Abstract
Ubiquitylation of histone H2B and/or a component of the system that ubiquitylates H2B is required for methylation of histone H3 at lysine 4 (H3K4) in yeasts and probably in humans. In this study, the single ubiquitylation site was mapped to conserved lysine 115 of the C-terminal region of histone H2B in the single-cell model organism Tetrahymena thermophila. In strains lacking H2B ubiquitylation, H3K4 methylation was not detectably affected. As in other organisms, the E2 ubiquitin-conjugating enzyme Ubc2 and the E3 ubiquitin ligase Bre1 were required for H2B ubiquitylation. However, neither enzyme was required for H3K4 methylation. These studies argue that, in T. thermophila, the histone ubiquitylation mechanism is not required for H3K4 methylation, demonstrating that different organisms can speak different languages in the “cross-talk” among post-translational modifications on different histones.
Introduction
In the nuclei of eukaryotic cells, DNA is highly compacted into chromatin, a nucleoprotein complex, whose major protein components are the histones. The basic unit of chromatin is the nucleosome core in which 146 base pairs of double-stranded DNA are wrapped in ∼1.75 left-handed superhelical turns around an octamer containing two molecules of each of the four conserved core histones H2A, H2B, H3, and H4 (1, 2). A fifth, less conserved linker histone H1, associates with a variable length of linker DNA between nucleosome cores. Site-specific histone post-translational modifications, such as acetylation, methylation, phosphorylation, and ubiquitylation, are correlated with diverse chromatin functions, including DNA replication, DNA repair, chromatin assembly, gene transcription, and other chromatin-based processes (3, 4). It has been suggested that at least some distinct histone modifications act sequentially or in combination to form what has been variously referred to as a histone, epigenetic, or nucleosome “code” that is read by other proteins or protein complexes to alter chromatin structure and regulate distinct downstream events (5, 6). Deciphering this code, which is likely to have complex redundant, combinatorial, and multivalent features, is essential for understanding the in vivo function of the diverse post-translational modifications on histones.
Ubiquitin is a 76-amino acid globular protein that is conserved in eukaryotes. Unlike proteins that are targeted by polyubiquitylation to the proteosome for degradation, histones are reversibly modified by covalent attachment of one ubiquitin or a short ubiquitin chain. The bulk of histone ubiquitylation occurs on chromatin by the addition of a single ubiquitin molecule via an isopeptide linkage to a specific lysine residue in the C-terminal tail of histones H2A and H2B (7, 8). To a lesser extent, histones H1 (9), H3, and H4 (10, 11) also can be ubiquitylated in vivo. Ubiquitylation on different histones has distinct functions (12).
A number of recent studies have focused on ubiquitylation of H2B. In the budding yeast Saccharomyces cerevisiae, H2B is monoubiquitylated at lysine 123 by an evolutionarily conserved E2 ubiquitin-conjugating enzyme, Rad6/Ubc2 (13). Initial reports indicated that Ubc2-mediated H2B monoubiquitylation is a prerequisite for methylation of histone H3 at lysine 4 and lysine 79 by the histone methyltransferases Set1 and Dot1, respectively (14–17), providing the first evidence of “cross-talk” between different post-translational modifications on different histones. Bre1 was identified as an E3 ubiquitin ligase that associates with Rad6/Ubc2 and directs it for H2B ubiquitylation (18, 19). In budding yeast, Ubc2-Bre1-mediated H2B ubiquitylation is highly dynamic, and the sequential ubiquitylation and deubiquitylation of H2B helps to establish the appropriate level of H3K43 methylation and regulates transcriptional initiation (20, 21). Ubc2-Bre1-mediated H2B ubiquitylation is dependent on the transcriptional elongation complex Paf1 and promotes efficient RNA polymerase II elongation (22–24). In human cells, H2B ubiquitylation is also involved in transcriptional activation and appears to affect H3K4 methylation, as shown by knockdown of human orthologs of yeast BRE1 (25, 26). It has been shown that Rad6/Ubc2 is also required for H2B ubiquitylation and efficient H3K4 di- and trimethylation in human cells (27). In the fission yeast Schizosaccharomyces pombe, Ubc2-Bre1-mediated H2B monoubiquitylation is conserved and is required for H3K4 methylation (28, 29). The apparent conservation of the relationship between H2B ubiquitylation and H3K4 methylation in yeasts and humans has led to the general belief that this pattern of cross-talk is likely to be universal. However, a recent study (30) has called some of the initial results on budding yeast into question. It was shown that in S. cerevisiae, H3K4 methylation is not solely dependent on H2B ubiquitylation and that elimination of the H2B ubiquitylation site, combined with an additional mutation on H2B, which was not identified in previous studies, are required to eliminate H3K4 methylation. This study also confirmed that BRE1 is required for H3K4 methylation in S. cerevisiae, suggesting that components of the H2B ubiquitylation mechanism but not ubiquitylation itself were essential for H3K4 methylation. In addition, in both budding and fission yeast, H2B ubiquitylation has functions in transcription that are independent of H3K4 methylation (28, 29). H2B ubiquitylation in fission yeast also functions in cell growth, nuclear structure, and heterochromatin derepression by unknown mechanisms that are independent of H3K4 methylation, and H2B ubiquitylation regulates the expression of more genes than does H3K4 methylation (28).
In this study, the single ubiquitylation site in Tetrahymena thermophila histone H2B was mapped to the conserved lysine 115 of its C-terminal region. In mutant strains lacking H2B ubiquitylation, H3K4 methylation is not detectably affected. Tetrahymena orthologs of Ubc2 and Bre1 were identified as the conserved E2 and E3 enzymes specific for H2B ubiquitylation at Lys115. Neither enzyme is required for H3K4 methylation. These studies strongly suggest that, unlike in yeasts and mammals, neither Ubc2-Bre1-mediated H2B ubiquitylation nor the major components of the H2B ubiquitylation mechanism are required for H3K4 methylation in Tetrahymena, demonstrating that different organisms speak different languages and use different components in the cross-talk between post-translational modifications on different histones.
EXPERIMENTAL PROCEDURES
Tetrahymena Strains and Culture Conditions
T. thermophila wild-type strains B2086, CU428, and CU427 were kindly provided by P. J. Bruns (Cornell University, Ithaca, NY). Strains used in this study are listed in Table 1. Cells were grown in super proteose peptone medium (1% proteose peptone, 0.1% yeast extract, 0.2% glucose, 90 μm EDTA ferric sodium salt) at 30 °C (31). For conjugation, midlogarithmic phase growing cultures of different mating types were washed, starved (16–24 h at 30 °C), and mated in 10 mm Tris-HCl (pH 7.5) (32).
TABLE 1.
