Abstract
Ricin A-chain (RTA) and saporin-L1 (SAP) catalyze adenosine depurination of 28S rRNA to inhibit protein synthesis and cause cell death. We present the crystal structures of RTA and SAP in complex with transition state analogue inhibitors. These tight-binding inhibitors mimic the sarcin–ricin recognition loop of 28S rRNA and the dissociative ribocation transition state established for RTA catalysis. RTA and SAP share unique purine-binding geometry with quadruple π-stacking interactions between adjacent adenine and guanine bases and 2 conserved tyrosines. An arginine at one end of the π-stack provides cationic polarization and enhanced leaving group ability to the susceptible adenine. Common features of these ribosome-inactivating proteins include adenine leaving group activation, a remarkable lack of ribocation stabilization, and conserved glutamates as general bases for activation of the H2O nucleophile. Catalytic forces originate primarily from leaving group activation evident in both RTA and SAP in complex with transition state analogues.
Keywords: ricin toxin A-chain, saporin-L1, sarcin-ricin loop, depurination
Ricin A-chain (RTA) is the catalytic subunit of ricin, a Centers for Disease Control and Prevention category B bioterrorism agent derived from Ricinus communis seeds. RTA catalyzes the depurination of an invariant adenosine residue, A4234, within the GA4234GA tetraloop motif of the highly conserved sarcin–ricin loop of eukaryotic 28S rRNA (1). Clinical trials have exploited the toxicity of RTA in RTA-antibody constructs to kill leukemia and lymphoma cells (e.g., RTA conjugated to anti-CD22) (2–5). Side effects limit the utility of RTA immunotoxins (6, 7). Targeted inhibitors against ribosome-inactivating proteins (RIPs) could improve immunotoxin cancer therapies by rescuing normal cells following toxin treatment.
Saporin-L1 (SAP), a homologue of RTA from Saponaria officinalis (soapwort) leaves, exhibits N-glycohydrolase activities on 80S ribosomes, poly(A) RNA, and other cellular DNA and RNAs (8–12). SAP releases multiple adenines from ribosomes, whereas RTA shows exquisite specificity.
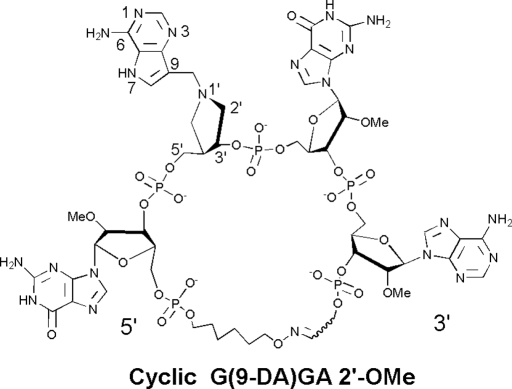
Truncated oligonucleotide constructs of the ribosomal sarcin–ricin loop are RTA and SAP substrates (13–16). In addition to hairpin stem–loop structures, RTA hydrolyzes adenine from cyclic GAGA loops that possess a 5′- to 3′-covalently closed synthetic linker (17, 18) (Fig. 1).
Fig. 1.
The structure of cyclic G(9-DA)GA 2′-OMe. 9-DA is a transition state mimic of 2′-deoxyadenosine. Atomic numbering for 9-DA follows that for purine nucleosides.
Transition state analysis of RTA-mediated depurinations established that hydrolysis of adenine involves a ribocation intermediate, followed by attack of an activated water. Adenine activation is a major driving force for RTA catalysis (19, 20). Efficient catalysis by RTA requires the invariant Glu-177 and Arg-180 residues (21–25). RTA and other RIPs have evolved to become near-perfect catalysts for mammalian ribosomes (21, 26–28). Here, we use transition state analogues to establish the catalytic site features contributing to this remarkable catalytic activity.
RTA transition state structures have guided the design and synthesis of potent RIP inhibitors (17, 29). Inhibitors for RTA and SAP include 1′-aza-sugars with a nonhydrolyzable 9-deazaadenine to mimic the elevated pKa of the leaving group (15). Dissociative transition states are characterized by increased ribosyl–adenine distance. A methylene bridge between aza-sugar and adenine groups in transition state analogues serves to mimic the dissociative transition state geometry (Fig. 1). RTA is active on mammalian ribosomes at physiological pH but is active on small RNAs and inhibited by transition state analogues only at low pH values (17).
In contrast to RTA, SAP depurinates synthetic oligonucleotides and mammalian ribosomes at physiological pH. Consequently, SAP binds transition state analogue inhibitors in both stem–loop and linear oligonucleotide geometries with low nanomolar affinity and is effective at protecting ribosomes from SAP at physiological pH (15). Cyclic oxime G(9-DA)GA 2′-OMe, linear trinucleotide G(9-DA)Gs3 2′-OMe, and dinucleotide s3(9-DA)Gs3 2′-OMe inhibitors inhibit SAP with slow-onset dissociation constant (Ki*) values of 3.9 to 7.5 nM (15) (Fig. 1 and SI).
The RIPs crystallize readily, but a sustained problem in the field has been the lack of structures with catalytic significance. Of the almost 100 crystal structures of RIPs listed in the Protein Data Bank, none of the N-ribohydrolases contain an oligonucleotide stem–loop structural analogue or tight-binding inhibitors. The present work provides unique information in the context of transition state analogue inhibitors.
Results
SAP and RTA Structures.
