Abstract
Although remission rates for metastatic melanoma are generally very poor, some patients can survive for prolonged periods following metastasis. We used gene expression profiling, mitotic index (MI), and quantification of tumor infiltrating leukocytes (TILs) and CD3+ cells in metastatic lesions to search for a molecular basis for this observation and to develop improved methods for predicting patient survival. We identified a group of 266 genes associated with postrecurrence survival. Genes positively associated with survival were predominantly immune response related (e.g., ICOS, CD3d, ZAP70, TRAT1, TARP, GZMK, LCK, CD2, CXCL13, CCL19, CCR7, VCAM1) while genes negatively associated with survival were cell proliferation related (e.g., PDE4D, CDK2, GREF1, NUSAP1, SPC24). Furthermore, any of the 4 parameters (prevalidated gene expression signature, TILs, CD3, and in particular MI) improved the ability of Tumor, Node, Metastasis (TNM) staging to predict postrecurrence survival; MI was the most significant contributor (HR = 2.13, P = 0.0008). An immune response gene expression signature and presence of TILs and CD3+ cells signify immune surveillance as a mechanism for prolonged survival in these patients and indicate improved patient subcategorization beyond current TNM staging.
Keywords: gene expression analysis, immune response, TNM staging, tumor infiltrating leukocytes
Melanoma is the deadliest form of skin cancer, and its incidence is on the rise (1–3). Treatment options for advanced melanoma are limited and rarely curative. While 5 year survival for stage III melanoma patients can reach up to 69% depending on the patient subcategory, the reported survival for stage IV disease is rarely longer than a year (3). Although long-term survival for patients with advanced melanoma is low despite currently available therapies, some patients can survive for prolonged periods with metastatic disease. The ability to predict survival in metastatic melanoma with greater accuracy could improve current treatment decisions and aid in the design of new therapies that might be tailored to specific subgroups of patients. The majority of innovative and improved prediction models, however, are geared toward evaluating the metastatic potential of primary tumors, as opposed to evaluating the progression potential of metastatic disease. It would potentially be useful to biologically subclassify melanoma that has already metastasized, beyond the use of the conventional Tumor, Node, Metastasis (TNM) staging, into categories that more accurately predict patient survival (4).
The use of gene expression profiling has yielded an enormous amount of information leading to the definition of molecular signatures for a wide variety of tumor types (5–7). For breast cancer, gene expression profiles are already in use to classify tumors biologically in ways that impact decisions regarding the most appropriate form of treatment (8, 9). For melanoma, gene expression profiling has been used to establish molecular signatures of disease progression. This has been done by comparing normal skin to benign nevi and to primary and metastatic melanomas (10, 11). Here, we use gene expression profiling to define molecular signatures of different subsets of advanced melanoma associated with differing survival potential. We observe that expression of genes associated with immune response and cell division are related to survival, and explore the measurement of mitotic index (MI), tumor infiltrating leukocytes (TILs) and CD3+ cells in histologic sections of metastatic lesions as simple predictors of patient postrecurrence survival.
Results
Gene Expression Profiling of Metastatic Melanoma Lesions Identifies Genes Associated with Survival.
To evaluate the association between gene expression profiles and survival in patients with metastatic melanoma, we evaluated 44 metastatic melanoma tissue samples from 38 patients who were followed clinically for a median of 20 months (2–38 months range) after excision of the metastatic lesion. Thirty-nine of the tumor samples were taken from patients with stage III disease, with 5 samples from patients with stage IV disease (Table S1). We evaluated the association of gene expression profiles of patient tumors and survival based on time from excision of the metastatic lesion to last follow-up or death. Using the Significant Analysis of Microarrays (SAM) (12) with a false discovery rate (FDR) (13) of 5.34% and filtering for at least a 1.5 fold change in expression between patients with prolonged survival (>1.5 years) compared to those with shorter survival, we identified a set of 266 genes (Dataset S1) that are significantly associated with postrecurrence survival.
To gain insight into the functional classes of these 266 genes, we analyzed them using the National Institute of Allergy and Infectious Diseases/National Institutes of Health Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resource 2008 (Table S2). In the group of patients with prolonged survival, the top functional annotation cluster for up-regulated transcripts was “immune system process” (top enrichment score of 13.42, representing 40 transcripts). The transcripts included those encoding MHC class II molecules (HLA-DOB, HLA-DPB1), T cell-associated molecules (ICOS, CD3d, ZAP70, TRAT1, TARP, GZMK, LCK, CD27), chemokines, chemokine receptors and adhesion molecules (CD11A, CXCL13, CCL19, CCR7, VCAM1, AMICA1) and a number of other innate and adaptive immune response molecules (CD79A, LTB, CLEC4G, CLECL1, FCER1A, IKZF1, TAP1, IRF1, IRF8, GBP2, IL4R, IL2RG, IKZF1, C3, MYADM, TLR10, NLRC5, FCAMR, BTLA, NLRC3, CD48). The up-regulation of immune system transcripts in metastatic lesions of patients with longer survival suggests that the immune response may keep tumor growth and metastasis in check in these patients.
