Abstract
Clostridium difficile is a major and growing problem as a hospital-associated infection that can cause severe, recurrent diarrhea. The mechanism by which the bacterium colonizes the gut during infection is poorly understood but undoubtedly involves protein components within the surface layer (S-layer), which play a role in adhesion. In C. difficile, the S-layer is composed of two principal components, the high and low molecular weight S-layer proteins, which are formed from the post-translational cleavage of a single precursor, SlpA. In the present study, we demonstrate that a recently characterized cysteine protease, Cwp84 plays a role in maturation of SlpA. Using a gene knock-out approach, we show that inactivation of the Cwp84 gene in C. difficile 630ΔErm results in a bacterial phenotype in which only immature, single chain SlpA comprises the S-layer. The Cwp84 knock-out mutants (CDΔCwp84) displayed significantly different colony morphology compared with the wild-type strain and grew more slowly in liquid medium. SlpA extracted from CDΔCwp84 was readily cleaved into its mature subunits by trypsin treatment. Addition of trypsin to the growth medium also cleaved SlpA on CDΔCwp84 and increased the growth rate of the bacterium in a dose-dependent manner. Using the hamster model for C. difficile infection, CDΔCwp84 was found to be competent at causing disease with a similar pathology to the wild-type strain. The data show that whereas Cwp84 plays a role in the cleavage of SlpA, it is not an essential virulence factor and that bacteria expressing immature SlpA are able to cause disease.
Introduction
Clostridium difficile, a Gram-positive, anaerobic bacterium is a frequent cause of hospital-associated infections (1). C. difficile infection (CDI)2 is primarily associated with the use of broad-spectrum antibiotics that suppress the normal gut flora allowing the bacterium to colonize the large intestine. Symptoms of CDI range from mild, self-limiting diarrhea to life-threatening colitis (1). Cases of CDI have risen significantly over the past 10 years, which is caused in part to the appearance of more virulent strains of the bacterium (2). In the United Kingdom, there were more than 50,000 cases in 2007 with over 8,000 associated deaths. In the United States, the mortality rate of CDI has increased from 5.7 per million in 1999 to 23.7 per million in 2004, and it has been described as the silent epidemic causing more deaths than all other intestinal diseases combined (3).
Two large protein cytotoxins, Toxins A and B, are recognized as the principal virulence factors of the bacterium that cause extensive damage to the gut wall during infection (4). These toxins mediate their action via disruption of the cell cytoskeleton by the glucosylation of the Rho family of GTPases. Recently, it has been shown that these toxins contain a cysteine protease domain, which appears to play a role in releasing the glucosylation domain to mediate its disruptive activity within the cell (5). Whereas the protein toxins of C. difficile undoubtedly play a key role in the progression of CDI, the mechanism by which C. difficile establishes and maintains its niche within the gut during infection is poorly understood. As has been found for other Gram-positive bacteria, it almost certainly involves the complex orchestration of a range of virulence factors, which may include toxins, enzymes, adhesins, and contributions from flagellae and fimbriae (6). Previous studies have begun to identify potential candidates that may play a role in C. difficile gut colonization, but the key determinants of this aspect of pathogenesis are still unclear (7–10). Many of these components are likely to reside among the complex array of proteins that make up the surface layer of the bacterium.
The outermost layer of C. difficile consists of a paracrystalline S-layer with a regularly arranged lattice structure (11, 12). C. difficile is unusual in that the S-layer components: the high molecular weight and low molecular weight S-layer proteins (HMW SLP and LMW SLP) are encoded by a single gene (slpA), which is post-translationally cleaved into its component SLPs (13). These have been found to vary in both size and amino acid sequence among strains (13, 14). Recent structural studies suggest that the HMW and LMW SLPs form an elongated complex in which the two polypeptides are arranged end-to-end (15). Distributed within the S-layer are various other surface proteins, which are attached to the cell envelope in a variety of ways including both covalent and noncovalent modes of attachment (6). Analysis of the C. difficile genome reveals the presence of at least 28 SLP paralogs containing a similar cell binding motif (Pfam PF04122), which may be displayed on the bacterial cell surface among the major S-layer proteins (16).
These, although minor components, are likely to play key roles in pathogenesis such as cell signaling, matrix degradation, and adherence. Studies on the 66-kDa Cwp66 suggest a role in the adhesion of C. difficile to epithelial cells (9). Cwp84 is a C. difficile surface protein that has recently been shown to possess cysteine protease activity and which may have a role in the degradation of extracellular matrices (7). This enzyme, which appears to be displayed on the bacterial cell surface, exhibited degrading activity against fibronectin, laminin, and vitronectin. That these and other minor putative surface components are expressed on the bacterial cell surface during human CDI has been confirmed by the presence of antibodies in patient sera (17).
Recently, targeted gene knock-out systems for the genus Clostridium have been successfully developed, which, in combination with in vivo models for CDI, provide a powerful set of tools to study the pathogenesis of the disease (18, 19). Preparation of Toxin A and B knock-out mutants has demonstrated the importance of Toxin B in the pathogenesis of CDI (20). The ClosTron gene knock-out system, which utilizes a retargeted mobile group II intron from the ltrB gene of Lactococcus lactis (Ll.LtrB), has been shown to provide stable, reproducible insertional inactivation of specific genes (19). In the present study, we used the ClosTron system to further understand the role of the cysteine protease Cwp84 in the pathogenesis of C. difficile. We demonstrate that this enzyme plays a key role in the maturation of the SlpA precursor protein into its component HMW and LMW SLPs. We have also characterized the mutant bacterium and assessed its competence to cause CDI.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Their Growth
Escherichia coli TOP10 (Invitrogen) was used for cloning and plasmid propagation and E. coli CA434 as the conjugative donor strain employed in the creation of C. difficile mutants (21). These strains were cultured aerobically at 37 °C in L broth or L agar (BioMerieux), containing chloramphenicol (25 μg/ml). The C. difficile 630ΔErm strain (22) and C. difficile knock-out mutants were grown at 37 °C in an atmosphere of 10% H2, 10% CO2, and 80% N2 in a Don Whitley MG1000 anaerobic workstation. Working stocks of C. difficile 630ΔErm and derived mutants were maintained on Anaerobic Agar with Horse Blood (AAB; Oxoid) and sBHI (23) + 5 μg/ml erythromycin, respectively. Fecal samples from euthanized animals were taken by colectomy and C. difficile cultured using alcohol shock and plated onto C. difficile selective medium (E&O Labs) and grown as above.