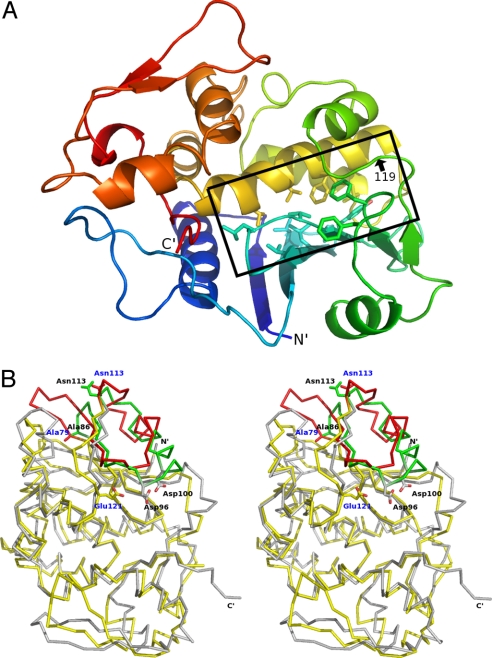
SAP and RTA (30% identity) share conserved tertiary and secondary structural elements. SAP is a monomer assembled into 2 partially segregated domains: an N-terminal, 6-stranded, mixed β-sheet with a short intervening α-helix (residues 1–119) and a C-terminal α-helical cluster that is followed by a 2-stranded antiparallel β-sheet (residues 120–257) (Fig. 2A). Nonpolar side chains project from the N-terminal β sheet (Leu-56, Val-63, Leu-65, Val-67, Val 72, Val-74, Tyr-77, Phe-91, and Phe-94) and from the C-terminal α-helical cluster (Val-151, Phe-164, Leu-165, Val-168, and Val-172), providing hydrophobic character to the interdomain active site cleft. The C-terminal, 2-stranded, β-sheet is structurally distinct in SAP and RTA. This region involves substrate access to the catalytic site and is implicated in the substrate specificities for SAP and RTA (30). The structural comparison between SAP and RTA also indicates a difference in the N-terminal segment formed by the short α-helix and 2 preceding β-strands (SAP 79–113 and RTA 86–113) (Fig. 2B).
Fig. 2.
Protein folds of RTA and SAP. (A) SAP structure is depicted in a ribbon diagram. The N-terminus is blue, with a color change to green (residues 1–119), yellow, and red, progressing to the C-terminus (residues 120–257). The position of residue 119 is indicated. The interface between N-terminal and C-terminal domains is highlighted in the black box, and the hydrophobic side chains in the interface region are shown. (B) Stereo-view of the overlaid Cα traces of RTA (gray) and SAP (yellow) is shown. The side chains are shown for Ala-79, Asn-113, and Glu-121 of SAP (blue labels) and Ala-85, Asp-96, Asp-100, and Asn-113 of RTA (black labels). Residues 79–113 of SAP and residues 85–113 of RTA are shown in red and green, respectively, to highlight a region that differs. Both structures are from inhibitor-bound complexes (3HIW and 3HIO).
Transition State Analogues.
Cyclic G(9-DA)GA 2′-OMe contains the tetranucleotide sequence of the GAGA sarcin–ricin loop but with the ricin-susceptible adenosine replaced by DADMe-Immucillin-A (9-DA), a transition state mimic (Fig. 1). The loop structure is maintained by a covalent oxime linker with geometry similar to stem–loop substrates (17). This cyclic oligonucleotide contains 5 phosphodiester bridges that are designed to be resistant to phosphodiesterases. 2′-O-Methylation prevents phosphodiesterase action and the 2′-deoxy of 9-DA provides chemical stability. 9-DA is a ribocation transition state mimic of the N-ribosyl hydrolase reactions catalyzed by RTA and proposed for SAP (15). Cyclic G(9-DA)GA 2′-OMe is an imperfect transition state mimic because it inhibits RTA with a competitive inhibition constant (Ki) of 300 nM at pH 4.0, binding only 320-fold tighter than substrate in the same construct (17). SAP inhibition by cyclic G(9-DA)GA 2′-OMe gives a Ki* of 3.9 nM at pH 7.7, binding 40,000-fold tighter than substrate. RTA action on ribosomes is not inhibited by these analogues at neutral pH values (15). Linear transition state analogues, including the trinucleotide G(9-DA)Gs3 2′-OMe (Ki = 7.5 nM) and dinucleotide s3(9-DA)Gs3 2′-OMe (Ki = 6.4 nM), inhibit SAP but not RTA (15) (Fig. S1). Both G(9-DA)Gs3 2′-OMe and s3(9-DA)Gs3 have 3 phosphodiester groups. These linear nanomolar inhibitors of SAP did not inhibit RTA at concentrations up to 100 μM in substrate competition assays, and therefore exhibit >104-fold specificity for SAP. Of these inhibitors, only cyclic G(9-DA)GA 2′-OMe was a significant RTA inhibitor and only at low pH (near 4.0).
Structure of RTA with Cyclic G(9-DA)GA 2′-OMe.
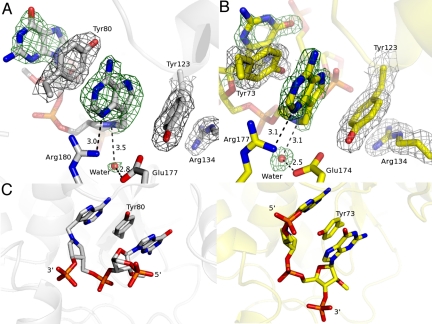
The final 2mFo-DFc electron density map contoured at the 1σ-level for the cyclic G(9-DA)GA 2′-OMe-RTA complex exhibited well-defined electron density for the 9-DA base, the methylene bridge of 9-DA, the preceding guanosine base, and the 5 phosphate diester groups of the cyclic inhibitor. Weak electron density was observed around the 9-DA N1′-aza-sugar analogue, especially around the position equivalent to the 5′-ribosyl carbon. Electron density was missing for the third guanine, the fourth adenine, and the oxime linker region, implying disorder of these substructures. The 9-deazaadenyl base of 9-DA caused a surprising reorganization of the catalytic site to form a continuous quadruple π-stack with the first guanine of the inhibitor and Tyr-80 and Tyr-123 residues of RTA (Fig. 3A). Tyr-123 also interacts with Arg-134 through a cation-π interaction and shares a hydrogen bond with the catalytic site Glu-177.
