.08 | 0.432 | 2.16 | 0.177 | 2.33 | 0.177 | 2.31 | 0.177 |
Mutation frequencies (substitutions per nucleotide) were calculated by scoring the mutations present in sequences of molecular clones relative to their consensus sequence. One substitution was considered to compute the maximum mutation frequencies if no mutations were scored. Repeated mutations were scored only once for each isolate and genomic region. The consensus sequence was obtained by direct sequencing of RT-PCR products.
Shannon entropy values were calculated using the following formula: −Σi [(pi × ln pi)/ln N], in which pi is the frequency of each sequence in the mutant spectrum and N is the total number of sequences compared (54).
189 molecular clones (129,886 nt) were sequenced.
191 molecular clones (117,767 nt) were sequenced.
189 molecular clones (108,297 nt) were sequenced.
186 molecular clones (107,322 nt) were sequenced.
ND, not determined.
Amplification targets and oligonucleotides.
Conserved sequences in the ToCV genome were identified after multiple-sequence alignment, with the ClustalX program (50), of the criniviruses (GenBank accession numbers are in parentheses) ToCV (Florida isolate, AY903447 and AY903448; AT80/99 Spain isolate, DQ983480 and DQ136146), LIYV (U15440, U15441), Strawberry pallidosis-associated virus (AY488137, AY488138), Beet pseudo yellows virus (AY330918, AY330919), CYSDV (AY242077, AY242078), Sweet potato chlorotic stunt virus (AJ428554, AJ428555), and Potato yellow vein virus (AJ557128, AJ557129, AJ508757) and the closterovirus Beet yellow virus (X73476). Conserved amino acid domains of the ToCV proteins RdRp, Hsp70h, CP, and CPm were identified using the ANAGram protein function assignment program (http://jaguar.genetica.uma.es/anagram.htm) (41). The sequences of oligonucleotide primers designed within conserved genomic regions of RNA1 were as follows (nucleotide numbers are according to the AT80/99 Spain ToCV isolate [30, 31]): RdRp (from 6664 to 7426; 763 nt), primers MA396 (forward; 5′-TGGTCGAACAGTTTGAGAGC-3′) and MA397 (reverse; 5′-TGAACTCGAATTGGGACAGA-3′). Primer sequences targeting conserved genomic regions of RNA2 were as follows: Hsp70h (from 1171 to 1827; 657 nt), primers MA394 (forward; 5′-CCGGCTGATTACAAGTCTGG-3′) and MA395 (reverse; 5′-CTCTTGTGCATGGAGCATTG-3′); CP (from 4456 to 5072; 617 nt), primers MA461 (forward; 5′-ACATCTCTCATTCCGGCTAATC-3′) and MA462 (reverse; 5′-TACAGTTCCTTGCCCTCGTTAC-3′); and CPm (from 6503 to 7119; 617 nt), primers MA392 (forward; 5′-TAAGGTCCAAACCGAAGTGG-3′) and MA393 (reverse; 5′-AAAGCTGACTCGTGCTCACA-3′).
Viral-RNA amplification, cloning, and sequencing.
RNAs were amplified by two-step reverse transcription (RT)-PCR as follows. One microliter of RNA was thawed at 70°C for 3 min, and after it was cooled on ice, 8 pmol of reverse primer was added, and the mixture was incubated at 65°C for 5 min and cooled on ice. A mixture containing 2.5 U of avian myeloblastosis virus reverse transcriptase (Promega) and 10 U of recombinant RNasin RNA inhibitor (Promega) was added to the primer mixture in a final reaction volume of 20 μl. The RT reaction was done at 37°C for 45 min, followed by incubation at 94°C for 5 min. Five microliters of the first-strand reaction mixture was used in a touchdown PCR in a volume of 50 μl with 1.25 U of Pfu DNA polymerase and 32 pmol of reverse and forward primers. Touchdown PCR proceeded with an initial denaturation at 95°C for 2 min, followed by 20 cycles at 95°C for 1 min, 60°C for 30 s (with a decrease of 0.5°C per cycle), and 72°C for 2 min and finally 10 additional cycles at an annealing temperature of 45°C. To ensure that the small quantity of template did not produce a bottleneck, we used cDNA only from RNA samples that had yielded positive products in a 1/10 dilution. Ten microliters of PCR product was treated with 10 U of exonuclease I and 2 U of shrimp alkaline phosphatase (both from Fermentas) and was directly sequenced to obtain the consensus sequence. The PCR products were cloned into the pCR-BluntII-TOPO vector (Invitrogen) and transferred into Escherichia coli Top10 electrocompetent cells (Invitrogen). Plasmids from 9 to 20 positive colonies for each ORF and isolate were amplified with ϕ29 DNA polymerase (TempliPhi amplification kit; Amersham) by following the manufacturer's protocol and were then sequenced. The basal mutation frequency (or experimental error), which was determined after sequencing 22 molecular clones of a T7 runoff transcript of the CP region, was 1.47 × 10−4 (two mutations found in 13,574 nt sequenced). The observed error was comparable to that reported for T7 or SP6 RNA polymerase (0.5 × 10−4 or 1.34 × 10−4 misincorporations per copied nucleotide, respectively [21, 43]). These numbers of misincorporations would generate 0.67 or 1.82 mutations, respectively.
Sequence analysis.
Multiple-sequence alignments for each field isolate and genomic region were performed using ClustalX with the default parameters. A standard nonparametric one-way run test, available in SPSS 14.0 statistical software (SPSS Inc.), was performed in order to analyze the distribution of mutations as a function of genome position. The acceptability of amino acid changes identified in each genomic region was evaluated following the structure-genetic (SG) matrix of Feng et al. (17). In this matrix, structural similarities, as well as probabilities of amino acid changes, are assigned values between 0 and 6; drastic amino acid changes are given a value of 0, whereas synonymous replacements are given a value of 6.
Mutation frequencies were calculated by scoring different mutations (repeated mutations were counted only once) relative to the consensus sequence divided by the total number of nucleotides sequenced (12). Shannon entropy values were calculated using the following formula: −Σi [(pi × ln pi)/ln N], in which pi is the frequency of each sequence in the mutant spectrum and N is the total number of sequences compared (54). The calculated Shannon entropy values range from 0 (all sequences are identical) to 1 (all sequences are different).
Alignments were used to estimate pairwise genetic distances by Kimura's two-parameter method (23) implemented with MEGA version 3.1 (26) (available at http://www.megasoftware.net). Standard deviations were calculated by the bootstrap method with 1,000 repeats (37). Pairwise synonymous substitutions per synonymous site (dS) and nonsynonymous substitutions per nonsynonymous site (dN) were calculated according to the Pamilo-Bianchi-Li method based on Kimura's two-parameter model (27, 37, 39). Phylogenetic relationships were inferred by the neighbor-joining method available in MEGA3. The robustness of evolutionary relationships was assessed by 1,000 bootstrap replicates (37). The pairwise nucleotide identity profile of the putative recombinant molecule found in the RdRp region of sample AT80/99 was determined with SimPlot (29).
