Abstract
Totipotent stem cells have the potential to differentiate into every cell type. Renewal of totipotent stem cells in the germline and cellular differentiation during early embryogenesis rely upon posttranscriptional regulatory mechanisms. The Caenorhabditis elegans RNA binding protein, MEX-3, plays a key role in both processes. MEX-3 is a maternally-supplied factor that controls the RNA metabolism of transcripts encoding critical cell fate determinants. However, the nucleotide sequence specificity and requirements of MEX-3 mRNA recognition remain unclear. Only a few candidate regulatory targets have been identified, and the full extent of the network of MEX-3 targets is not known. Here, we define the consensus sequence required for MEX-3 RNA recognition and demonstrate that this element is required for MEX-3 dependent regulation of gene expression in live worms. Based on this work, we identify several candidate MEX-3 targets that help explain its dual role in regulating germline stem cell totipotency and embryonic cell fate specification.
Keywords: embryogenesis, nematode, RNA-binding protein, post-transcriptional regulation, maternal effect
Totipotent stem cells have the capacity to differentiate into an entire organism. Multicellular organisms maintain a population of totipotent stem cells in the germline to assure reproductive potential (1). These stem cells give rise to the gametes that eventually produce the next generation. The gametes that arise from these cells encode all of the information necessary to pattern the development of a new organism. This information is carried in the form of DNA, epigenetic marks, and cytoplasmic components. All of this information must be established and maintained in totipotent germline stem cells.
After fertilization, totipotency is lost in most cellular lineages as tissues and organs begin to differentiate. In many organisms, cell-fate specification occurs early during embryogenesis at a time when the organism's genome is transcriptionally quiescent (1, 2). Thus, posttranscriptional regulation of maternally and paternally supplied gene products provides the basis for the loss of totitpotency and cell fate specification.
In the nematode Caenorhabditis elegans, the conserved dual KH (hnRNP K homology) domain protein, MEX-3, is required for both maintaining totipotency in the germline and cell fate specification in the early embryo (Fig. S1A) (3, 4). Worms with a homozygous null mex-3 mutation have a fully penetrant maternal-effect lethal phenotype, resulting in embryos that fail to undergo body morphogenesis and produce excess muscle and hypodermal cells (3). Furthermore, these embryos produce between three and six cells that resemble germline progenitor cells, as opposed to two in a wild-type embryo. In addition, worms that are mutant for mex-3 and a second KH domain protein, gld-1, are sterile with a germline tumor that contains numerous transdifferentiated cells forming a cell mass similar to a germline teratoma (4).
In the germline, MEX-3 protein is expressed in distal germ cells and maturing ooctyes (3). Upon fertilization, cytoplasmic MEX-3 is present throughout the entire embryo and then becomes restricted, predominantly to the anterior founder cell (AB) at the end of the two-cell stage (Fig. S1 B–D). After the four-cell stage, MEX-3 begins to disappear from the embryo. A small amount of MEX-3 remains in the posterior germline lineage, where it localizes to RNA-rich bodies including germ granules in early embryos and CAR-1/CGH-1 granules in late oocytes (3, 5–7).
A family of related human proteins (hMex-3A–D and TINO) are differentially recruited to RNA granules involved in posttranscriptional regulatory mechanisms (8, 9). In addition to a region of MEX-3 homology, these factors contain a carboxy-terminal RING finger domain and numerous phosphorylation sites. Qualitative experiments show that hMex-3A, -3B, and -3C bind directly to RNA homopolymers, whereas hMex-3D/TINO binds to a sequence that contains AU-rich elements (8, 9). The precise sequence specificity and affinity of the MEX-3 family has not been defined.
Because MEX-3 contains conserved RNA-binding domains, it likely exerts a role in development at the posttranscriptional level. Consistent with this idea, several lines of evidence indicate that MEX-3 regulates the spatial and temporal translation of two key targets, pal-1 and nos-2. PAL-1 is a Caudal-like homeodomain protein required to specify the posterior blastomere C (10). Its protein expression pattern is anticorrelated with that of MEX-3 in early embryos, as expected if MEX-3 negatively regulates pal-1 translation. PAL-1 expression is not observed until the four-cell stage, where it accumulates in the nuclei of the posterior blastomeres. mex-3 mutant embryos show ectopic expression of PAL-1, and the anterior blastomeres take on a C-like fate resulting in excess muscle in the anterior (3). The gene encoding the Nanos homolog NOS-2 is also dependent upon MEX-3 for its protein expression pattern (11). This protein is required for the proper development of primordial germ cells and their incorporation into the somatic gonad. In the early embryo, NOS-2 is not observed until the 16-cell stage in the germline precursor P4. However, in mex-3 mutant embryos, NOS-2 is expressed ectopically throughout the entire embryo (11).
Translational reporter experiments suggest that MEX-3 is regulating its targets in a 3′UTR dependent manner (10, 11). An RNA reporter containing the 3′UTR of pal-1 fused to lacZ is translated in early embryos in a pattern that matches endogenous PAL-1 (10). In the absence of MEX-3, ectopic expression of the reporter is observed in oocytes and anterior blastomeres. Furthermore, recent work demonstrates that MEX-3 represses nos-2 mRNA translation through its 3′UTR (11). Using a GFP::H2B translational reporter transgene, several cis-regulatory elements within the 3′UTR required for the spatial and temporal control of nos-2 mRNA translation (subA–E) were identified (12). Both in vitro and in vivo experiments suggest that MEX-3 represses nos-2 translation by interacting with a repeat sequence present within the regulatory elements subB and subC (11), yet the precise specificity determinants remain unknown.
