Abstract
Protein phosphorylation is dynamically regulated in eukaryotic cells via modulation of the enzymatic activity of kinases and phosphatases. Like phosphorylation, acetylation has emerged as a critical regulatory protein modification that is dynamically altered in response to diverse cellular cues. Moreover, acetyltransferases and deacetylases are tightly linked to cellular signaling pathways. Recent studies provide clues about the mechanisms utilized to regulate acetyltransferases and deacetylases. The therapeutic value of deacetylase inhibitors suggests that understanding acetylation pathways will directly impact our ability to rationally target these enzymes in patients. Recently discovered mechanisms which directly regulate the catalytic activity of acetyltransferases and deacetylases provide exciting new insights about these enzymes.
The Dynamic Regulation of Acetyltransferases and Deacetylases
In both the plant and animal kingdoms, protein function is often regulated via the post-translational addition of covalent modifications. The best characterized of these modifications is the phosphorylation of serine, threonine, and tyrosine residues. Phosphorylation, like other modifications, can dynamically regulate a broad range of protein functions. Consequences of phosphorylation include changes in sub-cellular localization, changes in protein protein interactions, alterations in protein half-life, and changes in the catalytic activity of proteinaceous enzymes. Research from the past decade indicates that the post-translational acetylation of lysine residues is also of broad importance in regulating eukaryotic protein function.
Kinase and phosphatase activity often responds to cellular stimuli, including engagement of growth factor receptors, changes in nutrient demand and metabolic activity, or genotoxic insults, in turn phosphorylating many down-stream targets thereby allowing the cell to properly respond to its environment. Many proteins, both histones and non-histone proteins (Table 1), are targeted by acetyltransferases and deacetylases (still often referred to as HATs and HDACs owing to the early characterization of histones as substrates). Recent evidence from a number of groups indicates that, like kinases, the enzymatic activity of acetyltransferases and deacetylases might be dynamically regulated and that this dynamic regulation can play an intricate role in modulating the response to cellular stimuli. Several mechanisms are known to regulate these enzymes: changes in subcellular localization, ubiquitylation-mediated degradation, changes in interactions with regulatory partners or cofactors, alterations in the cellular pools of acetyl-CoA1, and biophysical changes to the enzymes themselves that directly alter their catalytic activity. Many of these regulatory mechanisms have been discussed in recent reviews 2–4. Very recently a number of exciting examples have come to light in which acetyltransferases and deacetylases are regulated by this final mechanism, i.e. direct biophysical alterations intrinsic to the enzymes themselves. A number of examples of this direct control of enzymatic activity will be discussed here, including (i) control of p300 acetyltransferase activity by phosphorylation, acetylation, and sumoylation; (ii) modulation of histone deacetylase 1 (HDAC1) activity via p300-mediated acetylation which affects transcriptional activation programs; (iii) activation of Tat-interacting protein of 60kDa (TIP60) through direct sumoylation in response to DNA damage, as well as through altered interaction with the interacting partner, activating transcription factor 2 (ATF2); (iv) sirtuin 1 (SIRT1) enzymatic control by cell-cycle dependant phosphorylation and DNA damage induced sumoylation. In addition, we will discuss how these mechanism of direct regulation are linked to specific cellular processes including responses to genotoxic stress and malignant transformation. Although we have attempted to focus solely on mechanisms which directly alter intrinsic catalytic activity, these mechanisms are often linked to some of the other regulatory mechanisms mentioned above. For example, changes in Tip60 subcellular localization might be linked to altered post-translational modifications that in turn directly control enzymatic activity5 (Figure 1). We expect that many similar examples will emerge in future studies as the identification of critical protein acetylation events continues to grow at a remarkable rate.
Table 1.
Representative examples of substrates for acetyltransferases and deacetylases
| Acetyltransferases | ||
|---|---|---|
| Classical Name | Substrates (partial list) | Reference |
| CBP (KAT3A) | H2A; H2B; HDAC1; p53 | [18] |
| p300 (KAT3B) | H2A; H2B; HDAC1; p53; GATA-1a; p73 | [34,65,66,67] |
| Tip60 (HTATIP or KAT5) | H4, H2A, H3 not H2B; p53,; ATM | [46,53,54,49] |
| Deacetylases | ||
| Classical Name | Substrates (partial list) | |
| HDAC1 | p53 AR; MyoDc; E2F-1 | [68,69,70,71] |
| SIRT1 | p300; p53; ARb; PARP1d | [29,72–74,75,76] |
GATA binding protein 1
Androgen Receptor
MyoD- myogenic differentiation 1
poly (ADP-ribose) polymerase 1
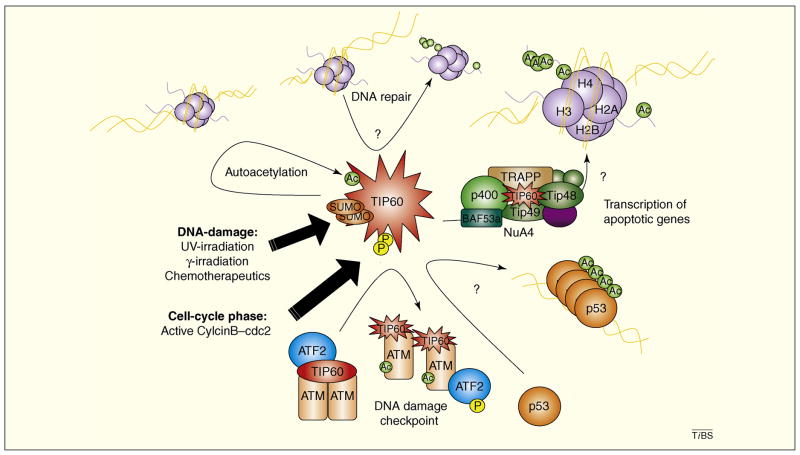
Figure 1. Points at which Tip60 enzymatic activity might modulate biological responses to cellular events.