A hierarchical analysis of molecular variance (AMOVA) was used to evaluate the distribution of the genetic diversity, based on the type and frequency of every sequence in each isolate and on the likelihood that the distribution was random; the AMOVA was performed using Arlequin 3.01 (16). Total variance was partitioned into variance components due to within-isolate and between-isolate variance. Genetic differentiation between ToCV subpopulations was estimated with the F statistic (55), and its significance was tested by nonparametric permutation analysis based on 10,000 repetitions (16).
RESULTS
Distribution of genetic variability within field isolates of ToCV.
To determine the genetic composition of ToCV isolates, RNA was extracted from 13 tomato plants infected with ToCV, and four genomic regions in the RdRp (RNA1), Hsp70h, CP, and CPm (RNA2) ORFs were amplified. Between 9 and 20 clones for each genomic region and isolate were sequenced. Mutations (base substitutions, indels, and nonsense mutations) were computed only once per isolate after comparison of each sequence with its corresponding consensus sequence. The genetic variabilities of the four genomic regions together accounted for a total of 165 mutations (identified in 755 clones corresponding to 463,272 nt sequenced), represented in Fig. 1 (see the supplemental material for a list of mutations and amino acid replacements). Base substitutions in all 13 isolates were distributed throughout the genome of ToCV, and insertions were found only in the RdRp and Hsp70h regions, whereas deletions were found in the four genomic regions. The RdRp region accumulated the highest number of mutations per nucleotide position (representing 42% of the total), whereas Hsp70h, CP, and CPm harbored 25%, 16%, and 18% of the total computed mutations, respectively. An overall χ2 test indicated significant differences in the frequencies of mutation for the four regions (χ2 = 14.64; df = 3; P = 0.0022). Pairwise χ2 tests corrected using the sequential Bonferroni method also revealed that the number of mutations per nucleotide position was significantly higher in the RdRp region than in the other regions.
Mutations shared by several isolates were found in the RdRp region and, to a lesser extent, in the Hsp70h and the CP regions, but they were not detected in the CPm region. The distribution of mutations after a standard nonparametric one-way run test was shown to be random in the genomic regions RdRp, Hsp70h, and CPm (P = 0.81, 0.13, and 0.90, respectively). In contrast, mutations in the CP region were not randomly distributed (P < 0.01), suggesting the existence of mutational hot spots. Altogether, these results indicate that the accumulation of mutations was not evenly distributed among the four genomic regions of ToCV that were analyzed.
Types of mutations and acceptability of amino acid changes.
The types of mutations predominant in field isolates of ToCV were studied by pooling the changes belonging to each of the four genomic regions. Percentages with respect to the number of mutations within each genomic region and with respect to the total number of mutations found in all genomic regions jointly were calculated (Table 1). Considering each genomic region independently, the most abundant mutations were base substitutions (82.9 to 94.2%); insertions were found only in the RdRp (1.4%) and Hsp70h (2.4%) regions, whereas deletions were present in the four regions and especially in the Hsp70h (14.6%) and CP (11.5%) regions. In general, transitions were more common than transversions, except in the CP region, where transversions amounted to 52%. U→C transitions were the most frequent base substitutions in the RdRp, Hsp70h, and CPm regions, followed by G→A. G→U transversions (27%) were the most common changes within the CP region. Moreover, C→G transversions were the rarest nucleotide substitutions and were found only in the CP region. The abundance of transversions over transitions in the CP region suggests that negative selection suppressed the number of transitional changes, because polymerases in general have misincorporation tendencies that favor transitions. When mutations from all genomic regions were pooled, transitions accounted for 66.4% whereas transversions accounted for 33.6%. Insertions and deletions represented 1.2% and 8.5% of the total number of mutations, respectively, and most were situated in U- or A-rich regions (data not shown). Nonsense mutations were found only in the RdRp (4.3%) and Hsp70h (11.5%) regions.
TABLE 1.
Numbers (and percentages) of different types of mutations found within 13 field isolates of ToCV
| Type of mutation | RNA1 |
RNA2 |
Total |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RdRp |
Hsp70h |
CP |
CPm |
|||||||
| n | % | n | % | n | % | n | % | n | % | |
| A→C | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 2 | 6.9 | 2 | 1.2 |
| A→G | 6 | 8.7 | 1 | 2.4 | 1 | 3.8 | 3 | 10.4 | 11 | 6.7 |
| A→U | 4 | 5.8 | 1 | 2.4 | 2 | 7.7 | 1 | 3.5 | 8 | 4.9 |
| G→A | 10 | 14.5 | 6 | 14.6 | 2 | 7.7 | 5 | 17.2 | 23 | 13.8 |
| G→C | 3 | 4.3 | 1 | 2.4 | 1 | 3.8 | 1 | 3.4 | 6 | 3.6 |
| G→U | 3 | 4.3 | 3 | 7.3 | 7 | 27.0 | 3 | 10.4 | 16 | 9.7 |
| C→A | 3 | 4.3 | 1 | 2.4 | 0 | 0.0 | 1 | 3.5 | 5 | 3.0 |
| C→G | 0 | 0.0 | 0 | 0.0 | 1 | 3.8 | 0 | 0.0 | 1 | 0.6 |
| C→U | 11 | 15.9 | 6 | 14.6 | 2 | 7.7 | 3 | 10.4 | 22 | 13.3 |
| U→A | 0 | 0.0 | 1 | 2.4 | 1 | 3.8 | 1 | 3.5 | 3 | 1.8 |
| U→C | 20 | 29.0 | 12 | 29.3 | 6 | 23.1 | 5 | 17.2 | 43 | 26.1 |
| U→G | 5 | 7.2 | 2 | 4.9 | 0 | 0.0 | 2 | 6.9 | 9 | 5.5 |
| Substitutions | 65 | 94.2 | 34 | 82.9 | 23 | 88.5 | 27 | 93.1 | 149 | 90.3 |
| Insertions | 1 | 1.4 | 1 | 2.4 | 0 | 0.0 | 0 | 0.0 | 2 | 1.2 |
| Deletions | 3 | 4.3 | 6 | 14.6 | 3 | 11.5 | 2 | 6.9 | 14 | 8.5 |
| Total | 69 | 100 | 41 | 100 | 26 | 100 | 29 | 100 | 165 | 100 |
| Transitions | 47 | 72.3 | 25 | 72.7 | 11 | 48.0 | 16 | 59.3 | 99 | 66.4 |
| Transversions | 18 | 27.7 | 9 | 27.3 | 12 | 52.0 | 11 | 40.7 | 50 | 33.6 |
| Nonsynonymous | 23 | 35.4 | 25 | 74.3 | 15 | 65.2 | 18 | 67.0 | 81 | 54.4 |
| Synonymous | 42 | 64.6 | 9 | 25.7 | 8 | 34.8 | 9 | 33.0 | 68 | 45.6 |
| Stop codons | 3 | 4.3 | 0 | 0.0 | 3 | 11.5 | 0 | 0.0 | 6 | 3.6 |
To assess whether selective constraints could be acting on the viral genomic RNA, synonymous and nonsynonymous mutations were computed for the four genomic regions. As indicated in Table 1, nonsynonymous mutations were more abundant in the regions belonging to RNA2 (from 65.2 to 74.3%), suggesting that selective constraints act at the RNA level. In the RdRp region located in RNA1, however, the number of synonymous changes was higher, indicating that in this case selection could be operating at the protein level instead.