Gene Knock-out and Characterization of Mutants
The ClosTron gene knock-out system developed by Heap et al. (19) was used for insertional knock-out of cwp84 (CD2787). The algorithm available on the Targetron Design Site was used to identify intron insertion sites within cwp84, one of which, 347 bp from the start codon, ultimately generated mutant designated CDΔCwp84347. The PCR primers, designed by the algorithm (Table 1) were synthesized (Eurofins MWG Operon) and used to assemble a 353-bp product by splicing overlap extension PCR, which would facilitate intron retargeting. The PCR product was excised from a 1% agarose gel and purified using the QIAquick gel extraction kit (Qiagen), then cloned into pMTL007-EC2 via HindIII and BsrGI, and the constructs transformed into E. coli TOP10. Sequence confirmation of purified plasmid constructs was performed using pMTL007-specific primers CspFdx-F1 and pMTL007-R1 (supplemental Table S1). The derivative pMTL007 plasmid construct was transformed, via electroporation, into the conjugative donor E. coli CA434 and then transferred via conjugation into the C. difficile 630ΔErm (24). Successful transconjugants and subsequent integrants were selected for by streaking onto C. difficile selective medium (E&O Labs) with the addition of thiamphenicol (15 μg/ml) and then on sBHI agar containing erythromycin (5 μg/ml), respectively. PCR was performed using primers flanking cwp84 (Table 1) to demonstrate the integration of the Ll.LtrB intron. PCR using the Erm-RAM primers (supplemental Table S1) confirmed the Ermr phenotype was caused by the splicing of the group I intron from the group II intron following integration and not a spontaneous Ermr mutant.
TABLE 1.
SOE and other PCR primers for mutant construction and analysis
| IBS | EBS1D | EBS2 | |
|---|---|---|---|
| cwp84–347 348a | AAAAAAGCTTATAATTATCCTTAGAAAACGACCAGGTGCGCCCAGATAGGGTG | CAGATTGTACAAATGTGGTGATAACAGATAAGTCGACCAGCATAACTTACCTTTCTTTGT | TGAACGCAAGTTTCTAATTTCGATTTTTTCTCGATAGAGGAAAGTGTCT |
| EBS Universal: | CGAAATTAGAAACTTGCGTTCAGTAAAC | ||
| Cwp84_F | TGAGCTAGCGCAGAAAACCATAAAACTCTAGATG | ||
| Cwp84_R | ATACTCTTTTAGGAAAATAGGAATTCAC | ||
| CD1751_F | GTGAAAAAATTTACTTC | ||
| CD1751_R | CTATTTAGCACTTTTTA |
Two additional cwp84 knock-out mutants were also constructed using the above protocol. For these, intron insertions were made at 677 and 2054 bp in cwp84, and the resulting mutants designated CDΔCwp84677 and CDΔCwp842054, respectively. A knock-out mutant of the Toxin A gene was also constructed, for control purposes, in which the intron was inserted at position 980 of the Toxin A gene (tcdA) to give the mutant CDΔToxinA980. Sequences of PCR primers used for construction of these mutants are provided in supplemental Table S1.
Analysis of the CD1751 Gene
Genomic DNA was extracted from stationary phase C. difficile 630ΔErm and CDΔCwp84347 using Qiagen DNeasy Blood and Tissue kit. PCR amplification of full-length CD1751 was performed using the primers shown in Table 1.
Cwp84 mRNA
Total RNA was extracted with Qiagen RNeasy mini kit as per manufacturer's instructions from stationary phase C. difficile 630ΔErm and CDΔCwp84347. To isolate mRNA and remove residual genomic DNA contamination, the total RNA was treated with mRNA-ONLY™ Prokaryotic mRNA Isolation kit (Epicenter Biotech) then TURBO DNA- free™ kit (Ambion). Reverse transcription PCR (RT-PCR) using primers amplifying cwp84 either pre-intron insertion site or flanking the intron insertion site (supplemental Table S1) was performed using SuperScript III One-Step RT-PCR System with Platinum® TaqDNA (Invitrogen).
Extraction and Characterization of SLPs
SLP extraction was essentially as described (10, 13) using low pH glycine extraction. Briefly, the harvested cell pellets were washed once with 10 mm HEPES containing 100 mm NaCl, pH 7.4, resuspended in 0.2 m glycine pH 2.2 and incubated for 30 min at 22 °C. Cells were then removed by centrifugation, and the supernatant neutralized with 2 m Tris-HCl, pH 9.0. Extracted SLPs were dialyzed into 50 mm HEPES containing 0.15 m NaCl, pH 7.4 and stored at −80 °C if not used directly.