Ectopic expression of NOS-2 in the mex-3 mutant likely explains the presence of extra germline precursor cells in terminal mutant embryos. Likewise, ectopic expression of PAL-1 likely explains the presence of excess muscle. However, the mex-3 mutant phenotype, and its role in maintaining germline totipotency, suggests that MEX-3 regulates additional mRNA targets. To determine the nucleotide binding specificity of MEX-3 and identify new candidate regulatory targets, we set out to define the determinants of MEX-3 binding and map the cis-acting elements within nos-2 and pal-1 required for MEX-3 regulation.
Results
Identification of a High Affinity MEX-3 RNA Aptamer.
To better characterize how MEX-3 recognizes its mRNA targets we performed an affinity elution-based in vitro selection experiment (SELEX) (Fig. S2A) (13–15). With a starting pool of ssRNA comprised of 30 nucleotides of randomized sequence, RNA target sequences were selected by immobilizing the RNA binding domain of MEX-3 fused to the C terminus of maltose binding protein (MBP) on amylose resin. To monitor the progress of the selection, an EMSA was performed, where fluorescently labeled RNA samples from either pool 0, pool 4, or pool 7 were equilibrated with varying concentrations of MEX-3 and resolved on a native polyacrylamide slab gel (Fig. S2B). After seven rounds of selection and enrichment, a pool of RNA that binds with high affinity to MEX-3 was identified.
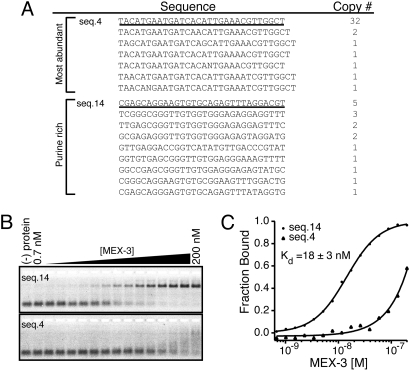
To analyze the sequences present within pool 7, the cDNA was cloned, and DNA from individual transformants was sequenced. Of 69 recovered sequences, 56 segregate into two major classes (Fig. 1A). The remaining sequences display no obvious similarity (Fig. S2C). The first group is comprised of 39 highly-related sequences with only 0–3 variable nucleotides between them. The other main group consists of 17 sequences that are highly purine-rich. To test whether MEX-3 binds with high affinity to either of these sequence classes, EMSA experiments were performed with the most abundant RNA sequence from each group. Quantitative analysis reveals that MEX-3 binds to the purine-rich example (seq.14) with high affinity (Kd,app = 18 ± 3 nM) (Fig. 1 B and C). In contrast, MEX-3 binds with lower affinity to the RNA example (seq.4) from the most abundant group (Kd,app = 160 ± 6 nM). It is not clear why seq.4 and its variants dominate the in vitro selection yield. To test if MEX-3 binds with high affinity to any of the unrelated RNAs, EMSA experiments were performed with two representative sequences. Neither sequence binds to MEX-3 with high affinity (Fig. S2D). Because seq.14 binds to MEX-3 with 9-fold higher affinity than seq.4, we chose to investigate seq.14 in more detail.
Fig. 1.
In vitro selection of MEX-3 RNA aptamers. (A) The two main sequence classes recovered after seven rounds of selection are shown. The first class is the most abundant with 39 sequences and the second class is purine-rich. Underlined are the examples tested for binding activity. The copy number of each clone is listed next to its sequence. (B) An EMSA of MEX-3 bound to seq.14 and seq.4 RNA. (C) A graph of MEX-3 versus fraction bound of seq.14 and seq.4 RNA is plotted and fit with the Hill equation. The calculated apparent equilibrium dissociation constant (Kd,app) for seq.14 is reported as the average ± one standard deviation of at least three replicates.
Identification of a 12-Nucleotide Element Sufficient for MEX-3 Binding.
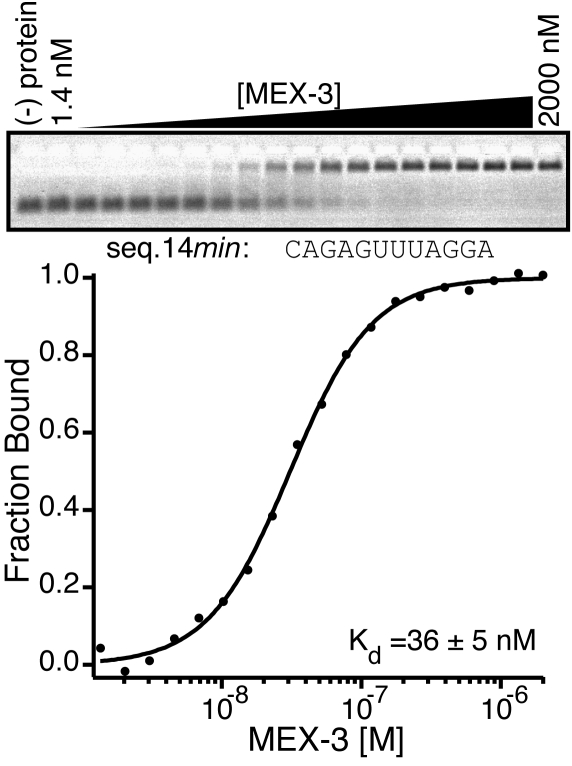
To determine the minimal sequence required for binding, truncation analysis was performed where either the 5′ or 3′-end of seq.14 was shortened by three nucleotide fragments (Fig. S3). The binding affinity of MEX-3 to these sequences was determined by EMSA. As many as 15 bases from the 5′-end (Kd,app < 43 nM) or six bases from the 3′-end (Kd,app < 21 nM) can be removed without a dramatic reduction in affinity. The results identify the region that is essential for recognition by MEX-3 (Fig. S3, shaded region). Based on these results, we designed a 12-nucleotide fragment (seq.14min) that encompasses this region and tested its ability to interact with MEX-3. Indeed, MEX-3 binds to this sequence with high affinity (Kd,app = 36 ± 5 nM) (Fig. 2). These experiments reveal the minimal region of the aptamer required for high affinity MEX-3 binding.