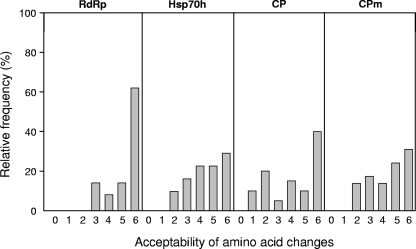
The relative frequencies and acceptabilities of amino acid changes according to the SG matrix of Feng et al. (17) for all four genomic regions are illustrated in Fig. 2. While the Hsp70h and CPm regions exhibited similar distributions with relative frequencies inversely proportional to acceptability values, the CP region allowed more severe amino acid changes (values 1 and 2), indicating a higher tolerance in this region. Conversely, most of the amino acid changes in the RdRp region (60%) were of value 6 (i.e., synonymous), meaning that, despite being the most variable coding region, RdRp does not tolerate drastic amino acid changes.
FIG. 2.
Acceptabilities of nucleotide substitutions identified in each genomic region, determined according to the SG system (17), and their relative frequencies. The values range from 0 (most drastic change) to 6 (synonymous change).
Quasispecies structure of ToCV intraisolate populations.
Considering the intraisolate genetic variability observed in the ToCV field samples under study, estimations of mutation frequency, Shannon entropy, and average genetic distance were used to characterize each mutant spectrum. The mutation frequency values within each genomic region for the 13 isolates of ToCV were calculated. As shown in Table 2, the variation of mutation frequency values depended on the particular genomic region, being highest for the RdRp region and ranging from 0.9 × 10−4 substitutions per nucleotide for isolate AT45/02 to 22.7 × 10−4 substitutions per nucleotide for isolate AT80/99. For the three regions of RNA2, mutation frequency values (the number of substitutions per nucleotide) ranged from <1.08 × 10−4 to 6.07 × 10−4 for the Hsp70h region, from <1.16 × 10−4 to 5.82 × 10−4 for the CP region, and from <1.08 × 10−4 to 6.12 × 10−4 for the CPm region. These results are in agreement with mutation frequency values obtained for several animal and plant RNA viruses (12, 42, 44).
The proportion of different genomes in a mutant distribution was measured by calculating the Shannon entropy. Very different degrees of heterogeneity and complexity of the ensemble of ToCV RNA molecules in each genomic region for each isolate were found, as shown in Table 2. Shannon entropy values ranged from 0.089 to 0.706 in the RdRp region, from 0 to 0.477 in the Hsp70h region, from 0 to 0.429 in the CP region, and from 0 to 0.347 in the CPm region. These results indicate that the heterogeneity and complexity of ToCV varied greatly, not only between isolates, but also among the four genomic regions analyzed within the same isolate. They also show that, in general, the RdRp region presents the most variable and heterogeneous mutant spectra.
The genetic complexity of ToCV field isolates was estimated by calculating pairwise genetic distances within isolates using Kimura's two-parameter method. Average genetic distances (d) for each genomic region in each field isolate are shown in Table 3. The maximum value of d estimated for the RdRp region was 2.89-fold higher than that for the Hsp70h and CP regions and 3.89-fold higher than the maximum value of d for the CPm region. Furthermore, the measure of dispersion of d between mutant spectra around the mean (i.e., the coefficient of variation [CV]) among the four regions was highest for RdRp (CV = 1.106, 0.697, 0.986, and 0.775 for RdRp, Hsp70h, CP, and CPm, respectively). The CV value of >1 indicates that d for RdRp has a “high-variance” distribution of data points. However, d values obtained for each genomic region after pooling all sequences from all mutant spectra (referred as total in Table 3) were very similar, with the largest difference being 4.88-fold higher for RdRp in relation to the Hsp70h region. These results indicate, first, that RdRp is by far the most diverse genomic region of ToCV intrahost populations and, second, that the great differences in d values between isolates within each genomic region might be underestimated if total d is employed.
TABLE 3.
Averages of estimated pairwise genetic distances (d) by Kimura's two-parameter method for the different genomic regions in 13 field isolates of ToCV
| Isolate | RNA1 |
RNA2 |
||||||
|---|---|---|---|---|---|---|---|---|
| RdRp |
Hsp70h |
CP |
CPm |
|||||
| d | SE | d | SE | d | SE | d | SE | |
| AT1/97 | 0.00358 | 0.00142 | 0.00206 | 0.00116 | 0.00024 | 0.00021 | 0.00070 | 0.00038 |
| AT3/98 | 0.00046 | 0.00031 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00124 | 0.00046 |
| AT24/99 | 0.00037 | 0.00026 | 0.00106 | 0.00053 | 0.00000 | 0.00000 | 0.00021 | 0.00020 |
| AT80/99 | 0.00803 | 0.00213 | NDb | ND | ND | ND | ND | ND |
| AT198/00 | 0.00251 | 0.00061 | 0.00065 | 0.00036 | 0.00022 | 0.00021 | 0.00070 | 0.00038 |
| AT230/00 | 0.00034 | 0.00032 | 0.00098 | 0.00038 | 0.00131 | 0.00084 | 0.00044 | 0.00030 |
| AT205/00 | 0.00087 | 0.00037 | 0.00070 | 0.00040 | 0.00037 | 0.00025 | 0.00000 | 0.00000 |
| AT36/02 | 0.00056 | 0.00031 | 0.00082 | 0.00039 | 0.00062 | 0.00035 | 0.00023 | 0.00022 |
| AT45/02 | 0.00018 | 0.00018 | 0.00061 | 0.00035 | 0.00138 | 0.00064 | 0.00070 | 0.00040 |
| AT31/03 | 0.00056 | 0.00030 | 0.00128 | 0.00087 | 0.00042 | 0.00030 | 0.00023 | 0.00023 |
| AT138/03 | 0.00020 | 0.00019 | 0.00022 | 0.00022 | 0.00175 | 0.00128 | 0.00110 | 0.00062 |
| AT204/04 | 0.00056 | 0.00031 | 0.00022 | 0.00021 | 0.00047 | 0.00033 | 0.00023 | 0.00023 |
| AT379/04 | 0.00111 | 0.00044 | 0.00082 | 0.00039 | 0.00024 | 0.00022 | 0.00023 | 0.00024 |
| Totala | 0.00650 | 0.00143 | 0.00133 | 0.00042 | 0.00206 | 0.00086 | 0.00277 | 0.00118 |
Estimated pairwise genetic distances by Kimura's two-parameter method after all the viral variants from the different isolates within each genomic region were pooled.