Genes that were down-regulated in patients with prolonged survival belonged to multiple functional annotation clusters, with the top 2 enrichment scores of 1.61 and 1.17 representing about 10 transcripts involved in (but not limited to) “cell cycle phase,” “M phase,” “cofactor binding,” “cell division,” “cytoskeleton,” and “aminotransferase” (Table S2). Genes in this category —which could be more broadly characterized as “cell proliferation”—included ANLN, PDE4D, CDK2, CXCL1, CGREF1, NUSAP1, and SPC24. The up-regulation of genes associated with cell division in patients with high mortality risk, suggests that higher rates of mitosis within metastatic lesions is associated with more rapid tumor growth and spread of metastatic disease.
Tumor Infiltrating Leukocytes and Tumor Cell Mitoses Are Predictive of Patient Survival.
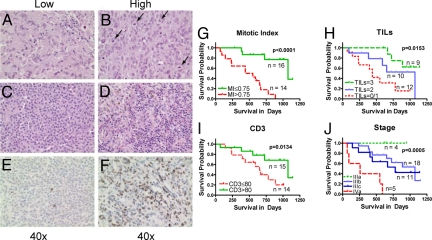
To determine if a simple, independent method could demonstrate an association of immune or proliferative parameters with patient survival, we examined histological sections from the same pathology specimens used for gene expression analysis. We quantified 3 different parameters—TILs, MI and CD3+ T cell count (CD3), and assessed whether any or all of these were independently associated with survival (Fig. 1 A–F).
Fig. 1.
Metastatic melanoma patient survival differ based on MI, TILs, and CD3 cell count. All available tissue specimens used for gene chip hybridization were also examined for the presence of mitoses; (A) reflects low and (B) reflects high levels of mitosis with bottom left corner showing a magnified section of the slide. They were also examined and scored for presence of TILs. (C) shows a representative view of low and (D) shows a high level of TILs. Paraffin embedded samples were also stained for CD3; (E) shows low levels and (F) shows high levels of CD3+ cells present in the melanoma sample. Kaplan-Meier survival curves for groups based on MI (G, P < 0.0001), TILs (H, P = 0.0163), CD3 cell count (I, P = 0.0134), and stage at recurrence/metastasis (J, overall P = 0.0006, but the separation of IIIb and IIIc is not significant P = 0.59).
We divided the patients into 3 groups based on the prevalence of TILs within their tumor (<25% TILs, 25 to 50% TILs, and >50% TILs as assessed by percentage of the lesion area represented by leukocytes, see methods). In addition, using the median value as the cutoff point, we divided the patients into 2 groups each based on CD3 count (lower and higher than 80 CD3+ cells per 10 High Power Fields (HPFs) and MI (lower and higher than 0.75 Mitoses per HPF, Table S3). Median survival estimates along with the 95% confidence intervals for these groups are provided in Table S3. Shown in Fig. 1 G, H, I, and J are Kaplan-Meier survival curves for the groups defined by MI, TILs, CD3 counts and TNM stage at the time of surgery, respectively. All 3 histological parameters were significantly associated with survival: patients with lower MI survived significantly longer (P < 0.0001, log rank test) as did patients with higher TIL indices (P = 0.0163) and CD3 counts (P = 0.0134). Please note that due to some missing histological specimens, certain figures and tables have differing specimen numbers (see Methods).
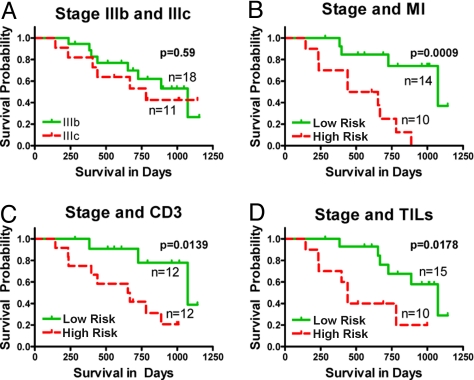
TNM staging in our cohort was effective in separating patients with stage IIIA (n = 4) and stage IV (n = 5) disease by survival (P = 0.0006, log rank test). However, the vast majority (n = 29) of patients in the cohort had stage IIIB or IIIC disease, and here TNM staging showed no differential association with survival (P = 0.59) (Figs. 1J, and 2A).
Fig. 2.
MI, CD3 counts, and TILs aid staging of IIIb and IIIc patients in predicting their survival. Patients with staging of IIIb and IIIc are represented in (A). Their survival capabilities cannot be distinguished using only staging (P = 0.59). By incorporating MI, CD3, and TILs (B–D) in the model, it is possible to improve the ability to separate stage IIIb/IIIc patients based on their survival (P = 0.0009, P = 0.0139, and P = 0.0178, respectively).