Tetrahymena strains used in this study
| Strain | Mic genotypea | Mac genotypea | Mac phenotypeb |
|---|---|---|---|
| B2086 | WTc | WT | II |
| CU428 | mpr1-1/mpr1-1 | WT | mp-s, VII |
| CU427 | chx1-1/chx1-1 | WT | cy-s, VI |
| B*VI | Genetically unfunctional mic (star) | WT | VI |
| B*VII | Genetically unfunctional mic (star) | WT | VII |
| hDΔHTB-86B4A6 | htb1-1::neo2/htb1-1::neo2; htb2-1::neo2/htb2-1::neo2 | WT | pm-s |
| hDΔHTB-86D2A2 | htb1-1::neo2/htb1-1::neo2; htb2-1::neo2/htb2-1::neo2 | WT | pm-s |
| HTB1 | htb1-1::neo2/htb1-1::neo2; htb2-1::neo2/htb2-1::neo2 | HTB1/htb1-1::neo2; htb2-1::neo2/htb2-1::neo2 | pm-r |
| htb1-K115R | htb1-1::neo2/htb1-1::neo2; htb2-1::neo2/htb2-1::neo2 | htb1-2K115R/htb1-1::neo2; htb2-1::neo2/htb2-1::neo2 | pm-r |
| htb1-K111R | htb1-1::neo2/htb1-1::neo2; htb2-1::neo2/htb2-1::neo2 | htb1-3K111R/htb1-1::neo2; htb2-1::neo2/htb2-1::neo2 | pm-r |
| htb1-K(111, 115)R | htb1-1::neo2/htb1-1::neo2; htb2-1::neo2/htb2-1::neo2 | htb1-4K(111, 115)R/htb1-1::neo2; htb2-1::neo2/htb2-1::neo2 | pm-r |
| ΔHHT3ΔHHT4 | hht3-1::neo2, hht4-1::bsr1/hht3-1::neo2, hht4-1::bsr1 | hht3-1::neo2, hht4-1::bsr1/hht3-1::neo2, hht4-1::bsr1 | pm-r, bs-r |
| HHT3 | hht1-1::neo2/hht1-1::neo2; hht2-1::neo2, hhf2-1::neo2/hht2-1::neo2, hhf2-1::neo2; hhf1-1::neo2/hhf1-1::neo2 | hht1-1::neo2/hht1-1::neo2; HHT3, HHF2/hht2-1::neo2, hhf2-1::neo2; hhf1-1::neo2/hhf1-1::neo2 | pm-r |
| hht2-K4Q | hht1-1::neo2/hht1-1::neo2; hht2-1::neo2, hhf2-1::neo2/hht2-1::neo2, hhf2-1::neo2; hhf1-1::neo2/hhf1-1::neo2 | hht1-1::neo2/hht1-1::neo2; hht2-2K4Q, HHF2/hht2-1::neo2, hhf2-1::neo2; hhf1-1::neo2/hhf1-1::neo2 | pm-r |
| ΔUBC2 | ubc2-1::neo2/ubc2-1::neo2 | ubc2-1::neo2/ubc2-1::neo2 | pm-r |
| ΔBRE1 | bre1-1::neo2/bre1-1::neo2 | bre1-1::neo2/bre1-1::neo2 | pm-r |
a Only showing disrupted or mutant alleles.
b Including drug phenotype and mating type.
c WT, wild type.
Making HTB1 and HTB2 Knock-out Constructs
The HTB1 gene was cloned from Tetrahymena genomic DNA, and plasmid pHTB1 was constructed. It is a pBluescript KS(+) (Stratagene) derivative and contains 1.7 kb of HTB1 5′-flanking sequence, the HTB1 coding region, and 2.2 kb of the HTB1 3′-flanking sequence. Plasmid p4T2-1 (33) is a pBluescript KS(+) derivative and contains the neo2 cassette, which consists of 0.3 kb of 5′-flanking sequence of the HHF1 gene (encoding histone H4); the neo gene coding region, which confers resistance to paromomycin when expressed in Tetrahymena; and 0.3 kb of the 3′-flanking sequence of the BTU2 gene (encoding β-tubulin). To make the HTB1 knock-out construct (supplemental Fig. S2A), a 1.7-kb EcoRI-BstXI restriction fragment of the HTB1 5′-flanking sequence from pHTB1 was inserted into the EcoRV site of plasmid p4T2-1. A 1.4-kb fragment of the HTB1 3′-flanking sequence (from the TGA stop codon to a SpeI site) was PCR-amplified from pHTB1 and inserted into the SpeI site of p4T2-1. The final construct, referred to as pHTB1-KO, was digested with KpnI and SacI, and a 4.5-kb insert containing HTB1 5′-flanking sequence, the neo2 cassette, and HTB1 3′-flanking sequence was released and used for germ line transformation. All constructs were verified by sequencing.
The HTB2 gene was cloned from Tetrahymena genomic DNA, and plasmid pHTB2 was constructed. It is a pBluescript KS(+) (Stratagene) derivative and contains 1.7 kb of HTB2 5′-flanking sequence, the HTB2 coding region, and 2.4 kb of HTB2 3′-flanking sequence. To make the HTB2 knock-out construct (supplemental Fig. S2B), a 1.7-kb fragment of the HTB2 5′-flanking sequence (from a HindIII site to 33 bp upstream of the ATG start codon) was PCR-amplified from pHTB2 and inserted into the EcoRV site of p4T2-1. A 2.4-kb fragment of the HTB2 3′-flanking sequence (from the TGA stop codon to an EcoRI site) was PCR-amplified from pHTB2 and inserted into the SmaI site of p4T2-1. The final construct, referred to as pHTB2-KO, was digested with XhoI and SpeI, and a 5.5-kb insert containing HTB2 5′-flanking sequence, neo2 cassette, and HTB2 3′-flanking sequence was released and used for germ line transformation. All constructs were verified by sequencing.
Creation of HTB1 or HTB2 Single Knock-out Heterokaryons
To obtain germ line transformants in which a single allele in the micronucleus had been knocked out, B2086 and CU428 cells were mated and transformed at 2.5–3.5 h postmixing with either the HTB1 or HTB2 knock-out construct using the PDS-1000/He biolistic particle delivery system (Bio-Rad) as described previously (34). Germ line knock-out homozygous heterokaryon strains, in which both copies of either the HTB1 or HTB2 gene are replaced by the neo2 cassette in the germ line micronucleus but retain wild-type genes in the somatic macronucleus, were then created as described previously (35).
Creation of HTB1 and HTB2 Double Knock-out Heterokaryons
HTB1 and HTB2 double knock-out heterokaryon strains hΔΔHTB-86B4A6 and hΔΔHTB-86D2A2 (Table 1) were created by crossing the HTB1 and HTB2 single knock-out heterokaryon strains as described previously (35).
Making HTB1 Somatic Rescue Constructs and Creation of HTB1 Somatic Rescue Strains
PCR-directed mutagenesis was performed as described previously (36) on pHTB1-M, which is identical to pHTB1 except that it contains a slightly shorter 3′-flanking sequence of HTB1. At a specific residue, the codon for lysine was changed to arginine. The resulting plasmids or pHTB1-M were digested with PstI and XhoI, and the 4.1-kb inserts containing 1.7 kb of HTB1 5′-flanking sequence, the wild-type or mutated HTB1 gene, and 2.1 kb of HTB1 3′-flanking sequence were released and used for somatic rescue transformation. All constructs were verified by sequencing.