ND, not determined.
Thus, ToCV intraisolate variability in tomato field samples is characterized by the high heterogeneity and genetic complexity of RNA virus populations. High mutation frequencies, Shannon entropy values, and average genetic distances estimated from ToCV mutant spectra in four genomic regions indicate that ToCV exists as viral quasispecies in field tomato isolates.
Direction of selective forces on the ToCV genome.
To address whether positive or negative selection is shaping ToCV populations, pairwise synonymous substitutions per synonymous site (dS) and nonsynonymous substitutions per nonsynonymous site (dNS) were estimated using the Pamilo-Biachi-Li method and the dNS/dS ratio, when its calculation was possible. As shown in Table 4, the dNS/dS ratio for each quasispecies and genomic region, estimated individually, ranged from 0.037 to 2.241, suggesting different constraints on sequence change depending on both the field isolate and the genomic region analyzed. Thus, the RdRp and CPm regions contained mutant spectra subjected to negative selection (dNS/dS < 1) and to positive selection (dNS/dS > 1). In both the Hsp70h and CP regions; however, purifying or negative selection seemed to be acting on every quasispecies. Average dNS/dS ratios for the genomic regions were between 0.265 and 0.768, suggesting that ToCV quasispecies in all four genomic regions would be under negative selection. These data contrasted greatly with the dNS/dS ratios obtained after pairwise analysis of pooled sequences from each genomic region (indicated as the total in Table 4). In this case, the RdRp (dNS/dS = 0.110) and CP (dNS/dS = 0.115) regions would have similar stringent constraints on amino acid change, indicating the effect of purifying selection. On the other hand, Hsp70h (dNS/dS = 1.364) would be subjected to strong positive selection and CPm (dNS/dS = 1.071) to nearly neutral selection.
TABLE 4.
Analysis of diversity for nonsynonymous (dNS) and synonymous (dS) positions in different genomic regions of 13 field isolates of ToCV
| Isolate | RNA1 |
RNA2 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RdRp |
Hsp70h |
CP |
CPm |
|||||||||
| dNS | dS | dNS/dSa | dNS | dS | dNS/dS | dNS | dS | dNS/dS | dNS | dS | dNS/dS | |
| AT1/97 | 0.00035 | 0.00945 | 0.037 | 0.00228 | 0.00000 | 0.00000 | 0.00000 | 0 | 0.00076 | 0.00062 | 1.214 | |
| AT3/98 | 0.00038 | 0.00069 | 0.553 | 0.00000 | 0.00000 | 0 | 0.00000 | 0.000000 | 0 | 0.00134 | 0.00111 | 1.211 |
| AT24/99 | 0.00000 | 0.00110 | 0 | 0.00140 | 0.00000 | 0.00000 | 0.00000 | 0 | 0.00033 | 0.00000 | - | |
| AT80/99 | 0.00151 | 0.02510 | 0.060 | NDd | ND | ND | ND | ND | ND | ND | ND | ND |
| AT198/00 | 0.00118 | 0.00942 | 0.125 | 0.00063 | 0.00079 | 0.804 | 0.00026 | 0.00000 | 0.00054 | 0.00182 | 0.297 | |
| AT230/00 | 0.00000 | 0.00325 | 0 | 0.00078 | 0.00114 | 0.687 | 0.00037 | 0.00316 | 0.117 | 0.00025 | 0.00059 | 0.431 |
| AT205/00 | 0.00114 | 0.00051 | 2.241 | 0.00079 | 0.00000 | 0 | 0.00043 | 0.00000 | 0 | 0.00000 | 0.00000 | 0 |
| AT36/02 | 0.00074 | 0.00000 | 0.00068 | 0.00251 | 0.270 | 0.00057 | 0.00000 | 0.00000 | 0.00000 | 0 | ||
| AT45/02 | 0.00000 | 0.00055 | 0 | 0.00093 | 0.00000 | 0.00078 | 0.00233 | 0.335 | 0.00070 | 0.00071 | 0.989 | |
| AT31/03 | 0.00052 | 0.00174 | 0.301 | 0.00027 | 0.00000 | 0.00000 | 0.00061 | 0 | 0.00038 | 0.00000 | - | |
| AT138/03 | 0.00023 | 0.00000 | 0.00036 | 0.00335 | 0.108 | 0.00105 | 0.00304 | 0.345 | 0.00027 | 0.00457 | 0.059 | |
| AT204/04 | 0.00074 | 0.00000 | 0.00059 | 0.00065 | 0.909 | 0.00055 | 0.00000 | 0.00025 | 0.00059 | 0.423 | ||
| AT379/04 | 0.00113 | 0.00055 | 2.060 | 0.00000 | 0.00200 | 0 | 0.00027 | 0.00000 | 0.00038 | 0.00000 | - | |
| Meanb | 0.768 | 0.555 | 0.265 | 0.660 | ||||||||
| Totalc | 0.0021 | 0.0190 | 0.110 | 0.0015 | 0.0011 | 1.364 | 0.0007 | 0.0061 | 0.115 | 0.0030 | 0.0028 | 1.071 |
Pairwise synonymous substitutions per synonymous site (dS) and nonsynonymous substitutions per nonsynonymous site (dNS) calculated according to the Pamilo-Bianchi-Li method based on Kimura's two-parameter model.
Estimated dNS/dS values per genomic region were computed as the arithmetic mean of the dNS/dS values for each isolate.
Estimated dNS/dS values per genomic region after pooling all the viral variants from the different isolates within each genomic region.
ND, not determined.
Phylogenetic analysis of mutant spectra: evidence for a low frequency of recombination.
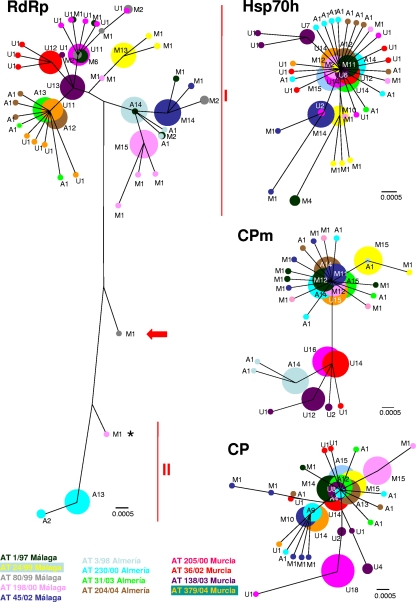
The phylogenetic relationships of sequences belonging to quasispecies of genomic regions RdRp, Hsp70h, CP, and CPm were deduced from multiple-sequence alignments by the neighbor-joining method. Figure 3 illustrates unrooted trees representing phylogeny within the four genomic regions. The composition, frequency, and phylogenetic relationship of the sequences found in each quasispecies determined the different shapes of the trees. The inferred tree for the RdRp region showed the maximum bifurcation, whereas the Hsp70h tree exhibited the lowest branching of all four regions. Phylogenetic trees of the CP and CPm regions displayed similar divergence levels, as determined by the analogous distributions of their mutant spectra. Interestingly, in the RdRp region, phylogenetic distances were higher because of sequences that differed by 10 base substitutions, indicated as types I and II. Even though most quasispecies in this region showed consensus sequences of type I, with closely related but nonidentical master sequences, a type II consensus sequence was found in sample AT230/00. However, a type II variant, immersed in an ensemble of type I RNA molecules, was detected in sample AT198/00. This sequence lacked two mutations present in the AT230/00 consensus and was positioned at an intermediate distance between type I and type II RNAs. Furthermore, a putative recombinant RNA molecule was found in sample AT80/99 (Fig. 3). The 5′ moiety of the amplified RdRp fragment was identical to consensus sequence AT230/00 (type II), and the 3′ moiety was identical to consensus sequence AT80/99 (type I). Although SimPlot analysis strongly suggests a recombinant origin for this molecule (not shown), it remains possible that convergence of sequences in this genomic region could be responsible for the observed difference between the moieties.
