Summary
Thylakoid membranes have a unique complement of proteins, most of which are synthesized in the cytosol, imported into the stroma, and translocated into thylakoid membranes by specific thylakoid translocases. Known thylakoid translocases contain a core multi-spanning, membrane-integrated subunit which is also nuclear-encoded and imported into chloroplasts before being integrated into thylakoid membranes. Thylakoid translocases play a central role in determining the composition of thylakoids, yet the manner by which the core translocase subunits are integrated into the membrane is not known. We used biochemical and genetic approaches to investigate integration of the core subunit of the chloroplast Tat translocase, cpTatC, into thylakoid membranes. In vitro import assays show that cpTatC correctly localizes to thylakoids if imported into intact chloroplasts, but it does not integrate into isolated thylakoids. In vitro transit peptide processing and chimeric precursor import experiments suggest that cpTatC possesses a stroma-targeting transit peptide. Import time course and chase assays confirmed that cpTatC targets to thylakoids via a stromal intermediate, suggesting that it might integrate through one of the known thylakoid translocation pathways. However, chemical inhibitors to the cpSecA-cpSecY and cpTat pathways did not impede cpTatC localization to thylakoids when used in import assays. Analysis of membranes isolated from Arabidopsis thaliana mutants lacking cpSecY or Alb3 showed that neither is necessary for cpTatC membrane integration or assembly into the cpTat receptor complex. These data suggest the existence of another translocase, possibly one dedicated to the integration of chloroplast translocases.
Keywords: TatC, protein translocation, thylakoid biogenesis, SecY2, Twin arginine, membrane protein integration
Introduction
Most of what is known about thylakoid protein translocation comes from studies of developed chloroplasts. Following import into the plastid, all nuclear-encoded thylakoid proteins studied to date are inserted into thylakoids from the plastid stroma by one of four conserved translocation pathways, all of which are evolutionarily derived from a bacterial endosymbiont (Schuenemann, 2007). Some membrane proteins integrate into thylakoids by an unassisted or ‘spontaneous’ mechanism. The remaining known membrane proteins and lumenal proteins are inserted by translocases. The cpSRP-FtsY-Alb3 translocase integrates a class of light harvesting membrane proteins, the cpSecA-cpSecYE translocase transports globular lumenal proteins and integrates some membrane proteins, and the cpTat translocase transports folded lumenal proteins and integrates a very limited number of membrane proteins (Schuenemann, 2007). The cpSecYE and SRP subunits also appear to co-translationally integrate at least one plastid-encoded membrane protein (Zhang et al., 2001; Nilsson and van Wijk, 2002). Little is known about how membrane-bound translocase subunits integrate into thylakoid membranes. Some translocase subunits, e.g. cpSecE and the cpTat subunits Hcf106 and Tha4, ‘spontaneously’ integrate into the membrane (Steiner et al., 2002; Fincher et al., 2003). In fact, Hcf106 and Tha4 are found in the envelope as well as the thylakoids, as might be expected of proteins that integrate spontaneously. However, the mechanism of integration of the multi-spanning core subunits cpSecY, Alb3, and cpTatC, is completely unknown.
We have begun to examine the targeting pathway of the core subunits. The present work focuses on cpTatC because we have an efficient biochemical assay for its localization. When incubated with intact chloroplasts, pcpTatC is imported, integrated into thylakoids, and assembled into the cpTat receptor complex (Fincher et al., 2003). In contrast to spontaneously integrating membrane proteins, membrane integration of cpTatC appears to require a translocase because neither the precursor nor the mature form of cpTatC integrates into isolated thylakoids under any experimental conditions examined (Fincher et al., 2003). Here, we present evidence that cpTatC targets to thylakoid membranes via a stromal intermediate, and that cpTatC membrane integration is not altered by competition with precursors of the cpSec and cpTat pathways. Furthermore, cpTatC was found integrated and assembled into the cpTat receptor complex in membranes isolated from Arabidopsis seedlings that carry knockout mutations in genes encoding Alb3 and cpSecY. These data suggest cpTatC integrates via a translocase other than those already known to exist at thylakoid membranes. The possible involvement of two new candidate translocases will be discussed.
Results
pcpTatC possesses a stroma-targeting transit peptide
cpTatC is synthesized as a precursor (pcpTatC) with a 50-residue amino-terminal targeting peptide that is cleaved upon import into chloroplasts (Mori et al., 2001). Two experiments were conducted to determine if pcpTatC possesses a stroma-targeting transit peptide. First, radiolabeled pcpTatC was incubated with isolated stroma to determine if its transit peptide is cleaved by the stromal transit peptidase (Richter and Lamppa, 1999). Isolated stroma cleaved pcpTatC to a protein the size of mature cpTatC (mcpTatC), similar to cleavage of the precursor to the small subunit of ribulose bisphosphate carboxylase oxygenase (pSSU) to mature size SSU (mSSU) (Figure 1a). Reduction in size was due to amino-terminal processing because stroma had no effect on in vitro translated mcpTatC. Processing was conducted by the stromal peptidase because ortho-phenanthroline, an inhibitor of the stromal transit peptidase, prevented processing of pcpTatC as well as pSSU (Figure 1a).
Figure 1.
Precursor to cpTatC possesses a stromal targeting transit peptide. (a) Radiolabeled pSSU, pcpTatC, and mature cpTatC (mcpTatC) were each incubated with isolated stroma and 3 mM PMSF with or without 10 mM 1,10-phenanthroline (OP), as shown above the panel, at 25°C, for 30 minutes. Samples were analyzed using SDS-PAGE and fluorography. (b) Scheme illustrating swapping of the pSSU and pcpTatC transit peptides. Numbers above constructs represent the number of amino acids from the translation start site. (c) Radiolabeled pcpTatC, SSUtpcpTatC, pSSU, and cpTatCtpSSU were each incubated with chloroplasts in a protein import assay (Experimental Procedures) for 20 minutes. Chloroplasts were treated with 100 μg/mL thermolysin and then re-purified, washed, lysed, and fractionated. Translation product (Tp) equivalent to 5% of each assay, chloroplasts (Cp), stroma (S), membranes (M), and thermolysin-treated membranes (+T) were analyzed using SDS-PAGE and fluorography. The positions of molecular weight markers are shown at left.
In a second experiment, the transit peptides and small portions of the mature domain of pSSU and pcpTatC were swapped (Figure 1b, S1b), and the chimeric precursor proteins were assayed for localization in a chloroplast import assay. As shown in Figure 1c, pcpTatC is imported into chloroplasts, processed to mature size, and localized primarily to the membranes. Integration into the membranes is shown by the appearance of characteristic degradation products upon treatment of the membranes with thermolysin (Mori et al., 2001). The same import and localization pattern was obtained for SSUtpcpTatC, which was directed by the known stroma-targeting SSU transit peptide, except that import was more efficient. Similarly, the cpTatC transit peptide directed cpTatCtpSSU import into the stroma, albeit much less efficiently than the homologous transit peptide. These results demonstrate that cpTatC possesses a stroma-targeting transit peptide.
The cpTatC non-conserved stromal domain is not necessary for targeting to thylakoid membranes or assembly into the 700 kD cpTat receptor complex
Precursors that possess stroma-targeting transit peptides do not necessarily reach the chloroplast stroma. For example, the outer envelope translocase subunit TOC75 also has a cleavable stroma-targeting transit peptide, but a second peptide sequence stops transfer before TOC75 reaches the stroma (Inoue and Keegstra, 2003). The possibility that cpTatC possesses a similar membrane retention domain was examined by looking for sequence elements that might serve as localization signals. Targeting signals are frequently extra peptides that have reduced sequence conservation among orthologous proteins as they do not constitute functional domains of the protein (Bruce, 2001). Mature cpTatC proteins possess long hydrophilic amino-terminal extensions before the first transmembrane domain (Mori et al., 2001), whereas bacterial TatC proteins have short hydrophilic amino termini (Buchanan et al., 2002). Alignment of cpTatC proteins from a variety of plant species revealed a conserved region constituting transmembrane domains, soluble loops, and 29 residues amino-proximal to the first transmembrane domain. Most sequence divergence was found in regions that represented transit peptides and the first ~ 50 residues of the amino terminus of the mature protein (Figure S1a). To assess its potential role in targeting, the non-conserved region (NC) was deleted from SSUtpcpTatC, producing SSUtpΔNCcpTatC (Figure 2a, S1b). This precursor imported into chloroplasts, localized to thylakoids, and produced the same degradation products as wild type pcpTatC (Figure 2b). In thylakoids, cpTatC is present in a ~700 kD receptor complex that can be detected by Blue Native PAGE (BN-PAGE) (Cline and Mori, 2001). Imported ΔNCcpTatC also assembled into a large receptor complex, but with a reduced molecular weight (Figure 2b). The lower molecular weight would be expected if all of the cpTatC subunits in the complex were ΔNCcpTatC, although that remains to be determined. Treating the digitonin-solubilized membrane extract with Hcf106 antibodies before BN-PAGE caused a band shift to higher molecular weight, verifying that ΔNCcpTatC is assembled with Hcf106 in the cpTat receptor complex (data not shown). Thus, the non-conserved stromal domain of cpTatC is not necessary for targeting of cpTatC to thylakoid membranes or assembly into the 700 kD cpTat receptor complex.