Fig. 3.
Catalytic site geometries for cyclic inhibitor G(9-DA)GA 2′-OMe bound to RTA and SAP. (A) π-Stacking interaction between the inhibitor and RTA is shown. The 2mFo-DFc electron density map and the inhibitor- and water nucleophile-omit mFo-DFc map were drawn in gray at a contour level of 1σ and in green at a contour level of 3σ, respectively. Glu-177 and Arg-180 are shown. (B) π-Stacking interaction between the inhibitor and SAP is shown. The 2mFo-DFc electron density map and the inhibitor- and water nucleophile-omit mFo-DFc map were drawn in gray at a contour level of 1σ and in green at a contour level of 3σ, respectively. Glu-174 and Arg-177 are indicated. (C) Comparison of inhibitor-bound RTA (Left, gray) and inhibitor-bound SAP (Right, yellow). The tyrosine involved in π-stacking is indicated. Distances in A and B are in angstroms.
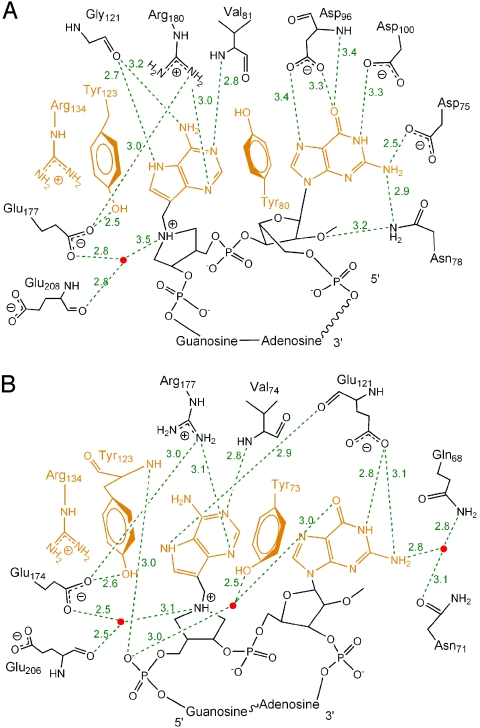
In addition to the π-stacking interaction, the N1, N6, and N7 groups of 9-DA participate in a hydrogen-bonding network with the Val-81 amide and the Gly-121 carbonyl (Fig. 4B). The guanidino nitrogen of Arg-180 is 3.0 Å from N3 of 9-DA. The first guanine of cyclic G(9-DA)GA 2′-OMe is tethered within a hydrogen-bonding network that includes the amide and/or carboxylate side chains of Asn-78 (2.9 Å), Asp-75 (2.5 Å), Asp-96 (3.3 Å and 3.4 Å), Asp-100 (3.3 Å), and the Asp-96 amide (3.4 Å).
Fig. 4.
Two-dimensional map of the cyclic transition state analogue inhibitor bound to the active sites of RTA (A) and SAP (B). The purines and residues involved in π-stacking are highlighted in orange. Water molecules are drawn as red dots. The hydrogen bonds are depicted as green dashed lines. The interaction between phosphodiester groups and the RIPs is shown in Figs. S2 and S3. The hydrogen bonds are depicted in green dashed lines (Å).
RTA has been proposed to use electropositive surface-exposed arginine and lysine residues to recognize the backbone phosphates of RNA tetraloop substrates (31). However, only the Arg-258 guanidino group interacts with the second and fourth phosphodiester groups (Fig. S2). Other interactions to the phosphodiesters are backbone-mediated and include (i) the Arg-213 amide and the third phosphodiester and (ii) water-bridged hydrogen bonds between the Asn-201 carbonyl oxygen and the third inhibitor phosphodiester. The first and fifth phosphodiesters of cyclic G(9-DA)GA 2′-OMe have no direct interactions with RTA.
Structure of SAP with Cyclic G(9-DA)GA 2′-OMe.
The final 2mFo-DFc electron density map contoured at the 1σ-level for the complex of SAP with cyclic G(9-DA)GA 2′-OMe exhibited well-defined electron density for the 9-DA base, the third guanosine moiety, and 4 (1–4) of the 5 phosphodiester links. Diffuse electron density was observed for the first guanine, the fourth adenosine, and the oxime linker. Two SAP molecules in the crystallographic asymmetrical unit are packed so that the first guanines of 2 SAP-inhibitor complexes have potential for overlap. We optimized the placement of the first guanine with respect to the coordinates of the first and second phosphates and its proximity to the adjacent 9-DA inhibitor group to avoid structural overlap. Contacts between SAP and the cyclic G(9-DA)GA 2′-OMe inhibitor were similar to those between SAP and the dinucleotide inhibitor s3(9-DA)Gs3 2′-OMe, and this comparison proved useful in assigning the geometry of the cyclic complex (Figs. S3–S6).
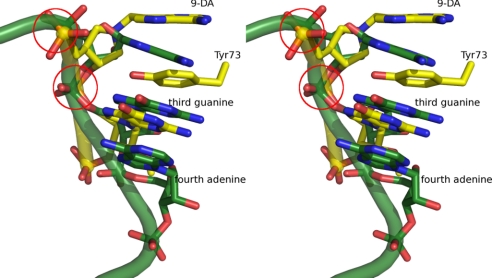
Cyclic G(9-DA)GA 2′-OMe bound to SAP revealed a quadruple π-stack at the catalytic site containing the 9-deazaadenyl of 9-DA, the third guanosine base from the inhibitor and residues Tyr-73 and Tyr-123 (Fig. 3B). This is remarkably similar to the π-stacking observed for inhibitor-bound RTA; however, in SAP, the equivalent tyrosine residues (Tyr-80 and Tyr-123 in RTA) interact with the third guanosine and 9-DA of cyclic G(9-DA)GA 2′-OMe. Thus, the 5′- to 3′-orientation of the SAP-bound inhibitor is reversed with respect to its placement in the RTA active site. Another noteworthy difference between RTA and SAP catalytic sites is the side chain of Tyr-73. In SAP, Tyr-73 is situated between 2 purine rings, and is displaced by 47° in relation to Tyr-80 in inhibitor-bound RTA. This geometry allows hydrogen bond formation with a water molecule that bridges Tyr-73, the second phosphodiester, and the third guanosine of the inhibitor (Figs. 3 and 4).