Tip60 (red) enzymatic activity is altered in response to cellular events including genomic insults and cell-cycle state. These physiological events modulate Tip60 acetyltransferase activity by both the addition of modifications such as sumoylation (orange) and phosphorylation (yellow), as well as through modifications to interacting partners, e.g. ATF2 (blue) or by modulating interaction with the multi-protein NuA4 acetyltransferase complex. As a result, Tip60 enzymatic activity might play an integral part in facilitating (i) activation of the DNA damage checkpoint through ATM (gold) activation and ATF2 displacement (ii) DNA repair through the exchange of histones (purple) at sites of damage (iii) support of transcription through acetylation (green) of histones H2A and H4 and the transcription factor p53 (orange), which can facilitate the expression of pro-apoptotic target genes. As demonstrated by the question marks, the role of Tip60 enzymatic modulation in these pathways is yet to be formally demonstrated.
Regulation of p300/CBP catalytic activity: acetylation and transcription
A variety of mechanisms regulate p300 and its paralog, CREB binding protein (CBP). These two proteins are highly conserved and share many over-lapping functions; however, they also play unique roles during specific physiological processes (reviewed in 6). p300/CBP contain several conserved domains including (i) three cysteine-histidine (CH)-rich domains and a KIX domain, which are important for interactions with a number of cellular transcription factors, including p53, E2F1, and RelA (also known as p65)6; (ii) the bromodomain, common to p300, CBP and other eukaryotic acetyltransferases, which is believed to facilitate interactions with acetylated lysine residues; and (iii) the centrally located and highly conserved acetyltransferase domain.
p300/CBP phosphorylation
A number of signaling pathways that activate kinase cascades trigger phosphorylation of p300 and CBP and can have profound effects on their function7–9. Cell-cycle dependent CBP/p300 phosphorylation, mediated by the cyclin E cyclin dependent kinase 2 (CDK2) complex, was one of the first reported examples of acetyltransferase regulation by enzymatic activity modulation 10. This phosphorylation stimulates acetyltransferase activity, which presumably increases histone acetylation at target promoters and facilitates CBP/p300-dependent transactivation of genes required for progression through S-phase 10. The role of p300/CBP in the transactivation of these cell cycle genes might result from the alteration of histone acetylation levels; however p300-mediated acetylation of non-histone substrates likely also plays a role.
p300/CBP autoacetylation
Early in vitro assays using the p300 and CBP acetyltransferase domains uncovered autoacetylation activity 11, 12, 13. However, it was more than a decade after their initial cloning that the regulatory role of p300 and CBP autoacetylation was first investigated at a mechanistic level. Thompson et al. dissected the function of p300 autoacetylation, specifically focusing on the enzyme’s catalytic activity 14. Recombinant hypoacetylated p300 protein was generated and subjected to biochemical analysis, which led to the identification of a novel lysine-rich activation loop within the enzyme. The unacetylated form of this loop could decrease p300 enzymatic activity by acting as an inhibitory pseudosubstrate. Upon autoacetylation the loop changes confirmation and no longer inhibits p300 enzymatic activity. This mechanism is reminiscent of one used by some tyrosine kinases 15. These kinases also contain regulatory loops that suppress activity when hypophosphorylated; by contrast, phosphorylation by either upstream kinases or via autophosphorylation renders the kinases fully active. Further characterization of the function for the p300 activation loop was done in mammalian cells by comparing the enzymatic activity of wild-type p300 to that of a mutant that had a deletion of the activation loop (p300Δ) 16. The p300Δ protein was more active than its wild-type counterpart in both its ability to acetylate the non-histone substrate p73 and to transactivate genes in the context of a luciferase reporter. The authors went on to establish the physiological relevance of their in vitro observations by validating the acetylation of p300 at K1499 in vivo.
Further work by this group characterized the mechanisms of acetylation within the activation loop 16. They found that efficient “auto”-acetylation could occur in trans via an intermolecular interaction with other p300 molecules; however, acetylation of this loop by PCAF, a member of a distinct acetyltransferase family, was minimal 17. Furthermore the group identified five acetylation sites within the activation loop whose modification precedes the more robust autoacetylation of the protein. Others have independently verified that several of the autoacetylated lysines are modified on endogenous CBP as well 18. This evolutionary conservation suggests that this mechanism of acetyltransferase regulation is likely of fundamental importance in higher eukaryotes. Intriguingly, the TIP60 and PCAF acetyltransferases can also undergo autoacetylation 19–21, despite the lack of primary sequence conservation with p300. It is tempting to speculate that autoacetylation might be a mode of regulation for many acetyltransferases. Such a regulatory mode is especially relevant in light of very recent evidence: the catalytic cores of these distinct families of acetyltransferases share similar structures, despite the fact that they lack primary sequence conservation22–24. Another intriguing aspect of the common autoacetylation exhibited by mammalian acetyltransferases is evident when one considers that many of these enzymes also contain bromodomains that are capable of interacting with acetylated lysines. The lysine residues subject to autoacetylation might themselves be recognized by these bromodomains in order to facilitate additional inter- or intra-molecular homotypic regulatory interactions.