FIG. 3.
Phylogenetic relationships among sequences of RdRp, Hsp70h, CP, and CPm regions of the ToCV genome, inferred by the neighbor-joining method from multiple-sequence alignments. Each individual sequence is represented by a circle with a different color for each isolate. The circle sizes are proportional to the number of identical sequences in each node and are indicated with a number. Sequences belonging to each isolate are identified with a color code, as indicated at the bottom, and a letter: A for the isolates from Almería, M for the isolates from Málaga, and U for the isolates from Murcia. I, RNA1 type I sequences; II, RNA1 type II sequences. The bar below each tree indicates 0.0005 nucleotide substitutions per site. The asterisk indicates the type II sequence found in sample AT198/00. The arrow indicates the recombinant sequence found in sample AT80/99.
An AMOVA test was used to determine whether genetic variation represented in the phylogenetic trees of each genomic region was a reflection of variation within isolates or between isolates. The results indicated that the observed variability in the genomic regions RdRp (81.8%), CP (71.8%), and CPm (82.5%) was due mainly to differences between populations (Table 5). Quasispecies from the RdRp region displayed different master sequences surrounded by their own mutant spectrum. In the CP and CPm regions, most of the mutant spectra shared the same master sequence, but the extent of variation within each spectrum was very narrow, accounting for the higher diversity between isolates than within isolates. In the Hsp70h region, most variation (61.6%) came from within populations because quasispecies in this region shared the master sequence but displayed distinctive mutant spectra.
TABLE 5.
AMOVA for haplotype distributions in different genomic regions of ToCV
| Region | Source of variation | df | Sum of squares | Variance components | % of variation | FSTa | P |
|---|---|---|---|---|---|---|---|
| RNA1 | |||||||
| RdRp | Among isolates | 12 | 353.888 | 2.01357 | 81.8 | 0.8182 | <0.001 |
| Within isolates | 175 | 78.288 | 0.44736 | 18.2 | |||
| Total | 187 | 432.176 | 2.46093 | ||||
| RNA2 | |||||||
| Hsp70h | Among isolates | 11 | 34.447 | 0.17886 | 38.4 | 0.3840 | <0.001 |
| Within isolates | 179 | 51.369 | 0.28698 | 61.6 | |||
| Total | 190 | 85.817 | 0.46584 | ||||
| CP | Among isolates | 11 | 80.974 | 0.45628 | 71.8 | 0.7183 | <0.001 |
| Within isolates | 177 | 31.671 | 0.17893 | 28.2 | |||
| Total | 188 | 112.646 | 0.63521 | ||||
| CPm | Among isolates | 11 | 121.581 | 0.70363 | 82.5 | 0.8252 | <0.001 |
| Within isolates | 174 | 25.930 | 0.14902 | 17.5 | |||
| Total | 185 | 147.511 | 0.85265 |
FST, F statistic.
DISCUSSION
Knowledge about the structure and genetic content of natural plant virus populations is limited. Despite the growing number of studies of virus diversity, few of them address the genetic characterization of populations that plant viruses establish within their hosts. However, there is increasing evidence that intrahost mutant spectra in both plant DNA and RNA viruses exist as virus quasispecies and that such quasispecies play a key role in virus adaptability and pathogenesis (11, 20). Virus quasispecies are subjected to drastic and frequent changes of environment and repeated bottlenecks that result in fitness loss due to the accumulation of deleterious mutations (6, 13), although a virus population may lose or gain fitness depending on the initial fitness of the population and the size of the bottleneck (38). Understanding virus quasispecies structure and behavior is fundamental for the design of effective control strategies because viral quasispecies are reservoirs of emerging viral variants (10, 11, 20, 34).
In this work, mutant spectra of 13 field isolates of ToCV sampled from 1997 to 2004 have been characterized by nucleotide sequencing of 755 molecular clones. Four genomic regions, located in two separately encapsidated genomic segments, have been sequenced, amounting to a total of 463,272 nt. Sequence determination of molecular clones has been documented to be a valid experimental approach for characterizing quasispecies from natural infections of bacterial, plant, and animal RNA viruses (1, 44). Our results have shown uneven distributions of mutations in the four genomic regions analyzed, strongly suggesting that representative regions of the viral genome should be considered to obtain rigorous estimates of mutation frequencies and genetic distances between sequences within mutant clouds. The need for analysis of several genomic regions has also been emphasized in previous studies of the variability of animal and plant RNA viruses (1, 18, 33). Some genomic regions of plant viruses can exhibit extremely low variation (48) and give an erroneous idea of virus genetic variability.
Genetic variation in a quasispecies can be overestimated because of experimental error introduced during the process of copying and amplification of viral RNA. This error may arise from the limited amounts of initial template and the use of low-fidelity polymerases (12). In the present study, such bias in the determination of intrahost virus population nucleotide heterogeneity was prevented by observing the following precautions for RT-PCR amplification. First, RNAs were transcribed and cloned only from RNA that yielded amplification products after a 1:10 dilution (i.e., the quantity of template was not limiting). Second, a Pfu high-fidelity DNA polymerase was used during PCR amplification. Third, the reaction conditions were those reported to favor detection of most of the viral variants (19).