As expected, the progeny of two HTB double knock-out heterokaryon strains were not viable because they lacked both genes for H2B, an essential, major component of the nucleosome core. Somatic rescue strains were created by transforming mating double knock-out heterokaryon strains hΔΔHTB-86B4A6 and -86D2A2 at late conjugation (20–24 h postmixing) by biolistic transformation with a wild-type or mutated copy of the HTB1 gene and selecting paromomycin-resistant progeny (34).
Cloning the Tetrahymena UBC2 Gene and Creation of UBC2 Knock-out Strains
A gene (TTHERM_00550720) encoding a putative Ubc2 was identified in the Tetrahymena Genome Database (available on the World Wide Web) using S. cerevisiae Rad6/Ubc2 as the query and named UBC2. A 3.0-kb HindIII-HincII fragment containing 0.8 kb of 5′-flanking sequence, the 456 bp UBC2 coding region, and 1.7 kb of 3′-flanking sequence was amplified by PCR from genomic DNA and cloned into the pBluescript KS(+) vector (Stratagene). The construct, named pUBC2, was verified by sequencing.
To make the UBC2 knock-out construct pUBC2-KO, the 456-bp UBC2 coding region in pUBC2 was replaced by the neo2 cassette (33). pUBC2-KO was digested with HindIII and SpeI to release a 4.0-kb ubc2-1::neo2 insert, which was used for germ line transformation. Germ line knock-out homozygous heterokaryon strains, in which both copies of the UBC2 gene are replaced by the neo2 cassette in the germ line micronucleus but retain wild-type genes in the somatic macronucleus, were then created as described previously (35). The UBC2 knock-out homozygous homokaryon strains (ΔUBC2), in which ubc2-1::neo2 had completely replaced all micronuclear and macronuclear UBC2 copies, were created by mating UBC2 germ line knock-out heterokaryon strains and selecting their paromomycin-resistant progeny. The ΔUBC2 strains were verified by PCR.
Cloning the Tetrahymena BRE1 Gene and Creation of BRE1 Knock-out Strains
A gene (TTHERM_00790530) encoding a putative Bre1 was identified in the Tetrahymena Genome Database and named BRE1. A 5.7-kb fragment containing 1.2 kb of 5′-flanking sequence, the 3.2-kb BRE1 coding region, and 1.3 kb of 3′-flanking sequence was amplified by PCR from genomic DNA and cloned into the pBluescript KS(+) vector (Stratagene). The construct, named pBRE1, was verified by sequencing.
To make the BRE1 knock-out construct, the BRE1 coding region, except for 80 bp of 3′ sequence, in pBRE1 was replaced by the neo2 cassette (33). The final construct, referred to as pBRE1-KO, was digested with KpnI and SacI, and a 4.0-kb bre1-1::neo2 insert was released and used for germ line transformation. Germ line knock-out homozygous heterokaryon strains, in which both copies of the BRE1 gene are replaced by the neo2 cassette in the germ line micronucleus but retain wild-type genes in the somatic macronucleus, were then created as described previously (35). The BRE1 knock-out homozygous homokaryon strains (ΔBRE1), in which bre1-1::neo2 had completely replaced all micronuclear and macronuclear BRE1 copies, were created by mating BRE1 germ line knock-out heterokaryon strains and selecting their paromomycin-resistant progeny. The ΔBRE1 strains were verified by PCR.
PCR Analysis
Total genomic DNA was isolated from cells of wild-type, ΔUBC2, and ΔBRE1 strains, as described previously (37), and used for PCR. The PCR products were separated on agarose gels. The primers used for the UBC2 locus were UBC2-04 (5′-GAATTAAACTTACTTATGTCTATC-3′), UBC2-seqC (5′-GAACGAGTGATTGAACTTG-3′), and neo-sense (5′-GAATGGGCTGACCGCTTCCTCGTGC-3′). The primers used for the BRE1 locus were BRE1-18 (5′-TTTACAGATAGATTGATTGAGTG-3′), BRE1-19 (5′-TTCCATATCTATCTATCTATCTTG-3′), and neo-253 (5′-CAGCCAGTCCCTTCCCGCTTCAGTGAC-3′).
Growth Analysis
Cells from each strain were used to inoculate 50 ml of super proteose peptone medium at starting densities of 1 × 104 cells/ml. Cultures were grown at 30 °C with vigorous shaking (150 rpm). Cells were counted by a Coulter counter (model ZBI, Coulter Electronics, Inc.) at frequent intervals. Growth data were plotted using Cricket Graph III (Computer Associates). Doubling times were calculated using the linear portion of the logarithmic growth curve.
Fluorescence Microscopy
Cells from each strain were grown to 2–3 × 105 cells/ml and fixed in 1.8% formaldehyde at room temperature, followed by staining with 0.4 μg/ml of the DNA-specific dye 4′,6-diamidine-2-phenylindole dihydrochloride (Roche Applied Science). Macro- and micronuclei were visualized using an Olympus BH-2 fluorescence microscope. Images were recorded using a CFW-1310 M Grayscale digital camera (Scion Corp.) and processed using Adobe Photoshop.
Nuclear Isolation and Histone Extraction
Macronuclei were isolated from growing cells as described previously (31) and extracted with 0.2 m H2SO4 for 12–18 h at 4 °C, and the acid-soluble material was precipitated with 20% trichloroacetic acid (38).
CNBr Cleavage
Acid-extracted histones of isolated macronuclei were incubated with 1 mg/ml CNBr in 70% formic acid for 24 h at room temperature in the dark (39). For control, the samples were incubated with 70% formic acid only.
Protein Electrophoresis and Western Blotting
∼15–20 μg of acid-extracted macronuclear histones were separated by 15% SDS-PAGE (40) and transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore) using a Trans-Blot SD semidry electrophoretic transfer cell (Bio-Rad). Blots were blocked in 5% nonfat dry milk in TBST1 (100 mm Tris-HCl (pH 7.5), 0.9% NaCl, 0.1% Tween 20) for 1 h at room temperature and incubated overnight at 4 °C with primary rabbit anti-ubiquitin antiserum (Sigma) diluted 1:1,000 in blocking buffer. Membranes were washed in TBST1 and incubated with a 1:5,000 dilution of goat anti-rabbit immunoglobulin (IgG) horseradish peroxidase-conjugated secondary antibody (Sigma) in 1% bovine serum albumin/TBST1 for 1 h at room temperature. Blots were processed using the Western Lightning chemiluminescence reagent (PerkinElmer Life Sciences) and exposed to Blue Sensitive Autoradiographic Film (Marsh Bio Products, Inc.). Before reprobing with other antisera, blots were incubated for 30 min at 50 °C in stripping buffer (62.5 mm Tris-HCl (pH 6.8), 2% SDS, 100 mm β-mercaptoethanol). The stripped blots were blocked in blocking buffer and probed with rabbit antisera made against Tetrahymena histones diluted in 1% bovine serum albumin/TBST1. Blots were then processed as described above for the anti-ubiquitin antiserum, except that a 1:20,000 dilution of the secondary antibody was used. The following histone antisera were used: anti-H2B (1:15,000 dilution), anti-H2A (1:20,000 dilution), anti-H4 (1:4,000 dilution) antisera (provided by Dr. C. David Allis, Rockefeller University, New York, NY) and anti-H2A.Z antiserum (1:10,000 dilution) (41).