Similar to the RTA cyclic G(9-DA)GA 2′-OMe complex, the π-stacked purines of the SAP-bound cyclic inhibitor participate in an extensive hydrogen-bonding network with surrounding residues (Fig. 4B). N1, N6, and N7 of the 9-deazaadenyl base (9-DA) share hydrogen bonds with the Glu-121 backbone carbonyl and the Val-74 amide. The guanidino nitrogen of Arg-177 (equivalent to Arg-180 in RTA) is 3.1 Å from N3 of 9-DA. The third guanosine base forms 2 hydrogen bonds with the Glu-121 side chain and interacts, via a bridging water molecule, with the side chains of Gln-68 and Asn-71. The second phosphodiester of cyclic G(9-DA)GA 2′-OMe shares hydrogen bonds with the Tyr-123 backbone amide, the Tyr-233 phenyl hydroxyl group, and the Asn-207 amide side chain (Fig. S3). The third and fourth phosphodiesters form hydrogen bonds with the Trp-209 amide and the Arg-214 guanidino group, respectively. The first and fifth phosphodiesters of cyclic G(9-DA)GA 2′-OMe do not interact with catalytic site groups of SAP.
Catalytic Water.
Structures of inhibitor-bound RTA and SAP contain water molecules at distances of 3.5 Å and 3.1 Å, respectively, from the N1′-position of the 9-DA 1′-aza-sugar. This water is appropriately positioned for nucleophilic attack at the C1′-position of adenosine and is within hydrogen-bonding distance of the conserved carboxylate groups of RTA (Glu-177) and SAP (Glu-174), candidates for the role of general base in the catalytic mechanism (Fig. 3 A and B). The active site glutamates of RTA and SAP (Glu-177 and Glu-174) are 4.1 Å and 3.3 Å, respectively, from N1′ (pKa ≈8) of 9-DA, forming weak ion pairs. These glutamates are in a better position to accept a proton from water than to stabilize the developing ribocation known to form at the transition state of RTA (20).
Discussion
Leaving Group Interactions.
In the acid-catalyzed solvolysis of adenosine, protonation of adenine N7 and N1 forms an adenine dication causing loss of the N-ribosidic bond (20). Formation of an adenine cation facilitates extraction of bonding electrons from the ribosyl group to the adenine leaving group. At the transition state, N-ribosidic bond separation occurs by cationic character in both the adenine leaving group and the ribocation. Anionic adenine would not readily separate from the ribocation without water participation. Following transition state passage, the ribocation is neutralized by migration of the ribocation anomeric carbon to the nucleophilic water. In the complexes shown here, adenine leaving group activation is readily identified. RTA and SAP contacts both favor formation of adenyl cations. Stabilization of N7-protonated adenines occurs via backbone carbonyl oxygens (Gly-121 in RTA and Glu-121 in SAP) with N7 proton donation presumably from solvent. Face-to-face aromatic π-stacking of active site tyrosines (Tyr-123 and Tyr-80 in RTA, Tyr-123 and Tyr-73 in SAP) with the adenine base also elevates the pKa of the adenine, and thereby facilitates leaving group activation (32). At one end of the π-tetra-stack in both enzymes is a guanidine cation (Arg-134 in both RTA and SAP), adding more cation character through these groups. Polarization of N1 of the scissile adenines toward the transition state is provided by proton sharing with backbone amides (Val-81 in RTA, Val-74 in SAP). In conjugated ring systems, cationic polarization at any site facilitates electron pull from the cationic adenine toward the ribosyl group. At N3 of the adenine in both RIPs, a guanidine cation is in position to protonate N3 (Arg-180 in RTA, Arg-177 in SAP). These contacts to adenine support an adenine cation on the path to transition state formation. Mutagenesis studies of RTA have suggested that protonation of adenine N3 by invariant Arg-180 is critical for leaving group activation (22–25).
Structural studies of RTA with the dinucleotide ApG (PDB ID code: 1ApG) also suggested leaving group activation by π-stacking interactions between the adenine ring, Tyr-80, and Tyr-123; hydrogen-bonding interactions between N7 and Gly-121; transition state stabilization by N3 protonation with Arg-180; and amide backbone interactions between N6 and N1 with Val-81 (22, 23). The interactions around the 9-deazaadenyl base of cyclic G(9-DA)GA 2′-OMe in complexes with RTA and SAP are similar but also include the extended π-stacking interactions.
The Water Nucleophile.
The catalytic water is not apparent in earlier reports of the crystal structure of RTA, and Glu-177 or Arg-180 was considered to be a candidate base for activation (32, 33). In the RTA- and SAP-cyclic G(9-DA)GA 2′-OMe inhibitor complexes, the 9-deazaadenyl bases share a common binding orientation, the 1-aza-sugar groups are aligned in the same plane, and a water molecule occupies a position near N1′ appropriate for nucleophilic attack (Fig. 3C). The Arg-180 guanidino group of RTA and Arg-177 of SAP are positioned to polarize or protonate adenine N3 but are not in appropriate positions to interact with the nucleophilic water. Glu-177 (RTA) and Glu-174 (SAP) carboxylates, situated 2.8 Å (2.5 Å in SAP) from the water molecule, apparently act as general bases (Fig. S7).
Inhibitor Binding to SAP.