Fig. 2.
A 12-nucleotide element sufficient for MEX-3 binding. An EMSA experiment showing MEX-3 bound to seq.14min RNA (Upper) with a plot of MEX-3 versus fraction bound of seq.14min RNA (Lower). The Kd,app is reported on the graph.
Determination of the MEX-3 Consensus Sequence.
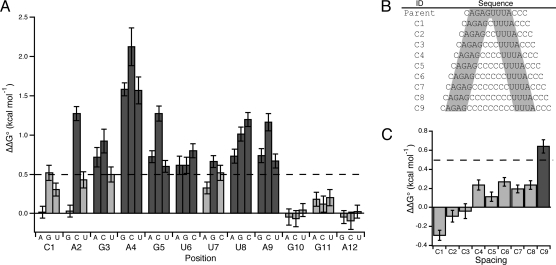
To determine the MEX-3 consensus sequence and analyze the thermodynamic contribution of each base, EMSA was used to measure the change in standard free-energy change (ΔΔG°) of every single point mutation of seq.14min RNA (Fig. 3A). Mutation of eight positions causes a significant reduction in binding affinity (positions 2–9; ΔΔG > 0.5 kcal mol−1), whereas mutation of four positions has little or no effect on binding (positions 1, 10–12; ΔΔG < 0.5 kcal mol−1). The cutoff of 0.5 kcal mol−1 represents a twofold reduction in binding affinity at 20 °C. Based on this quantitative assay, we define the MEX-3 recognition element (MRE) as (A/G/U)(G/U)AGU(U/A/C)UA. Analysis of this sequence reveals that five positions have a fixed nucleotide specificity, whereas three positions are partially degenerate allowing one or more base substitution at each position. The first four bases are predominantly purine-rich and the last four bases are AU-rich. MEX-3 contains two KH domains, and the typical binding surface of this motif is a four-nucleotide element with variable specificity (16–20). Therefore, we predict that each KH domain in MEX-3 recognizes a four-base half-site within the MRE.
Fig. 3.
Mutagenesis of seq.14min RNA. (A) This sequence was mutated systematically and the Kd,app was determined for every single point mutation. Each mutant was compared with the Kd,app of seq.14min to calculate the change in standard free energy change (ΔΔG°). The bars represent the ΔΔG° for each point mutation indicated on the x-axis. Base substitutions that have greater than a twofold loss in binding affinity are shown by dark gray bars (dashed line, ΔΔG° > 0.5 kcal mol−1), whereas mutations that have less than a twofold loss in binding affinity are shown by light gray bars. The error was propagated from the standard deviation of seq.14min and the respective point mutation. (B) The parent sequence containing the eight-nucleotide consensus with a background of cytodine is shown followed by nine spacing mutants (C1–C9). The shaded nucleotides represent each half-site within the MEX-3 consensus sequence. (C) The Kd,app of each spacing mutant was compared with the Kd,app of the parent sequence to calculate ΔΔG°. The spacing mutant that has greater than a twofold loss in binding affinity is shown by the dark gray bar (dashed line, ΔΔG° > 0.5 kcal mol−1).
To test this hypothesis, a series of spacing mutants were made where 0–9 cytidines were inserted between positions G5 and U6 (Fig. 3B). A background of cytidine was used, because this base is not tolerated in the MRE with the exception of one position (U7). These experiments reveal that MEX-3 binds to a bipartite recognition element that can tolerate as many as eight nucleotides in between each specificity determinant (Fig. 3C). Based on this data, we redefine the MRE as (A/G/U)(G/U)AGN(0–8)U(U/A/C)UA.
MEX-3 Binds Specifically to MREs Present in nos-2 and pal-1 Transcripts.
Next, we asked if the regulatory targets of MEX-3 (nos-2 and pal-1) harbor an MRE within their 3′UTR. A search of the 3′UTR of nos-2 and pal-1 reveals that both transcripts contain two copies of the MRE (Fig. S4A). In the case of nos-2, the MREs are present within the previously defined regulatory elements of subB and subC (Fig. S4A), suggesting that the MRE is a functional cis-regulatory sequence.