Mass Spectrometry and Data Processing
These analyses were conducted at the Centre for Proteomic Research, University of Southampton. Excised protein bands were subjected to in situ trypsin digestion using previously described methods (25). The resulting peptides were separated by nano-reverse phase liquid chromatography, using a Dionex C18 PepMap, 3 μm, 100 Å (150 mm × 75 μm, i.d.) column, and electrosprayed into a quadrupole time-of-flight tandem mass spectrometer. All data were acquired using a Q-tof Global Ultima (Waters Ltd) fitted with a nanoLockSpray™ source. A survey scan was acquired from m/z 375 to 1800 with the switching criteria for MS to MS/MS including ion intensity and charge state. The collision energy used to perform MS/MS was varied according to the mass and charge state of the eluting peptide. All MS/MS spectra were automatically processed using MASCOT distiller (Matrix Science) and subsequently searched against a FASTA-formatted listing of protein sequences predicted from the C. difficile 630 genome sequences using the search algorithm MASCOT ver2.2. The following parameters were used: parent mass tolerance was 150 ppm, fragment mass tolerance 0.25 Da, carbamidomethylation was set as a fixed modification, the oxidation of methionine as a variable modification and a maximum of one missed cleavage was allowed. The significance threshold for search results was set at p < 0.05, which indicates identity or extensive homology.
Preparation of C. difficile Spores
C. difficile 630ΔErm and CDΔCwp84347 were cultured in sBHI for a total of 18 days. Samples were taken at days 10 and 18 to quantitate sporulation efficiency. A portion (300 μl) of 48 h C. difficile starter culture was spread onto AAB plates and incubated at 37 °C for 10 days. After incubation, the bacterial lawn was scraped off and transferred to 15 ml of sterile DMEM and centrifuged for 20 min at 2000 × g. The resulting pellet was washed twice by resuspension into 12 ml of DMEM followed by centrifugation and then resuspended into DMEM and heat shocked 62 °C for 40 min. The resulting crude spore suspension was aliquoted and stored at −80 °C.
Animal Model for C. difficile Infection
Syrian hamsters (80–100 g) were housed (2 per cage) in isolator cages fitted with air filters on their lids to minimize contamination between groups. Groups of 14 hamsters were divided into a test subgroup of 10 and control subgroup of 4 animals. All 14 hamsters were weighed and administered clindamycin (2 mg in 0.2 ml of sterile H2O) by the orogastric route on day 0. On day 2, hamsters in the test subgroup were challenged with 200 colony-forming units of C. difficile spores in 0.2 ml of DMEM, given orogastically. Control subgroup animals received just DMEM. All animals were weighed daily and monitored 6 times/day for 12 days for disease symptoms that included diarrhea, weight loss, lethargy, and tender abdomen (26, 27). Hamsters were scored on a 0–3 scale for disease symptoms, and animals in advanced stages of disease that had become immobile were euthanized. Fecal samples of euthanized animals were taken by colectomy, and C. difficile cultured as described above.
RESULTS
Production of CDΔCwp84347
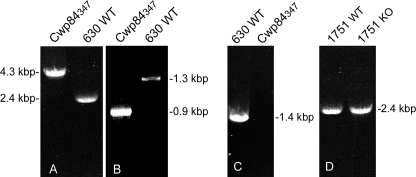
Of 28 potential sites for Ll.LtrB intron integration into cwp84, an insertion site between base pairs 347 and 348, which is close to the putative active site region of the Cwp84 cysteine protease (8), was chosen for the initial investigation. C. difficile 630ΔErm, transconjugants with the inserted intron were derived by selection for erythromycin resistance. PCR, using primers flanking cwp84, was used to confirm intron insertion in the Erm-resistant C. difficile clone gDNA. Fig. 1 shows amplification of the cwp84 gene from C. difficile 630ΔErm and CDΔCwp84347 yielded PCR products consistent with correct insertion of the intron. In addition to Erm resistance being demonstrated phenotypically, confirmatory PCR of the ErmRAM region was also performed (Fig. 1). This integration was also found to be stable through several passages of the bacterium. Transcriptional analysis of the cwp84 gene using RT-PCR showed expression of the transcript in stationary phase cells from wild-type C. difficile 630ΔErm and complete absence of a similar size product from CDΔCwp84347. Analysis of the CD1751 gene was also undertaken, because this shows significant homology (70.4%) to the cwp84 gene and encodes another putative C. difficile surface-associated cysteine protease. PCR analysis of CD1751 in CDΔCwp84347 showed this gene to be unmodified by the gene knock-out procedure (Fig. 1).
FIGURE 1.
A, confirmation of intron integration in cwp84 gene by PCR. Lane 1, 4.3-kbp product from CDΔCwp84347. Lane 2, 2.4-kbp product from C. difficile 630ΔErm. B, demonstration of erythromycin retrotransposition-activated marker (Erm-RAM) self-splicing, an event strictly coupled to group II intron integration. Lane 1, 900-bp product from CDΔCwp84347. Lane 2, 1300-bp product from native pMTL007-EC2 shuttle vector. C, RT-PCR of cwp84 mRNA flanking intron integration site in stationary phase cultures. Lane 1, C. difficile 630ΔErm, RT+; lane 2, CDΔCwp84347, RT+. D, PCR confirmation that CD1751 (a cysteine protease with homology to Cwp84) is unaffected by the knock-out of cwp84. Lane 1, C. difficile 630ΔErm; lane 2, CDΔCwp84347.