Figure 2.
The cpTatC non-conserved stromal domain is not necessary for cpTatC localization. (a) Scheme illustrating deletion of the cpTatC non-conserved region (NC region). Numbers above constructs represent the number of amino acids from the translation start site. (b) Radiolabeled pcpTatC and SSUtpΔNCcpTatC were incubated with chloroplasts in a protein import assay (Experimental Procedures) for 20 minutes. Chloroplasts were treated with 100 μg/mL thermolysin, re-purified, washed, lysed, and fractionated into stroma and membranes. Translation product (Tp) equivalent to 5% of each assay, chloroplasts (Cp), stroma (S), membranes (M), and thermolysin-treated membranes (+T) were analyzed using SDS-PAGE and fluorography. Membranes were also dissolved in 1% digitonin and analyzed using Blue Native PAGE and fluorography. The positions of molecular weight markers (Mr), or monomeric (440 kD) and dimeric (880 kD) ferritin for BN-PAGE, are shown at left.
pcpTatC import and protease accessibility assays reveal a stromal mcpTatC
A small percentage of imported cpTatC is always found in the soluble fraction of chloroplasts recovered from import assays (Figure 2b, lane 3). A protease accessibility assay was performed to test whether the soluble cpTatC was stromal or located in the inter-envelope membrane space (Jackson et al., 1998). When incubated with intact chloroplasts, thermolysin digests exposed outer envelope proteins, but it cannot penetrate the outer envelope membrane (Figure 3b). Trypsin can cross the outer envelope to access the intermembrane space, allowing digestion of TOC75, but it cannot cross the inner envelope membrane, preventing digestion of TIC110 (Figure 3b). Radiolabeled pcpTatC was imported into chloroplasts and the chloroplasts were then treated with buffer, trypsin, or thermolysin. After treatment, repurified chloroplasts were fractionated into soluble and membrane fractions. The percentage of soluble cpTatC from the trypsin treated chloroplasts was essentially the same as that from the thermolysin treated chloroplasts, indicating that the soluble mcpTatC is located in the stroma rather than the inter-envelope membrane space (Figure 3a).
Figure 3.
Soluble cpTatC is located in the stroma, not the envelope intermembrane space. (a) Radiolabeled pcpTatC was incubated with chloroplasts in a protein import assay (Experimental Procedures) for 10 minutes. Recovered chloroplasts were treated with IB (−), trypsin, or thermolysin. Proteases were inhibited before chloroplast re-isolation, lysis, and fractionation. Translation product (Tp) equivalent to 5% of each assay, stroma (S), and membrane (M) fractions were analyzed using SDS-PAGE and fluorography. Gel band extraction was used to quantify radiolabeled cpTatC in each chloroplast fraction for each treatment. Numbers below bands represent the relative percentages of cpTatC found in each fraction. (b) Chloroplasts from IB- (−), trypsin-(Tr), and thermolysin-treated (Th) samples were also analyzed by SDS-PAGE and immunoblotting with antibodies to TOC75 (applied at 1:2500) or TIC110 (applied at 1:5000). The positions of molecular weight markers (Mr) are shown at left.
pcpTatC targets to thylakoid membranes via a stromal intermediate
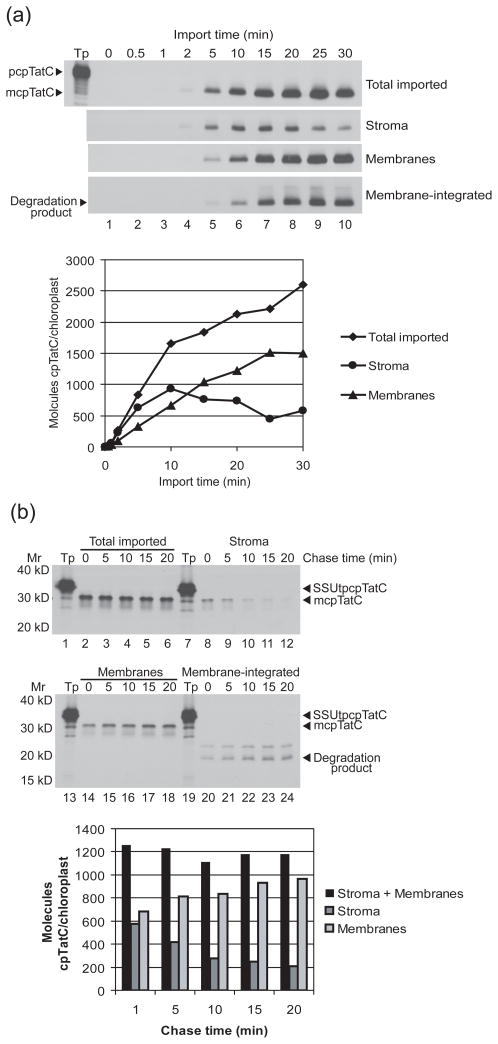
In order to determine whether the stromal mcpTatC is a stromal intermediate or a dead-end import product, a rapid stopping time course of a protein import assay was conducted (Reed et al., 1990). During the first five minutes of import, mcpTatC accumulated in the stroma at a rate similar to import into chloroplasts (Figure 4a), after which its accumulation slowed and then declined. Initially, membrane-associated mcpTatC accumulated more slowly, but it continued to accumulate after stromal mcpTatC accumulation leveled off and declined. Similar kinetics of accumulation were obtained from three independent experiments (Figure S2). These import kinetics are similar to those exhibited by thylakoid proteins that are known to target via stromal intermediates (Reed et al., 1990; Cline et al., 1992).
Figure 4.
cpTatC targets to thylakoid membranes via a stromal intermediate. (a) Radiolabeled pcpTatC was incubated with chloroplasts in an import time course assay (Experimental Procedures). At the times indicated, samples of the import reaction were transferred to 3.3 mM HgCl2 to terminate import and localization. Chloroplasts in each sample were treated with 100 μg/mL thermolysin, repurified, brought to equal chlorophyll concentration, lysed, and fractionated into membranes and stroma. Translation product (Tp) equivalent to 5% of each assay, chloroplasts (Total imported), stroma, membranes, and thermolysin-treated membranes (Membrane-integrated) were analyzed using SDS-PAGE and fluorography. Gel band extraction was used to quantify radiolabeled cpTatC in each chloroplast fraction at each time point. Plotted are molecules of cpTatC per chloroplast estimated from DPMs in bands and chlorophyll content of each sample for each fraction and time point. (b) Chloroplasts and radiolabeled SSUtpcpTatC (Tp) were incubated in a import chase time course assay (Experimental Procedures). After 5 minutes of import, chloroplasts were diluted, treated with thermolysin, repurified, resuspended to the original import reaction volume, and placed back in import reaction conditions. At time points, samples of chloroplasts were re-isolated, washed, lysed, and fractionated. Chloroplasts (‘Total imported’), stroma, membranes, and thermolysin-treated membranes (‘Membrane-integrated’) were analyzed using SDS-PAGE and fluorography. Radiolabeled cpTatC in each chloroplast fraction was quantified as above. Plotted are the molecules of cpTatC per chloroplast of stroma, membrane, and sum of stroma and membrane cpTatC at each time point. The positions of molecular weight markers (Mr) are shown at left.