Regulating p300/CBP catalytic activity via autoacetylation during transcriptional initiation
It is clear that autoacetylation can control p300 activity, but what is not yet clear from this discussion is whether autoacetylation occurs constitutively, or whether it can be dynamically modulated in response to physiological stimuli. p300 is recruited to promoters by transcriptional activators such as p53 25. Once recruited, p300 acetylates histones, an important event for transcriptional activation. Interestingly, the presence of p300 can be inhibitory to transcription and only after p300 is released can TFIID and other components of the basal transcriptional machinery assemble 26. This finding suggests that p300 recruitment and release are both necessary for maximal transcription to proceed. How might this occur? Evidence from Black et al. supports a model in which the autoacetylation of p300 induces a conformational change that directly affects its ability to interact with Mediator, a multi-protein complex that plays critical roles in the early stages of transcriptional activation 26. p300 is recruited to the promoter in a hypoacetylated “inactive” state (Figure 2). Once at the promoter, p300, via its enhanced catalytic activity, acetylates both histones and itself, causing a conformational change that allows it to be released from Mediator, thereby permitting subsequent assembly of the basal transcriptional machinery. The authors conclude that p300 creates an autocatalytic switch whereby p300–promoter association activates the enzyme, thus facilitating p300 autoacetylation and subsequent release. Autoacetylation might be triggered by the simultaneous recruitment of multiple p300 molecules to the same locus. If autoacetylation can occur in trans, as suggested by in vitro data 26, this might represent the mechanistic switch needed to initiate p300’s role in transcription. Unfortunately, this model fails to explain how the first autoacetylation event can occur among a promoter- bound cluster of non-acetylated/inactive p300 molecules. It is possible that an as yet unrecognized steric effect of promoter recruitment or oligomer formation initiates p300 autoacetylation. Alternatively, the low levels of acetylation activity inherent to the non-acetylated p300 isoform might be sufficient to trans autoacetylate other p300 molecules when present at high local concentration on the promoter. Such a feed-forward mechanism could then trigger the activation of the full complement of available p300 molecules.
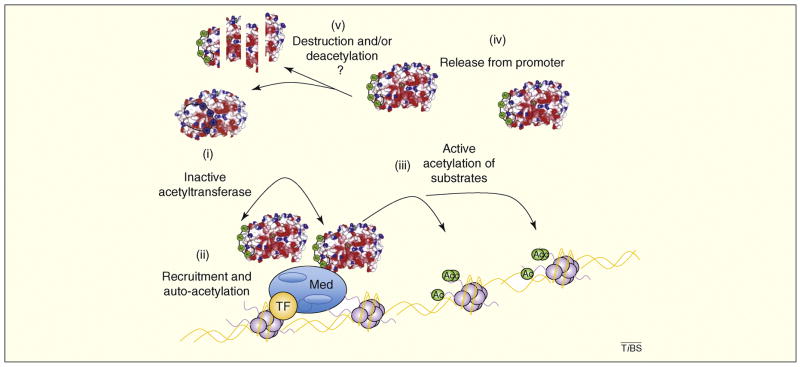
Figure 2. A catalytic switch activates p300/CBP at target promoters.
p300 and CBP each contain an unstructured positively charged loop which can inhibit their catalytic activity and is thought to modulate their transcriptional coactivator function. (i) Before the loop is acetylated the positively charged domain remains tightly associated with the enzymatic cleft, blocking activity. (ii) Upon recruitment to a target promoter, multiple p300/CBP molecules are in close proximity and can autoacetylate and fully activate each other. (iii) Once the acetyltransferases are fully active, they can acetylate targets including histones (purple), transcription factors (TF; orange) and possibly other components of the transcriptional machinery which are necessary for robust transcriptional activation of the targeted gene. (iv) Autoacetylation also allows for p300/CBP release from the promoter, which might be a necessary step for the continued assembly of the basal transcriptional machinery. (v) p300 and CBP are now in an active, highly acetylated state. The mechanism by which they are returned to the unacetylated state is unclear, but it could involve degradation and subsequent production of new protein or activation loop deacetylation. Mediator (Med): blue.
Competition between acetylation and sumoylation modulates p300 activity
A region between amino acids 1004–1045 within p300 is repressive to the transcriptional activation of some p300-responsive genes27. This region has been designated the cell cycle regulatory domain 1 (CRD1), because the repressive nature of this region can be de-repressed by the expression of p21 (also called cyclin-dependent kinase inhibitor 1A; CDKN1A) in a CRD1-dependent manner 27. More recently, sumoylation and acetylation sites were identified within the p300 CRD1 region 28, 29. Interestingly, data suggests that the sumoylated region of the p300 CRD1 domain is important for the SIRT1-mediated repression of a subset of genes that use p300 as a coactivator. Biochemical analysis revealed that acetylation and sumoylation might target the same lysines within p300, suggesting that the repressive effects of SIRT1 could stem from its ability to deacetylate these sites, thereby allowing SUMO1 conjugation and suppression of p300 activity29. Microarray analysis identified a subset of genes which utilize p300 as a coactivator and that are also repressed by SIRT1 in a p300-dependent manner; some of these genes correlate with metabolism and cellular differentiation, known SIRT1-associated pathways29. Therefore, it is intriguing to consider that the switch in CRD1 modification between sumoylation and acetylation might enable differential regulation of p300/CBP-mediated gene expression. Both the acetyltransferase domain and the CRD1 domain are highly modified regions of p300 and CBP and seem to modulate the complex role that p300/CBP play in gene activation and the cellular decision to proliferate or arrest.