Morphology and Growth Characteristics of CDΔCwp84347
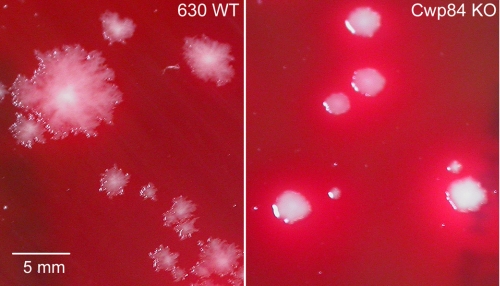
Grown on non-selective solid medium, CDΔCwp84347 showed significantly different colony morphology compared with wild-type C. difficile 630ΔErm. Whereas the latter showed a characteristic irregular, translucent appearance, CDΔCwp84347 formed more regular circular colonies, which were creamy white in appearance (Fig. 2). In liquid medium, CDΔCwp84347 grew more slowly than wild-type C. difficile 630ΔErm, and at higher culture ODs had a propensity to aggregate and form a precipitate. The slower growth of CDΔCwp84347 was not a general consequence of the knock-out process as a mutant strain containing a tcdA (Toxin A) knock-out grew at a similar rate to the wild-type strain (Table 2). CDΔCwp84347 retained its ability to sporulate, albeit at reduced efficiency compared with the wild type. The level of sporulation, estimated from viable counts before and after heat-shock treatment, was determined to be 0.64 ± 0.03% for 18-day cultures of CDΔCwp84347 compared with 7.18 ± 3.45% for wild-type C. difficile 630ΔErm. Colonies produced from CDΔCwp84347 spore preparations were shown to retain the intron insertion within the cwp84 gene, providing further evidence of the stability of the mutation.
FIGURE 2.
Difference in colony morphology of wild-type C. difficile 630ΔErm and CDΔCwp84347 grown on solid phase medium.
TABLE 2.
Growth rates of C. difficile mutants
Growth rates were measured on the linear portion of the growth curve (n = 3).
| C. difficile strain | Growth rate | WT |
|---|---|---|
| A600 h−1 | % | |
| C. difficile 630ΔErm | 0.29 ± 0.011 | 100 |
| CDΔCwp84347 | 0.12 ± 0.007 | 42 |
| CDΔToxinA980 | 0.26 ± 0.018 | 90 |
| C. difficile 630ΔErm + trypsina | 0.30 ± 0.002 | 103 |
| CDΔCwp84347 + trypsin (μg/ml) | ||
| 0 | 0.12 ± 0.002 | 42 |
| 1 | 0.13 ± 0.008 | 43 |
| 3 | 0.16 ± 0.003 | 55 |
| 10 | 0.17 ± 0.003 | 58 |
| 100 | 0.21 ± 0.018 | 71 |
a Trypsin added to growth medium at 100 μg/ml.
Analysis of Surface Layer Proteins
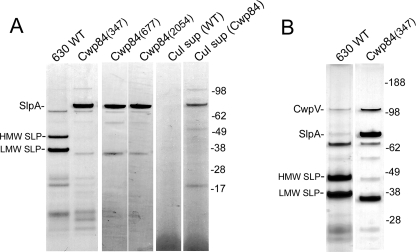
Previous studies have demonstrated that the major SLPs of C. difficile can be conveniently extracted by several methods including low pH buffer washing (10, 13). SDS-PAGE of SLPs extracted from wild-type C. difficile 630ΔErm by this method showed a characteristic band pattern (12, 13) in which the two principal SLPs, derived from the post-translational cleavage of the single precursor, SlpA, appeared as bands of ∼37 and 48 kDa, corresponding to the LMW SLP and HMW SLP, respectively (Fig. 3A). In contrast, these bands were absent in SLP extracts of CDΔCwp84347 in which a new prominent band of ∼84 kDa was evident by SDS-PAGE analysis. This band was subjected to shotgun proteomic analysis (GeLC-MS/MS) using in-gel tryptic digestion. Mascot individual ion scores of >47 for 13 individual peptides positively identified the 84-kDa band as the SlpA precursor protein, which has a predicted molecular mass of 76 kDa (supplemental Fig. S1). N-terminal sequence analysis of the SlpA precursor protein band gave the amino acid sequence ATTGT, which is identical to that of the mature LMW SLP and demonstrated that the signal peptide sequence had been removed from the precursor protein. These data show that in the CDΔCwp84347 mutant, the maturation of the SlpA precursor protein is incomplete, in that following removal of the signal peptide, cleavage into the HMW and LMW SLPs fails to occur. Another characteristic of CDΔCwp84347 is that the SlpA precursor protein was evident in the growth medium in 24-h cultures (Fig. 3A) unlike wild-type C. difficile 630ΔErm, where there was no evidence of any SLPs being released under similar growth conditions.
FIGURE 3.
A, analysis of extracted SLPs from wild-type C. difficile 630ΔErm, CDΔCwp84347, CDΔCwp84677, and CDΔCwp842054. Lanes 5 and 6, 24-h culture supernatant fluids from C. difficile 630ΔErm (Cul sup WT), CDΔCwp84347 (Cul sup Cwp84), respectively. B, analysis of extracted SLPs by silver-stained SDS-PAGE from wild-type C. difficile 630ΔErm and CDΔCwp84347.
Additional Cwp84 Gene Knock-out Mutants
To demonstrate conclusively that knock-out of the Cwp84 gene resulted in a bacterial phenotype expressing the single chain SlpA precursor, two further cwp84 mutants, CDΔCwp84677 and CDΔCwp842054, were constructed using different intron insertion sites at 677 and 2054 bp, respectively. These mutants were characterized in a similar manner to CDΔCwp84347, which confirmed both insertion of the intron and absence of Cwp84 mRNA product. For both mutants, analysis of the extracted SLPs showed the presence of the single chain SlpA precursor and complete absence of the HMW and LMW SLP bands (Fig. 3A).