Cyclic oligonucleotides serve as efficient substrates for RTA and SAP because they mimic the folded GAGA loop of larger stem–loop substrates (15, 17). SAP structures with inhibitors show a quadruple π-stacking interaction between 9-DA, the third guanine, and 2 RIP active site tyrosines. Superposition of the SAP π-stacked guanosine of cyclic G(9-DA)GA 2′-OMe with the third guanosine of the GAGA tetraloop structure obtained for a 27-nucleotide mimic of the sarcin–ricin loop (PDB ID code: 1Q9A) produced a similar geometry for the 5′- and 3′-phosphate diesters of 9-DA with a small deviation in the position of the 3′-phosphate of the third guanosine (Fig. 5) (34). Crystallographic and NMR structures for free GAGA tetraloops report sheared guanine–adenine base pairs between the first guanine and fourth adenine and a triple purine stack among the last 3 nucleotides (34–37). This spatial arrangement of the GAGA tetraloop permits intercalation of SAP Tyr-73 between adenine and guanine bases. Likewise, the 9-deazaadenyl base at the edge of the triple purine stack in bound inhibitor enables Tyr-123 of SAP to insert on the outside of the equivalent tetraloop substrate without disrupting the tetraloop internal purine stack. In addition, SAP interacts with a face of the third guanine. Thus, the π-stacking interaction between the third guanine and fourth adenine of the tetraloop substrate remains similar to that of unbound stem loops.
Fig. 5.
Stereo-view of the superimposed inhibitors bound to SAP (yellow, PDB ID code 3HIW) and an unbound GAGA tetraloop (green, PDB ID code 1Q9A). The partial nucleic acid backbone of the inhibitor (yellow) and GAGA tetraloop (green) is shown. Tyr-73 and 9-DA and the third guanosine are depicted in yellow. The last 3 nucleotides of the unbound GAGA tetraloop are shown in green. The 5′- and 3′-phosphate diesters of 9-DA are highlighted in the red circles.
RTA and SAP Exhibit Distinct Modes of Inhibitor Binding.
RTA and SAP employ conserved structurally equivalent residues to recognize 9-DA and the third guanine (SAP) or first guanine (RTA) of cyclic G(9-DA)GA 2′-OMe. Despite similarities in the π-stacked geometries of the RIP-inhibitor complexes, the RTA and SAP inhibitor-binding modes are reversed. The different 3′- to 5′-polymer alignment implies that these RIPs exhibit divergent binding specificities. Distinct conformations are observed for the phosphates of cyclic inhibitor bound to RTA and to SAP (Fig. 3C). The cyclic inhibitor bound to RTA requires the first guanine to orient into a face-to-face contact with the RTA-bound adenine to form the quadruple π-stack. The orientation of this guanine is accompanied by separation of the sheared guanine–adenine base pair (between the first guanine and the fourth adenine), apparent in comparing the crystallographic and NMR structures for the sarcin–ricin loop GAGA tetraloop (34–37).
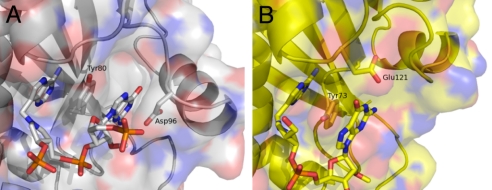
The molecular basis for the distinct binding mechanisms in RTA and SAP apparently arises from structural differences in the N-terminal region, which includes the fourth/fifth β-strands and a short α-helix (SAP 79–113 and RTA 86–113) (Fig. 2). Asp-96 of RTA projects from a loop between this helix and the fifth β-strand, and hydrogen bonds to guanine from the side (Fig. 6A). In contrast, SAP uses Glu-121 (a Gly in RTA), located on the loop between the N-terminal β-sheet and C-terminal α-helical cluster, to form hydrogen bonds to guanine (Fig. 6B). In order for RTA to bind substrate in the mode observed for SAP-inhibitor complex, the π-stacking interaction between the third guanine and fourth adenine of the GAGA tetraloop must be broken and the fourth adenine, which forms a sheared guanine–adenine base pair with the first guanine, requires reorientation to accommodate the N-terminal helix for hydrogen bonds between Asp-96 and Asp-100 to the third guanine (Fig. 4).
Fig. 6.
Cyclic inhibitor in the active sites of RTA and SAP. (A) Surface of RTA is shown relative to the active site. Tyr-80, Asp-96, 9-DA, and the first guanosine of the inhibitor are shown. Negative, positive, and neutral charges are shown in red, blue, and gray, respectively. (B) Surface of SAP is drawn to illustrate the active site. Tyr-73, Glu-121, 9-DA, and the third guanosine of the inhibitor are shown. The negative, positive, and neutral charges are drawn in red, blue, and yellow, respectively.
Inhibitor Conformation in RTA.
Crystals of the RTA-inhibitor complexes were also produced with the linear inhibitors G(9-DA)Gs3 2′-OMe and s3(9-DA)Gs3 2′-OMe (Fig. S1). Clear electron density was only observed for the 9-DA groups of these bound inhibitors. The disordered hydroxypyrrolidine and guanosine groups in these structures indicate stable interactions only to the scissile adenine analogue. Crystallization of RTA with cyclic G(9-DA)GA 2′-OMe caused the crystallographic C-axis to decrease by 5 Å and revealed the π-stacked guanine moiety of the inhibitor. This guanine moiety participates in 6 hydrogen bonds with surrounding residues, 3 of which involve Asp-96 (Fig. 4A). A previous report of RTA with bound ApG (PDB ID code: 1APG) lacked the guanine-to-Tyr-80 π-interaction and the hydrogen-bonding interactions with Asp-96. The conformation difference at Asp-96 (the rsmd for the Cα of Asp-96 is 1.73 Å, and the overall rmsd for the Cα is 0.64 Å) establishes a rearrangement relative to bound ApG.