An import-chase assay was conducted to determine if stromal cpTatC is the direct precursor to thylakoid-integrated cpTatC (Reed et al., 1990; Li and Schnell, 2006). After a brief period of pSSUtpcpTatC import, all remaining precursor was removed from the chloroplast surface by treatment with thermolysin. Chloroplasts were returned to import reaction conditions, and samples were removed at subsequent time points to follow the sorting of imported protein between the internal chloroplast compartments (Figure 4b). pSSUtpcpTatC was used in this experiment instead of pcpTatC, because the SSU transit peptide imparts increased import efficiency upon cpTatC (Figure 1c), and produces proportionally more stromal mcpTatC at early time points. The data show that cpTatC chases from stroma to thylakoids. Similar results were obtained from three independent chase experiments (data not shown). Taken together, the results of the time course and chase experiments indicate that cpTatC targets to thylakoid membranes via a stromal intermediate.
cpTatC is neither integrated by the cpTat nor the cpSecA/cpSecYE pathways
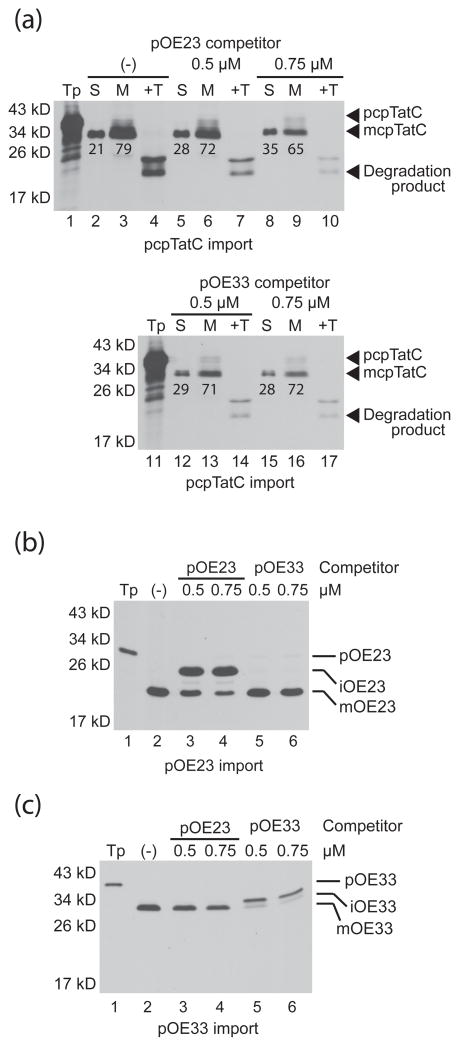
cpTatC targeting to thylakoid membranes via a stromal intermediate suggests that a conserved thylakoid translocase might serve as the cpTatC integrase (See Discussion). We used in organello competition to determine if cpTatC is integrated either by the cpSec or cpTat pathways (Cline et al., 1993). In organello competition involves pre-importing chemical quantities of either a cpSec or cpTat pathway precursor into chloroplasts before adding radiolabeled test precursor to the assay. The TOC-TIC import apparatus imports chemical amounts of precursors much faster than the cpSec or cpTat apparatus can transport them from the stroma, which results in amounts of stromal intermediate that saturates the thylakoid translocases. Radiolabeled stromal intermediate accumulates if both stromal species use the same thylakoid translocase. As shown in Figure 5a, pre-import of the cpTat precursor pOE23 or the cpSec pathway precursor pOE33 neither caused accumulation of stromal cpTatC nor a decrease in membrane associated cpTatC. The efficacy and specificity of the competition is shown in Figure 5b and 5c for control proteins: radiolabeled pOE23, which accumulates as intermediate only when competing with pre-imported pOE23, and radiolabeled pOE33, which accumulates as intermediate only after pre-import of pOE33.
Figure 5.
In organello competition indicates that cpTatC integration does not proceed via the cpTat or cpSec pathways. Chloroplasts were incubated with 5 mM ATP, 1.5 mM DTT, and either 0.3 M urea (−) or unlabeled bacterially expressed pOE23 or pOE33 (competitor) at the final concentrations depicted above the panels for 7 min in the light. Radiolabeled pcpTatC (a), pOE23 (b), or pOE33 (c) was added to the reaction mixtures and the import reaction allowed to continue for an additional 15 minutes. Chloroplasts from pcpTatC reactions were recovered, lysed, and fractionated into stroma (S) and membranes (M). An aliquot of the membrane fraction was treated with thermolysin (+T). Samples were analyzed by SDS-PAGE and fluorography. Chloroplasts from import reactions with radiolabeled pOE23 or pOE33 were recovered and directly analyzed. Gel band extraction was performed to quantify the relative percentages of stromal and membrane-associated cpTatC, which are represented by numbers below the bands in panel (a). The positions of molecular weight markers (Mr) are shown at left.
Other inhibitors were applied to import assays: 1) azide to inhibit the ATPase domain of cpSecA and 2) ionophores to dissipate the proton gradient, which is required by the cpTat and which stimulates the cpSecA/cpSecYE and SRP/Alb3 pathways. Neither treatment had a significant effect on cpTatC targeting to thylakoids (Figures S3, S4).
Neither cpSecY nor Alb3 are necessary for cpTatC integration
In bacteria, SecYE and YidC, the bacterial orthologs to cpSecYE and Alb3, operate with other translocase subunits in a modular fashion, e.g. SRP can target precursors to SecYE or YidC, and SecA is not always required to integrate membrane proteins through SecYE (Fandl et al., 1988; Valent et al., 1998; Scotti et al., 1999; Bloois et al., 2004). In order to more directly determine if Alb3 or cpSecY are necessary to integrate cpTatC, membranes were isolated from Arabidopsis seedlings carrying knockout mutations in genes that encode either Alb3 or cpSecY (Sundberg et al., 1997; Skalitzky, 2006). Chloroplasts could not be analyzed from mutant seedlings, which contain grossly deformed plastids with few internal membranes. The isolated membranes were tested for the presence, membrane integration, and assembly status of cpTatC. Thylakoids isolated from wildtype Arabidopsis were also analyzed to help identify translocase-specific and non-specific bands. As shown in Figure 6a, cpTatC was detected in membranes from both mutants. cpTatC in mutant membranes was resistant to alkaline carbonate extraction (Figure 6c) (Mori et al., 2001), indicating that it is integrally associated with the membranes. Moreover, cpTatC was found assembled into the 700 kD Tat receptor complex in membranes from mutant seedlings (Figure 6b). Samples of membranes from mutant seedlings were also queried using antibodies that detect core translocase subunits Alb3, cpSecY, or Alb4 (Figure 6a, 6c). Neither Alb3 nor cpSecY were detected in membranes isolated from each respective mutant (Figure 6a). The minor band migrating slightly below the Alb3 band in the alb3 membranes is a non-specific band that is extracted by alkaline carbonate (Figure 6c). Interestingly Alb4 and cpSecY were present in the alb3 membranes and Alb3 and Alb4 were present in the secy membranes (Figure 6a). Alkaline carbonate extraction (Figure 6c) verified that Alb3, cpSecY, and Alb4 were integrally associated with the wild-type and mutant membranes, although Alb3 was frequently low in abundance in alkaline extraction experiments with secy membranes. We occasionally observed a double banding pattern when detecting cpSecY (Figure 6a); the lower band may result from some partial degradation during isolation. In addition to showing that cpTatC does not require either cpSecY or Alb3 for integration, these findings raise important questions regarding the integration machinery for other translocase subunits Alb3, Alb4, and cpSecY.
Figure 6.
Neither Alb3 nor SecY are necessary for cpTatC integration. (a) Arabidopsis seedlings were germinated on MS media, grown, and harvested as described (Experimental Procedures). Membranes were isolated from seedlings and samples of equal mass tissue were analyzed by SDS-PAGE and immunoblotting. (b) Isolated membranes were also dissolved in 2% digitonin and analyzed by BN-PAGE and immunoblotting using antibodies to atcpTatC. (c) Membranes were subjected to alkaline carbonate (-CO3) or mock extraction (IB). Supernatants (S) and membrane pellets (P) from each extraction were analyzed using SDS-PAGE and immunoblotting. Thylakoids (Thy) prepared from isolated Arabidopsis chloroplasts were analyzed in the far right lanes of each SDS-PAGE gel to provide reference bands for the other lanes. (*) Non-specific bands. All antibodies were applied at 1:5000.