Effect of cwp84 Knock-out on the Maturation of Surface Protein CwpV
In C. difficile 630, the cwpV gene (CD0514) encodes a predicted surface protein of 167 kDa. Recently, it has been shown by analysis of surface protein extracts that CwpV is processed into two fragments: a smaller fragment of ∼40 kDa derived from the N-terminal region and a larger fragment of ∼120 kDa derived from its C-terminal region (28). Silver-stained SDS-PAGE of SLPs extracted from wild-type C. difficile 630ΔErm and CDΔCwp84347 both showed the presence of a band of ∼115 kDa (Fig. 3B). Proteomic analysis of the 115-kDa band from CDΔCwp84347 identified this band as the 120-kDa C-terminal fragment of CwpV (supplemental Fig. S2). These data suggest that the maturation of CwpV is not affected by knock-out of cwp84.
Effect of Trypsin Treatment on the SlpA Precursor
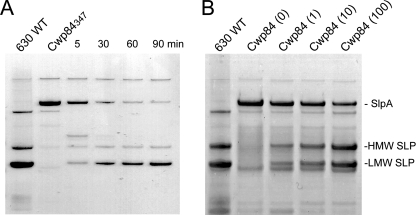
Previous studies have demonstrated that maturation of the SlpA precursor involves cleavage at the Ser-345—Ala-346 bond within the sequence: ETKSANDT. Because there is a lysine residue close to the natural site of cleavage within the SlpA precursor, we assessed the ability of trypsin to generate analogs of the LMW and HMW SLPs. In initial experiments, the effect of trypsin on SLPs extracted from CDΔCwp84347 by low pH treatment was assessed. Treatment with a low concentration of trypsin (1 μg/ml) resulted in the disappearance of the 84-kDa band and concomitant appearance of two bands of similar size to the HMW and LMW SLP bands from wild-type C. difficile 630ΔErm (Fig. 4). GeLC-MS/MS analysis confirmed the identity of the 48- and 37-kDa bands as the HMW and LMW SLPs, respectively (data not shown). In addition, N-terminal sequencing of the 48-kDa band gave the expected sequence for the trypsin-derived product (supplemental Fig. S1). Similar results were obtained when higher trypsin concentrations (5 and 10 μg/ml) were used. The LMW SLP appeared relatively resistant to trypsin digestion, whereas the HMW SLP appeared to be slowly degraded (Fig. 4A).
FIGURE 4.
A, cleavage of extracted SlpA with trypsin. Cleavage of SLP extracts from CDΔcwp84347 with 1 μg/ml trypsin (DPCC-treated, bovine pancreas; Sigma T1005) in 50 mm HEPES pH 7.4 containing 0.15 m NaCl at 22 °C for various times. B, extracted SLPs from 24-h cultures of CDΔcwp84347 grown in sBHI supplemented with 0, 1, 10, and 100 μg/ml trypsin. Prior to low pH extraction of SLPs, cultures were washed with 10 mm HEPES containing 100 mm NaCl, pH 7.4, and 0.5 mg/ml trypsin inhibitor and then extracted in 0.2 m glycine, pH 2.2, buffer containing 0.1 mg/ml trypsin inhibitor.
Effect of Trypsin Treatment on the Growth and SLPs of CDΔCwp84347
Because the extracted SlpA precursor protein is rapidly cleaved in solution by trypsin, we assessed whether or not the protein displayed on the surface of the bacterium was assessable to cleavage. CDΔCwp84347 was grown in medium supplemented with various concentrations of trypsin followed by washing with medium containing trypsin inhibitor. Fig. 4B shows SLPs extracted from cells treated in this manner compared with the SLP extracts from the untreated wild type and CDΔCwp84347 cultures. N-terminal sequences (SANDT and ATTGT) of the prominent bands, which appeared as a result of trypsin treatment, confirmed their identity as the HMW and LMW SLPs, respectively. The data show that >50% of the SlpA precursor protein is cleaved on cells grown in the presence of the highest trypsin concentration used. Compared with trypsin treatment of the extracted SlpA, the HMW SLP appeared more resistant to trypsin degradation, which may reflect its proposed orientation within the S-layer in which the HMW SLP is the lesser exposed of the two subunits (15). Addition of trypsin to the CDΔCwp84347 growth medium significantly increased its growth rate in a dose-dependent manner (Table 2), consistent with the cleavage of SlpA (Fig. 4B). The presence of trypsin in the growth medium also eliminated the propensity of CDΔCwp84347 to aggregate, which provides further evidence that the SlpA precursor protein making up the S-layer is accessible to the protease in the medium.
Ability of CDΔCwp84347 to Cause Infection
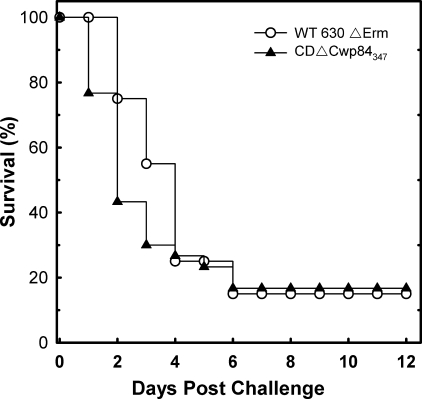
To assess the capacity of CDΔCwp84347 to cause CDI, the hamster model was used, as it is widely recognized as the best model currently available (26). This disease model parallels most of the recognized characteristics of the human disease, and hamsters can readily be made susceptible to CDI by treatment with a broad spectrum antibiotic such as clindamycin. After administration of clindamycin, test groups were challenged 48-h later by orogastric administration of 2 × 102 spore-forming units produced from either CDΔCwp84347 or wild-type C. difficile 630ΔErm. Of 30 hamsters (3 test groups of 10) challenged with CDΔCwp84347, 83.3% succumbed to severe disease within 6-days postchallenge compared with 85% challenged with C. difficile 630ΔErm (20 hamsters, 2 test groups of 10). Both strains displayed typical symptoms of CDI (diarrhea, weight loss, lethargy). Fig. 5 shows combined survival plots for these data, which were analyzed with a log rank test to assess the relative hazard of succumbing to disease. The difference in median survival time (with 95% confidence intervals) of 2 days for CDΔCwp84347 and 4 days for C. difficile 630ΔErm was found not to be statistically different (p value of 0.218). Fecal samples were removed from euthanized animals by colectomy and C. difficile isolated and analyzed. PCR of the cwp84 gene (as described in Fig. 1) demonstrated that all C. difficile isolates from the CDΔCwp84347 test groups contained the expected intron (4.3-kbp product), whereas all isolates from the wild-type group gave a 2.4-kbp product consistent with the wild-type gene. The data demonstrate that the CDΔCwp84347 is capable of causing CDI in the hamster model at a similar rate of onset as the wild-type strain.