Alternative Substrate Binding Modes for RIPs.
Inhibitors bound to SAP and RTA exhibit opposite phosphate diester polarity. The phosphodiester structures of unbound sarcin–ricin GAGA tetraloops differ from the geometry of inhibitor bound to RTA. RTA slowly depurinates the fourth adenine of the GAGA tetraloop substrate after the more specific second adenine has been hydrolyzed (17). The phosphodiester geometry of inhibitor bound to RTA is similar to that of a GAGA tetraloop construct bound to the ribotoxin restrictocin (PDB ID code: 1JBT), although base recognition by this RIP does not use π-stacking (38). Restrictocin, a ribonuclease, binds to the third guanosine and fourth adenosine of the GAGA tetraloop and unfolds the loop and hydrolyzes the phosphodiester bond between the third guanosine and fourth adenosine (28). The geometry of quadruple π-stacked guanine and 9-deazaadenyl of RTA-bound inhibitor resembles that of the third and fourth bases of the restrictocin-bound GAGA tetraloop.
Oligonucleotide Base Hydrolases.
Base hydrolytic reactions similar to the plant-depurinating RIPs are common in the DNA repair enzymes. DNA glycosylases initiate base excision repair by recognizing and cleaving mutagenic base lesions. Human uracil DNA glycosylase (UDG), human 3-methyladenine DNA glycosylase (AAG), human 8-oxoguanine glycosylase (OGG), and Methanobacterium thermoformicicum thymine DNA glycosylase (TDG) represent 4 major classes of DNA glycosylases that have been structurally characterized (39–42). Like RTA and SAP, human UDG and human AAG are single-domain proteins with a relatively small DNA-interaction surface. Human OGG and bacterial TDG contain multiple domains in which the active sites are located at domain junctions. Human UDG recognizes mismatched deoxyuridine by hydrogen bonding to every polar atom of uracil and by a favorable face-to-face π-stacking interaction between uracil and Phe-158 (39). Human AAG uses a confined pocket to fit the methyladenine base between Tyr-127 and His-136 and achieves the binding specificity by tight π-electron donor–acceptor interactions between the aromatic side chains and the cationic alkyl base (40). Human OGG specifically recognizes 8-oxo-guanine by hydrogen bonding to O6, N1, and protonated N7 of 8-oxo-guanine and sandwiching the base between Phe-319 and Cys-253 (41). Bacterial TDG from M. thermoformicicum tethers N2, N3, and O4 of thymine via hydrogen bonds without π-stacking interactions, whereas the C5 methyl group fits into a hydrophobic cleft (42). These DNA repair glycosylases interact only with the single base of the mismatched nucleotide, and none of them bind to oligonucleotides using the quadruple π -stack interactions found in RTA and SAP.
Conclusions
RIPs RTA and SAP bind to oligonucleotide transition state mimics with 2 active site tyrosine residues and an arginine to form a polarized quadruple π-stack with the adenine analogue (9-deazaadenyl) and its adjacent guanine of the cyclic inhibitor G(9-DA)GA 2′-OMe. The bound 9-deazaadenyl and guanine bases are tethered in a hydrogen-bonding network with the active site residues. Leaving group interactions from these RIPs are captured by the 9-deazaadenyl group. Distinct phosphodiester directional binding mechanisms observed in inhibitor-bound RTA and SAP may reflect 2 distinct modes of GAGA tetraloop recognition in the RIPs.
SAP is distinguished from RTA by its ability to bind tightly to trinucleotide [G(9-DA)Gs3 2′-OMe] and dinucleotide [s3(9-DA)Gs3 2′-OMe] transition state analogue inhibitors. Their binding modes are the same as that for bound cyclic G(9-DA)GA 2′-OMe. RTA prefers a folded GAGA tetraloop for binding and catalysis, whereas SAP can efficiently bind and catalyze linear GAGA sequences. SAP is catalytically efficient with synthetic (and ribosomal) substrates at neutral pH, whereas RTA depurinates small stem loops only at acidic pH.
Potent inhibitors of RIP proteins may have application as immunotoxin rescue therapeutics by inhibiting unwanted RIP activity in the blood. Structure-based design of RIP inhibitors has focused on the adenine-binding pocket (43, 44). Crystal structures of cyclic G(9-DA)GA 2′-OMe bound to RTA and SAP establish that the adjacent guanine base of the GAGA tetraloop (first for RTA or third for SAP) substantially contributes to these previously undescribed RIP-binding mechanisms. The transition state analogues described here capture this mechanism and establish an unprecedented polarized tetra-π-stack-binding mechanism at the catalytic sites of 2 representative RIPs.
Materials and Methods
Materials and methods are provided in SI Text and include (i) preparation of proteins, (ii) crystallization details, (iii) data collection, (iv) structure determination, and (v) model refinement summary in Table S1. The structure factors and atomic coordinates of SAP Y73A, apo SAP, dinucleotide-bound SAP, trinucleotide-bound SAP, cyclic inhibitor-bound SAP, and cyclic inhibitor-bound RTA have been deposited to the Protein Data Bank with the ID codes 3HIQ, 3HIS, 3HIT, 3HIV, 3HIW, and 3HIO, respectively.
Supplementary Material
Acknowledgments.
We thank Dr. P. Tyler of Industrial Research Ltd. for providing 9-DA used in this study. We thank Dr. H. Nika and the Laboratory of Macromolecular Analysis and Proteomics of the Albert Einstein College of Medicine of Yeshiva University for mass spectrum analysis of fragmented SAP. This work was submitted in partial fulfillment of the Ph.D. degree program for M.B.S., Albert Einstein College of Medicine, Yeshiva University. Data on x-ray diffraction for this study were measured at Beamline X29A of the National Synchrotron Light Source. Financial support of Beamline X29A of the National Synchrotron Light Source comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the U.S. Department of Energy and from the National Center for Research Resources of the National Institutes of Health. This work was supported by National Institutes of Health research Grant CA072444 and National Institutes of Health Medical Science Training Program Grant T32-CM00728.