Discussion
Cavalier-Smith classified cell membranes into two categories: ‘genetic’ and ‘derived’ (Cavalier-Smith, 2000). Genetic membranes always arise by growth and division, and they are continuously maintained through the germ line from cell to daughter cell. Genetic membranes acquire proteins through the action of specific translocases and frequently synthesize their own polar lipids. Derived membranes obtain lipids and proteins from genetic membranes by membrane flow, e.g. vesicles. The endoplasmic membrane and plastid inner envelope membrane are examples of genetic membranes, whereas the tonoplast and Golgi membranes are derived. The thylakoid membrane of plant chloroplasts has features of both genetic and derived membranes. In mature chloroplasts, thylakoids take up their unique complement of proteins through a set of specific translocases (Cline and Theg, 2007; Schuenemann, 2007) and thylakoids are passed to daughter chloroplasts by division (Cran and Possingham, 1972). Conversely, during chloroplast biogenesis, thylakoids are thought to arise from the inner envelope by membrane invagination and fission (Carde et al., 1982; Benning et al., 2006; Kobayashi et al., 2007). This poses the question. “At what point do inner plastid envelope invaginations become thylakoids?” The key may lie in determining when, where, and how thylakoid translocases are inserted into the membranes because they play a central role in determining the protein composition of the thylakoid membrane and lumen.
cpTatC, cpSecY, and Alb3 are thylakoid translocase proteins derived from the cyanobacterial progenitor of chloroplasts (Dalbey and Kuhn, 2000). In bacteria, the orthologous proteins are co-translationally integrated into the cytoplasmic or thylakoid membranes by SRP and SecY/E/G and/or YidC, similar to other multi-spanning membrane proteins. Co-translational insertion insures that highly hydrophobic proteins are not exposed to the aqueous environment where they would aggregate. As the chloroplast evolved, genes encoding thylakoid translocases were relocated to the nucleus, precluding a co-translational mode of integration and necessitating alternate strategies to integrate these proteins into the membrane. One strategy might integrate cpTatC into the inner envelope during plastid import by a novel insertion mechanism. Such a strategy is used by a class of mitochondrial inner membrane proteins and at least two chloroplast inner envelope proteins (Brink et al., 1995; Knight and Gray, 1995; Sirrenberg et al., 1996). cpTatC might then be relocated to thylakoids by membrane flow. Another possibility would be to import cpTatC to the stroma allowing it to enter a conserved, but altered, translocation pathway. The localization of LHC apoproteins is an example of this latter strategy. preLHC apoproteins are imported into the plastid stroma where they bind cpSRP, a modified SRP adapted to post-translational insertion, which targets them to a conserved FtsY receptor before subsequent integration via Alb3.
Our studies show that cpTatC possesses a stromal-targeting transit peptide, appears to lack an envelope retention domain, and proceeds to thylakoids via a stromal intermediate. This suggests that cpTatC is integrated by a conserved translocase, either directly into the thylakoid membrane or, alternatively, into the inner envelope membrane. In that regard, a small amount of envelope-associated cpTatC was generally detected in protein import assays (Fincher et al., 2003), although it was not technically feasible to determine if envelope-associated cpTatC is an intermediate or an off-pathway product.. At the time of this study, three conserved translocases were known: cpSecA/cpSecY/E, cpSRP/cpFtsY/Alb3, and cpTat, all of which are thylakoid localized in mature chloroplasts (Fincher et al., 2003). cpSecA is homologous to SecA, an ATP-powered translocation motor required to move large hydrophilic domains across the membrane through a SecYE channel (Andersson and Heijne, 1993), and cpSRP and cpFtsY are involved in targeting precursors to Alb3 (Moore et al., 2000). A number of genetic studies have ruled out certain accessory subunits from being involved in cpTatC integration. The cpTat substrate OE23 was correctly localized to the thylakoid lumen in maize seedlings that lack cpSecA (Voelker et al., 1997). Mature size OE23 was also detected in membranes isolated from Arabidopsis plants that carry knockout mutations in SRP subunits cpSRP54 and SRP43 (Hutin et al., 2002). The chloroplast Tat pathway was indirectly ruled out by the fact that in vitro-integrated Tha4 complemented cpTat transport function of maize thylakoids from a Tha4 null mutant line (Fincher et al., 2003). Our biochemical studies verify that cpTatC does not employ the cpSec and cpTat translocases in the known configurations.
Asakura et al. previously detected cpSecY and cpTatC in protein extracts from maize seedlings harboring a knockout mutation in the gene encoding FtsY; and more recently, cpTatC and cpSecY were detected in protein extracts from Arabidopsis alb3 and ftsy mutants (Asakura et al., 2004; Asakura et al., 2008). We extend these findings by showing that cpTatC is integrated into membranes and assembled into the cpTat receptor complex in the absence of Alb3 or cpSecY. These findings are particularly relevant because in E. coli, indirect evidence suggests that TatC is integrated in a SecYE-dependent manner (Yi et al., 2003).
We also found that cpSecY and Alb4 were integrated in membranes from alb3 mutant seedlings and Alb3 and Alb4 were integrated in membranes from cpsecy mutant seedlings. This is contrary to evidence from E. coli that suggests the Alb3 homolog, YidC, is integrated in a Sec and SRP dependent manner (Koch et al., 2002). We cannot rule out the possibility that Alb3 alone integrates Alb3 and that cpSecY/E integrates cpSecY; however, it is clear from our results that cpTatC is not integrated by any of the known translocases, implying the existence of an uncharacterized plastid translocase. Although the translocase may be novel, it is more likely to be conserved because cpTatC is derived from the bacterial endosymbiont, which originally integrated this multi-spanning protein from the topologically equivalent (i.e. to stroma) cytoplasm.
Two new conserved candidate cpTatC integrases were recently discovered: Alb4 and SecY2 (Gerdes et al., 2006; Skalitzky, 2006). Alb4 is homologous to Alb3 and is located in the thylakoid membranes in chloroplasts. SecY2 possesses a predicted transit peptide, and when incubated with intact chloroplasts, is imported, processed to a smaller protein, and is recovered with the membrane fraction (unpublished results of the authors). The general importance of Alb4 is unclear as anti-sense RNA and T-DNA insertion lines failed to show a striking phenotype (Gerdes et al., 2006). On the other hand, Arabidopsis that are homozygous for knockout mutations in SecY2 are embryo lethal. In addition, promoter swapping experiments between cpSecY and SecY2 showed that these two proteins perform non-redundant functions (Skalitzky, 2006). Certainly this second plastid SecY is a candidate for insertion/translocation of proteins not accommodated by the known translocases. This arrangement is demonstrated by certain gram positive bacteria which possess an additional SecY that translocates a specific subset of proteins (Siboo et al., 2008). Considering the essential nature of the plastid SecY2, deciphering its function will require a conditional mutation or an inducible silencing approach, which we are now pursuing.
Experimental Procedures
Construction of Precursors
Sp6 transcription clones for mature size cpTatC (mcpTatC) and precursors to cpTatC (pcpTatC), the small subunit of RuBisCO (pSSU), atAlb4, atAlb3, psAlb3 (previously called cpOxa1p), and the 23 kD (pOE23) and 33 kD (pOE33) subunits to the oxygen-evolving complex were each previously described (Anderson and Smith, 1986; Cline et al., 1993; Moore et al., 2000; Mori et al., 2001; Gerdes et al., 2006). All novel restriction sites were inserted into constructs via Quickchange Mutagenesis (Stratagene). Reciprocal swaps between the transit peptides and mature domains of SSU and cpTatC were made as follows. MluI restriction sites were inserted twenty and eight amino acids after the stromal transit peptidase cleavage sites of pSSU and pcpTatC respectively, and the reciprocal chimeric precursors constructed by sub-cloning of restriction fragments (Figure S1b). In order to prepare cpTatC precursors lacking the non-conserved amino terminal domain, a BssHII restriction site was inserted into pcpTatC 47 residues after the stromal transit peptidase cleavage site. Precursors containing either the cpTatC or SSU transit peptide and cpTatC lacking the non-conserved amino terminus (ΔNCcpTatC) were prepared by an MluI, BssHII double restriction digest and subsequent ligation of fragments. The junction in SSUtpΔcpTatC consisted of amino acids 1–79 from pSSU and residues 198–353 from cpTatC. The TatCtpΔNCcpTatC fusion lacked pcpTatC residues 101–197.