To assess if any of the 3 histologic parameters, CD3 count, MI, or TILs, could significantly improve upon the ability of TNM staging in predicting postrecurrence survival, we fitted 3 multivariable Cox regression models. Each model involved one of these predictors (CD3 count or MI or TILs) and TNM stage as independent variables and postrecurrence survival as the dependent variable (Table 1 A–C). Adding any of the 3 histologic parameters significantly improved upon the ability of TNM stage to predict survival: MI was the strongest contributor (HR = 2.13, P = 0.0008) followed by CD3 count (HR = 0.80, P = 0.0022) and TILs (HR = 0.26, P = 0.0067). Using these models, we divided the patients into “low” and “high” risk groups using the median hazard ratio as a cut-off point. Kaplan-Meier survival curves of low and high risk groups among stage IIIB/IIIC patients based on each of these 3 models are shown in Fig. 2 B–D. For comparison, Kaplan-Meier survival curves for stage IIIB/IIIC patients based on TNM stage alone are provided in Fig. 2A. Adding any of the 3 parameters to TNM stage resulted in the ability to segregate stage IIIB and IIIC patients into high and low risk groups with significantly different survival probabilities. The median survival times were 1073 days in the low-risk group (95% confidence interval, 1073 to “not reached”) and 496 days in the high-risk group (95% confidence interval, 237 to “not reached”) based on the model with TNM and MI as predictors. Out of 15 patients in stage IIIb, 11 segregated into low risk and 4 into high risk. In the case of 9 IIIc patients, 3 segregated into low risk and 6 into high risk (Table 2). The clinical characteristics of the low and high risk groups predicted by the model with TNM and MI as predictors are provided in Table 2.
Table 1.
Hazard ratios (HRs) and 95% Confidence Intervals (CIs) for TNM stage, MI, CD3 count, and TIL based on 3 multivariable Cox proportional hazards models of postrecurrence survival
| A | HR (95% CI) | p value |
|---|---|---|
| TNM stage* IIIA/IIIB vs. IIIC/IV | 2.05 (0.76, 5.54) | 0.16 |
| Mitotic Index | 2.13 (1.38, 3.32) | 0.0008 |
| B | ||
| TNM stage* IIIA/IIIB vs. IIIC/IV | 1.82 (0.70, 4.74) | 0.22 |
| TILs | 0.26 (0.10, 0.69) | 0.0067 |
| C | ||
| TNM stage* IIIA/IIIB vs. IIIC/IV | 1.27 (0.46, 3.56) | 0.64 |
| CD3 count | 0.80 (0.70, 0.92) | 0.0022 |
All three histologic parameters, MI, CD3, and TILs, add to the ability of TNM stage in predicting postrecurrence survival. A, HRs and 95% CIs for TNM stage and MI based on a Cox regression model involving these variables. B, HRs and 95% CIs for TNM stage and TIL based on a Cox regression model involving these variables. C, HRs and 95% CIs for TNM stage and CD3 count based on a Cox regression model involving these variables.
*Due to small sample size, TNM stage was dichotomized (IIIA/B vs. IIIC/IV). The other three variables were dichotomized for convenience using medians as cut-off points.
Table 2.
Clinical characteristics of low and high risk groups predicted based on a Cox multivariable regression model of survival since recurrence/metastasis (R/M) with MI, and TNM stage as predictors (N = 30)
| Low risk (N = 16) | High risk (N = 14) | Statistical test | ||
|---|---|---|---|---|
| Sex | Female | 5 (31%) | 6 (43%) | Fisher's P value = 0.71 |
| Male | 11 (69%) | 8 (57%) | ||
| Age at recurrence | Mean = 65 (SD = 18) | Mean = 59 (SD = 21) | Wilcoxon rank sum P = 0.38 | |
| CD3 cell count | ≤80 | 4 (25%) | 10 (83%) | Fisher's P value = 0.0063 |
| > 80 | 12 (75%) | 2 (17%) | ||
| missing | 0 | 2 | ||
| Mitotic index | ≤0.75 | 15 (94%) | 1 (7%) | Fisher's P value < 0.0001 |
| > 0.75 | 1 (6%) | 13 (93%) | ||
| TILs index | 0–25% | 4 (25%) | 8 (57%) | Fisher's P value = 0.22 |
| 25–50% | 6 (37.5%) | 4 (29%) | ||
| 50–100% | 6 (37.5%) | 2 (14%) | ||
| Stage at recurrence/metastasis | IIIA | 2 (12.5%) | 0 | Fisher's P value = 0.0137 |
| IIIB | 11 (69%) | 4 (29%) | ||
| IIIC | 3 (19%) | 6 (43%) | ||
| IV | 0 | 4 (29%) | ||
| Radiation | Yes | 4 (25%) | 2 | Fisher's P value = 0.67 |
| No | 12 (75%) | 10 | ||
| Missing | 0 | 2 | ||
| Immunotherapy | Yes | 0 | 2 (17%) | Fisher's P value = 0.17 |
| No | 16 (100%) | 10 (83%) | ||
| Missing | 0 | 2 | ||
| Chemotherapy | Yes | 8 (50%) | 3 (25%) | Fisher's P value = 0.25 |
| No | 8 (50%) | 9 (75%) | ||
| Missing | 0 | 2 |
We then used an independent cohort of patients to see if these observations could be validated. We analyzed 52 additional metastatic melanoma samples taken from 25 stage IIIb and 27 stage IIIc patients. MI of the patients in the validation cohort was significantly lower than that of the patients in the original cohort studied (P = 0.0176, Fig. S1a) while postrecurrence survival was longer, although not statistically significant (P = 0.10, Fig. S1b). TNM stage was a significant predictor of survival in the validation cohort (P = 0.003). MI alone was not significant in additionally separating survival in the validation cohort. To examine this further, we then combined the 2 cohorts into an expanded cohort of 90 patient samples; using the original MI cutoff of 0.75, we were able to separate the patients into high and low risk groups with significantly different survival (Fig. S1c, P < 0.0001). For stage IIIb/c patients in the combined cohort, multivariate Cox proportional hazards model showed that MI was a more important predictor of survival [HR = 3.08, 95% CI: (1.38, 6.90), P = 0.0062] than TNM stage [HR = 2.13, 95% CI: (1.02, 4.47), P = 0.05]. Furthermore, TIL frequency was a significant predictor of survival in IIIc patients in the validation cohort (Fig. S1d, P = 0.0197) but not for stage IIIb patients.