Suppression of HDAC1 enzymatic activity by p300-mediated acetylation influences transcriptional regulation
Histone acetylation is commonly associated with active transcription, whereas histone deacetylation is typically linked to gene repression. Paradoxically, it has been known for some time that a small subset of genes requires deacetylase activity for their transcription30, 31. Although many of these deacetylase-activated genes might be indirect downstream targets, it appears that the transcription of at least some is directly activated by these normally repressive enzymes. For example, research in 1990 demonstrated that the addition of the deacetylase inhibitor butyrate to cells would inhibit glucocorticoid receptor (GR)-mediated mouse mammary tumor virus (MMTV) transcription 32. Subsequent studies established that this inhibition happens very rapidly, occurring within five minutes of hormone addition 33. Additionally, cells treated with the deacetylase inhibitor trichostatin A (TSA) exhibit impaired transcription, but display minimal changes in histone acetylation levels. This finding led to the suggestion that, in this system, a non-histone substrate might be the relevant deacetylase target. Using specific deletion mutants of the MMTV promoter region, the authors showed this HDAC-dependant GR-mediated transcription is correlated with the proximal promoter and specifically by the region including the TATA box. Therefore the relevant non-histone target(s) is likely a component of the basal transcriptional machinery; however, the specific target(s) remain unknown. These fundamental early studies set the stage for more recent advances in our understanding of HDAC regulation, including those described below.
The Hager group, in an additional study of GR-mediated MMTV transcription, identified HDAC1 as the specific enzyme responsible for the effects elicited by the broad spectrum deacetylase inhibitors34. Furthermore, they found that HDAC1 activity is required for active MMTV transcription and that HDAC1 acetylation regulates its activity, which then modulates the transcriptional activity from the MMTV promoter 34. Following the addition of a GR-agonist, the MMTV reporter undergoes biphasic activation and subsequent repression, a series of events which is correlated with polymerase II occupancy at the promoter 35. Hager’s group sought to identify acetylated proteins that associated with the GR at different times, in the hope that doing so might provide insight into the biphasic MMTV activation 34. Interestingly, upon agonist addition, HDAC1, although present at all time points, is specifically acetylated during the phase of transcriptional decline. Further analysis also revealed that HDAC1 acetylation correlates with p300 recruitment to the GR complex. A series of analyses revealed that, indeed, p300 directly acetylates HDAC1, thereby rendering HDAC1 nearly inactive, as measured by in vitro deacetylation of core histone H4. Mass spectrometry identified six p300-dependent acetylated lysines within HDAC1. Of these lysines, two (K218 and K220) lie within the enzyme’s catalytic core and the other four (K432, K438, K439 and K441) are located within the C-terminal region. To test whether acetylation of these lysines can influence HDAC1 enzymatic activity, point mutations were introduced which resulted in the conversion of all six lysines to either arginine (mimicking an unacetylated site; 6K/R) or glutamine (mimicking an acetylated site; 6K/Q). Analysis of these HDAC1 mutants further bolstered the hypothesis that acetylation blocks HDAC1 activity, as the 6K/Q mutant had minimal activity towards histones. By contrast, the 6K/R mutant displayed wild-type deacetylase activity. Interestingly, K218 and K220, which are located in the HDAC1 core catalytic domain, are predicted to be in close proximity to the HDAC1 monovalent ion binding pocket, a domain critical for deacetylase activity. This finding led to the hypothesis that these two residues might be the important targets of p300-mediated HDAC1 inactivation. However, a more focused set of HDAC1 mutants which express lysine to glutamine alterations in only one or a few of the acetylated lysines revealed that modification of the C-terminal lysine residues was more important for HDAC1 inactivation and that the core catalytic domain lysine targets contribute only minimally to HDAC1 regulation.
Hager and colleagues propose an intriguing model for GR-mediated transcription, in which ligand addition facilitates HDAC1 recruitment to the MMTV promoter34. They posit that the nonacetylated (active) form of HDAC1 deacetylates an unknown, non-histone target that is necessary for rapid and highly active transcription. After a longer period of hormone stimulation, however, an acetyltransferase, e.g. p300, is recruited to the GR complex and acetylates HDAC1, thus suppressing its catalytic activity. This event leads to a second, less-active phase of GR-mediated transcription. Many questions regarding this intricate acetylation-deacetylation cascade remain unanswered. What is the non-histone target deacetylated by HDAC1? Does the recruitment of an acetyltransferase result in less transcription solely through HDAC1 acetylation or can this acetyltransferase also acetylate the same target that HDAC1 deacetylates? Is there a deacetylase that can target HDAC1 and reverse this repressive affect? Indeed HDAC1 cannot auto-deacetylate in vitro 34, leaving a few possibilities: another deacetylase might reverse the acetylation mark, HDAC1 could require a specific co-factor, missing in the in vitro reaction, for auto-deacetylation, or the cellular concentration of acetylated and non-acetylated HDAC1 might be modulated solely through protein turnover. Finally, do other cell signaling networks also utilize such modulation of HDAC1 activity to fine-tune their transcriptional regulatory targets? Literature examining this biochemical mechanism is limited to the GR-activated MMTV transcriptional system. However, growing evidence from genome wide profiling studies in multiple organisms points to a role for HDACs as positive regulators of transcription at at least a few loci 36–38, thereby suggesting that the role of HDAC1 in GR-meditated transcription is not an anomaly, but rather a precursor example of many other instances where the dynamic regulation of acetyltransferases and deacetylases might influence gene expression. It will be interesting to see how this non-canonical role for HDAC1 as a transcriptional activator unfolds. It is clear that a more detailed analysis of the dynamic acetylation-deacetylation of both histone and non-histone substrates is necessary to further our understanding of the role played by acetylation in regulating gene expression.