Nucleotide sequence encoding the first 126 amino acids of Arabidopsis pcpTatC was amplified from a cDNA clone by a polymerase chain reaction using primers flanked with NdeI and BamHI restriction sites. The PCR product was sub-cloned in pET-14b (Novagen) to produce an N-terminal 6x histidine-tagged fusion protein (pET14b-atcpTatC).
Preparation of radiolabeled precursors
RNA transcripts were produced by transcription with SP6 polymerase (Promega) and translated with a homemade wheat germ translation system in the presence of 3[H]-leucine (Cline, 1986). Where indicated, in vitro-coupled transcription, translation with wheat germ TnT (Promega and NEN Life Science Products) was performed following the manufacture’s guidelines. Translation products were diluted with one volume of 60 mM leucine in 2X import buffer (IB, 1X = 50 mM HEPES/KOH, pH 8.0, 0.33 M sorbitol) prior to use unless otherwise indicated in the figure legend.
Preparation of bacterially expressed proteins and antibodies
pOE23 and pOE33 from pea were each expressed in E. coli, and purified as inclusion bodies as previously described (Cline et al., 1993). Inclusion bodies were dissolved in 10 M urea, 10 mM DTT before use in in organello competition experiments. The amino-terminal 126 amino acids from atpcpTatC were expressed in E. coli (strain BL21) from the pET-14b plasmid and purified as inclusion bodies as above. Inclusion bodies were dissolved in 6 M Urea and subjected to Ni-affinity purification according to manufacturers instructions (GE Healthcare), with the modification that all buffers contained 6 M Urea. Affinity-purified atcpTatC peptide was used for the production of antibodies in rabbits (Cocalico Biologicals). Antibodies to psAlb3 have been described (Mori et al., 2001). We found that the psAlb3 antibodies react with the atAlb4 protein but not the atAlb3 protein (Figure S5). psAlb3 antibodies were used to detect Alb4 in immunoblots (Figure 6). Antibodies to atcpSecY (Schuenemann et al., 1999), atAlb3 (Moore et al., 2000), and atAlb4 (Gerdes et al., 2006) have been described.
Preparation of chloroplasts, stromal extract, and total cell membranes
Peas (Pisum sativum L. cv. Laxton’s Progress 9 Improved) used for chloroplast isolation were grown as described (Cline, 1986). Intact chloroplasts were isolated from 9- to 10-day old pea seedlings and were resuspended in IB at 1 mg/mL of chlorophyll. For preparation of stromal extract, chloroplasts were lysed hypotonically by resuspension in 10 mM HEPES/KOH, pH 8.0 and incubation on ice for 10 minutes followed by centrifugation at 150,000 x g for 30 minutes at 2 C to pellet the membranes. Chlorophyll concentrations were determined according to Arnon (Arnon, 1949). Arabidopsis seed harboring a knockout mutation in the gene encoding Alb3 were obtained from the Arabidopsis Biological Resource Center (Stock# CS16) (Rhee et al., 2003). The cpSecY knockout mutant (Ws ecotype) contains a T-DNA insertion in the third exon and was previously designated scy1-2 (Skalitzky, 2006). Arabidopsis seeds were sterilized, plated to media, and grown for 2 or 4 weeks (20°C, 16 hour photoperiod, 100 μE/m2/s of light) before total membrane or intact chloroplast isolation (Smith et al., 2003). Media used to grow alb3 mutant seedlings contained 20 mg/L hygromycin. Pale alb3 or secy mutant seedlings were harvested from plates for total membrane isolation, using a method adapted from (Schaller et al., 1995). Equal masses of tissue from wildtype and mutant Arabidopsis seedlings were subjected to probe homogenization (PT10-35, Kinematica GmBH) in ice-cold membrane extraction buffer (30 mM Tris pH 8, 20% glycerol, 5 mM EDTA, 5mM EGTA, 1mM PMSF) and filtration through Miracloth (Calbiochem). Filtrate underwent centrifugation at 150,000 xg, for 30 minutes, at 2°C. Membrane pellets were resuspended in equal volumes of IB.
Chloroplast import and thylakoid protein integration assays
Radiolabeled precursor proteins were incubated with isolated chloroplasts (0.33 mg/mL chlorophyll), 5 mM MgATP, and IB, in 120 μE of light in a 25°C water bath for times specified in figure legends. After import, samples were treated with 100 μg/mL thermolysin on ice for 30 minutes. Proteolysis was stopped by addition of 0.5 M EDTA to a final concentration of 10 mM, and chloroplasts were re-isolated by centrifugation through 35% Percoll, 5 mM EDTA, in IB. Intact chloroplasts were washed, resuspended to equal concentrations of chlorophyll, then analyzed using SDS-PAGE. Alternatively, chloroplasts were lysed hypotonically by resuspension in 10 mM HEPES/KOH, pH 8.0 and incubation on ice for 10 minutes. Lysed chloroplasts were analyzed using SDS-PAGE and/or fractionated into membranes and stroma by differential centrifugation (12,000 xg, 10 minutes, 4°C). Membranes were resuspended in IB and analyzed using SDS-PAGE, and/or combined with 100 μg/mL thermolysin on ice for 30 minutes. Proteolysis was stopped by addition of 10 mM EDTA before analysis by SDS-PAGE and fluorography.
For import time course assays, reaction samples were rapidly stopped with 3.3 mM HgCl2 and analyzed as described (Reed et al., 1990). Chase time course assays were done similar to (Li and Schnell, 2006). Chloroplasts and MgATP were pre-incubated for 10 minutes, at 25°C, in 120 μE of light. SSUtpcpTatC precursor was then added and incubation continued for 5 minutes. Chloroplasts were diluted five-fold in ice-cold IB and treated with 200 μg/mL thermolysin on ice for 15 minutes to remove surface-bound precursor. Chloroplasts were then washed, resuspended to the original reaction volume in IB, 5 mM MgATP and incubated in the light at 25°C. At time points, samples of chloroplasts were taken for re-isolation by centrifugation through 35% Percoll cushions. Intact chloroplasts were lysed, quantified, adjusted to equal chlorophyll concentrations, and fractionated. Chloroplasts, fractions, and thermolysin-treated membranes were analyzed by SDS-PAGE and fluorography. Radiolabelled proteins were extracted from excised gel bands and quantified by scintillation counting (Cline, 1986).
Protease accessibility assay
Radiolabeled pcpTatC was imported into chloroplasts for 10 minutes. Aliquots of the import reaction were treated with 200 μg/mL trypsin (Sigma, 6300 BAEE units/mg), 100 μg/mL thermolysin, or IB for 30 minutes on ice (Li and Schnell, 2006). Thermolysin and trypsin were inhibited by a 10 minute incubation on ice in the presence of 10 mM EDTA or trypsin inhibitors (1 mM PMSF, 0.05 mg/mL TLCK, 0.1 mg/mL soybean trypsin inhibitor, and 2 μg/mL aprotinin). Chloroplasts were re-isolated, washed, quantified, adjusted to equal chlorophyll concentration, lysed, and fractionated in the presence of all respective protease inhibitors. Stroma and membrane fractions were analyzed using SDS-PAGE and fluorography as well as by immunoblotting with antibodies to TOC75 or TIC110.