FIGURE 5.
Virulence of C. difficile 630ΔErm and CDΔCwp84347 in hamsters. Data show time from challenge to severe disease/death in the hamster model for CDI. For CDΔCwp84347, the data represent three test groups of 10 animals and for C. difficile 630ΔErm, two test groups of 10 animals.
DISCUSSION
Over the past 10 years, infection by C. difficile has become a serious problem within health care environments, and the bacterium has achieved superbug status causing and contributing to many thousands of deaths within the developed world (29). Many aspects of CDI remain unclear, in particular the mechanisms by which the bacterium establishes and maintains its niche within the gut. The study of such aspects of the disease process is of paramount importance for the development of new therapeutics and prophylactics. It is only comparatively recently that it has become possible to generate stable, targeted gene knock-outs in C. difficile (19). Combined with proteomics and in vivo models for CDI, such techniques provide powerful tools with which to study disease pathogenesis. In the present study, we describe the knock-out of cwp84, a gene encoding a putative surface protein Cwp84 and demonstrate that this cysteine protease plays a key role in the maturation of the surface protein layer.
Insertional knock-out of cwp84 at three different locations led to a bacterial phenotype in which the S-layer, normally composed of the HMW and LMW SLPs, contained the single SlpA precursor polypeptide. Successful knock-out of the cwp84 gene was confirmed by both PCR analysis of the gene and absence of mRNA. Whereas the ClosTron gene knock-out system offers a high degree of stringency with respect to gene targeting to minimize the risk of promiscuous intron insertion, we scanned the C. difficile genome to ensure that the chosen Cwp84 target sites were unique. In addition, that gene knock-out via three different intron insertion sites resulted in the same phenotypic outcome makes it highly unlikely that this was the result of inactivation of genes other than cwp84.
The SlpA precursor polypeptide expressed by CDΔCwp84347 was found to lack its signal polypeptide, which indicates that signal peptide removal and cleavage into its subunits are two distinct events. Removal of the signal peptide by a signal peptidase during passage through the cell membrane followed by cleavage into the HMW and LMW SLP by a cell wall peptidase would appear to be the logical chain of events. To date only Cwp84 and the protease encoded by the CD1751 gene have been implicated as surface layer-associated cysteine proteases. The presence of Cwp84 within S-layer proteins has been demonstrated, and the enzyme has been shown to contain cysteine protease activity toward a range of extracellular matrix proteins including fibronectin, laminin, and vitronectin. The putative CD1751 cysteine protease displays 63.2% amino acid homology with Cwp84 but as yet has not been characterized. PCR analysis of the CD1751 gene in CDΔCwp84 demonstrated that it was unaffected by the knock-out procedure, and no integration of the intron had occurred. That there was no evidence of the HMW and LMW SLP bands in surface protein extracts from CDΔCwp84347 suggests that CD1751 has no role in the cleavage of the SlpA precursor. In addition, analysis of the genome of a virulent C. difficile strain (R20291, Stoke Mandeville) shows CD1751 to be deleted, providing further evidence against any significant role in pathogenesis.
Collectively, the above data provide strong evidence that Cwp84 plays a key role in maturation of the SlpA precursor polypeptide into HMW and LMW SLPs. Interestingly, the maturation of CwpV, a surface-associated protein that is also cleaved into two fragments (28) was unaffected by cwp84 knock-out, which suggests a different/additional processing mechanism for this surface protein.
Purified SLPs frequently retain their ability to form a regular two-dimensional array in suspension and on a variety of surfaces including liposomes. Based on the accessibility of SLP subunits to antibodies and location of the putative cell wall-binding motifs, which are contained within the HMW SLP, an orientation for the S-layer in which the LMW SLP forms the outermost layer has been proposed (14, 30). Recently, this model has been supported by structural data in which small-angle x-ray scattering analysis was used to analyze a complex of the HMW and LMW SLPs (15). These data showed the complex to be an elongated structure in which a C-terminal region of the LMW SLP interacts with an N-terminal region of the HMW SLP. In CDΔCwp84347, it is unlikely that the single chain SlpA precursor would be able to form a similar complex, with perhaps implications on the packing and assembly of the S-layer. It is therefore not surprising that CDΔCwp84347 had markedly different morphology and growth characteristics compared with wild-type C. difficile 630ΔErm. CDΔCwp84347 grew more slowly and had a tendency to clump and form string-like precipitates, which also suggests a change in the property of the bacterial surface layer with an increased degree of intercell adherence. A significant finding is the presence of the SlpA precursor polypeptide in the growth medium during culture of CDΔCwp84347. This suggests a weakened attachment via the cell wall-binding motifs, because the mature SLP subunits were absent from the medium of wild-type C. difficile 630ΔErm. Overall, the above observations suggest that the structural integrity of the S-layer is compromised within CDΔCwp84347, which is possibly caused by the inability of the SlpA precursor polypeptide to form a properly locked lattice structure.