Our results showed that ToCV populations in a natural host consisted of heterogeneous and complex mutant distributions. These distributions contain a most abundant master sequence surrounded by a mutant spectrum of closely related variants. Within the family Closteroviridae, of which ToCV is a member, there have been few studies of intraisolate genetic diversity in field isolates. In perennial plants, like citrus and grapevine, the variability of the closterovirus CTV (25, 47) and the ampeloviruses GLRaV-1 and GLRaV-3 (24, 51) have been analyzed by Single-strand conformation polymorphism. Estimates of mutation frequencies, however, have not been previously presented for criniviruses or for members of the other two genera in the family Closteroviridae, Closterovirus and Ampelovirus. Our results give a clear picture of the extent of genomic heterogeneity and complexity that the crinivirus ToCV exhibits in its primary natural host, tomato. For the most variable region, RdRp, if only mutations from the predominant RNA1 sequences (i.e., type I variants) in isolates AT80/99 and AT198/00 are considered, mutation frequencies are reduced to 15.8 × 10−4 and 4.8 × 10−4, respectively. Even so, mutation frequencies for the RdRp region are higher than those for the Hsp70h, CP, and CPm regions located in RNA2. For the CP region of the crinivirus CYSDV, genetic diversity (d) within three field isolates has been documented to be from 1.9 × 10−4 to 7.3 × 10−4 (46). In the case of Closteroviridae-infected perennial plants, these figures may be up to 1,000-fold higher. For instance, intraisolate diversity of CTV in citrus reached 0.142, with diversity in this case estimated as the genetic distance between two haplotypes (defined by the single-strand conformation polymorphism pattern) (25). For the Hsp70h region of GLRaV-1 in grapevine, diversity values obtained from sequencing data were from 2.0 × 10−3 to 3.6 × 10−2, although these figures were obtained from comparison of six or fewer clones (24). The lower level of genetic variation found in ToCV than in CTV and GLRaV-1 might be caused in part by differences in host plants and/or modes of transmission, as suggested for CYSDV (46). CTV and GLRaV-1 have perennial hosts that can be infected for decades, whereas tomato, the primary host for ToCV, is an annual, and infections generally do not last for more than 180 days before the crop ends. ToCV is transmitted only by its whitefly vectors, whereas CTV and GLRaV-1 are also transmitted by vegetative propagation, which could relax selective constraints related to insect transmission.
In the RdRp region, coexistence of phylogenetically distant sequences was detected. In this region, two types of RNA 1, namely, types I and II, have been found (G. Lozano, J. Navas-Castillo, unpublished results). While mutant variants in most ToCV isolates analyzed in this study were closely related to type I, all sequences belonging to isolate AT230/00 displayed type II-related sequences. Isolate AT198/00, however, was found to have one type II variant coexisting in a virus spectrum dominated by type I variants. A similar situation has been described for CTV isolates, in which mutant spectra harbor two divergent master sequences (47). A mutant virus with the potential to become dominant may remain as a minority in the population because of the suppressive effect of the mutant spectrum and can be retained by complementation (8, 12). In an evolving quasispecies, bottlenecks can isolate genomes that are able to reinitiate an infection (4). In nature, intrahost ToCV populations are subjected to repeated whitefly inoculations, and it is likely that the low-frequency type II variant might have been introduced into a tomato plant previously infected by the type I sequences. However, maintenance of this type II variant in that mutant spectrum (and the same applies to deleterious nonsense mutations found in RdRp and CP mutant spectra) can be explained by lack of negative or purifying selection and the modulating effect of the mutant spectrum surrounding it, supporting the view that the quasispecies as a whole, and not individual genomes, is the target of selection (40, 52). Similarly, the cucumovirus Tomato aspermy virus maintains in its mutant spectrum a cell-to-cell movement-defective mutant by complementation, providing another example of the ability of plant virus quasispecies to keep variants with low fitness (32).
Recombination in RNA viruses is regarded as a mechanism that compensates for the accumulation of deleterious mutations and that helps to maintain genomic diversity (35). Thus, the large genomes of closteroviruses in particular are at high risk, and recombination might allow regeneration of functional genomes from deleterious ones. Nonhomologous recombination, indicated by the generation of defective RNAs, as well as homologous recombination, has been reported in natural isolates of CTV (3, 53, 56). In the last study (56), which was carried out in a single sweet orange plant through a combination of genome-wide resequencing analysis and deep sequencing of selected genomic regions, an extraordinarily large proportion (17.6%) of the sequenced molecules were recombinants derived from the three predominant genotypes. Although the search for recombinant molecules was not the aim of our work, the current study suggests that recombination is not frequent in ToCV, in contrast to closteroviruses, since we detected only one putative recombinant molecule. The low abundance of recombinant molecules, however, might indicate strong negative selection against those variants. In the particular case of the AT80/99 RdRp recombinant molecule, most mutations located in its type II sequence were synonymous, suggesting that this particular variant might have escaped from purifying selection.
Our results for all 13 isolates showed that quasispecies of regions belonging to RNA2 were the more restricted in genetic variation. Hsp70h and CPm regions displayed identical or very closely related master sequences and an abundance of nonsynonymous over synonymous changes, which might indicate functional constraints. In addition, structural constraints acting on the viral RNA might greatly limit sequence changes in these regions. Other restraints could include RNA folding into secondary structures, control of replication and translation, and protection against degradation (49).
From our results it is clear that, of all regions analyzed, the CP region was the most restricted with respect to sequence variation. There are several reasons for this. First, all isolates presented closely related, if not identical, master sequences, and their surrounding mutant spectra were extremely invariant. Second, this region was the only one where mutations were not distributed randomly according to the results of the run test. Third, transversions outnumbered transitional nucleotide substitutions in the CP region, suggesting purifying selection. Fourth, synonymous mutations were more abundant than nonsynonymous nucleotide substitutions in the CP region. Nonetheless, amino acid change for this protein may be subjected to greater purifying selection for the maintenance of the structure and stability of viral capsids and by interaction with the vector (7). On the other hand, quasispecies of the RdRp region were the most variable, although most of the substitutions were synonymous. Although this suggests that selection might not be acting at the nucleotide level, we cannot rule out the possibility that specific amino acid changes may be driving evolution. In this region (as well as in the CP region), nonsense mutants coding for truncated proteins were detected, suggesting that they may be maintained by complementation with viable mutants. For both RNA1 and RNA2, no geographical differences can be inferred from phylogenetic analyses.
One possible explanation for the relatively high mutation frequencies, high Shannon entropy, and high diversity values in the RdRp region is that the RdRp region is located in RNA1, which is specialized for replication, while the other regions studied are in RNA2. ToCV is a two-component virus, which means that during infection genomic segments 1 and 2 are separately encapsidated. Virions containing RNA1 inside the cell are able to decapsidate and replicate autonomously even in the absence of virions containing RNA2. However, to be encapsidated and to be able to spread within the plant, coinfection with RNA2 must occur. On the other hand, RNA2 must coincide with RNA1 to be replicated and encapsidated. More rounds of RNA1 replication before meeting RNA2 would mean more chances to accumulate mutations, resulting in imbalances of mutations between the segments. Asynchronous accumulation of RNA1 and RNA2 of the crinivirus LIYV upon coinfection of protoplasts has been reported (59). LIYV RNA1 progeny, both genomic and subgenomic RNAs, was detected in protoplasts as early as 12 h postinoculation (p.i.) and accumulated to high levels by 24 h p.i. In contrast, RNA2 progeny were not readily detected until ca. 36 h p.i.