In organello competition assay
In organello competition for the cpTat and cpSec pathways was conducted essentially as described (Cline et al., 1993). Briefly, unlabeled inclusion bodies of pOE23 or pOE33 were dissolved in 10 M urea, 10 mM DTT for 1 hr at 37 °C. Chloroplasts, 5 mM MgATP, and 1.5 mM DTT were then incubated with the unlabeled competitors for 7 minutes in light at 25 °C in order to accumulate the stromal intermediates iOE23 and iOE33, respectively. Competitors were aliquotted from stocks, such that the final competitor concentration was either 0.5 or 0.75 μM, or no competitor, and the urea concentration was 0.3 M in all assays. Radiolabeled precursors pcpTatC, pOE23, or pOE33 were then added (1/6 volume) and the incubation continued for an additional 15 minutes. Intact chloroplasts were recovered from assays by centrifugation through Percoll cushions. Recovered chloroplasts from the radiolabeled pOE23 and pOE33 assays were analyzed directly. Chloroplasts from cpTatC assays were lysed in 10 mM HEPES/KOH pH 8.0, 10 mM MgCl2 buffer, the membranes recovered by centrifugation at 3,200 x g for 8 minutes and the resulting supernatant centrifuged at 100,000 x g for 20 minutes to obtain stroma. An aliquot of the membrane fraction at ~1 mg Chl per mL IB was treated with 150 μg/mL thermolysin per mL for 40 minutes at 4 °C. Proteolysis was terminated with EDTA (10 mM final concentration), the membranes recovered by centrifugation and washed with IB, containing 14 mM EDTA.
Electrophoresis
cpTatC-containing samples destined for SDS-PAGE were incubated in sample buffer (0.1M Tris-HCl pH6.8, 8M urea, 5% SDS, 20% glycerol, 10% β-mercaptoethanol) for one hour at room temperature before electrophoresis, to prevent cpTatC aggregation. Samples destined for Blue Native PAGE were prepared as described (Gerard and Cline, 2007), except that 2% digitonin was included in solubilization buffers used to prepare Arabidopsis membranes. Gels were processed for fluorography or analyzed by immunoblotting (Cline, 1986; Cline and Mori, 2001). Immuno-labeled proteins were visualized by using the ECL method (Amersham Biosciences). Radiolabeled proteins were extracted from dried gel slices and quantified by scintillation counting (Cline, 1986). The number of molecules of mcpTatC, iOE23, or mOE23 were calculated from the dpm of an extracted band, the specific activity of the leucine used in the translations, the number of leucine residues per molecule, and the efficiency of radiolabeled leucine incorporation during precursor synthesis in vitro. Leucine residues for each molecule were derived from amino acid sequence data. Gel band extraction and quantitative immunoblotting were each used to quantify precursors from in vitro translation reactions (Fincher et al., 2003). Leucine incorporation efficiency was determined by comparing the amount of radiolabeled precursor to the amount of total precursor produced from an in vitro synthesis reaction. Chloroplasts per import assay sample were calculated from the chlorophyll concentration and the number of chloroplasts per microgram of chlorophyll, which was typically about 1 × 106.
Supplementary Material
Identification and modification of the cpTatC N-terminal stromal nonconserved domain. (a) The Pisum cpTatC precursor peptide sequence was used to BLAST all available plant gene indices at The Institute for Genomic Research (Quackenbush et al., 2001; Tsai et al., 2001; Pertea et al., 2003). Multalin was used to compare peptide sequences from the BLAST result (Corpet, 1988). The non-conserved region of Pisum cpTatC begins with amino acids ‘-CFAV-’ and ends with amino acids ‘–RSAI-.’ The remainder of the Pisum cpTatC peptide sequence, and that for each homolog, are omitted. (b) MluI and BssHII restriction sites were inserted in pSSU and pcpTatC. Restriction-based cloning was used to swap the transit peptides between each precursor and to delete the non-conserved stromal region from the SSUtpcpTatC precursor. Straight lines run adjacent the SSU (above text) and cpTatC (below text) transit peptides, and jagged lines run adjacent the cpTatC non-conserved region.
cpTatC targets to thylakoid membranes via a stromal intermediate. (a) Three import time course experiments were conducted (Experimental Proceedures), and radiolabeled cpTatC in each chloroplast fraction was quantified. Plotted are the percentages of stroma- and membrane-associated cpTatC relative to total imported cpTatC at each time point.
pcpTatC targeting is unaffected by protonophores. (a) Radiolabeled pOE23 or pcpTatC was incubated with chloroplasts, that were pretreated with (Nig/Val) or without (−) 0.5 μM nigericin and 1 μM valinomycin, in a protein import assay (Supplemental Procedures) for 20 minutes. Chloroplasts were treated with 100 μg/mL thermolysin and repurified. Chloroplasts from pcpTatC import reactions were lysed, and fractionated. Chloroplasts from pOE23 import reactions and total imported cpTatC (T), stroma (S), membrane (M), and thermolysin-treated membrane fractions (+T) were analyzed using SDS-PAGE and fluorography. Gel band extraction was used to quantify radiolabeled intermediate OE23 (iOE23), and mature OE23 (mOE23) and cpTatC (mcpTatC) in each chloroplast fraction for each treatment. Numbers below bands represent the relative percentages of cpTatC or OE23 found in each fraction. Translation products (Tp). The positions of molecular weight markers (Mr) are shown at left.
pcpTatC targeting is unaffected by azide. Radiolabeled pcpTatC, pOE33, or pOE23 was incubated with chloroplasts, that were pretreated without (−) or with 5 mM or 10 mM sodium azide, in a protein import assay (Supplemental Procedures) for 15 minutes. Chloroplasts from pcpTatC import reactions were repurified, lysed, and fractionated as described in Supplemental Procedures. Chloroplasts from pOE23 and pOE33 import reactions and total imported cpTatC (T), stroma (S), membrane (M), and thermolysin-treated membrane fractions (+T) were analyzed using SDS-PAGE and fluorography. Gel band extraction was used to quantify radiolabeled mature cpTatC (mcpTatC) in each chloroplast fraction for each treatment. Numbers below bands represent the relative percentages of cpTatC found in each fraction. Translation products (Tp).
Antibodies that detect psAlb3 protein, cross-react with atAlb4, but not atAlb3. (a) Radiolabeled precursors to atAlb3, atAlb4, and psAlb3 were translated in vitro and analyzed by SDS-PAGE and fluorography or immunoblotting with antibodies to psAlb3 (applied at 1:40000). The positions of molecular weight markers (Mr) are shown at left. (b) Isolated Arabidopsis thylakoids were analyzed by SDS-PAGE and immunoblotting with antibodies to psAlb3 (applied at 1:5000) and atAlb4 (applied at 1:5000). The positions of molecular weight markers (Mr) are shown between lanes.
Acknowledgments
We would like to thank Dr. Danja Schunemann for an SP6 transcription clone for precursor to Arabidopsis Alb4 and for antibodies to Arabidopsis Alb4. We thank Dr. Neil Hoffman and Dr. Ralph Henry for antibodies to Arabidopsis cpSecY and Alb3 and Ralph Henry for an SP6 transcription clone for the precursor to Arabidopsis Alb3. We also thank Dr. Danny Schnell for antibodies to TOC75 and TIC110 and the Arabidopsis Biological Resource Center for alb3 mutant seed. The authors appreciate critical review of the manuscript by Jose Celedon and Xianyue Ma. This work was supported in part by National Institutes of Health grant R01 GM46951 to K. Cline and by a NIH Biotechnology Training Grant to JH Harwood.
Footnotes
Arabidopsis seed stocks: CS16 (ABRC) and secy1-2
Additional supporting information may be found in the online version of this article: Figures S1 through S5 and associated experimental procedures and references.