Footnotes
The authors declare no conflict of interest.
Data Deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org, [PDB ID codes 3HIQ (SAP Y73A), 3HIS (SAP), 3HIT (SAP + s3(9-DA)Gs 2′-OMe), 3HIV (SAP + G(9-DA)Gs3 2′-OMe), 3HIW (SAP + cyclic G(9-DA)GA 2′-OMe), and 3HIO (RTA + cyclic G(9-DA)GA 2′-OMe)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0911606106/DCSupplemental.
References
- 1.Endo Y, Mitsui K, Motizuki M, Tsurugi K. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J Biol Chem. 1987;262:5908–5912. [PubMed] [Google Scholar]
- 2.Amlot PL, et al. A phase I study of an anti-CD22-deglycosylated ricin A chain immunotoxin in the treatment of B-cell lymphomas resistant to conventional therapy. Blood. 1993;82:2624–2633. [PubMed] [Google Scholar]
- 3.Messmann RA, et al. A phase I study of combination therapy with immunotoxins IgG-HD37-deglycosylated ricin A chain (dgA) and IgG-RFB4-dgA (Combotox) in patients with refractory CD19(+), CD22(+) B cell lymphoma. Clin Cancer Res. 2000;6:1302–1313. [PubMed] [Google Scholar]
- 4.Sausville EA, et al. Continuous infusion of the anti-CD22 immunotoxin IgG-RFB4-SMPT-dgA in patients with B-cell lymphoma: A phase I study. Blood. 1995;85:3457–3465. [PubMed] [Google Scholar]
- 5.Senderowicz AM, et al. Complete sustained response of a refractory, post-transplantation, large B-cell lymphoma to an anti-CD22 immunotoxin. Ann Intern Med. 1997;126:882–885. doi: 10.7326/0003-4819-126-11-199706010-00005. [DOI] [PubMed] [Google Scholar]
- 6.Baluna R, Coleman E, Jones C, Ghetie V, Vitetta ES. The effect of a monoclonal antibody coupled to ricin A chain-derived peptides on endothelial cells in vitro: Insights into toxin-mediated vascular damage. Exp Cell Res. 2000;258:417–424. doi: 10.1006/excr.2000.4954. [DOI] [PubMed] [Google Scholar]
- 7.Soler-Rodriguez AM, Ghetie MA, Oppenheimer-Marks N, Uhr JW, Vitetta ES. Ricin A-chain and ricin A-chain immunotoxins rapidly damage human endothelial cells: Implications for vascular leak syndrome. Exp Cell Res. 1993;206:227–234. doi: 10.1006/excr.1993.1142. [DOI] [PubMed] [Google Scholar]
- 8.Ferreras JM, et al. Distribution and properties of major ribosome-inactivating proteins (28 S rRNA N-glycosidases) of the plant Saponaria officinalis L. (Caryophyllaceae) Biochim Biophys Acta. 1993;1216:31–42. doi: 10.1016/0167-4781(93)90034-b. [DOI] [PubMed] [Google Scholar]
- 9.Stirpe F, et al. Ribosome-inactivating proteins from the seeds of Saponaria officinalis L. (soapwort), of Agrostemma githago L. (corn cockle) and of Asparagus officinalis L. (asparagus), and from the latex of Hura crepitans L. (sandbox tree) Biochem J. 1983;216:617–625. doi: 10.1042/bj2160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbieri L, et al. Polynucleotide:adenosine glycosidase activity of ribosome-inactivating proteins: Effect on DNA, RNA and poly(A) Nucleic Acids Res. 1997;25:518–522. doi: 10.1093/nar/25.3.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbieri L, Valbonesi P, Gorini P, Pession A, Stirpe F. Polynucleotide: Adenosine glycosidase activity of saporin-L1: Effect on DNA, RNA and poly(A) Biochem J. 1996;319(Pt 2):507–513. doi: 10.1042/bj3190507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbieri L, Valbonesi P, Govoni M, Pession A, Stirpe F. Polynucleotide:adenosine glycosidase activity of saporin-L1: Effect on various forms of mammalian DNA. Biochim Biophys Acta. 2000;1480:258–266. doi: 10.1016/s0167-4838(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 13.Amukele TK, Roday S, Schramm VL. Ricin A-chain activity on stem-loop and unstructured DNA substrates. Biochemistry. 2005;44:4416–4425. doi: 10.1021/bi0474362. [DOI] [PubMed] [Google Scholar]
- 14.Chen XY, Link TM, Schramm VL. Ricin A-chain: Kinetics, mechanism, and RNA stem-loop inhibitors. Biochemistry. 1998;37:11605–11613. doi: 10.1021/bi980990p. [DOI] [PubMed] [Google Scholar]
- 15.Sturm MB, Schramm VL. Transition state analogues rescue ribosomes from saporin-L1 ribosome inactivating protein. Biochemistry. 2009;48:9941–9948. doi: 10.1021/bi901425h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gluck A, Endo Y, Wool IG. Ribosomal RNA identity elements for ricin A-chain recognition and catalysis. Analysis with tetraloop mutants. J Mol Biol. 1992;226:411–424. doi: 10.1016/0022-2836(92)90956-k. [DOI] [PubMed] [Google Scholar]
- 17.Sturm MB, Roday S, Schramm VL. Circular DNA and DNA/RNA hybrid molecules as scaffolds for ricin inhibitor design. J Am Chem Soc. 2007;129:5544–5550. doi: 10.1021/ja068054h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allerson CR, Verdine GL. Synthesis and biochemical evaluation of RNA containing an intrahelical disulfide crosslink. Chem Biol. 1995;2:667–675. doi: 10.1016/1074-5521(95)90030-6. [DOI] [PubMed] [Google Scholar]
- 19.Chen XY, Berti PJ, Schramm VL. Transition-state analysis for depurination of DNA by ricin A-chain. J Am Chem Soc. 2000;122:6527–6534. [Google Scholar]
- 20.Chen XY, Berti PJ, Schramm VL. Ricin A-chain: Kinetic isotope effects and transition state structure with stem-loop RNA. J Am Chem Soc. 2000;122:1609–1617. [Google Scholar]