∼10 μg of CNBr-cleaved macronuclear histones were separated by 15% SDS-PAGE and transferred as above, except that Immobilon-PSQ polyvinylidene difluoride membranes (Millipore) were used for blotting. For the rabbit anti-trimethyl- histone H3 Lys4 (anti-H3K4me3) antiserum (Abcam), which was preabsorbed with 1 μg/ml dimethyl-histone H3 Lys4 (H3K4me2) peptide (Abcam) for 1 h at 37 °C, blots were blocked in 5% bovine serum albumin in TBST2 (TBST1 with Tween 20 at 0.5%) for 1 h at room temperature and incubated overnight at 4 °C with the primary rabbit antiserum diluted 1:5,000 in the blocking buffer. Membranes were washed in TBST2 and incubated with 1:10,000 dilution of goat anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody (Sigma) in the blocking buffer for 1 h at room temperature. Blots were then processed as above. For the rabbit anti-monomethyl-histone H3 Lys4 (anti-H3K4me1) and anti-H3K4me2 antisera (Upstate), blots were blocked in 5% nonfat dry milk in TBST1 and incubated with the primary rabbit antiserum (1:2,000 dilution for anti-H3K4me1 antiserum, 1:5,000 dilution for anti-H3K4me2 antiserum) in the blocking buffer. Membranes were then incubated with a 1:5,000 dilution of goat anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody (Sigma) in the blocking buffer. Blots were stripped and reprobed with the anti-H3 antiserum (1:6,000 dilution) as described above for the other histone antisera.
Microarray Analysis
In each experiment, total RNAs were extracted from wild-type CU428 and mutant strains at a density of 2 × 105 cells/ml and purified further with Qiagen RNeasy columns. Gene expression microarray analysis was performed as described (42).
RESULTS
The Two Tetrahymena H2B Genes Function Redundantly
In Tetrahymena, histone H2B.1 and H2B.2 (encoded by HTB1 and HTB2, respectively) are non-allelic variants of H2B that differ in three amino acids (26) (supplemental Fig. S1). HTB single knock-out homozygous homokaryon strains, ΔHTB1 and ΔHTB2, in which either the HTB1 or HTB2 gene, respectively, was disrupted in both macronuclear and micronuclear genomes, were viable and grew normally (data not shown), demonstrating that neither of two HTB genes is essential. HTB double knock-out heterokaryon strains were created, in which both copies of HTB1 and HTB2 genes were replaced by the neo2 cassette in the germ line micronucleus while wild-type genes were retained in the somatic macronucleus. When two HTB double knock-out heterokaryon strains with different mating types were mated, all of their exconjugant progeny failed to grow. The progeny of HTB double knock-out heterokaryons could be rescued by somatic transformation with the wild-type HTB1 gene, demonstrating that the growth defect was due to the absence of any copies of the HTB genes in the newly developed macronuclei of these exconjugant progeny. Therefore, like the other non-allelic genes encoding variants of the major core histones H2A, H3, and H4 (43, 44), the two genes encoding H2B are functionally redundant in T. thermophila.
Lysine 115 Is the Only Residue of H2B That Can Be Ubiquitylated in Tetrahymena
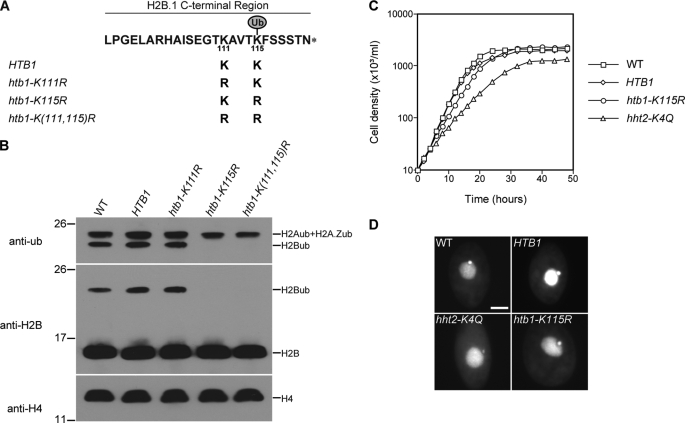
There are only two lysine residues, Lys111 and Lys115, in the Tetrahymena H2B C-terminal region. Lys115 corresponds to a conserved C-terminal lysine residue that is the only ubiquitylation site of H2B in mammals (45), S. cerevisiae (13), S. pombe (28, 29), and Arabidopsis (46). To identify the ubiquitylation site(s) in Tetrahymena H2B and eliminate the possibility that, in the absence of the preferred H2B ubiquitylation site, the other lysine residue could be ubiquitylated, single or double lysine to arginine (which cannot be ubiquitylated) mutations were introduced (Fig. 1A). The wild-type HTB1 gene and three mutated copies of the HTB1 gene, htb1-K111R, htb1-K115R, and htb1-K(111,115)R, were used to rescue the progeny of mating between two HTB1 and HTB2 double knock-out heterokaryons that would otherwise die due to the absence of a functional gene encoding H2B in their macronucleus. All three mutated HTB1s could rescue the progeny, suggesting that neither Lys111 nor Lys115 is essential.
FIGURE 1.
Identification of Lys115 as the only ubiquitylation site of histone H2B and demonstration that H2B ubiquitylation is not required for vegetative growth in Tetrahymena. A, C-terminal sequences of proteins encoded by the wild-type and mutant HTB1 genes used to rescue HTB knock-out strains. The two lysines in the C-terminal region of H2B.1 were changed, either separately or together, to arginines, which cannot be ubiquitylated. B, acid-extracted macronuclear histones of wild-type (WT) CU428, HTB1, and three htb1 mutant strains were analyzed by Western blotting using anti-ubiquitin (anti-ub) and anti-H2B antisera. Anti-H4 antiserum was used to monitor histone loading. The slower migrating band stained with anti-H2B antiserum in the middle panel and the faster band stained with anti-ubiquitin antiserum in the upper panel (from which the unstained lower portion has been removed) co-migrate, enabling them to be identified as ubiquitylated H2B (H2Bub). Molecular mass markers (in kDa) are indicated on the left. C, growth curves of wild-type (WT) CU428, HTB1, htb1-K115R, and hht2-K4Q strains. The doubling times are as follows: wild type, 2.3 h; HTB1, 2.2 h; htb1-K115R, 2.9 h; hht2-K4Q, 4.3 h. D, small micronuclei were observed in hht2-K4Q but not in htb1-K115R cells. Wild-type CU428, HTB1, htb1-K115R, and hht2-K4Q cells were stained with 4′,6-diamidine-2-phenylindole dihydrochloride. Scale bar, 10 μm.
Monoubiquitylated H2B can be easily distinguished from unmodified H2B by its slow migration on one-dimensional SDS-PAGE gels (Fig. 1B). In htb1-K115R and htb1-K(111,115)R, but not htb1-K111R mutant strains, monoubiquitylated H2B was not detectable (Fig. 1B), demonstrating that Lys115 is the only ubiquitylation site on histone H2B in Tetrahymena.