We then asked if MEX-3 binds specifically to the MREs present within the nos-2 and pal-1 3′-UTRs. To address this question, EMSA was performed to determine the binding affinity of MEX-3 to the five known regulatory elements of nos-2 (subA–E) (Fig. S4B) (12) and to a 21-nucleotide fragment of the pal-1 UTR that harbors one MRE. We anticipated that MEX-3 would bind to nos-2 subB, nos-2 subC, and the fragment from the pal-1 UTR, but not to nos-2 subA, subD, or subE sequences that lack the MRE. As expected, MEX-3 binds with high affinity to nos-2 subC (Kd,app = 40 ± 5 nM) (Fig. S4B) and the pal-1 3′UTR fragment (Kd,app = 25 ± 4 nM) (Fig. S4B) and with low affinity to nos-2 subA, subD, and subE (Kd,app > 200 nM). Both nos-2 subC and pal-1 MRE RNA reveal two shifted species, which is likely due to additional partial binding sites present within the sequences. We were surprised to observe that MEX-3 binds with low affinity to nos-2 subB (Kd,app = 420 ± 20). Because the MRE is near the 3′-end of the sequence, we suspected that the fluorescein label might be disrupting the predicted interaction. A longer variant (subB+) was prepared that includes seven additional nucleotides downstream. Consistent with our hypothesis, MEX-3 binds to this RNA with fourfold higher affinity (Kd,app = 106 ± 2 nM) (Fig. S4B). The data reveal that MREs are present within both MEX-3 regulatory targets, and that MEX-3 binds to them with high affinity.
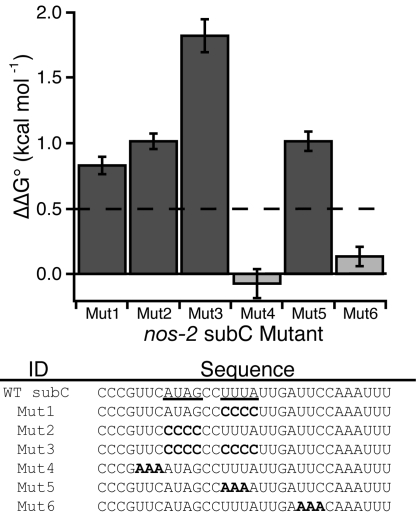
To verify that the MRE is essential for the interaction of MEX-3 with its mRNA targets, a series of mutations of the nos-2 subC element was prepared, and the ability of MEX-3 to bind to each was determined (Fig. 4). Each half-site was mutated such that either AUAG or UUUA was changed to CCCC. In each case, the binding affinity is significantly reduced (mut1, mut2; ΔΔG > 0.5 kcal mol−1). The binding affinity was then tested when both half-sites were mutated to CCCC and a dramatic reduction in binding is observed (mut3; ΔΔG > 1.5 kcal mol−1). When the half-site UUUA is mutated to AAAA, a loss in affinity is observed similar to mut1 or mut2 (mut5; ΔΔG > 0.5 kcal mol−1). Control mutations outside of the MRE have no significant change in relative binding affinity (mut4, mut6; ΔΔG < 0.5 kcal mol−1). Together, the data show that the MRE is required for the high affinity interaction between MEX-3 and nos-2 subC.
Fig. 4.
Mutagenesis of the nos-2 subC regulatory element. (Upper) The bar graph represents the ΔΔG° calculated from the Kd,app of each mutant sequence compared with the Kd,app of wild-type subC RNA. Mutations that have greater than a twofold loss in binding affinity are shown by the dark gray bars (dashed line, ΔΔG° > 0.5 kcal mol−1). (Lower) The sequences tested for MEX-3 binding are shown. The MRE half-sites are underlined in the wild-type sequence, and the mutations made are shown in bold (Mut1–6).
The Expression Pattern of a nos-2 3′UTR Reporter Depends upon the MRE.
To validate the relevance of the MRE consensus in vivo, we examined the expression pattern of a nos-2 3′UTR reporter, where specific mutations are made to both MRE sites. We used MosSCI (21) to generate single copy transgenic strains encoding green fluorescent protein (GFP) fused to histone H2B with the nos-2 3′UTR downstream (Fig. 5A). The pie-1 promoter was used in each transgene to drive germline transcription. Based on our biochemical assessment of the interaction between MEX-3 and RNA containing an MRE, the first half-site ATAG was mutated to CCCC in each MRE present within the nos-2 3′UTR (MREmut). We chose to mutate these nucleotides because this half-site has the most dramatic effect on MEX-3 binding (Fig. 4). The UUUA half site is located within a conserved repeat element that has previously been shown to be required for MEX-3 dependent regulation (11, 12).
Fig. 5.