Prevalidated Gene Expression Predictor of Survival in Metastatic Melanoma.
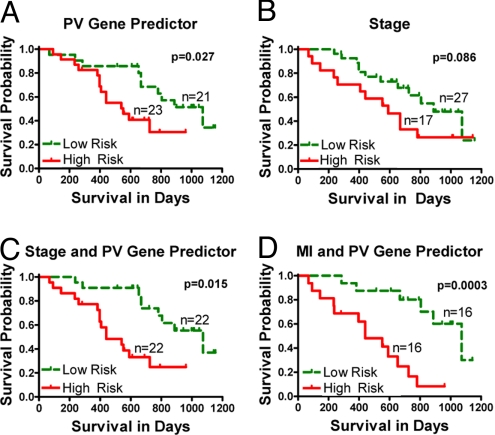
To test if gene expression signatures bear predictive prognostic potential in metastatic melanoma, we derived a gene expression predictor of survival using principal component analysis (PCA) (14) applied to the genes selected by SAM as described in the methods section. We used the method of prevalidation (PV) to derive the gene expression predictor and to compare its prediction accuracy to that of MI, TILs, CD3 cell count and TNM stage (15, 16). Kaplan-Meier survival curves of low-risk and high-risk groups predicted by the PV gene expression predictor are shown in Fig. 3A. The survival in the 2 groups was significantly different (log rank P = 0.027) indicating that gene expression profiles can predict survival in metastatic melanoma.
Fig. 3.
Gene signature (PV) and MI are capable of improving current outcome prediction model through machine learning. Predicted high-risk and low-risk groups obtained using (A) prevalidated gene expression predictor (P = 0.027), (B) Stage alone (P = 0.086), (C) combination of Stage and prevalidated gene predictor (P = 0.015) and (D) combination of MI and prevalidated gene predictor (P = 0.0003).
To confirm this observation using a different learning method, we tested the metastatic melanoma expression data using the Support Vector Machine algorithm (17, 18), with and without PCA. We obtained the best performance using the top 50 genes determined using the signal-to-noise ratio gene selection method, with measurements decorrelated using PCA: 78.57% sensitivity, 71.43% specificity, and 81.38% area under the ROC curve (AUC) (Table S4).
As an additional confirmatory method, we then tested our gene signature (Dataset S1) on recently published test samples (n = 29) that were completely independent of our study (19). This data set was very similar to ours as it contained relative mRNA levels of metastatic melanoma lesions from patients with mostly stage IIIb and IIIc disease, with time to recurrence as one of the study variables. We observed 61.54% sensitivity, 62.50% specificity, and 70.67% AUC when we applied our list of 266 genes (only 137 of which were present on their chips) to their data set. For comparison purposes, we performed the same signal-to-noise-ratio method described above but this time using their data set for both training and testing, reporting the best results using the top 20 genes: 69.23% sensitivity, 68.75% specificity, and 70.67% AUC. Comparing these 2 sets of results indicates that close to the maximal predictability power was achieved using the initial selection of genes from our data set despite extremely different platforms that the 2 datasets were generated on. This confirms the potential of metastatic melanoma gene expression profiles to predict patient outcome.
Metastatic Melanoma Risk Predictor.