References
- Anderson S, Smith SM. Synthesis of the small subunit of ribulose-bisphosphate carboxylase from genes cloned into plasmids containing the SP6 promoter. Biochem J. 1986;240:709–715. doi: 10.1042/bj2400709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson H, Heijne Gv. Sec dependent and sec independent assembly of E. coli inner membrane proteins: the topological rules depend on chain length. EMBO J. 1993;12:683–691. doi: 10.1002/j.1460-2075.1993.tb05702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiology. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Hirohashi T, Kikuchi S, Belcher S, Osborne E, Yano S, Terashima I, Barkan A, Nakai M. Maize Mutants Lacking Chloroplast FtsY Exhibit Pleiotropic Defects in the Biogenesis of Thylakoid Membranes. Plant Cell. 2004;16:201–214. doi: 10.1105/tpc.014787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Kikuchi S, Nakai M. Non-identical contributions of two membrane-bound cpSRP components, cpFtsY and Alb3, to thylakoid biogenesis. The Plant Journal. 2008 doi: 10.1111/j.1365-313X.2008.03659.x. [DOI] [PubMed] [Google Scholar]
- Benning C, Xu C, Awai K. Non-vesicular and vesicular lipid trafficking involving plastids. Current Opinion in Plant Biology. 2006;9:241–247. doi: 10.1016/j.pbi.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Bloois Ev, Haan GJ, Gier J-Wd, Oudega B, Luirink J. F1F0 ATP synthase subunit c is targeted by the SRP to YidC in the E. coli inner membrane. FEBS Letters. 2004;576:97–100. doi: 10.1016/j.febslet.2004.08.069. [DOI] [PubMed] [Google Scholar]
- Brink S, Fischer K, Klosgen RB, Flugge UI. Sorting of Nuclear-encoded Chloroplast Membrane Proteins to the Envelope and the Thylakoid Membrane. J Biol Chem. 1995;270:20808–20815. doi: 10.1074/jbc.270.35.20808. [DOI] [PubMed] [Google Scholar]
- Bruce BD. The paradox of plastid transit peptides: conservation of function despite divergence in primary structure. Biochemica et Biophysica Acta - Molecular Cell Research. 2001;1541:2–21. doi: 10.1016/s0167-4889(01)00149-5. [DOI] [PubMed] [Google Scholar]
- Buchanan G, Leeuw Ed, Stanley NR, Wexler M, Berks BC, Sargent F, Palmer T. Functional complexity of the twin-arginine translocase TatC component revealed by site-directed mutagenesis. Molecular Microbiology. 2002;43:1457–1470. doi: 10.1046/j.1365-2958.2002.02853.x. [DOI] [PubMed] [Google Scholar]
- Carde JP, Joyard J, Douce R. Electron Microscopic Studies of Envelope Membranes from Spinach Plastids. Biology of the Cell. 1982;44:315–324. [Google Scholar]
- Cavalier-Smith T. Membrane heredity and early chloroplast evolution. Trends in Plant Science. 2000;5:174–182. doi: 10.1016/s1360-1385(00)01598-3. [DOI] [PubMed] [Google Scholar]
- Cline K. Import of proteins into chloroplasts: membrane integration of a thylakoid precursor protein reconstituted in chloroplast lysates. Journal of Biological Chemistry. 1986;261:14804–14810. [PubMed] [Google Scholar]
- Cline K, Henry R, Li C-J, Yuan J. Pathways and Intermediates for the Biogenesis of Nuclear Encoded Thylakoid Proteins. Research in Photosynthesis. 1992;III:149–156. [Google Scholar]
- Cline K, Henry R, Li C, Yuan J. Multiple pathways for protein transport into or across the thylakoid membrane. The EMBO Journal. 1993;12:4105–4114. doi: 10.1002/j.1460-2075.1993.tb06094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Mori H. Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. Journal of Cell Biology. 2001;154:719–730. doi: 10.1083/jcb.200105149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Theg SM. The Sec and Tat Protein Translocation Pathways in Chloroplasts. In: Dalbey R, Koehler C, Tamanoi F, editors. The Enzymes. Vol. 25. Elsevier; 2007. pp. 1–25. [Google Scholar]
- Cran D, Possingham J. Two forms of division profile in spinach chloroplasts. Nature New Biology. 1972;235:142. [Google Scholar]
- Dalbey RE, Kuhn A. Evolutionarily Related Insertion Pathways of Bacterial, Mitochondrial, and Thylakoid Membrane Proteins. Annual Review of Cell and Developmental Biology. 2000;16:51–87. doi: 10.1146/annurev.cellbio.16.1.51. [DOI] [PubMed] [Google Scholar]
- Fandl JP, Cabelli R, Oliver D, Tai PC. SecA suppresses the temperature-sensitive SecY24 defect in protein translocation in Escherichia coli membrane vesicles. PNAS. 1988;85:8953–8957. doi: 10.1073/pnas.85.23.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher V, Dabney-Smith C, Cline K. Functional assembly of thylakoid deltapH-dependent/Tat protein transport pathway components in vitro. European Journal of Biochemistry. 2003;270:4930–4941. doi: 10.1046/j.1432-1033.2003.03894.x. [DOI] [PubMed] [Google Scholar]
- Gerard F, Cline K. The Thylakoid Proton Gradient Promotes an Advanced Stage of Signal Peptide Binding Deep within the Tat Pathway Receptor Complex. J Biol Chem. 2007;282:5263–5272. doi: 10.1074/jbc.M610337200. [DOI] [PubMed] [Google Scholar]
- Gerdes L, Bals T, Klostermann E, Karl M, Philippar K, Hunken M, Soll J, Schunemann D. A Second Thylakoid Membrane-associated Alb3/OxaI/YidC Homologue Is Involved in Proper Chloroplast Biogenesis in Arabidopsis thaliana. J Biol Chem. 2006;281:16632–16642. doi: 10.1074/jbc.M513623200. [DOI] [PubMed] [Google Scholar]
- Hutin C, Havaux M, Carde JP, Kloppstech K, Meiherhoff K, Hoffman N, Nussaume L. Double mutation cpSRP43/cpSRP54 is necessary to abolish the cpSRP pathway required for thylakoid targeting of the light-harvesting chlorophyll proteins. The Plant Journal. 2002;29:531–543. doi: 10.1046/j.0960-7412.2001.01211.x. [DOI] [PubMed] [Google Scholar]
- Inoue K, Keegstra K. A polyglycine stretch is necessary for proper targeting of the protein translocation channel precursor to the outer envelope membrane of chloroplasts. The Plant Journal. 2003;34:661–669. doi: 10.1046/j.1365-313x.2003.01755.x. [DOI] [PubMed] [Google Scholar]
- Jackson DT, Froehlich JE, Keegstra K. The Hydrophilic Domain of Tic110, an Inner Envelope Membrane Component of the Chloroplastic Protein Translocation Apparatus, Faces the Stromal Compartment. Journal of Biological Chemistry. 1998;273:16583–16588. doi: 10.1074/jbc.273.26.16583. [DOI] [PubMed] [Google Scholar]
- Knight JS, Gray JC. The N-Terminal Hydrophobic Region of the Mature Phosphate Translocator Is Sufficient for Targeting to the Chloroplast Inner Envelope Membrane. The Plant Cell. 1995;7:1421–1432. doi: 10.1105/tpc.7.9.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H. Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. PNAS. 2007;104:17216–17221. doi: 10.1073/pnas.0704680104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch HG, Moser M, Schimz KL, Mueller M. The Integration of YidC into the Cytoplasmic Membrane of Escherichia coli Requires the Signal Recognition Particle, SecA and SecYEG. Journal of Biological Chemistry. 2002;277:5715–5718. doi: 10.1074/jbc.C100683200. [DOI] [PubMed] [Google Scholar]
- Li M, Schnell DJ. Reconstitution of protein targeting to the inner envelope membrane of chloroplasts. Journal of Cell Biology. 2006;175:249–259. doi: 10.1083/jcb.200605162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M, Harrison MS, Peterson EC, Henry R. Chloroplast Oxa1p Homolog Albino3 Is Required for Post-translational Integration of the Light Harvesting Chlorophyll-binding Protein into Thylakoid Membranes. J Biol Chem. 2000;275:1529–1532. doi: 10.1074/jbc.275.3.1529. [DOI] [PubMed] [Google Scholar]
- Mori H, Summer EJ, Cline K. Chloroplast TatC plays a direct role in thylakoid ΔpH-dependent protein transport. FEBS Letters. 2001;501:65–68. doi: 10.1016/s0014-5793(01)02626-6. [DOI] [PubMed] [Google Scholar]
- Nilsson R, van Wijk KJv. Transient interaction of cpSRP54 with elongating nascent chains of the chloroplast-encoded D1 protein; ‘cpSRP54 caught in the act’. FEBS Letters. 2002;524:127–133. doi: 10.1016/s0014-5793(02)03016-8. [DOI] [PubMed] [Google Scholar]