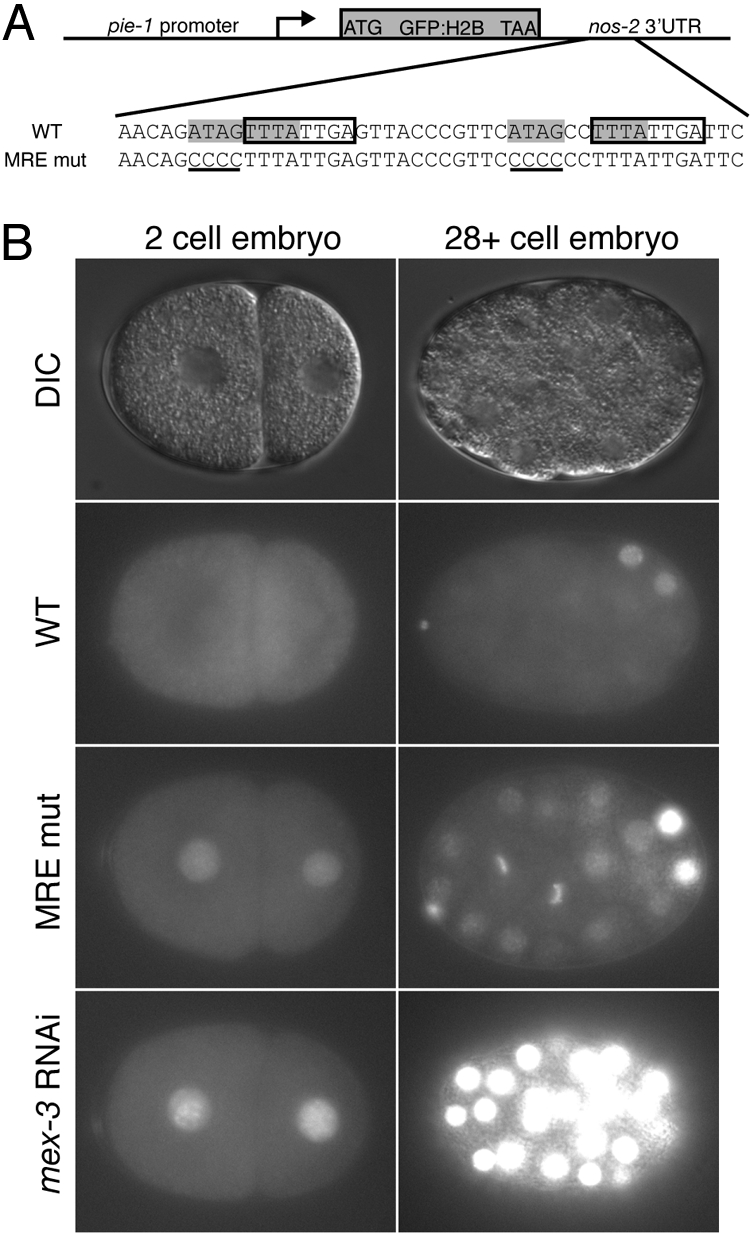
Validation of the MRE within the regulatory target nos-2. (A Upper) A schematic of the nos-2 3′UTR reporter transgene. A pie-1 promoter was used to drive germline specific transcription. (Lower) The region of the nos-2 3′UTR containing both MRE sites is shown. Each MRE is highlighted in gray. The conserved octamer is surrounded by a box, which was reported previously to be required for translation (12). Half of each MRE site was mutated to CCCC (shown in bold) to make the nos-2 MREmut reporter construct. (B) Expression of the GFP::H2B nos-2 3′UTR reporter constructs in early embryos. DIC images of 2-cell-stage embryos (Left) and later-staged (28+cells) embryos (Right) are shown. The GFP::H2B reporter expression pattern is compared between the nos-2 3′UTR, nos-2 MREmut 3′UTR, and the nos-2 3′UTR reporter in mex-3 RNAi embryos.
The expression pattern of the reporter harboring the wild-type nos-2 3′UTR matches the previously reported endogenous NOS-2 expression pattern (12, 22) (Fig. 5B). The reporter is absent in early embryos and is only observed in the germline precursor cells in older embryos. In stark contrast, the transgenic reporter that contains the nos-2 MREmut 3′UTR is observed both in early embryos and in all cells of embryos at approximately the 28-cell stage (Fig. 5B). Thus, mutation of the MREs disrupts both the spatial and temporal regulation of reporter expression. A similar pattern is observed with the wild-type reporter when mex-3 mRNA is depleted by RNAi (Fig. 5B). The results demonstrate that both MEX-3 and the MREs are required to appropriately pattern the expression of the reporter transgene.
It is interesting to note that reduction of MEX-3 by RNAi appears to have a more dramatic effect on nos-2 reporter expression than mutating its cis-acting response elements within the nos-2 3′-UTR (Fig. 5B). A possible explanation for this observation is that residual binding of MEX-3 to nos-2 3′UTR mutants leads to partial repression, because only part of the MRE is disrupted in each case. There may also be other specificity determinants from SELEX that we have not distinguished. Another possibility is that MEX-3 might regulate other trans-acting factors that feed back to repress nos-2 translation. Alternatively, other proteins that bind to the nos-2 3′UTR may facilitate MEX-3 binding, and this interaction has not been fully disrupted. Our data cannot distinguish between these possibilities.
Candidate MEX-3 Regulatory Targets Based on the MRE.
To identify new candidate MEX-3 targets, we used the MRE consensus as a pattern to search annotated C. elegans transcripts for potential MEX-3 binding sites. MREs throughout the entire genome were identified using the pattern matching tool PATSCAN (23). Because 3′UTRs are the primary determinant of spatial gene expression within the germline, we cross-referenced the predicted MREs with 3′UTR annotations from release WS190 of the C. elegans genome (24). Of 10,802 genes with an annotated 3′UTR, 2,834 (26.2%) contain at least one MRE in their 3′UTR (Table S1). To identify candidate MEX-3 targets required for embryogenesis, we filtered the results to include only transcripts present in 1–8 cell embryos (25) and that result in an embryonic lethal phenotype when knocked-down by RNAi (26). Based on this analysis, 214 candidate MEX-3 targets were identified (7.5% of all MRE-containing 3′UTRs, Fig. S5).