To see if PV gene expression predictor could add to the predictive power of TNM staging, we performed a multivariate Cox proportional hazards model with survival since surgery as a dependent variable, and TNM stage and PV gene expression predictor as independent variables. The PV gene expression predictor was significant (HR = 2.71, P = 0.03), and TNM stage was borderline significant (HR = 2.06, P = 0.08). Shown in Fig. 3 B and C are Kaplan-Meier survival curves for low and high risk groups predicted using models with stage at R/M only and with PV gene expression predictor and TNM stage together. This model segregated 11 stage IIIb and 6 stage IIIc tissue samples into high risk group, while putting 12 IIIb and 6 IIIc tissue samples into low risk group. Using gene expression analysis of metastatic melanoma patient samples, we are able to add to the predictive power of TNM stage as TNM stage alone was not able to separate patients with stage IIIB and IIIC (Table S5A).
We then performed a multivariate Cox proportional hazards model with survival since surgery as a dependent variable and stage, MI and PV gene expression predictor as independent variables (CD3 and TILs were not used as they were less predictive than MI). MI was the most significant predictor (HR = 2.54, P = 0.0002), but the PV gene expression predictor was also significant (HR = 3.64, P = 0.019) while stage was not (HR = 1.64, P = 0.30). When we removed the stage from the model, both MI (HR = 2.53, P = 0.0001) and PV gene expression predictor (HR = 3.91, P = 0.013) were still significant. Kaplan-Meier estimated survival curves for low-risk and high-risk groups predicted using this final model are shown in Fig. 3D. Table S5B shows the clinical characteristics of the patients according to the risk groups obtained using this best model. The rates of postrecurrence survival at 2 years (i.e., 730 days) in the low-risk and high-risk groups were 70% [95% CI is (49%, 100%)] and 14% [95% CI is (4.8%, 57%)], respectively. The median survival times were 1,073 days in the low-risk group (95% CI, 805 to “not reached”) and 440 days in the high-risk group (95% CI, 237 to “not reached”). The survival in the 2 groups was significantly different (log rank P = 0.0003).
Using gene expression analysis of metastatic melanoma patient samples, we are able to add to the predictive power of TNM staging, since stage alone was not able to separate patients with stage IIIB and IIIC disease. However, in our hands the best way to enhance survival prediction was by quantifying the MI, which has the added benefit of being much easier to perform than gene expression analysis. Thus MI provides a relatively simple and effective way to further differentiate a patient's ability to fight metastatic melanoma, either used alone or in combination with gene expression analysis.
Discussion
A number of studies analyzing human cancers have shown the importance of the immune response in the equilibrium state of primary neoplasia, but the importance of the immune system in keeping metastatic disease in check is less well understood (20–22). In melanoma, these types of studies have been heavily weighted toward stage I and stage II disease (20, 23). One study, however, has shown a correlation between TILs in resected lymph node metastases and patient survival (24). Similarly, studies of metastatic colorectal cancer, ovarian cancer, and follicular lymphoma have all demonstrated a better prognosis linked to the presence of infiltrating immune cells within tumor lesions (21, 22, 25). Only one other study has examined stage III melanoma by gene expression profiling and that study also linked up-regulation of certain genes associated with the immune system (e.g., HLA-E, PILRA, GTPBP2, IGKC) to time to tumor progression (19) and patient survival. However, that study did not directly address the influence of MI, TILs, or gene signatures on the improvement of TNM staging.
Despite these findings, the evaluation of the presence of leukocytes within metastatic lesions as a potentially easy and predictive tool of patient prognosis has not been sufficiently explored. This is possibly due to the conflicting studies that have shown both beneficial and detrimental effects of their presence (20–22). Here we show that, based on evaluation of TILs, CD3, and mRNA expression levels in the tumor, there is a comprehensive immune response in the tumors of stage III patients who survive for longer periods of time. We find an array of immune parameters among which are chemokines and adhesion molecules like CXCL13, CCL19, CCR7, VCAM1, and AMICA1 whose presence suggests active recruitment of the immune system into tumor sites. Establishing mechanisms underlying immune cell recruitment and activation at the molecular and cellular levels in metastatic lesions could be an important step toward advancement of immunotherapies in melanoma. For example, we detected higher levels of ICOS mRNA levels in the samples of patients who live longer, and the elevation of CD4+ICOShi IFN γ secreting T cells has been recently documented in the lesions of prostate cancer patients treated with anti-CTLA-4 antibody (26). Importantly, we were able to validate our gene signatures on an independent dataset from a study with a similar patient population that was published independently (19). Our data suggest that the immune response is in fact important in controlling advanced melanoma and indicate that its signature or quantification through TIL and CD3 counts can further subcategorize the staging system of recurrent tumors.
Another often forgotten and clinically underutilized parameter is MI. Its association with worse prognosis in melanoma has been examined (27), but in the current 6th edition of American Joint Committee on Cancer (AJCC) staging system it does not play a role (28), as a majority of the studies pertained to primary lesions. These studies have shown that MI in primary lesions is significantly associated with tumor thickness and ulceration that are the core determinants of the current staging system. However, MI will be included in the 7th edition of the AJCC staging system to address the classification of stage I melanoma (29). In our study of metastatic lesions, MI was the strongest indicator of patient survival and was the best single factor that improved current staging, significantly improving the separation between stage IIIB and IIIC patients, that we further validated by expanding patient samples with an additional 52 specimens. Our data support the use of MI in staging more advanced melanoma as well, following epidemiologic validation of this finding.