Understanding the mechanisms which regulate HDAC function has far-reaching clinical implications. Pharmacological HDAC inhibitors are being evaluated as treatments for a broad variety of diseases, and the first of these, vorinostat, was approved by the FDA in 2006 for treatment of Cutaneous T Cell Lymphoma. Studies targeting muscular dystrophy39, Huntington’s Disease40, Alzheimer’s Disease41, HIV infection42 and numerous other forms of cancer, are among the approximately 100 clinical trials involving HDAC inhibitors (http://clinicaltrials.gov/). Understanding more about how these enzymes are regulated by cellular signaling pathways (e.g. those activated by hormone treatment or chemotherapeutic strategies that induce DNA damage), is critical for our ability to use these inhibitors in a more efficacious manner.
TIP60 enzymatic activity changes in response to DNA damage
The MYST family of acetyltransferases is conserved from yeast to humans 16. Although the human MYST family comprises five members (TIP60, hMOF, HBO1, MOZ and MORF), TIP60 is the most thoroughly studied. Esa1p, a yeast MYST member associated with the NuA4 acetyltransferase complex43, is an important regulator of cell-cycle progression 44. TIP60, the mammalian homolog of Esa1p, was identified in an interaction screen that looked for proteins that associate with the HIV-1 Tat protein 45. TIP60 characterization identified it as an acetyltransferase with specific substrate specificity: it acetylates the core histones H2A, H3 and H4, but not H2B 46. TIP60 resides in a complex similar to the yeast Esa1p-containing NuA4 complex 47, although some of its functions might not require association with this complex. Like Esa1p, TIP60 is an important player in the DNA damage response and apoptotic program 47, 48. More recent studies have shed light on the role played by TIP60 in the DNA damage and apoptosis pathways via the identification of additional histone and non-histone substrates. For example, the ataxia telangiectasia mutated (ATM) kinase, which is a central kinase in the checkpoint and repair pathways initiated by double strand DNA lesions, is acetylated by TIP60 as well as by the closely related MYST family member, hMOF 49–50. Acetylation is a critical event in ATM activation and for downstream signaling events, including p53 phosphorylation and cell-cycle arrest. TIP60 is also necessary for chromatin remodeling; in this role it works within a large NuA4-related complex that acetylates and subsequently exchanges histones at sites of double stranded DNA breaks, a process so far best described in Drosophila melanogaster 51. TIP60 acts as a coactivator for transcription of stress responsive target genes. In some cases TIP60 works with p53, both acetylating histones when recruited by p53 to its target genes 52, and directly acetylating p53 53, 54, thereby facilitating active transcription of cell-cycle arrest and apoptotic gene programs. These examples point to a critical role for TIP60 in multiple steps during the DNA damage and apoptotic responses to genotoxic stress.
Multiple mechanisms for TIP60 regulation
As TIP60 involvement in cellular processes that lead to cell death is multifaceted, it is not surprising that TIP60 is subject to a complex network of regulation. At least two separate E3 ubiquitin ligases, hMDM2 and the Cullin3 ROC1 (also called Rbx1 or Hrt1) complex, can direct TIP60 degradation 55, 56. Both E3 ubiquitin ligases seem to be opposed by signaling from the DNA damage response as their interactions with TIP60 are lost following UV- or γ-irradiation, stresses which result in TIP60 accumulation. Furthermore, TIP60 can be regulated by changes in its sub-nuclear localization which occur in response to DNA damage. UV-irradiation triggers TIP60 sumoylation at K430 and K451. This event re-localizes TIP60 to promyelocytic leukemia (PML) bodies and correlates with increases in TIP60 function 5. The recruitment of TIP60 to PML bodies also facilitates p53 recruitment, p53-mediated p21 transcription and proper cell-cycle arrest. Interestingly, sumoylation of TIP60 also has a direct effect on the enzyme’s intrinsic catalytic activity.
TIP60 catalytic activity: one of many nodes in TIP60 regulation
The first report describing regulation of TIP60 acetyltransferase activity stemmed from the observation that TIP60 contains consensus sequences for phosphorylation by cell-cycle related kinases 57. Empirical studies demonstrated that TIP60 is phosphorylated on serine 86 and serine 90. Remarkably, substitution of these sites to alanine (to eliminate phosphorylation) rendered TIP60 a very weak acetyltransferase for histone substrates. These residues are phosphorylated by the cyclin-dependent kinase complex cyclinB–cdc2 (cell division cycle 2), and this phosphorylation might specifically enhance TIP60 activity during the G2/M phase of the cell cycle 57.