The htb1-K115R strain, lacking H2B ubiquitylation, grew a little more slowly than the wild-type strain (Fig. 1C) and had morphologically normal germ line micronuclei and somatic macronuclei when viewed in the light microscope (Fig. 1D). In contrast, hht2-K4Q cells (47), in which the only major H3 contains a K4Q mutation and thus cannot be methylated, showed severe defects in vegetative growth and micronuclear morphology, suggesting that H2B ubiquitylation may not be required for H3K4 methylation in Tetrahymena.
Both Major and Minor H3s Are Methylated at Lys4 in Tetrahymena
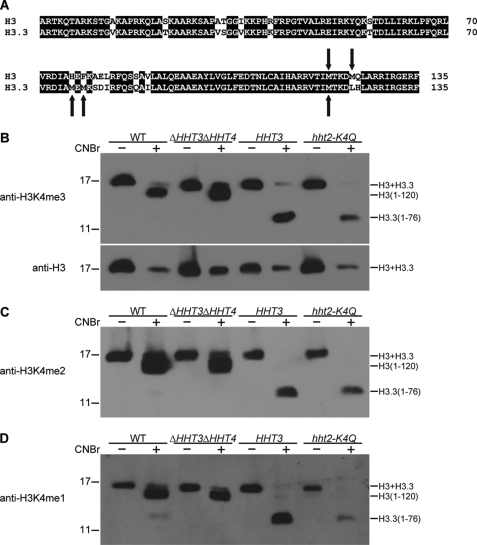
Tetrahymena has four genes encoding non-allelic variants of histone H3; HHT1 and HHT2 encode the same major H3 variant, and HHT3 and HHT4 encode minor variants H3.3 and H3.4, respectively, which cannot be separated by one-dimensional SDS-PAGE (48). Growing cells of Tetrahymena contain the major H3 and the minor H3.3. HHT4 is only expressed when HHT3 is knocked out. Both major and minor H3s are detected by antibodies specific for H3K4 methylation. Therefore, to determine whether H2B ubiquitylation is required for H3K4 methylation in Tetrahymena, both major and minor H3s need to be examined. The major H3 and the minor H3.3 have 2 and 3 methionine residues, respectively (Fig. 2A), only one of which is at the same position. CNBr cleaves proteins at methionine residues, so that, after CNBr cleavage, major and minor H3s generate different patterns of peptides, enabling the separation of the N-terminal fragments of major and minor H3 variants (39) and distinction of Lys4 methylation of major and minor H3s. Macronuclear histones from wild-type, ΔHHT3ΔHHT4, HHT2, and hht2-K4Q cells were incubated with CNBr and separated on SDS-polyacrylamide gel, followed by Western blotting with anti-H3K4me3 (Fig. 2B), anti-H3K4me2 (Fig. 2C), or anti-H3K4me1 (Fig. 2D) antisera to check the trimethylation, dimethylation, or monomethylation of major and minor H3s at Lys4, respectively (supplemental Fig. S3). Antisera specific for H3K4 methylation detected the 120-amino acid peptide (H3-(1–120)) of major H3 and the 76-amino acid peptide (H3.3-(1–76)) of minor H3.3, both of which contain lysine 4, indicating that both major and minor H3 variants can be methylated at Lys4 in Tetrahymena, although Lys4 methylation of major H3 is much more abundant than Lys4 methylation of minor H3.3 in wild-type cells. In ΔHHT3ΔHHT4 cells, both genes encoding minor H3s were knocked out (48), and antisera only detected Lys4 methylation of major H3. In HHT3 cells, minor H3.3 is the only H3 variant (48), and antisera only detected Lys4 methylation of minor H3.3. However, when minor H3.3 is the only H3 in the cells, its methylation is increased but does not appear to fully reach the wild-type level of Lys4 methylation of total H3s. In hht2-K4Q mutant cells, major H3 cannot be methylated at Lys4, and the level of Lys4 methylation of minor H3.3 increases, as in HHT3 cells. These results demonstrate that Lys4 methylation of major and minor H3s can be distinguished by CNBr cleavage.
FIGURE 2.
Major and minor H3s can be distinguished by CNBr cleavage. A, alignment of sequences of Tetrahymena major H3 and minor H3.3. The arrows indicate methionine residues that can be cleaved by CNBr. B, acid-extracted macronuclear histones from wild-type (WT) CU428 cells, from cells (ΔHHT3ΔHHT4) in which both minor H3s were knocked out and the only H3 in the cells is major H3, and cells (HHT3) containing only the minor H3.3 created by rescuing progeny of knock-out heterokaryons lacking all major H3s with the gene encoding H3.3, and hht2-K4Q cells in which the only major H3 contained a K4Q mutation. The histones were incubated without or with CNBr (1 mg/ml) for 24 h at room temperature and then were separated on a 15% SDS-polyacrylamide gel, followed by Western blotting with anti-H3K4me3 (preabsorbed with the H3K4me2 peptide to block nonspecific binding) and anti-H3 antisera. Bands corresponding to major H3, minor H3.3, major H3 peptide (H3-(1–120)), and minor H3.3 peptide (H3.3-(1–76)) are indicated on the right. Molecular mass markers (in kDa) are indicated on the left. C and D, the same analyses as in B were performed with anti-H3K4me2 or anti-H3K4me1 antisera, respectively.
Ubiquitylation of H2B Is Not Required for Methylation of H3 at Lysine 4
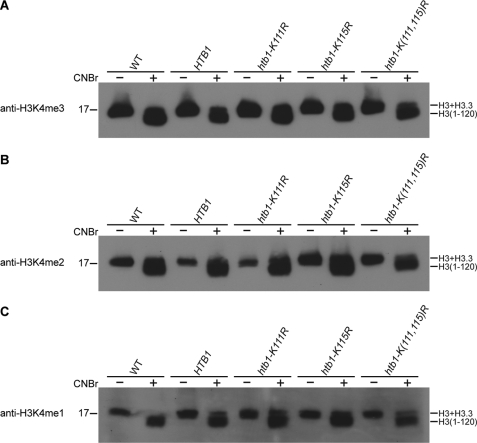
To investigate whether H2B ubiquitylation at Lys115 is required for H3K4 methylation of either H3 or H3.3, macronuclear histones of wild-type, HTB1, htb1-K111R, htb1-K115R and htb1-K(111,115)R strains were cleaved by CNBr and separated on SDS-polyacrylamide gel, followed by Western blotting with anti-H3K4me3, anti-H3K4me2, and anti-H3K4me1 antisera (Fig. 3, A–C). Lys4 methylation of neither major H3 nor minor H3.3 (visualized by overexposing these Western blots; data not shown) was abolished in any of these strains. Therefore, mutating the ubiquitylation site on H2B did not detectably affect H3K4 methylation. These results indicate that H2B ubiquitylation at Lys115 is not required for mono-, di-, or trimethylation of H3 at Lys4 in Tetrahymena, suggesting that the cross-talk of H2B ubiquitylation and H3K4 methylation observed in yeast and mammals may not be conserved for either major or minor H3s of Tetrahymena.