CDΔCwp84347 was assessed for its ability to cause CDI in the hamster disease model. The mutant strain was clearly competent at colonizing the hamster gut and caused a similar spectrum of symptoms to wild-type C. difficile 630ΔErm with a mean of fatality of 83.3% in the test groups. Bacterial isolates from the fecal samples taken from diseased animals confirmed CDΔCwp84347 as the causative strain and further illustrated the stability of the mutation. These data demonstrate first, that Cwp84 does not directly play a critical role in CDI pathogenesis in the animal model, and second, that bacteria expressing the SlpA precursor polypeptide are still competent at causing disease. With respect to the latter, one possibility is that the SlpA precursor polypeptide retains any key biological action(s) of the HMW and LMW SLPs. It has been shown that these SLPs have adhesive activity to cultured cell lines, and it is feasible that the domains mediating this activity are correctly folded within the SlpA precursor. Another possibility is that the SlpA precursor is matured via cleavage by the gut proteases. The SlpA precursor extracted from CDΔCwp84347 was shown to be highly sensitive to tryptic cleavage in vitro, giving rise to peptides indistinguishable in size on SDS-PAGE to the native HMW and LMW SLPs. More significantly, we showed that inclusion of trypsin in the CDΔCwp84347 growth medium led to a significant proportion of the cell associated SlpA precursor being cleaved. Thus, it is possible that a proportion of CDΔCwp84347 growing in the gut derives a mature S-layer via the action of gut proteases. Alignment of 14 SlpA sequences (31) at their putative cleavage sites suggests that the SlpA precursor could potentially be cleaved by the gut enzymes trypsin or chymotrypsin (Fig. 6).
FIGURE 6.
Alignment of predicted maturation cleavage sites within SlpA from 14 C. difficile ribotypes (31). Maturation of SlpA is generally predicted to occur C-terminal to a consensus motif TKS or TYX (31). Potential cleavage sites for trypsin or chymotrypsin are shown in bold.
Previous studies have shown that the cwp84 gene is highly conserved and that its expression, as well as other surface protein genes, is up-regulated in the presence of antibiotics, leading to an increase in bacterial adhesion (32). These observations led to speculation that the enzyme may play a role in the maturation of surface-associated adhesins. The data presented in the present study are entirely consistent with these findings, given the proposed role of the major S-layer proteins in cell adhesion. A direct role for Cwp84 as a colonization factor through the digestion of extracellular matrixes has also been proposed. Whereas the findings in the present study are not inconsistent with Cwp84 having additional roles involved in matrix degradation, they do demonstrate that the enzyme is not essential for CDI in the animal model, which could be because of redundancy of function within the surface-associated components.
It is interesting to note that Toxins A and B, the principal virulence factors of C. difficile, also rely on a cysteine protease in their actions. In the case of each of these toxins, an intrinsic cysteine protease activity mediates the release of a fragment containing a glucosyl transferase activity once the toxin has been internalized within the mammalian cell. In conclusion, the present study shows that the cysteine protease Cwp84 plays a critical role in the maturation of the S-layer of C. difficile. Whereas the CDΔCwp84347 mutant expressing the SlpA precursor polypeptide displays modified morphology and growth characteristics, it is still able to cause disease in animals. The CDΔCwp84 mutants created in the present study may also be of value to structural studies of the S-layer proteins, because they provide a potential source of purified SlpA precursor polypeptide.
Acknowledgments
We thank Nigel Minton and John Heap (University of Nottingham, UK), who developed the ClosTron gene knock-out system, for supplying us with the pMTL007 plasmid and C. difficile 630ΔErm strain. We would also like to thank Nigel Minton and Stephen Cartman (University of Nottingham, UK) for help in establishing the ClosTron system. We would like to acknowledge the excellent technical assistance of Paul Yeates, Nathan Wiblin, and Antony Rule in completing this study.
This work was supported by a Health Protection Agency studentship (to J. M. K.) and the United Kingdom Department of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- CDI
- C. difficile infection
- DMEM
- Dulbecco's modified Eagle's medium
- Erm
- erythromycin
- RT-PCR
- reverse transcription PCR
- SLP
- surface-layer proteins
- HMW
- high molecular weight
- LMW
- low molecular weight
- MS
- mass spectroscopy
- WT
- wild type
- SOE
- splice overlap extension.