We postulate that the progression of metastatic melanoma is manifested by the balance of uncontrolled proliferation (MI) and the comprehensive presence of the immune system (TILs, CD3, and the wide array of immune network molecules detected at the mRNA level). Whether the low proliferative capacity in certain patients allows them to develop an immune response or whether the immune system functions to control proliferation is not clear. Our data indicate that metastatic melanoma is biologically diverse and that there is a need to tailor clinical trials toward the molecular and cellular profile of each patient. Potentially, patients with an existent immune presence in the tumor lesions are more prone to further stimulation of T cells to fight the tumor burden. On the other hand, the biggest benefit from chemotherapy may be seen in the patients whose tumors have high mitotic rates. If so, then subcategorizing patients based on metastatic lesion immune cell infiltration and MI before clinical trial recruitment might yield much more profound results than seen so far.
Methods
Sample Population.
Under an Institutional Review Board approved protocol we enrolled the first 38 patients and collected 44 melanoma samples, since some patients had 2 or 3 recurrences. Patient median age at 1st recurrence was 62.5 with a range from 30 to 92. Sixty-three percent of patients were males and 37% were females. All of the patients underwent surgery, 32% received chemotherapy, 24% underwent radiation therapy, and 13% underwent immunotherapy. Eighteen percent of patients presented at stage I, 29% at stage II, 47% at stage III, and 3% at stage IV. For validation, we used data on an independent cohort of 29 patients available online at ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-TABM-403. Another independent cohort to validate our findings using MI, TILs, and CD3 consisted of 52 randomly selected samples from patients with stage IIIb and IIIc melanoma.
MI, CD3 Cell Count, and TILs.
We assessed TILs and MI in hematoxylin and eosin (H&E)-stained tissue sections and performed immunohistochemistry staining to assess tumor infiltrating CD3 positive cells. Since many of the tissue samples were from lymph node metastases, any lymphocytes in the vicinity of tumor borders were excluded. Tumor slides were examined by 2 pathologists who were both blinded to the patients' clinical data. MI was established by counting mitoses in 10 high power fields (HPF) per tumor section and then averaging the number by HPF (1.96 mm2). CD3 positive cells were counted only within the tumor at least 2 HFPs away from the tumors' interface with the normal lymph node parenchyma. CD3+ cells in 10 high power fields per tumor section were counted and that number is reported. On H&E stains we established presence of TILs and indexed to 4 categories (0 = 0–5%, 1 = 5–25%, 2 = 25–50% and 3 = 50%+) each showing the percentage of tumor section that was represented by TILs. As with CD3+ T cells, we looked only at the portion of tumor at least 2 HPFs away from the tumors' interface with the normal lymph node parenchyma. We hybridized 44 tissue samples from 38 patients to Genechips. However, MI, TILs, and CD3 were only available for 30, 31, and 29 of the 38 patients, respectively, with complete data on all 3 parameters available for a total of 28 patients. This explains the differing numbers in the tables. For example, n = 30 in Table 2 that describes a model based on MI. Table S5A describes the 44 samples (not patients) and Table S5B describes 32 samples (not patients).
Statistical Methods—Clinical Data Analysis.
The clinical data were summarized numerically and graphically to verify the normality assumption and for outlier detection. Box-Cox transformations were used to transform variables with deviations from normality, such as MI and CD3 cell count (30). The variable TILs were treated as ordered in the analysis. TNM stage was dichotomized in the analysis due to small sample size. For clinical data analysis, the unit of analysis was patient and not recurrence/metastasis. However, all reported results hold for per recurrence analysis. For each patient with multiple samples, the sample corresponding to the earliest recurrence/metastasis was used in the analysis. Cox proportional hazards model was used for prediction. The median estimated hazard ratio was used to divide the patients into low and high risk groups. All analyses were performed using the R language for statistical computing (31).
Gene Chip Processing.
Post surgery collected tissue was placed in RNAlater (Qiagen) at 4 °C overnight, then stored at −80 °C. Before whole RNA extraction (RNeasy Mini Kit, Qiagen), touch preparations were performed to ensure that the specimen obtained was mostly tumor tissue. RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies). Double stranded cDNA synthesis was performed using a SuperScript double-stranded cDNA synthesis kit from Invitrogen. In vitro transcription of biotin-labeled cRNA probes was done using an IVT labeling kit (Affymetrix). Fragmented biotin-labeled cRNA was hybridized on Affymetrix Human Genome U 133 Plus 2.0 chips, in the Rockefeller University Genomics Core laboratory.
Gene Chip Data Preprocessing.
The raw gene expression values were normalized using probe logarithmic error intensity estimate. Probes were grouped by their Unigene symbols and the median of expression levels of all probes in a group was taken to be the expression level of the transcript (32). This step resulted in the reduction of number of features from 54,675 to 23,940. The signals were then further quantile normalized (33).