To identify candidate MEX-3 targets that play a role in maintaining germline stem cell totipotency, we filtered our primary search results using microarray data that identify transcripts enriched in the germline (27). Of the germline enriched genes, 527 have at least one MRE (18.6% of all MRE-containing 3′UTRs) (Fig. S5). Because the role of MEX-3 in the germline is only revealed in the context of a mex-3 gld-1 double mutant (4), we rationalized that the most important targets might contain both MEX-3 and GLD-1 binding sites. To identify such transcripts, we searched the 3′UTRs of germline enriched genes for both the MRE and the STAR-binding element (SBE), recognized by GLD-1 (28) (Table S1). Based on this analysis, a total of 162 genes were identified as potential targets of both MEX-3 and GLD-1 (Fig. S5). Together, the results predict several candidate MEX-3 targets that play critical roles both in the embryo and in the germline (Table S1 and Table S2).
We wished to examine predicted unique targets in a functional assay. As a first step toward this goal, we determined the MEX-3 dependence of reporter expression in 10 transgenic lines harboring germline-specific GFP::H2B::3′UTR reporters containing at least one MRE. These strains represent a subset of a germline 3′UTR reporter library developed by Seydoux and colleagues (24). Worms were grown on either OP50 or mex-3 RNAi food, and the expression pattern of GFP was determined (Table S3). Two of the 10 (pal-1 and glp-1 3′-UTR reporter strains) demonstrated a change in reporter fluorescence upon mex-3 RNAi. pal-1 has previously been demonstrated to be a MEX-3 regulatory target and thus serves as a positive control (10). The glp-1 reporter shows an overall increase in GFP fluorescence and ectopic expression in the posterior of early embryos (2–4 cell), when mex-3 RNA is depleted (Fig. S6). There are two MEX-3 binding sites in a conserved region of the glp-1 3′-UTR. The binding sites surround previously characterized binding sites for the RNA-binding proteins POS-1 and GLD-1 (28, 29). The location of the binding sites suggests that regulation is direct and could potentially be influenced by the presence of other RNA-binding proteins. The results identify glp-1 as a new candidate MEX-3 regulatory target.
Discussion
Based on in vitro selection and biochemical experiments, we have defined the consensus MEX-3 recognition element (MRE: (A/G/U)(G/U)AGN (0–8)U(U/A/C)UA). The MRE is a bipartite element that consists of two four-nucleotide motifs separated by 0–8 nucleotides. MEX-3 contains two KH RNA binding domains. We predict that each motif binds specifically to one four-nucleotide half-site. Previous studies have shown that KH domains typically accommodate four nucleotides in their binding pockets, consistent with this hypothesis (16–20). Alignment of the KH domains between the human hMex-3 proteins and C. elegans MEX-3 reveal 79–81% sequence identity within the RNA-binding domain (8). Interestingly, part of the MRE is comprised of an AU-rich half-site which is proposed to be required for hMEX-3D/TINO RNA binding. We predict that the hMEX-3 proteins bind to RNA with similar specificity as their C. elegans homolog MEX-3.
The MRE is present in the 3′UTR of ≈26.2% of all genes in C. elegans. By filtering this list for transcripts expressed in the same tissues at the same time as MEX-3, we have narrowed the candidate targets to 214 transcripts in the embryo and 527 transcripts in the germline. The two previously characterized MEX-3 regulatory targets—nos-2 and pal-1—both contain two MRE sites in their 3′UTR. Mutating both sites in a nos-2 reporter leads to derepression of the transgene, as does reduction of MEX-3 by RNAi. Thus, the MRE is a critical cis-acting functional element.
Our search for candidate regulatory targets reveals several genes that play key roles during various stages of development. A number of these genes encode other RNA binding proteins, including cbp-3, puf-3, puf-6, puf-7, spn-4, mex-1, and gld-1 (28, 30–34). Interestingly, the mex-3 transcript also has two MREs within its own 3′UTR, suggesting that MEX-3 may regulate its own mRNA translation. Other candidate mRNA targets encode membrane proteins such as glp-1 and ooc-3. GLP-1 is a Notch homolog essential for mitotic proliferation of germ cells and maintenance of germline stem cells and the development of early blastomeres (35, 36). Our results suggest MEX-3 regulates this transcript, but more work is needed to show that regulation is direct. OOC-3 is a putative transmembrane protein localized to the endoplasmic reticulum and is required for the correct localization of the polarity determinants PAR-2 and PAR-3, two critical proteins that establish asymmetry in the early embryo immediately after fertilization (37, 38).
There are several candidate target genes that may contribute to the observed mex-3 mutant phenotype. The genes include ubc-9, pha-4, and tbx-2, all of which are required to specify ABa-derived pharyngeal muscle (39, 40). The 3′UTR of ubc-9 contains four MRE sites, whereas tbx-2 and pha-4 each have one. Similar to mex-3 mutant embryos, worms that lack these genes fail to produce pharyngeal tissue from the ABa blastomere.
Of particular interest is how MEX-3 functions to maintain totipotency in the germline. The unusual transdifferentiated germline teratoma phenotype is only observed in worms with mutations in both mex-3 and gld-1, a second KH-domain RNA binding protein (4). Moreover, recent evidence indicates that MEX-3 and PUF-8, a Puf-domain RNA binding protein, promote mitotic division of germ cells (41). These studies suggest that MEX-3 is playing redundant roles with other RNA binding proteins to ensure preservation and maintenance of germline stem cells. One potential mechanism to explain the redundancy is coregulation of the same mRNA target. A potential target that is recognized by both MEX-3 and GLD-1 is the mRNA encoding the putative transcription factor SOX-2. C. elegans SOX-2 is homologous to human SOX2, which is required for embryonic stem cell self-renewal and is one of the factors used to induce pluripotency in induced pluripotency stem cells (iPS) (42, 43). C. elegans sox-2 mRNA harbors one MRE and one SBE (GLD-1 binding sequence), consistent with the hypothesis that MEX-3 and GLD-1 play a redundant role in regulating SOX-2 expression.