FIGURE 3.
H2B ubiquitylation is not required for H3K4 methylation in Tetrahymena. Acid-extracted macronuclear histones from wild-type (WT) CU428, HTB1, htb1-K111R, htb1-K115R, and htb1-K(111,115)R strains were cleaved by CNBr and separated on 15% SDS-polyacrylamide gel, followed by Western blotting with anti-H3K4me3 (A), anti-H3K4me2 (B), or anti-H3K4me1 (C) antisera. Bands corresponding to major H3, minor H3.3, and major H3 peptide (H3-(1–120)) are indicated on the right. Bands corresponding to minor H3.3 peptide (H3.3-(1–76)) are not shown. Molecular mass markers (in kDa) are indicated on the left.
Ubc2 and Bre1 Are the Conserved E2 Ubiquitin Conjugation Enzyme and E3 Ubiquitin Ligase, Respectively, Specific for H2B Ubiquitylation in Tetrahymena
In addition to preventing ubiquitylation of H2B, mutating Lys115 to arginine might also disrupt some structural feature of Tetrahymena H2B that is required for the regulation of H3K4 methylation. As an alternative approach, we sought to identify and mutate the enzymes responsible for H2B ubiquitylation. A putative Tetrahymena Ubc2 gene (UBC2) was identified by searching the Tetrahymena Genome Database using S. cerevisiae Rad6/Ubc2 as the query. Southern blotting indicated that Tetrahymena UBC2 is a single copy gene (data not shown). Comparison of cDNA and genomic DNA sequences indicated that UBC2 contains no introns and encodes a putative protein of 151 amino acids (data not shown). The sequence alignment of Ubc2 orthologs of Tetrahymena, yeasts, and other eukaryotes (supplemental Fig. S4) demonstrates that Ubc2 proteins are highly conserved among diverse organisms. Phylogenetic analysis (supplemental Fig. S5) indicates that Tetrahymena Ubc2 is most closely related to the putative Ubc2 ortholog of another ciliate, Paramecium tetraurelia. UBC2 is expressed in growing, starved, and conjugating cells of Tetrahymena, especially strongly at 4–8 h postmixing in conjugation, after meiosis and during the earlier stages of nuclear differentiation (data not shown). To study the in vivo function of UBC2, UBC2 knock-out strains (ΔUBC2) were created, in which all micronuclear and macronuclear UBC2 copies were replaced by the neo2 cassette (supplemental Fig. S6A). Replacement in ΔUBC2 strains was confirmed by PCR (supplemental Fig. S6, B and C).
A putative Tetrahymena Bre1 gene (BRE1) was also identified by searching the Tetrahymena Genome Database using the RING finger domain sequence of S. cerevisiae Bre1 as the query. Southern blotting indicated that Tetrahymena BRE1 is a single copy gene (data not shown). Comparison of cDNA and genomic DNA sequences showed that BRE1 contains no introns and encodes a putative protein of 1075 amino acids (data not shown). Bre1 orthologs of Tetrahymena, yeasts, and other organisms (supplemental Fig. S7) contain a conserved C3HC4 RING finger domain in their C-terminal region, which is characteristic of a class of ubiquitin ligases (49). Other parts of Bre1s share low sequence homology (data not shown) and contain multiple predicted coiled-coil domains (18). Northern blotting failed to detect the mRNA of BRE1, but RT-PCR analysis showed that BRE1 is expressed in growing, starved, and conjugating cells of Tetrahymena (data not shown). To study the in vivo function of BRE1, BRE1 knock-out strains (ΔBRE1) were created, in which all micronuclear and macronuclear BRE1 copies were replaced by the neo2 cassette (supplemental Fig. S8A). Replacement in ΔBRE1 strains was confirmed by PCR (supplemental Fig. S8, B and C).
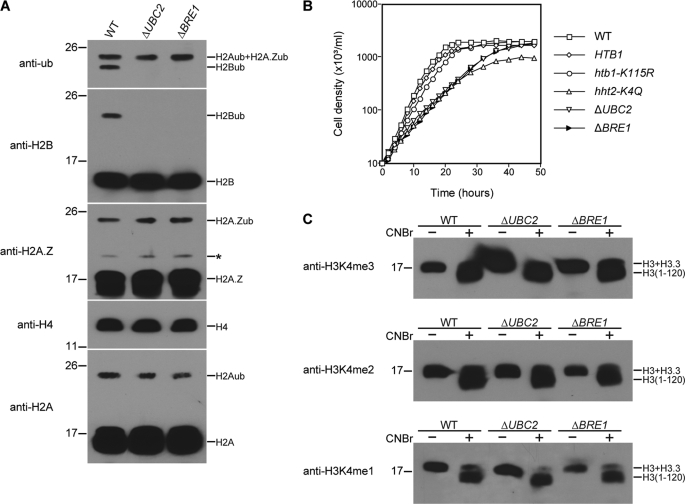
To study their in vivo function, complete knock-out strains of UBC2 and BRE1 were created, and the ubiquitylation levels of macronuclear histones of ΔUBC2 and ΔBRE1 cells were analyzed (Fig. 4A). In both ΔUBC2 and ΔBRE1 cells, H2B ubiquitylation was abolished, whereas ubiquitylation of major or minor H2As was not detectably affected. These results demonstrate that, as in other organisms, UBC2 and BRE1 in Tetrahymena encode the conserved E2 ubiquitin-conjugating enzyme and E3 ubiquitin ligase, respectively, that are specifically responsible for H2B ubiquitylation in vivo. ΔUBC2 and ΔBRE1 cells grew much more slowly than the wild-type strain (Fig. 4B), suggesting that UBC2 and BRE1 are required for normal vegetative growth of Tetrahymena. Importantly, this slow growth phenotype was not observed when H2B ubiquitylation was abolished by mutating Lys115, indicating that UBC2 and BRE1 in Tetrahymena may have functions in addition to H2B ubiquitylation.
FIGURE 4.
UBC2 and BRE1 are required for H2B ubiquitylation but not for H3K4 methylation in Tetrahymena. A, acid-extracted macronuclear histones of wild-type (WT) CU428, UBC2 knock-out (ΔUBC2), and BRE1 knock-out (ΔBRE1) strains were analyzed by Western blotting using anti-ubiquitin (anti-ub), anti-H2B, anti-H2A.Z, anti-H4, and anti-H2A antisera. Anti-H4 antiserum was used to monitor histone loading. The asterisk denotes an unknown protein that cross-reacts with anti-H2A.Z antiserum. Molecular mass markers (in kDa) are indicated on the left. B, growth curves of wild-type CU428, HTB1, htb1-K115, hht2-K4Q, ΔUBC2, and ΔBRE1 strains. The doubling times are as follows: wild type, 2.2 h; HTB1, 2.3 h; htb1-K115R, 2.6 h; hht2-K4Q, 4.1 h; ΔUBC2, 4.6 h; ΔBRE, 4.5 h. C, acid-extracted macronuclear histones of wild-type CU428, ΔUBC2, and ΔBRE1 strains were cleaved by CNBr and separated on 15% SDS-polyacrylamide gel, followed by Western blotting with anti-H3K4me3, anti-H3K4me2, or anti-H3K4me1 antisera. Bands corresponding to major H3 and minor H3.3 or major H3 peptide (H3-(1–120)) are indicated on the right. Bands corresponding to minor H3.3 peptide (H3.3-(1–76)) are not shown. Molecular mass markers (in kDa) are indicated on the left.