REFERENCES
- 1.Bartlett J. G. (2006) Ann. Intern Med. 145, 758–764 [DOI] [PubMed] [Google Scholar]
- 2.McDonald L. C., Killgore G. E., Thompson A., Owens R. C., Jr., Kazakova S. V., Sambol S. P., Johnson S., Gerding D. N. (2005) N. Engl. J. Med. 353, 2433–2441 [DOI] [PubMed] [Google Scholar]
- 3.Navaneethan U. (2008) Minerva Gastroenterol. Dietol. 54, 451–453 [PubMed] [Google Scholar]
- 4.Borriello S. P. (1998) J. Antimicrob. Chemother. 41, Suppl. C, 13–19 [DOI] [PubMed] [Google Scholar]
- 5.Egerer M., Giesemann T., Herrmann C., Aktories K. (2009) J. Biol. Chem. 284, 3389–3395 [DOI] [PubMed] [Google Scholar]
- 6.Desvaux M., Dumas E., Chafsey I., Hébraud M. (2006) FEMS Microbiol. Lett. 256, 1–15 [DOI] [PubMed] [Google Scholar]
- 7.Janoir C., Péchiné S., Grosdidier C., Collignon A. (2007) J. Bacteriol. 189, 7174–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savariau-Lacomme M. P., Lebarbier C., Karjalainen T., Collignon A., Janoir C. (2003) J. Bacteriol. 185, 4461–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waligora A. J., Hennequin C, Mullany P, Bourlioux P., Collignon A., Karjalainen T. (2001) Infect. Immun. 69, 2144–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright A., Wait R., Begum S., Crossett B., Nagy J., Brown K., Fairweather N. (2005) Proteomics 9, 2443–2452 [DOI] [PubMed] [Google Scholar]
- 11.Sára M., Sleytr U. B. (2000) J. Bacteriol. 182, 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerquetti M., Molinari A., Sebastianelli A., Diociaiuti M., Petruzzelli R., Capo C., Mastrantonio P. (2000) Microb. Pathog. 28, 363–372 [DOI] [PubMed] [Google Scholar]
- 13.Calabi E., Ward S., Wren B., Paxton T., Panico M., Morris H., Dell A., Dougan G., Fairweather N. (2001) Mol. Microbiol. 40, 1187–1199 [DOI] [PubMed] [Google Scholar]
- 14.Karjalainen T., Saumier N., Barc M. C., Delmée M., Collignon A. (2002) J. Clin. Microbiol. 40, 2452–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fagan R. P., Albesa-Jové D., Qazi O., Svergun D. I., Brown K. A., Fairweather N. F. (2009) Mol. Microbiol. 71, 1308–1322 [DOI] [PubMed] [Google Scholar]
- 16.Sebaihia M., Wren B. W., Mullany P., Fairweather N. F., Minton N., Stabler R., Thomson N. R., Roberts A. P., Cerdeño-Tárraga A. M., Wang H., Holden M. T., Wright A., Churcher C., Quail M. A., Baker S., Bason N., Brooks K., Chillingworth T., Cronin A., Davis P., Dowd L., Fraser A., Feltwell T., Hance Z., Holroyd S., Jagels K., Moule S., Mungall K., Price C., Rabbinowitsch E., Sharp S., Simmonds M., Stevens K., Unwin L., Whithead S., Dupuy B., Dougan G., Barrell B., Parkhill J. (2006) Nat. Genet. 38, 779–786 [DOI] [PubMed] [Google Scholar]
- 17.Wright A., Drudy D., Kyne L., Brown K., Fairweather N. F. (2008) J. Med. Microbiol. 57, 750–756 [DOI] [PubMed] [Google Scholar]
- 18.Carter G. P., Lyras D., Allen D. L., Mackin K. E., Howarth P. M., O'Connor J. R., Rood J. I. (2007) J. Bacteriol. 189, 7290–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heap J. T., Pennington O. J., Cartman S. T., Carter G. P., Minton N. P. (2007) J. Microbiol. Methods 70, 452–464 [DOI] [PubMed] [Google Scholar]
- 20.Lyras D., O'Connor J. R., Howarth P. M., Sambol S. P., Carter G. P., Phumoonna T., Poon R., Adams V., Vedantam G., Johnson S., Gerding D. N., Rood J. I. (2009) Nature 458, 1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purdy D., O'Keeffe T. A., Elmore M., Herbert M., McLeod A., Bokori-Brown M., Ostrowski A., Minton N. P. (2002) Mol. Microbiol. 46, 439–452 [DOI] [PubMed] [Google Scholar]
- 22.Hussain H. A., Roberts A. P., Mullany P. (2005) J. Med. Microbiol. 54, 137–141 [DOI] [PubMed] [Google Scholar]
- 23.Roberts A. K., Chierici R., Sawatzki G., Hill M. J., Volpato S., Vigi V. (1992) Acta Paediatr. 81, 119–124 [DOI] [PubMed] [Google Scholar]
- 24.Davis I, Carter G., Young M., Minton N. P. (2005) in Handbook on Clostridia (Duerre P. ed) pp. 37–49, CRC Press, Boca Raton [Google Scholar]
- 25.Shevchenko A., Jensen O. N., Podtelejnikov A. V., Sagliocco F., Wilm M., Vorm O., Mortensen P., Shevchenko A., Boucherie H., Mann M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambol S. P., Tang J. K., Merrigan M. M., Johnson S., Gerding D. N. (2001) J. Infect. Dis. 183, 1760–1766 [DOI] [PubMed] [Google Scholar]
- 27.Babcock G. J., Broering T. J., Hernandez H. J., Mandell R. B., Donahue K., Boatright N., Stack A. M., Lowy I., Graziano R., Molrine D., Ambrosino D. M., Thomas W. D., Jr. (2006) Infect. Immun. 74, 6339–6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emerson J. E., Reynolds C. B, Fagan R. P., Shaw H. A., Goulding D., Fairweather N. F. (2009) Mol. Microbiol. 74, 541–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brazier J. S. (2008) Br J. Biomed. Sci. 65, 39–44 [DOI] [PubMed] [Google Scholar]
- 30.Calabi E., Fairweather N. (2002) J. Bacteriol. 184, 3886–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eidhin D. N., Ryan A. W., Doyle R. M., Walsh J. B., Kelleher D. (2006) J. Med Microbiol. 55, 69–83 [DOI] [PubMed] [Google Scholar]
- 32.Denève C., Deloménie C., Barc M. C., Collignon A., Janoir C. (2008) J. Med. Microbiol. 57, 732–738 [DOI] [PubMed] [Google Scholar]