- Reed JE, Cline K, Stephens LC, Bacot KO, Viitanen PV. Early events in the import/assembly pathway of an integral thylakoid protein. European Journal of Biochemistry. 1990;194:33–42. doi: 10.1111/j.1432-1033.1990.tb19423.x. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcie-Hernandez M, Huala E, Lander G, Montoya M, Miller N, Mueller LA, Mundodi S, Reiser L, Tacklind J, Weems DC, Wu Y, Xu I, Yoo D, Yoon J, Zhang P. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Lamppa GK. Stromal Processing Peptidase Binds Transit Peptides and Initiates Their ATP-dependent Turnover in Chloroplasts. Journal of Cell Biology. 1999;147:33–44. doi: 10.1083/jcb.147.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Ladd AN, Lanahan MB, Spanbauer JM, Bleeker AB. The Ethylene Response Mediator ETR1 from Arabidopsis Form a Disulfide-linked Dimer. J Biol Chem. 1995;270:12526–12530. doi: 10.1074/jbc.270.21.12526. [DOI] [PubMed] [Google Scholar]
- Schuenemann D, Amin P, Hartmann E, Hoffman NE. Chloroplast SecY Is Complexed to SecE and Involved in the Translocation of the 33-kDa but Not the 23-kDa Subunit of the Oxygen-evolving Complex. J Biol Chem. 1999;274:12177–12182. doi: 10.1074/jbc.274.17.12177. [DOI] [PubMed] [Google Scholar]
- Schuenemann D. Mechanisms of protein import into thylakoids of chloroplasts. Biol Chem. 2007;388:907–915. doi: 10.1515/BC.2007.111. [DOI] [PubMed] [Google Scholar]
- Scotti PA, Valent QA, Manting EH, Urbanus ML, Driessen AJM, Oudega B, Luirink J. SecA Is Not Required for Signal Recognition Particle-mediated Targeting and Initial Membrane Insertion of a Nascent Inner Membrane Protein. J Biol Chem. 1999;274:29883–29888. doi: 10.1074/jbc.274.42.29883. [DOI] [PubMed] [Google Scholar]
- Siboo IR, Chaffin DO, Rubens CE, Sullam PM. Characterization of the Accessory Sec System of Staphylococcus aureus. Journal of Bacteriology. 2008;190:6188–6196. doi: 10.1128/JB.00300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg C, Bauer MF, Guiard B, Neupert W, Brunner M. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature. 1996;384:582–585. doi: 10.1038/384582a0. [DOI] [PubMed] [Google Scholar]
- Skalitzky CA. Botany. Madison: University of Wisconsin. Master of Science; 2006. Genetic Analysis of SCY1 and SCY2 Function in Arabidopsis thaliana; p. 62. [Google Scholar]
- Smith MD, Fitzpatrick L, Keegstra K, Schnell DJ. Vitro Analysis of Chloroplast Protein Import. Current Protocols in Cell Biology. 2003:16.11–16.21. doi: 10.1002/0471143030.cb1116s17. [DOI] [PubMed] [Google Scholar]
- Steiner JM, Kocher T, Nagy C, Loffelhardt W. Chloroplast SecE: evidence for spontaneous insertion into the thylakoid membrane. Biochemical and Biophysical Research Communications. 2002;293:747–752. doi: 10.1016/S0006-291X(02)00285-1. [DOI] [PubMed] [Google Scholar]
- Sundberg E, Slagter J, Fridborg I, Cleary S, Robinson C, Coupland G. ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell. 1997;9:717–730. doi: 10.1105/tpc.9.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent QA, Scotti PA, High S, Gier J-WLd, Heijne Gv, Lentzen G, Wintermeyer W, Oudega B, Luirink J. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R, Mendel-Hartvig J, Barkan A. Transposon-Disruption of a Maize Nuclear Gene, tha1, Encoding a Chloroplast SecA Homologue: In Vivo Role of cp-SecA in Thylakoid Protein Targeting. Genetics. 1997;145:467–478. doi: 10.1093/genetics/145.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L, Jiang F, Chen M, Cain B, Bolhuis A, Dalbey RE. YidC Is Strictly Required for Membrane Insertion of Subunits a and c of the F1F0ATP Synthase and SecE of the SecYEG Translocase. Biochemistry. 2003;42:10537–10544. doi: 10.1021/bi034309h. [DOI] [PubMed] [Google Scholar]
- Zhang L, Paakkarinen V, Suorsa M, Aro EM. A SecY Homologue Is Involved in Chloroplast-encoded D1 Protein Biogenesis. J Biol Chem. 2001;276:37809–37814. doi: 10.1074/jbc.M105522200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification and modification of the cpTatC N-terminal stromal nonconserved domain. (a) The Pisum cpTatC precursor peptide sequence was used to BLAST all available plant gene indices at The Institute for Genomic Research (Quackenbush et al., 2001; Tsai et al., 2001; Pertea et al., 2003). Multalin was used to compare peptide sequences from the BLAST result (Corpet, 1988). The non-conserved region of Pisum cpTatC begins with amino acids ‘-CFAV-’ and ends with amino acids ‘–RSAI-.’ The remainder of the Pisum cpTatC peptide sequence, and that for each homolog, are omitted. (b) MluI and BssHII restriction sites were inserted in pSSU and pcpTatC. Restriction-based cloning was used to swap the transit peptides between each precursor and to delete the non-conserved stromal region from the SSUtpcpTatC precursor. Straight lines run adjacent the SSU (above text) and cpTatC (below text) transit peptides, and jagged lines run adjacent the cpTatC non-conserved region.
cpTatC targets to thylakoid membranes via a stromal intermediate. (a) Three import time course experiments were conducted (Experimental Proceedures), and radiolabeled cpTatC in each chloroplast fraction was quantified. Plotted are the percentages of stroma- and membrane-associated cpTatC relative to total imported cpTatC at each time point.
pcpTatC targeting is unaffected by protonophores. (a) Radiolabeled pOE23 or pcpTatC was incubated with chloroplasts, that were pretreated with (Nig/Val) or without (−) 0.5 μM nigericin and 1 μM valinomycin, in a protein import assay (Supplemental Procedures) for 20 minutes. Chloroplasts were treated with 100 μg/mL thermolysin and repurified. Chloroplasts from pcpTatC import reactions were lysed, and fractionated. Chloroplasts from pOE23 import reactions and total imported cpTatC (T), stroma (S), membrane (M), and thermolysin-treated membrane fractions (+T) were analyzed using SDS-PAGE and fluorography. Gel band extraction was used to quantify radiolabeled intermediate OE23 (iOE23), and mature OE23 (mOE23) and cpTatC (mcpTatC) in each chloroplast fraction for each treatment. Numbers below bands represent the relative percentages of cpTatC or OE23 found in each fraction. Translation products (Tp). The positions of molecular weight markers (Mr) are shown at left.
pcpTatC targeting is unaffected by azide. Radiolabeled pcpTatC, pOE33, or pOE23 was incubated with chloroplasts, that were pretreated without (−) or with 5 mM or 10 mM sodium azide, in a protein import assay (Supplemental Procedures) for 15 minutes. Chloroplasts from pcpTatC import reactions were repurified, lysed, and fractionated as described in Supplemental Procedures. Chloroplasts from pOE23 and pOE33 import reactions and total imported cpTatC (T), stroma (S), membrane (M), and thermolysin-treated membrane fractions (+T) were analyzed using SDS-PAGE and fluorography. Gel band extraction was used to quantify radiolabeled mature cpTatC (mcpTatC) in each chloroplast fraction for each treatment. Numbers below bands represent the relative percentages of cpTatC found in each fraction. Translation products (Tp).
Antibodies that detect psAlb3 protein, cross-react with atAlb4, but not atAlb3. (a) Radiolabeled precursors to atAlb3, atAlb4, and psAlb3 were translated in vitro and analyzed by SDS-PAGE and fluorography or immunoblotting with antibodies to psAlb3 (applied at 1:40000). The positions of molecular weight markers (Mr) are shown at left. (b) Isolated Arabidopsis thylakoids were analyzed by SDS-PAGE and immunoblotting with antibodies to psAlb3 (applied at 1:5000) and atAlb4 (applied at 1:5000). The positions of molecular weight markers (Mr) are shown between lanes.