Significance Analysis of Microarrays.
SAM (12) was used to identify genes that are significantly associated with postrecurrence survival using time from recurrence to death (or censored) as the outcome variable. One thousand permutations of the data were used to estimate the FDR (13) and to select differentially expressed genes. Additionally, the patients were dichotomized into 2 groups: those with prolonged survival (>1.5 years) and those with “shorter survival” (<1.5 years). A 2-sample nonparametric comparison was used in SAM to identify genes that are differentially expressed between these 2 groups. The significant gene lists resulting from the 2 types of analyses (survival and 2-sample comparison) were then compared.
Prevalidated Gene Expression Predictor.
To derive a gene expression signature of postrecurrence survival, we used the method of PV (15, 16). PV outputs a prediction for each patient based on the model that is estimated without using that patient's data. We used per recurrence analysis because PV allowed us to reduce bias that might arise due to the dependence among multiple recurrences of the same patient. An 11-fold PV was used to construct a gene expression predictor of postrecurrence survival. The 44 samples were divided into 11 groups of 4 samples randomly, but in such a way that samples from the same patient were always grouped together to reduce bias. At each PV fold, one of the 11 groups of 4 samples was set aside as a test set and the remaining 40 samples were used as a training set. The training set was analyzed using SAM to select the top 3 up-regulated and top 3 down-regulated genes, resulting in an output of 6 top genes. We calculated the first principal component of the 6 genes in the training data. We fit the Cox proportional hazards model with the first principal component of the 6 genes in the training set as a predictor, and survival since metastatic excision as a dependent variable. Based on this model, we estimated hazard ratios for the training set and divided the test set cases into low risk and high risk, using the median training set hazard ratio as a cutoff point. This procedure was repeated 11 times, each time reserving a different set of 4 samples for the test set. Note that for each patient, the above PV procedure outputs a prediction based on the model that was estimated without using that patient's data and, therefore, no overfitting occurs. Varying the number of genes selected by SAM between 4 and 20 produced similar PV predictors. The resulting PV gene expression predictor was compared to the other clinical predictors in a multivariable Cox regression model.
Gene Selection via Signal-to-Noise Ratio.
To select the informative genes which should be included in the model, we used the signal-to-noise ratio (SNR), a feature selection method found to perform well in gene expression experiments (5, 18). The signal-to-noise ratio favors genes that have nonoverlapping distributions with far apart means. We experimented with the top 10, 30, 50, 100, 300, 500, and 1,000 genes, and used 1.5- and 2-fold change to further narrow down the set of candidate genes. We found that the best performance measures do not improve with the inclusion of more than the top 50 genes (Table S4). Although different methods were applied, the overlap between SAM genes and top 50 SNR genes is remarkably high.
Prediction, Performance Evaluation, and Estimation of Statistical Significance.
Due to their ability to handle datasets with a small number of highly dimensional examples with correlated features, support vector machines (SVM) are a popular supervised learning method to analyze gene expression data (17, 18). To estimate the prediction accuracy, we used leave-one-out cross-validation. Here one example is systematically held out and the model is built on all of the remaining examples and tested on the example which was hidden while the model was learned. We report the following performance measurements: prediction accuracy, sensitivity, specificity and AUC. In each leave-one-out iteration, values of Unigene features were normalized to have zero mean and unit variance using z-score normalization. In our experiments we report the results with and without the use of PCA. We set the amount of retained variance after performing PCA to 0.95.
Immunohistochemistry.
Immunohistochemistry was performed on formalin fixed, paraffin embedded tissues using mouse anti-human CD3, clone PS-1 (Ventana Medical Systems). In brief, sections were deparaffinized in xylene, rehydrated through graded alcohols and rinsed in distilled water. Heat induced epitope retrieval was performed in 10 mM citrate buffer pH 6.0 for 10 min in a 1200-Watt microwave oven at 90% power. CD3 was applied undiluted and incubated for 30 min. Primary antibody was detected with Ventana's biotinylated goat anti-mouse secondary followed by application of streptavidin-horseradish-peroxidase conjugate. The complex was visualized with 3,3 diaminobenzidene and enhanced with copper sulfate. Slides where washed in distilled water, counterstained with hematoxylin, dehydrated, and mounted with permanent media.
Supplementary Material
Acknowledgments.
We thank Dr Shalini Mulaparthi for her assistance in obtaining the pilot grant, Dr. Patrick Ott for clinical consultations and NYU IMCG for the help with the patient samples. This work was been supported by the National Institutes of Health Grants P30 CA016087–29 and R37 AI044628 (to N.B.), the Cancer Research Institute, New York University Cancer Center Biostatistics Shared Resource, the Emerald Foundation, and National Science Foundation Grant IIS-0447773 (to S.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905139106/DCSupplemental.