To determine whether H2B ubiquitylation was required for H3K4 methylation in wild-type (as opposed to mutated) Tetrahymena and whether the enzymes responsible for H2B ubiquitylation might themselves regulate H3K4 methylation through another pathway, we examined H3K4 methylation in the ΔUBC2 and ΔBRE1 cells. Macronuclear histones of ΔUBC2 and ΔBRE1 cells were cleaved by CNBr and analyzed by Western blotting with antibodies specific for tri-, di-, or monomethylation of H3K4. Lys4 methylation of major H3 (Fig. 4C) and minor H3.3 (data not shown) was not detectably affected in ΔUBC2 or ΔBRE1 cells. These results demonstrate that neither ubiquitylation of H2B nor UBC2 or BRE1 is required for H3K4 methylation in Tetrahymena.
Microarray Analysis
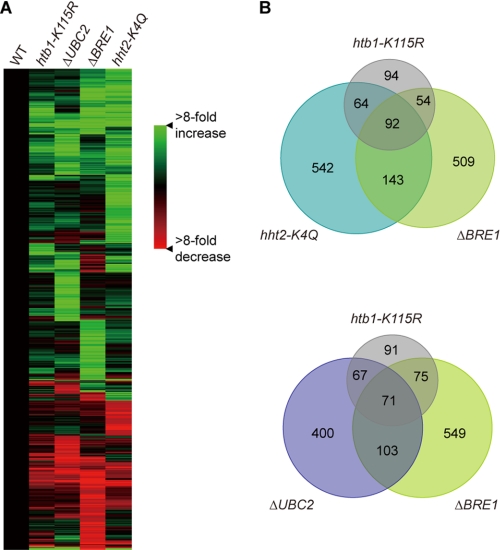
Microarray analyses were performed to compare the gene expression profiles of wild-type and mutant strains (Fig. 5 and supplemental Fig. S9). Although 92 genes showed similar changes in expression in htb1-K115R, ΔBRE1, and hht2-K4Q strains, unlike in yeast, these genes cannot be regulated by cross-talk between H2B ubiquitylation and H3K4 methylation. Only 71 genes were similarly affected in all three mutations (htb1-K115R, ΔUBC2, and ΔBRE1) that eliminated H2B ubiquitylation; many of these genes are likely to be regulated by H2B ubiquitylation. Consistent with our observations that mutation of the H3K4 site, knock-out of UBC2, or knock-out of BRE1 had greater effects on growth than mutation of the H2B ubiquitylation site, elimination of H3K4 methylation and knock-out of either of the ubiquitylation enzymes affects a broader spectrum of genes than elimination of H2B ubiquitylation. Taken together, the large differences in growth and in the number of genes whose expression is altered in the respective mutants argue strongly that UBC2 and BRE1 are involved in pathways other than H2B ubiquitylation and that UBC2 and BRE1 mainly function independently of each other.
FIGURE 5.
Microarray analysis of gene expression profiles of wild-type (WT) CU428, HTB1, htb1-K115, ΔUBC2, ΔBRE1, and hht2-K4Q strains. A, genes whose expression levels were altered by at least 4-fold in one or more mutant strains (supplemental Table S1) were clustered using a hierarchical clustering algorithm. All data sets are normalized to the values of a wild-type CU428 strain. B, Venn diagrams show overlap among htb1-K115, hht2-K4Q, ΔUBC2, and ΔBRE1 data sets. The number of genes whose expression level had over 4-fold change is indicated.
DISCUSSION
In this study, we have shown that, in Tetrahymena, mutant strains lacking H2B ubiquitylation could be created by rescuing the progeny of HTB double knock-out heterokaryons with a mutated HTB1 gene lacking the conserved ubiquitylation site, making Tetrahymena the only organism besides yeasts in which the in vivo function of H2B ubiquitylation has been studied directly by mutating the ubiquitylation site.
We have also shown that genes encoding Tetrahymena Ubc2 and Bre1 are the E2 ubiquitin-conjugating enzyme and E3 ubiquitin ligase, respectively, for H2B ubiquitylation, indicating that, as in yeasts and human cells, Ubc2-Bre1-mediated H2B ubiquitylation is conserved in Tetrahymena. It was suggested that, in human cells, H2B ubiquitylation requires UbcH6, which has E2 ubiquitin-conjugating enzyme activity in vitro (26). However, compared with the previously identified human HR6A and HR6B enzymes (50, 51), UbcH6 has less sequence homology to Ubc2s of S. cerevisiae and S. pombe (supplemental Figs. S4 and S5). Together with the evidence that Ubc2 is required for H2B ubiquitylation in Tetrahymena and yeasts, these results suggest that Ubc6H is not a true Ubc2 ortholog. A recent study (27) also showed that Rad6/Ubc2, not Ubc6H, is required for H2B ubiquitylation in human cells.
We have also shown that H3K4 mono-, di-, and trimethylation occur in Tetrahymena. However, although H2B ubiquitylation is required for H3K4 methylation in some strains of S. cerevisiae (15, 17), in S. pombe (28, 29), and probably in mammals (25, 26), none of the levels of H3K4 methylation were detectably affected in the HTB ubiquitylation mutant strain of Tetrahymena. Our study has shown that none of the levels of H3K4 methylation were detectably affected in UBC2 or BRE1 knock-out strains of Tetrahymena. These results strongly argue that, despite the conservation of the site of H2B monoubiquitylation and H3K4 mono-, di-, and trimethylation and the fact that the genes involved in the H2B ubiquitylation pathway are highly conserved, the cross-talk between Ubc2-Bre1-mediated H2B ubiquitylation and H3K4 methylation is not universally conserved among all organisms.
Acknowledgments
We thank P. J. Bruns (Cornell University) for cell strains, C. D. Allis (Rockefeller University) for providing the antibodies to Tetrahymena histones, and J. Bowen for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM021793.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S9.
- H3K4
- histone H3 lysine 4
- H3K4me3
- trimethyl-histone H3 lysine 4
- H3K4me2
- dimethyl-histone H3 lysine 4
- H3K4me1
- monomethyl-histone H3 lysine 4.