The results obtained from this in-depth study of the intraisolate genetic variability of ToCV should contribute to our understanding of the biology, epidemiology, and evolution of the rapidly growing list of viruses in the complex family Closteroviridae. This information is especially important because the viruses in this family infect a broad range of economically important crops.
Supplementary Material
Acknowledgments
This research was funded by grants AGL2001-0542 and AGL2004-06959-C04-01/AGR to J.N.-C. and BFU2007-65080BMC to A.G.-P. (Plan Nacional de I+D+I, Ministerio de Educación y Ciencia [MEC], Spain). G.L. was the recipient of an FPI fellowship from MEC. A.G.-P. was supported by a Ramón y Cajal contract from the Ministerio de Ciencia e Innovación (Spain) and the European Social Fund.
We thank Pedro Moreno and Bruno Gronenborn for their critical reading of the manuscript. Also, many thanks to Marta Montserrat for her statistical advice and Pablo Navas for his editing help.
Footnotes
Published ahead of print on 30 September 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Arias, A., E. Lázaro, C. Escarmís, and E. Domingo. 2001. Molecular intermediates of fitness gain of an RNA virus: characterization of a mutant spectrum by biological and molecular cloning. J. Gen. Virol. 82:1049-1060. [DOI] [PubMed] [Google Scholar]
- 2.Ayllón, M. A., C. López, J. Navas-Castillo, S. M. Garnsey, J. Guerri, R. Flores, and P. Moreno. 2001. Polymorphism of the 5′ terminal region of Citrus tristeza virus (CTV) RNA: incidence of three sequence types in isolates of different origin and pathogenicity. Arch. Virol. 146:27-40. [DOI] [PubMed] [Google Scholar]
- 3.Ayllón, M. A., C. López, J. Navas-Castillo, M. Mawassi, W. O. Dawson, J. Guerri, R. Flores, and P. Moreno. 1999. New defective RNAs from Citrus tristeza virus: evidence for a replicase driven template switching mechanism in their generation. J. Gen. Virol. 80:817-821. [DOI] [PubMed] [Google Scholar]
- 4.Bergstrom, C. T., P. McElhany, and L. A. Real. 1999. Transmission bottlenecks as determinants of virulence in rapidly evolving pathogens. Proc. Natl. Acad. Sci. USA 96:5095-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao, L. 1988. Evolution of sex in RNA viruses. J. Theor. Biol. 133:99-112. [DOI] [PubMed] [Google Scholar]
- 6.Chao, L. 1990. Fitness of RNA virus decreased by Muller's ratchet. Nature 348:454-455. [DOI] [PubMed] [Google Scholar]
- 7.Chare, E. R., and E. C. Holmes. 2004. Selection pressures in the capsid genes of plant RNA viruses reflect mode of transmission. J. Gen. Virol. 85:3149-3157. [DOI] [PubMed] [Google Scholar]
- 8.de la Torre, J. C., and J. J. Holland. 1990. RNA virus quasispecies populations can suppress vastly superior mutant progeny. J. Virol. 64:6278-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolja, V. V., J. F. Kreuze, and J. P. T. Valkonen. 2006. Comparative and functional genomics of closteroviruses. Virus Res. 117:38-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domingo, E. 2007. Virus evolution, p. 389-421. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 11.Domingo, E., C. Escarmis, L. Menéndez-Arias, C. Perales, M. Herrera, I. Novella, and J. J. Holland. 2008. Viral quasispecies: dynamics, interactions and pathogenesis, p. 87-118. In E. Domingo, C. Parrish, and J. Holland (ed.), Origin and evolution of viruses. Elsevier, Amsterdam, The Netherlands.
- 12.Domingo, E., V. Martín, C. Perales, A. Grande-Pérez, J. García-Arriaza, and A. Arias. 2006. Viruses as quasispecies: biological implications. Curr. Top. Microbiol. Immunol. 299:51-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duarte, E., D. Clarke, A. Moya, E. Domingo, and J. J. Holland. 1992. Rapid fitness losses in mammalian RNA virus clones due to Muller ratchet. Proc. Natl. Acad. Sci. USA 89:6015-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elena, S. F., P. Carrasco, J. A. Darós, and R. Sanjuán. 2006. Mechanisms of genetic robustness in RNA viruses. EMBO Rep. 7:168-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escarmís, C., E. Lazaro, and S. C. Manrubia. 2006. Population bottlenecks in quasispecies dynamics. Curr. Top. Microbiol. Immunol. 299:141-170. [DOI] [PubMed] [Google Scholar]
- 16.Excoffier, L., G. Laval, and S. Schneider. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, D. F., M. S. Johnson, and R. F. Doolittle. 1985. Aligning amino acid sequences: comparison of commonly used methods. J. Mol. Evol. 21:112-125. [DOI] [PubMed] [Google Scholar]
- 18.García-Arenal, F., and A. A. Fraile. 2008. Questions and concepts in plant virus evolution: a historical perspective, p. 1-14. In M. J. Roosinck (ed.), Plant virus evolution. Springer-Verlag, Berlin, Germany.
- 19.Grande-Pérez, A., G. Gómez-Mariano, P. R. Lowenstein, and E. Domingo. 2005. Mutagenesis-induced, large fitness variations with an invariant arenavirus consensus genomic nucleotide sequence. J. Virol. 79:10451-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland, J. J. 2006. Transitions in understanding of RNA viruses: a historical perspective. Curr. Top. Microbiol. Immunol. 299:371-401. [DOI] [PubMed] [Google Scholar]
- 21.Huang, J., L. G. Brieba, and R. Sousa. 2000. Misincorporation by wild-type and mutant T7 RNA polymerases: identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry 39:11571-11580. [DOI] [PubMed] [Google Scholar]
- 22.Hull, R. 2002. Matthew's plant virology. Academic Press, San Diego, CA.