In addition to the cell cycle-dependent regulation of its catalytic activity, the aforementioned ability of TIP60 to acetylate and thereby facilitate ATM kinase activation is regulated during the DNA damage response. Recently, the Ronai group demonstrated that ATF2 directly interacts with TIP60 and modulates its function at this step of the DNA damage response 56. ATF2 interacts with the TIP60 C-terminus, which contains the enzyme’s catalytic MYST domain and zinc finger region. This direct association between TIP60 and ATF2 represses TIP60 catalytic activity and also facilitates Cullin3–ROC1-mediated TIP60 degradation. After cells encounter DNA damage, TIP60 levels are no longer affected by the degradation complex. These data suggest that, prior to DNA damage, ATF2 maintains low TIP60 levels by facilitating TIP60 degradation, but that it has no effect on TIP60 protein levels after γ-irradiation. However, the authors elucidated a second function for ATF2 in regulating TIP60 following DNA damage. After DNA damage, TIP60 and ATF2 seem to be more loosely associated. Interestingly, ATF2 is an ATM substrate and the authors suggest that the ATF2 phosphorylation status might alter its interaction with TIP60. In this second state of the ATF2 TIP60 complex, ATF2 seems to be necessary for maximal TIP60 enzymatic activity because ATF2 depletion in damaged cells attenuates TIP60 acetyltransferase activity 56. These observations support the existence of a stress-responsive positive feedback loop between ATF2, TIP60 and ATM that facilitates the necessary cell-cycle arrest and/or apoptotic responses. This loop is connected by a complex series of interrelated post-translational modifications (eg, TIP60 phosphorylation, ATF2 phosphorylation and ATM acetylation).
In addition to ATF2-mediated regulation of TIP60 activity, sumoylation also modulates TIP60 activity5. As mentioned, following UV-irradiation, TIP60 is sumoylated, which alters the protein’s localization, targeting it to PML bodies. As well as changing its localization, Cheng et al. report a change in the catalytic activity of the protein. The authors generated different recombinant TIP60 plasmids that either block TIP60 sumoylation (TIP60-dm) or mimic a constitutively sumoylated TIP60 (TIP60-SUMO). Expression of these plasmids in HeLa cells revealed robust H2A acetylation following UV-irradiation in cells expressing either wild-type TIP60 (wt-TIP60) or TIP60-SUMO. By contrast, cells expressing TIP60-dm were unable to increase H2A acetylation. Additionally, TIP60 sumoylation directly influenced the intrinsic acetyltransferase activity in vitro. wt-TIP60 and TIP60-dm were expressed and affinity purified from 293T cells before and after UV-irradiation. In contrast to wt-TIP60, which was robustly activated by the DNA damage stimulus, as measured by acetylation of an H4 peptide, the activity of TIP60-dm was not significantly enhanced by UV-irradiation. Moreover, wt-TIP60 isolated from UV-irradiated cells that had been depleted of Ubc9, which is required for sumoylation, displayed catalytic activity as low as the non-sumoylatable TIP60-dm protein. These data suggest that TIP60 sumoylation influences both the localization and catalytic activity of the enzyme, thereby influencing its ability to mediate the response to DNA damage.
Consistent with its tight regulation during the response to genotoxic stress, changes in TIP60 expression and function have been identified in cancer, in both mouse models and human tumors 58, 59. For example, human prostate cancer samples display loss of TIP60, an event that has been correlated with its role in repressing the transcription of the metastasis gene KAI159. Human lymphomas and mammary tumors also display mono-allelic loss of TIP6059, implicating it as a tumor suppressor. Similarly, the progression of murine tumors driven by the MYC oncoprotein is decreased by the presence of TIP60 and even the loss of a single TIP60 allele is sufficient to enhance tumor growth 59. Together, these data suggest an important link between TIP60 levels or activity in the cell and the signals generated by the oncogene/DNA damage response pathways.
SIRT1 activity is regulated by phosphorylation and sumoylation
Sasaki et al. recently showed that in vitro SIRT1 activity can be regulated by its phosphorylation status. Specifically, SIRT1 interacts with and is directly phosphorylated by the CyclinB–CDK1 complex60. Although SIRT was known to be phosphorylated [63], this is the first evidence of a specific kinase targeting SIRT1. Additionally, Sasaki et al. identified two sites that were targeted by this cyclin-dependent kinase, threonine 530 and serine 540, and found that when substituted to alanines the resultant SIRT1 protein inhibits cellular proliferation 60. In vivo studies documenting the relevance of SIRT1 phosphorylation in different biological contexts will be both exciting and necessary for a full understanding of how SIRT1 phosphorylation might regulate deacetylase activity and thereby modulate the acetylation status of the various SIRT1 targets in the cells.
Another recent report characterized sumoylation-dependent regulation of SIRT1 deacetylase activity61. These authors identify SIRT1 lysine 734 as a sumoylated site and showed that sumoylation does not affect nuclear localization, but does abrogate SIRT1-mediated deacetylation of histone 4 lysine 16 and p53 lysine 382. Additionally, the mutant SIRT1 is defective for functionally repressing p53 transcriptional activity and apoptotic potential. To specifically investigate whether SIRT1 enzymatic activity is modulated by sumoylation, the authors performed an in vitro deacetylation assay on a p53 peptide, using recombinant SIRT1 which had either been previously sumoylated in vitro or left unmodified. Indeed, the authors found a two-fold increase in the deacetylase activity of sumoylated SIRT1. There was no change in the sumoylation levels or activity of the SIRT1-K734R mutant. The authors also showed that SUMO1/sentrin specific peptidase 1 (SENP1) interacts with and desumoylates SIRT1 in response to cellular stress, (eg, UV-irradiation and hydrogen peroxide treatment). SENP1 depleted cells contained more active, sumoylated SIRT1 and were more resistant to stress-induced apoptosis than their wild type counterparts. Yang et al. suggest an interesting “molecular switch” mechanism by which the stress-induced apoptotic potential of the cell is governed, in part, by SIRT1 sumoylation status. It is interesting to note that SENP1 can also desumoylate p53 and MDM262. Given the central role played by these multiple SENP1 substrates in diverse signaling pathways, additional study will be required to sort out their relative contributions. However, it is also appealing to imagine that a cell might use one protein, SENP1, to functionally modulate a pathway at multiple levels, e.g. by differentially regulating the sumoylation status and function of p53, MDM2 and SIRT1.