REFERENCES
- 1.Wolffe A. P. (1998) Chromatin: Structure and Function, 3rd Ed., Academic Press, Inc., San Diego [Google Scholar]
- 2.Kornberg R. D., Lorch Y. (1999) Cell 98, 285–294 [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 4.Berger S. L. (2007) Nature 447, 407–412 [DOI] [PubMed] [Google Scholar]
- 5.Ruthenburg A. J., Li H., Patel D. J., Allis C. D. (2007) Nat. Rev. Mol. Cell Biol. 8, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner B. M. (2007) Nat. Cell Biol. 9, 2–6 [DOI] [PubMed] [Google Scholar]
- 7.Goldknopf I. L., Busch H. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 864–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.West M. H., Bonner W. M. (1980) Nucleic Acids Res. 8, 4671–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pham A. D., Sauer F. (2000) Science 289, 2357–2360 [DOI] [PubMed] [Google Scholar]
- 10.Chen H. Y., Sun J. M., Zhang Y., Davie J. R., Meistrich M. L. (1998) J. Biol. Chem. 273, 13165–13169 [DOI] [PubMed] [Google Scholar]
- 11.Wang H., Zhai L., Xu J., Joo H. Y., Jackson S., Erdjument-Bromage H., Tempst P., Xiong Y., Zhang Y. (2006) Mol. Cell 22, 383–394 [DOI] [PubMed] [Google Scholar]
- 12.Weake V. M., Workman J. L. (2008) Mol. Cell 29, 653–663 [DOI] [PubMed] [Google Scholar]
- 13.Robzyk K., Recht J., Osley M. A. (2000) Science 287, 501–504 [DOI] [PubMed] [Google Scholar]
- 14.Briggs S. D., Xiao T., Sun Z. W., Caldwell J. A., Shabanowitz J., Hunt D. F., Allis C. D., Strahl B. D. (2002) Nature 418, 498. [DOI] [PubMed] [Google Scholar]
- 15.Dover J., Schneider J., Tawiah-Boateng M. A., Wood A., Dean K., Johnston M., Shilatifard A. (2002) J. Biol. Chem. 277, 28368–28371 [DOI] [PubMed] [Google Scholar]
- 16.Ng H. H., Xu R. M., Zhang Y., Struhl K. (2002) J. Biol. Chem. 277, 34655–34657 [DOI] [PubMed] [Google Scholar]
- 17.Sun Z. W., Allis C. D. (2002) Nature 418, 104–108 [DOI] [PubMed] [Google Scholar]
- 18.Hwang W. W., Venkatasubrahmanyam S., Ianculescu A. G., Tong A., Boone C., Madhani H. D. (2003) Mol. Cell 11, 261–266 [DOI] [PubMed] [Google Scholar]
- 19.Wood A., Krogan N. J., Dover J., Schneider J., Heidt J., Boateng M. A., Dean K., Golshani A., Zhang Y., Greenblatt J. F., Johnston M., Shilatifard A. (2003) Mol. Cell 11, 267–274 [DOI] [PubMed] [Google Scholar]
- 20.Henry K. W., Wyce A., Lo W. S., Duggan L. J., Emre N. C., Kao C. F., Pillus L., Shilatifard A., Osley M. A., Berger S. L. (2003) Genes Dev. 17, 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kao C. F., Hillyer C., Tsukuda T., Henry K., Berger S., Osley M. A. (2004) Genes Dev. 18, 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng H. H., Dole S., Struhl K. (2003) J. Biol. Chem. 278, 33625–33628 [DOI] [PubMed] [Google Scholar]
- 23.Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. (2003) J. Biol. Chem. 278, 34739–34742 [DOI] [PubMed] [Google Scholar]
- 24.Xiao T., Kao C. F., Krogan N. J., Sun Z. W., Greenblatt J. F., Osley M. A., Strahl B. D. (2005) Mol. Cell. Biol. 25, 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J., Hake S. B., Roeder R. G. (2005) Mol. Cell 20, 759–770 [DOI] [PubMed] [Google Scholar]
- 26.Zhu B., Zheng Y., Pham A. D., Mandal S. S., Erdjument-Bromage H., Tempst P., Reinberg D. (2005) Mol. Cell 20, 601–611 [DOI] [PubMed] [Google Scholar]
- 27.Kim J., Guermah M., McGinty R. K., Lee J. S., Tang Z., Milne T. A., Shilatifard A., Muir T. W., Roeder R. G. (2009) Cell 137, 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanny J. C., Erdjument-Bromage H., Tempst P., Allis C. D. (2007) Genes Dev. 21, 835–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zofall M., Grewal S. I. (2007) J. Biol. Chem. 282, 14065–14072 [DOI] [PubMed] [Google Scholar]
- 30.Foster E. R., Downs J. A. (2009) J. Cell Biol. 184, 631–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. (1975) in Methods in Cell Biology (Prescott D. M. ed) Vol. 9, pp. 311–327, Academic Press, Inc., New York: [DOI] [PubMed] [Google Scholar]
- 32.Bruns P. J., Brussard T. B. (1974) J. Exp. Zool. 188, 337–344 [DOI] [PubMed] [Google Scholar]
- 33.Gaertig J., Gu L., Hai B., Gorovsky M. A. (1994) Nucleic Acids Res. 22, 5391–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cassidy-Hanley D., Bowen J., Lee J. H., Cole E., VerPlank L. A., Gaertig J., Gorovsky M. A., Bruns P. J. (1997) Genetics 146, 135–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hai B., Gorovsky M. A. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1310–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pogulis R. J., Vallejo A. N., Pease L. R. (1996) in Methods in Molecular Biology (Trower M. K. ed) Vol. 57, pp. 167–176, Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 37.Gaertig J., Gorovsky M. A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 9196–9200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allis C. D., Glover C. V., Gorovsky M. A. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 4857–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glover C. V. (1979) Structure and Function of the Inner Histones of Tetrahymena Macronuclei: Partial Sequences and Histone-Histone Interactions Ph.D. thesis, University of Rochester, Rochester, NY [Google Scholar]
- 40.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 41.Stargell L. A., Bowen J., Dadd C. A., Dedon P. C., Davis M., Cook R. G., Allis C. D., Gorovsky M. A. (1993) Genes Dev. 7, 2641–2651 [DOI] [PubMed] [Google Scholar]
- 42.Miao W., Xiong J., Bowen J., Wang W., Liu Y., Braguinets O., Grigull J., Pearlman R. E., Orias E., Gorovsky M. A. (2009) PLoS ONE 4, e4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X., Gorovsky M. A. (1996) Nucleic Acids Res. 24, 3023–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y., Mochizuki K., Gorovsky M. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1679–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorne A. W., Sautiere P., Briand G., Crane-Robinson C. (1987) EMBO J. 6, 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sridhar V. V., Kapoor A., Zhang K., Zhu J., Zhou T., Hasegawa P. M., Bressan R. A., Zhu J. K. (2007) Nature 447, 735–738 [DOI] [PubMed] [Google Scholar]
- 47.Liu Y. (2003) Genetic Studies of RNA-mediated DNA Elimination in the Developing Macronuclei of Tetrahymena thermophila Ph.D. thesis, University of Rochester, Rochester, NY [Google Scholar]
- 48.Cui B., Liu Y., Gorovsky M. A. (2006) Mol. Cell. Biol. 26, 7719–7730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joazeiro C. A., Weissman A. M. (2000) Cell 102, 549–552 [DOI] [PubMed] [Google Scholar]
- 50.Koken M. H., Reynolds P., Jaspers-Dekker I., Prakash L., Prakash S., Bootsma D., Hoeijmakers J. H. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 8865–8869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koken M., Reynolds P., Bootsma D., Hoeijmakers J., Prakash S., Prakash L. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 3832–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]