Here, we reveal the requirements for RNA recognition by MEX-3 and identify cis-acting elements in the 3′UTR of its mRNA targets nos-2 and pal-1. Our results demonstrate that a large number of transcripts contain a MEX-3 recognition element within their 3′UTR, but caution that the MRE is not the sole determinant of MEX-3-dependent regulation. Our results reveal a number of candidate targets that could potentially explain the diverse roles of MEX-3 in regulating embryonic cell fate specification and maintaining totipotency within the germline. Further work is needed to define which candidate targets are the most important factors contributing to the mex-3 mutant phenotypes.
Methods
Protein Expression and Purification.
The sequence encoding amino acids 45–205 of MEX-3 were amplified from the corresponding ORFeome clone (Open Biosystems) and subcloned into pMal-c (New England Biolabs), a protein expression vector that encodes an N-terminal maltose-binding protein (MBP) tag. For details of the purification, refer to SI Materials and Methods.
In Vitro RNA Selection.
RNA library design and in vitro selection protocols were adapted from published protocols with a few modifications (15, 44). For experimental details refer to SI Materials and Methods.
Electrophoretic Mobility Shift Assay.
RNA transcripts or synthetic oligonucleotides (Integrated DNA Technologies) were 3′-end-labeled with fluorescein 5-thiosemicarbazide (Invitrogen) following the protocol described (45). Electrophoretic mobility shift experiments and data analysis were carried out as previously described with a few modifications (refer to SI Materials and Methods for changes) (45). Due to dissociation in the gel, the 5′ Frag3 RNA EMSA experiment was fit to a Langmuir isotherm to determine the apparent equilibrium dissociation constant.
Worm Strain Generation.
All transgenic worm strains were made using biolistic transformation (46, 47) or MosSCI (21). For cloning and experimental details refer to SI Materials and Methods.
Bioinformatics.
MREs were identified in release WS190 of the C. elegans genome using the pattern matching tool PATSCAN (23). Using a custom MySQL database, the predicted MREs were cross-referenced with the 3′UTR annotations for all transcripts from Wormbase release WS190 to identify genes with 3′UTR containing candidate MEX-3 binding sites. To identify genes that may be regulated by MEX-3, the results were filtered through genome-wide datasets describing genes expressed during one to eight cell embryos (25), genes required to complete embryogenesis (26), and genes whose expression is increased in the germline relative to an average of all tissues (27).
Supplementary Material
Acknowledgments.
The authors thank Dr. Geraldine Seydoux (Johns Hopkins University, Baltimore), Dr. Eric Jorgensen (University of Utah, Salt Lake City), Dr. Marian Walhout (University of Massachusetts Medical School, Worcester), and Dr. Craig Mello (University of Massachusetts Medical School, Worcester) for reagents; Dr. Mary Munson and Dr. Phillip Zamore for critical comments concerning the manuscript; and Dr. Nick Rhind for sharing equipment. This work was supported by National Institutes of Health Grant GM081422 (to S.P.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907916106/DCSupplemental.
References
- 1.Seydoux G, Braun RE. Pathway to totipotency: Lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 2.Farley BM, Ryder SP. Regulation of maternal mRNAs in early development. Crit Rev Biochem Mol Biol. 2008;43:135–162. doi: 10.1080/10409230801921338. [DOI] [PubMed] [Google Scholar]
- 3.Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell. 1996;87:205–216. doi: 10.1016/s0092-8674(00)81339-2. [DOI] [PubMed] [Google Scholar]
- 4.Ciosk R, DePalma M, Priess JR. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science. 2006;311:851–853. doi: 10.1126/science.1122491. [DOI] [PubMed] [Google Scholar]
- 5.Jud M., Razelun J., Bickel J., Czerwinski M., Schisa J. A. Conservation of large foci formation in arrested oocytes of Caenorhabditis nematodes. Dev Genes Evol. 2007;217:221–226. doi: 10.1007/s00427-006-0130-3. [DOI] [PubMed] [Google Scholar]
- 6.Jud MC, et al. Large P body-like RNPs form in C. elegans oocytes in response to arrested ovulation, heat shock, osmotic stress, and anoxia and are regulated by the major sperm protein pathway. Dev Biol. 2008;318:38–51. doi: 10.1016/j.ydbio.2008.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallo CM, Munro E, Rasoloson D, Merritt C, Seydoux G. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev Biol. 2008;323:76–87. doi: 10.1016/j.ydbio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Buchet-Poyau K, et al. Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res. 2007;35:1289–1300. doi: 10.1093/nar/gkm016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnini M, et al. Identification of TINO: A new evolutionarily conserved BCL-2 AU-rich element RNA-binding protein. J Biol Chem. 2004;279:20154–20166. doi: 10.1074/jbc.M314071200. [DOI] [PubMed] [Google Scholar]
- 10.Hunter CP, Kenyon C. Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell. 1996;87:217–226. doi: 10.1016/s0092-8674(00)81340-9. [DOI] [PubMed] [Google Scholar]