- 21.Robertus JD, Monzingo AF. The structure of ribosome inactivating proteins. Mini-Rev Med Chem. 2004;4:477–486. doi: 10.2174/1389557043403837. [DOI] [PubMed] [Google Scholar]
- 22.Monzingo AF, Robertus JD. X-ray analysis of substrate analogs in the ricin A-chain active site. J Mol Biol. 1992;227:1136–1145. doi: 10.1016/0022-2836(92)90526-p. [DOI] [PubMed] [Google Scholar]
- 23.Kim Y, Robertus JD. Analysis of several key active site residues of ricin A chain by mutagenesis and X-ray crystallography. Protein Eng. 1992;5:775–779. doi: 10.1093/protein/5.8.775. [DOI] [PubMed] [Google Scholar]
- 24.Kim Y, et al. Structure of a ricin mutant showing rescue of activity by a noncatalytic residue. Biochemistry. 1992;31:3294–3296. doi: 10.1021/bi00127a035. [DOI] [PubMed] [Google Scholar]
- 25.Frankel A, Welsh P, Richardson J, Robertus JD. Role of arginine 180 and glutamic acid 177 of ricin toxin A chain in enzymatic inactivation of ribosomes. Mol Cell Biol. 1990;10:6257–6263. doi: 10.1128/mcb.10.12.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sturm MB, Schramm VL. Detecting ricin: A sensitive luminescent assay for ricin A-chain ribosome depurination kinetics. Anal Chem. 2009;81:2847–2853. doi: 10.1021/ac8026433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsnes S, Fernandez-Puentes C, Carrasco L, Vazquez D. Ribosome inactivation by the toxic lectins abrin and ricin. Kinetics of the enzymic activity of the toxin A-chains. Eur J Biochem. 1975;60:281–288. doi: 10.1111/j.1432-1033.1975.tb21001.x. [DOI] [PubMed] [Google Scholar]
- 28.Korennykh AV, Piccirilli JA, Correll CC. The electrostatic character of the ribosomal surface enables extraordinarily rapid target location by ribotoxins. Nat Struct Mol Biol. 2006;13:436–443. doi: 10.1038/nsmb1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roday S, et al. Inhibition of ricin A-chain with pyrrolidine mimics of the oxacarbenium ion transition state. Biochemistry. 2004;43:4923–4933. doi: 10.1021/bi0498499. [DOI] [PubMed] [Google Scholar]
- 30.Savino C, Federici L, Ippoliti R, Lendaro E, Tsernoglou D. The crystal structure of saporin SO6 from Saponaria officinalis and its interaction with the ribosome. FEBS Lett. 2000;470:239–243. doi: 10.1016/s0014-5793(00)01325-9. [DOI] [PubMed] [Google Scholar]
- 31.Olson MA. Ricin A-chain structural determinant for binding substrate analogues: A molecular dynamics simulation analysis. Proteins. 1997;27:80–95. doi: 10.1002/(sici)1097-0134(199701)27:1<80::aid-prot9>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 32.Versees W, Loverix S, Vandemeulebroucke A, Geerlings P, Steyaert J. Leaving group activation by aromatic stacking: An alternative to general acid catalysis. J Mol Biol. 2004;338:1–6. doi: 10.1016/j.jmb.2004.02.049. [DOI] [PubMed] [Google Scholar]
- 33.Mlsna D, Monzingo AF, Katzin BJ, Ernst S, Robertus JD. Structure of recombinant ricin A chain at 2.3 A. Protein Sci. 1993;2:429–435. doi: 10.1002/pro.5560020315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Correll CC, Beneken J, Plantinga MJ, Lubbers M, Chan YL. The common and the distinctive features of the bulged-G motif based on a 1.04 A resolution RNA structure. Nucleic Acids Res. 2003;31:6806–6818. doi: 10.1093/nar/gkg908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Correll CC, et al. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc Natl Acad Sci USA. 1998;95:13436–13441. doi: 10.1073/pnas.95.23.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jucker FM, Heus HA, Yip PF, Moors EH, Pardi A. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J Mol Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- 37.Szewczak AA, Moore PB, Chang YL, Wool IG. The conformation of the sarcin/ricin loop from 28S ribosomal RNA. Proc Natl Acad Sci USA. 1993;90:9581–9585. doi: 10.1073/pnas.90.20.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X, Gerczei T, Glover LT, Correll CC. Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping. Nat Struct Biol. 2001;8:968–973. doi: 10.1038/nsb1101-968. [DOI] [PubMed] [Google Scholar]
- 39.Parikh SS, et al. Uracil-DNA glycosylase-DNA substrate and product structures: Conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc Natl Acad Sci USA. 2000;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc Natl Acad Sci USA. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 42.Mol CD, Arvai AS, Begley TJ, Cunningham RP, Tainer JA. Structure and activity of a thermostable thymine-DNA glycosylase: Evidence for base twisting to remove mismatched normal DNA bases. J Mol Biol. 2002;315:373–384. doi: 10.1006/jmbi.2001.5264. [DOI] [PubMed] [Google Scholar]
- 43.Carra JH, et al. Fragment-based identification of determinants of conformational and spectroscopic change at the ricin active site. BMC Struct Biol. 2007;7:72–82. doi: 10.1186/1472-6807-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller DJ, et al. Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition. J Med Chem. 2002;45:90–98. doi: 10.1021/jm010186s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.