Concluding remarks
Kinases and phosphatases are critical regulators of important cellular processes; accordingly, their enzymatic activity is tightly modulated in response to biological cues. Activating mutations in these enzymes have been implicated as causative in human cancer 63. Furthermore, inhibitors of these enzymes are effective treatments for some forms of cancer 64. Recent studies clearly demonstrate that acetyltransferases and deacetylases are similarly linked to signaling pathways and that they undergo enzymatic regulation analogous to kinases and phosphatases. It is also now clear that inappropriate acetylation of histone and non-histone targets plays an important role in a number of human diseases. Exciting advances in our understanding of the biochemical mechanisms that control the enzymatic activity of acetyltransferases and deacetylases, such as those highlighted here, promise to identify many new points of regulation where these pathways can be attacked with targeted therapies. It is only via the detailed mechanistic analysis of these enzymes that we will identify opportunities to implement these interventions in a rational manner.
References
- 1.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Legube G, Trouche D. Regulating histone acetyltransferases and deacetylases. EMBO Rep. 2003;4:944–947. doi: 10.1038/sj.embor.embor941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyteca S, et al. To die or not to die: a HAT trick. Mol Cell. 2006;24:807–808. doi: 10.1016/j.molcel.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Dokmanovic M, et al. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Z, et al. Functional characterization of TIP60 sumoylation in UV-irradiated DNA damage response. Oncogene. 2008;27:931–941. doi: 10.1038/sj.onc.1210710. [DOI] [PubMed] [Google Scholar]
- 6.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 7.Nakajima T, et al. The signal-dependent coactivator CBP is a nuclear target for pp90RSK. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 8.Liu YZ, et al. Nerve growth factor up-regulates the transcriptional activity of CBP through activation of the p42/p44(MAPK) cascade. J Biol Chem. 1998;273:32400–32407. doi: 10.1074/jbc.273.49.32400. [DOI] [PubMed] [Google Scholar]
- 9.Chawla S, et al. CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- 10.Ait-Si-Ali S, et al. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- 11.Ogryzko VV, et al. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 12.Martens JH, et al. Scaffold/matrix attachment region elements interact with a p300-scaffold attachment factor A complex and are bound by acetylated nucleosomes. Mol Cell Biol. 2002;22:2598–2606. doi: 10.1128/MCB.22.8.2598-2606.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamamori Y, et al. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 14.Thompson PR, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- 15.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 16.Thomas T, Voss AK. The diverse biological roles of MYST histone acetyltransferase family proteins. Cell Cycle. 2007;6:696–704. doi: 10.4161/cc.6.6.4013. [DOI] [PubMed] [Google Scholar]
- 17.Karanam B, et al. Kinetic and mass spectrometric analysis of p300 histone acetyltransferase domain autoacetylation. J Biol Chem. 2006;281:40292–40301. doi: 10.1074/jbc.M608813200. [DOI] [PubMed] [Google Scholar]
- 18.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 19.Creaven M, et al. Control of the histone-acetyltransferase activity of Tip60 by the HIV-1 transactivator protein, Tat. Biochemistry. 1999;38:8826–8830. doi: 10.1021/bi9907274. [DOI] [PubMed] [Google Scholar]
- 20.Patel JH, et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, et al. Adenovirus E1B 55-kilodalton oncoprotein inhibits p53 acetylation by PCAF. Mol Cell Biol. 2000;20:5540–5553. doi: 10.1128/mcb.20.15.5540-5553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trievel RC, et al. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc Natl Acad Sci U S A. 1999;96:8931–8936. doi: 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Y, et al. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat Struct Biol. 2002;9:862–869. doi: 10.1038/nsb849. [DOI] [PubMed] [Google Scholar]
- 24.Neuwald AF, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 25.Lill NL, et al. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 26.Black JC, et al. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 27.Snowden AW, et al. A novel transcriptional repression domain mediates p21(WAF1/CIP1) induction of p300 transactivation. Mol Cell Biol. 2000;20:2676–2686. doi: 10.1128/mcb.20.8.2676-2686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girdwood D, et al. P300 transcriptional repression is mediated by SUMO modification. Mol Cell. 2003;11:1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 29.Bouras T, et al. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 30.McKnight GS, et al. Butyrate and related inhibitors of histone deacetylation block the induction of egg white genes by steroid hormones. Cell. 1980;22:469–477. doi: 10.1016/0092-8674(80)90357-8. [DOI] [PubMed] [Google Scholar]
- 31.Lallemand F, et al. Direct inhibition of the expression of cyclin D1 gene by sodium butyrate. Biochem Biophys Res Commun. 1996;229:163–169. doi: 10.1006/bbrc.1996.1774. [DOI] [PubMed] [Google Scholar]
- 32.Bresnick EH, et al. Glucocorticoid receptor-dependent disruption of a specific nucleosome on the mouse mammary tumor virus promoter is prevented by sodium butyrate. Proc Natl Acad Sci U S A. 1990;87:3977–3981. doi: 10.1073/pnas.87.10.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulholland NM, et al. Inhibition of MMTV transcription by HDAC inhibitors occurs independent of changes in chromatin remodeling and increased histone acetylation. Oncogene. 2003;22:4807–4818. doi: 10.1038/sj.onc.1206722. [DOI] [PubMed] [Google Scholar]