- 23.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 24.Kominek, P., M. Glasa, and M. Bryxiova. 2005. Analysis of the molecular variability of Grapevine leafroll-associated virus 1 reveals the presence of two distinct virus groups and their mixed occurrence in grapevines. Virus Genes 31:247-255. [DOI] [PubMed] [Google Scholar]
- 25.Kong, P., L. Rubio, M. Polek, and B. W. Falk. 2000. Population structure and genetic diversity within California Citrus tristeza virus (CTV) isolates. Virus Genes 21:139-145. [DOI] [PubMed] [Google Scholar]
- 26.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 27.Li, W. H. 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36:96-99. [DOI] [PubMed] [Google Scholar]
- 28.Livieratos, I. C., E. Eliasco, G. Müller, R. C. L. Olsthoorn, L. F. Salazar, C. W. A. Pleij, and R. H. A. Coutts. 2004. Analysis of Potato yellow mosaic virus RNA: evidence for a tripartite genome and conserved 3′-terminal structures among members of the genus Crinivirus. J. Gen. Virol. 85:2065-2075. [DOI] [PubMed] [Google Scholar]
- 29.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lozano, G., E. Moriones, and J. Navas-Castillo. 2006. Complete nucleotide sequence of the RNA2 of the crinivirus tomato chlorosis virus. Arch. Virol. 151:581-587. [DOI] [PubMed] [Google Scholar]
- 31.Lozano, G., E. Moriones, and J. Navas-Castillo. 2007. Complete sequence of the RNA1 of a European isolate of tomato chlorosis virus. Arch. Virol. 152:839-841. [DOI] [PubMed] [Google Scholar]
- 32.Moreno, I. M., J. M. Malpíca, E. Rodríguez-Cerezo, and F. García-Arenal. 1997. A mutation in tomato aspermy cucumovirus that abolishes cell-to-cell movement is maintained to high levels in the viral RNA population by complementation. J. Virol. 71:9157-9162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moury, B., C. Desbiez, M. Jacquemond, and H. Lecoq. 2006. Genetic diversity of plant virus populations: Towards hypothesis testing in molecular epidemiology. Adv. Virus Res. 67:49-87. [DOI] [PubMed] [Google Scholar]
- 34.Moya, A., E. C. Holmes, and F. González-Candelas. 2004. The population genetics and evolutionary epidemiology of RNA viruses. Nat. Rev. Microbiol. 2:279-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy, P. D. 2008. Recombination in plant RNA viruses, p. 133-164. In M. J. Roosinck (ed.), Plant virus evolution. Springer-Verlag, Berlin, Germany.
- 36.Nee, S. 1987. The evolution of multicompartmental genomes in viruses. J. Mol. Evol. 25:277-281. [DOI] [PubMed] [Google Scholar]
- 37.Nei, M., and S. Kumar. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY.
- 38.Novella, I. S., S. F. Elena, A. Moya, E. Domingo, and J. J. Holland. 1995. Size of genetic bottlenecks leading to virus fitness loss is determined by mean initial population fitness. J. Virol. 69:2869-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pamilo, P., and N. O. Bianchi. 1993. Evolution of the zfx and zfy genes: rates and interdependence between the genes. Mol. Biol. Evol. 10:271-281. [DOI] [PubMed] [Google Scholar]
- 40.Perales, C., V. Martín, C. M. Ruiz-Jarabo, and E. Domingo. 2005. Monitoring sequence space as a test for the target of selection in viruses. J. Mol. Biol. 345:451-459. [DOI] [PubMed] [Google Scholar]
- 41.Pérez, A. J., G. Thode, and O. Trelles. 2004. AnaGram: protein function assignment. Bioinformatics 20:291-292. [DOI] [PubMed] [Google Scholar]
- 42.Pita, J. S., and M. J. Roosinck. 2008. Virus populations, mutation rates, and frequencies, p. 109-121. In M. J. Roosinck (ed.), Plant virus evolution. Springer-Verlag, Berlin, Germmany.
- 43.Pugachev, K. V., F. Guirakhoo, S. W. Ocran, F. Mitchell, M. Parsons, C. Penal, S. Girakhoo, S. O. Pougatcheva, J. Arroyo, D. W. Trent, and T. P. Monath. 2004. High fidelity of yellow fever virus RNA polymerase. J. Virol. 78:1032-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roossinck, M. J., and W. L. Schneider. 2006. Mutant clouds and occupation of sequence space in plant RNA viruses. Curr. Top. Microbiol. Immunol. 299:337-348. [DOI] [PubMed] [Google Scholar]
- 45.Rubio, L., J. Soong, J. Kao, and B. W. Falk. 1999. Geographic distribution and molecular variation of isolates of three whitefly-borne closteroviruses of cucurbits: lettuce infectious yellows virus, cucurbit yellow stunting disorder virus, and beet pseudo-yellows virus. Phytopathology 89:707-711. [DOI] [PubMed] [Google Scholar]
- 46.Rubio, L., Y. Abou-Jawdah, H. X. Lin, and B. W. Falk. 2001. Geographically distant isolates of the crinivirus Cucurbit yellow stunting disorder virus show very low genetic diversity in the coat protein gene. J. Gen. Virol. 82:929-933. [DOI] [PubMed] [Google Scholar]
- 47.Rubio, L., M. A. Ayllón, P. Kong, A. Fernández, M. Polek, J. Guerri, P. Moreno, and B. W. Falk. 2001. Genetic variation of Citrus tristeza virus isolates from California and Spain: evidence for mixed infections and recombination. J. Virol. 75:8054-8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider, W. L., and M. J. Roossinck. 2001. Genetic diversity in RNA virus quasispecies is controlled by host-virus interactions. J. Virol. 75:6566-6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simmonds, P., and D. B. Smith. 1999. Structural constraints on RNA virus evolution. J. Virol. 73:5787-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turturo, C., P. Saldarelli, D. Yafeng, M. Digiaro, A. Minafra, V. Savino, and G. P. Martelli. 2005. Genetic variability and population structure of Grapevine leafroll-associated virus 3 isolates. J. Gen. Virol. 86:217-224. [DOI] [PubMed] [Google Scholar]
- 52.Vignuzzi, M., J. K. Stone, J. J. Arnold, C. E. Cameron, and R. Andino. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vives, M. C., L. Rubio, A. Sambade, T. E. Mirkov, P. Moreno, and J. Guerri. 2005. Evidence of multiple recombination events between two RNA sequence variants within a Citrus tristeza virus isolate. Virology 331:232-237. [DOI] [PubMed] [Google Scholar]
- 54.Volkestein, M. V. 1994. Physical approaches to biological evolution. Springer-Verlag, Berlin, Germany.
- 55.Weir, B. S., and C. C. Cockerham. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358-1370. [DOI] [PubMed] [Google Scholar]
- 56.Weng, Z., R. Barthelson, S. Gowda, M. E. Hilf, W. O. Dawson, D. W. Galbraith, and Z. Xiong. 2007. Persistent infection and promiscuous recombination of multiple genotypes of an RNA virus within a single host generate extensive diversity. PloS ONE 2:e917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wintermantel, W. M., G. C. Wisler, A. G. Anchieta, H. Y. Liu, A. V. Karasev, and I. E. Tzanetakis. 2005. The complete nucleotide sequence and genome organization of tomato chlorosis virus. Arch. Virol. 150:2287-2298. [DOI] [PubMed] [Google Scholar]
- 58.Wisler, G. C., R. H. Li, H. Y. Liu, D. S. Lowry, and J. E. Duffus. 1998. Tomato chlorosis virus: a new whitefly-transmitted, phloem-limited, bipartite closterovirus of tomato. Phytopathology 88:402-409. [DOI] [PubMed] [Google Scholar]
- 59.Yeh, H. H., T. Tian, L. Rubio, B. Crawford, and B. W. Falk. 2000. Asynchronous accumulation of Lettuce infectious yellows virus RNAs 1 and 2 and identification of an RNA 1 trans enhancer of RNA 2 accumulation. J. Virol. 74:5762-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.