- 11.Jadhav S, Rana M, Subramaniam K. Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development. 2008;135:1803–1812. doi: 10.1242/dev.013656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Agostino I, Merritt C, Chen PL, Seydoux G, Subramaniam K. Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev Biol. 2006;292:244–252. doi: 10.1016/j.ydbio.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 13.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 14.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 15.Loughlin FE, et al. The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5′ splice site-like sequences. Proc Natl Acad Sci USA. 2009;106:5581–5586. doi: 10.1073/pnas.0802466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valverde R, Edwards L, Regan L. Structure and function of KH domains. Febs J. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 17.Makeyev AV, Liebhaber SA. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamarnik AV, Andino R. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J Virol. 2000;74:2219–2226. doi: 10.1128/jvi.74.5.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braddock DT, Baber JL, Levens D, Clore GM. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: Solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J. 2002;21:3476–3485. doi: 10.1093/emboj/cdf352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis HA, et al. Sequence-specific RNA binding by a Nova KH domain: Implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 21.Frokjaer-Jensen C, et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- 23.Dsouza M, Larsen N, Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- 24.Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130:889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- 26.Sonnichsen B, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 27.Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- 28.Ryder SP, Frater LA, Abramovitz DL, Goodwin EB, Williamson JR. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat Struct Mol Biol. 2004;11:20–28. doi: 10.1038/nsmb706. [DOI] [PubMed] [Google Scholar]
- 29.Farley BM, Pagano JM, Ryder SP. RNA target specificity of the embryonic cell fate determinant POS-1. RNA. 2008;14:2685–2697. doi: 10.1261/rna.1256708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lublin AL, Evans TC. The RNA-binding proteins PUF-5, PUF-6, and PUF-7 reveal multiple systems for maternal mRNA regulation during C. elegans oogenesis. Dev Biol. 2007;303:635–649. doi: 10.1016/j.ydbio.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Stumpf CR, Kimble J, Wickens M. A Caenorhabditis elegans PUF protein family with distinct RNA binding specificity. RNA. 2008;14:1550–1557. doi: 10.1261/rna.1095908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomes JE, et al. The maternal gene spn-4 encodes a predicted RRM protein required for mitotic spindle orientation and cell fate patterning in early C. elegans embryos. Development. 2001;128:4301–4314. doi: 10.1242/dev.128.21.4301. [DOI] [PubMed] [Google Scholar]
- 33.Mello CC, Draper BW, Krause M, Weintraub H, Priess JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. 1992;70:163–176. doi: 10.1016/0092-8674(92)90542-k. [DOI] [PubMed] [Google Scholar]
- 34.Jones AR, Francis R, Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol. 1996;180:165–183. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- 35.Hutter H, Schnabel R. glp-1 and inductions establishing embryonic axes in C. elegans. Development. 1994;120:2051–2064. doi: 10.1242/dev.120.7.2051. [DOI] [PubMed] [Google Scholar]
- 36.Hutter H, Schnabel R. Specification of anterior–posterior differences within the AB lineage in the C. elegans embryo: A polarising induction. Development. 1995;121:1559–1568. doi: 10.1242/dev.121.5.1559. [DOI] [PubMed] [Google Scholar]
- 37.Basham SE, Rose LS. Mutations in ooc-5 and ooc-3 disrupt oocyte formation and the reestablishment of asymmetric PAR protein localization in two-cell Caenorhabditis elegans embryos. Dev Biol. 1999;215:253–263. doi: 10.1006/dbio.1999.9447. [DOI] [PubMed] [Google Scholar]
- 38.Pichler S, et al. OOC-3, a novel putative transmembrane protein required for establishment of cortical domains and spindle orientation in the P(1) blastomere of C. elegans embryos. Development. 2000;127:2063–2073. doi: 10.1242/dev.127.10.2063. [DOI] [PubMed] [Google Scholar]
- 39.Roy Chowdhuri S, Crum T, Woollard A, Aslam S, Okkema PG. The T-box factor TBX-2 and the SUMO conjugating enzyme UBC-9 are required for ABa-derived pharyngeal muscle in C. elegans. Dev Biol. 2006;295:664–677. doi: 10.1016/j.ydbio.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Mango SE, Lambie EJ, Kimble J. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development. 1994;120:3019–3031. doi: 10.1242/dev.120.10.3019. [DOI] [PubMed] [Google Scholar]
- 41.Ariz M, Mainpal R, Subramaniam K. C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev Biol. 2009;326:295–304. doi: 10.1016/j.ydbio.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fong H, Hohenstein KA, Donovan PJ. Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells. 2008;26:1931–1938. doi: 10.1634/stemcells.2007-1002. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 44.Hori T, Taguchi Y, Uesugi S, Kurihara Y. The RNA ligands for mouse proline-rich RNA-binding protein (mouse Prrp) contain two consensus sequences in separate loop structure. Nucleic Acids Res. 2005;33:190–200. doi: 10.1093/nar/gki153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pagano JM, Farley BM, McCoig LM, Ryder SP. Molecular basis of RNA recognition by the embryonic polarity determinant MEX-5. J Biol Chem. 2007;282:8883–8894. doi: 10.1074/jbc.M700079200. [DOI] [PubMed] [Google Scholar]
- 46.Praitis V. Creation of transgenic lines using microparticle bombardment methods. Methods Mol Biol. 2006;351:93–107. doi: 10.1385/1-59745-151-7:93. [DOI] [PubMed] [Google Scholar]
- 47.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.