- 34.Qiu Y, et al. HDAC1 acetylation is linked to progressive modulation of steroid receptor-induced gene transcription. Mol Cell. 2006;22:669–679. doi: 10.1016/j.molcel.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 35.Becker M, et al. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 2002;3:1188–1194. doi: 10.1093/embo-reports/kvf244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reid G, et al. Multiple mechanisms induce transcriptional silencing of a subset of genes, including oestrogen receptor alpha, in response to deacetylase inhibition by valproic acid and trichostatin A. Oncogene. 2005;24:4894–4907. doi: 10.1038/sj.onc.1208662. [DOI] [PubMed] [Google Scholar]
- 37.Cho Y, et al. Individual histone deacetylases in Drosophila modulate transcription of distinct genes. Genomics. 2005;86:606–617. doi: 10.1016/j.ygeno.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Chang S, Pikaard CS. Transcript profiling in Arabidopsis reveals complex responses to global inhibition of DNA methylation and histone deacetylation. J Biol Chem. 2005;280:796–804. doi: 10.1074/jbc.M409053200. [DOI] [PubMed] [Google Scholar]
- 39.Berdeaux R, et al. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat Med. 2007;13:597–603. doi: 10.1038/nm1573. [DOI] [PubMed] [Google Scholar]
- 40.Thomas EA, et al. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington’s disease transgenic mice. Proc Natl Acad Sci U S A. 2008;105:15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green KN, et al. Nicotinamide restores cognition in Alzheimer’s disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J Neurosci. 2008;28:11500–11510. doi: 10.1523/JNEUROSCI.3203-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keedy KS, et al. A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J Virol. 2009;83:4749–4756. doi: 10.1128/JVI.02585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allard S, et al. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarke AS, et al. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamine J, et al. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J Biol Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- 47.Ikura T, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 48.Squatrito M, et al. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 49.Sun Y, et al. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta A, et al. Involvement of human MOF in ATM function. Mol Cell Biol. 2005;25:5292–5305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kusch T, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 52.Legube G, et al. Role of the histone acetyl transferase Tip60 in the p53 pathway. J Biol Chem. 2004;279:44825–44833. doi: 10.1074/jbc.M407478200. [DOI] [PubMed] [Google Scholar]
- 53.Sykes SM, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang Y, et al. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 55.Legube G, et al. Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. EMBO J. 2002;21:1704–1712. doi: 10.1093/emboj/21.7.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhoumik A, et al. Regulation of TIP60 by ATF2 modulates ATM activation. J Biol Chem. 2008;283:17605–17614. doi: 10.1074/jbc.M802030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lemercier C, et al. Tip60 acetyltransferase activity is controlled by phosphorylation. J Biol Chem. 2003;278:4713–4718. doi: 10.1074/jbc.M211811200. [DOI] [PubMed] [Google Scholar]
- 58.Kim JH, et al. Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature. 2005;434:921–926. doi: 10.1038/nature03452. [DOI] [PubMed] [Google Scholar]
- 59.Gorrini C, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 60.Sasaki T, et al. Phosphorylation regulates SIRT1 function. PLoS ONE. 2008;3:e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Y, et al. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9:1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen L, Chen J. MDM2-ARF complex regulates p53 sumoylation. Oncogene. 2003;22:5348–5357. doi: 10.1038/sj.onc.1206851. [DOI] [PubMed] [Google Scholar]
- 63.Kornek G, Selzer E. Targeted therapies in solid tumours: pinpointing the tumour’s Achilles heel. Curr Pharm Des. 2009;15:207–242. doi: 10.2174/138161209787002906. [DOI] [PubMed] [Google Scholar]
- 64.Pavlovsky C, et al. First-line therapy for chronic myeloid leukemia: Past, present, and future. Am J Hematol. 2009 doi: 10.1002/ajh.21380. [DOI] [PubMed] [Google Scholar]
- 65.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 66.Boyes J, et al. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 67.Costanzo A, et al. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol Cell. 2002;9:175–186. doi: 10.1016/s1097-2765(02)00431-8. [DOI] [PubMed] [Google Scholar]
- 68.Luo J, et al. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 69.Ito A, et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21:6236–6245. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaughan L, et al. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J Biol Chem. 2002;277:25904–25913. doi: 10.1074/jbc.M203423200. [DOI] [PubMed] [Google Scholar]
- 71.Mal A, et al. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 2001;20:1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langley E, et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 74.Luo J, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 75.Fu M, et al. Hormonal control of androgen receptor function through SIRT1. Mol Cell Biol. 2006;26:8122–8135. doi: 10.1128/MCB.00289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rajamohan SB, et al. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly (ADP-ribose) polymerase 1. Mol Cell Biol. 2009 doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]