References
- 1.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 2.Thompson JF, Scolyer RA, Kefford RF. Cutaneous melanoma. Lancet. 2005;365:687–701. doi: 10.1016/S0140-6736(05)17951-3. [DOI] [PubMed] [Google Scholar]
- 3.Fecher LA, Cummings SD, Keefe MJ, Alani RM. Toward a molecular classification of melanoma. J Clin Oncol. 2007;25:1606–1620. doi: 10.1200/JCO.2006.06.0442. [DOI] [PubMed] [Google Scholar]
- 4.Balch CM, Soong SJ. Predicting outcomes in metastatic melanoma. J Clin Oncol. 2008;26:168–169. doi: 10.1200/JCO.2007.13.8123. [DOI] [PubMed] [Google Scholar]
- 5.Golub TR, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 6.Mills K. Gene expression profiling for the diagnosis and prognosis of acute myeloid leukaemia. Front Biosci. 2008;13:4605–4616. doi: 10.2741/3026. [DOI] [PubMed] [Google Scholar]
- 7.van't Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–570. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 8.Henry NL, Hayes DF. Use of gene-expression profiling to recommend adjuvant chemotherapy for breast cancer. Oncology (Williston Park) 2007;21:1301–1309. discussion 1311, 1314–1319. [PubMed] [Google Scholar]
- 9.Nahleh ZA. Molecularly targeted therapy in breast cancer: The new generation. Recent Pat Anticancer Drug Discov. 2008;3:100–110. doi: 10.2174/157489208784638794. [DOI] [PubMed] [Google Scholar]
- 10.Haqq C, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci USA. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaeger J, et al. Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clin Cancer Res. 2007;13:806–815. doi: 10.1158/1078-0432.CCR-06-1820. [DOI] [PubMed] [Google Scholar]
- 12.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 14.Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. New York: Academic Press; 1979. [Google Scholar]
- 15.Hofling H, Tibshirani R. A study of pre-validation. Annals of Applied Statistics. 2008;2:643–664. [Google Scholar]
- 16.Tibshirani RJ, Efron B. Pre-validation and inference in microarrays. Stat Appl Genet Mol Biol. 2002;1 doi: 10.2202/1544-6115.1000. Article1. [DOI] [PubMed] [Google Scholar]
- 17.Brown MP, et al. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc Natl Acad Sci USA. 2000;97:262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furey TS, et al. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics. 2000;16:906–914. doi: 10.1093/bioinformatics/16.10.906. [DOI] [PubMed] [Google Scholar]
- 19.John T, et al. Predicting clinical outcome through molecular profiling in stage III melanoma. Clin Cancer Res. 2008;14:5173–5180. doi: 10.1158/1078-0432.CCR-07-4170. [DOI] [PubMed] [Google Scholar]
- 20.Piras F, et al. The predictive value of CD8, CD4, CD68, and human leukocyte antigen-D-related cells in the prognosis of cutaneous malignant melanoma with vertical growth phase. Cancer. 2005;104:1246–1254. doi: 10.1002/cncr.21283. [DOI] [PubMed] [Google Scholar]
- 21.Sato E, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA. 2005;102:18538–18543. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galon J, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 23.Clemente CG, et al. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer. 1996;77:1303–1310. doi: 10.1002/(SICI)1097-0142(19960401)77:7<1303::AID-CNCR12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 24.Mihm MC, Jr, Clemente CG, Cascinelli N. Tumor infiltrating lymphocytes in lymph node melanoma metastases: A histopathologic prognostic indicator and an expression of local immune response. Lab Invest. 1996;74:43–47. [PubMed] [Google Scholar]
- 25.Dave SS, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 26.Chen H, et al. Anti-CTLA-4 therapy results in higher CD4+ICOShi T cell frequency and IFN-gamma levels in both nonmalignant and malignant prostate tissues. Proc Natl Acad Sci USA. 2009;106:2729–2734. doi: 10.1073/pnas.0813175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Attis MG, Vollmer RT. Mitotic rate in melanoma: A reexamination. Am J Clin Pathol. 2007;127:380–384. doi: 10.1309/LB7RTC61B7LC6HJ6. [DOI] [PubMed] [Google Scholar]
- 28.Francken, et al. The prognostic importance of tumor mitotic rate confirmed in 1317 patients with primary cutaneous melanoma and long follow-up. Ann Surg Oncol. 2004;11:426–433. doi: 10.1245/ASO.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 29.Balch CM, Gershenwald JE, Soong S-J, Sober A, Kirkwood J. In: Cutaneous Melanoma. Balch CM, Houghton A, Sober A, Soong S-J, editors. St. Louis: Quality Medical Publishing; 2009. [Google Scholar]
- 30.Box GE, Cox DR. An analysis of transformations. J R Stat Soc B. 1964;26:211–246. [Google Scholar]
- 31.RDC Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 32.Pavlidis P, Lewis DP, Noble WS. Exploring gene expression data with class scores. Pac Symp Biocomput. 2002:474–485. [PubMed] [Google Scholar]
- 33.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.