Abstract
Triadin is a multiple proteins family, some isoforms being involved in muscle excitation-contraction coupling, and some having still unknown functions. To obtain clues on triadin functions, we engineered a triadin knock-out mouse line and characterized the physiological effect of triadin ablation on skeletal muscle function. These mice presented a reduced muscle strength, which seemed not to alter their survival and has been characterized in the present work. We first checked in these mice the expression level of the different proteins involved in calcium homeostasis and observed in fast muscles an increase in expression of dihydropyridine receptor, with a large reduction in calsequestrin expression. Electron microscopy analysis of KO muscles morphology demonstrated the presence of triads in abnormal orientation and a reduction in the sarcoplasmic reticulum terminal cisternae volume. Using calcium imaging on cultured myotubes, we observed a reduction in the total amount of calcium stored in the sarcoplasmic reticulum. Physiological studies have been performed to evaluate the influence of triadin deletion on skeletal muscle function. Muscle strength has been measured both on the whole animal model, using hang test or electrical stimulation combined with NMR analysis and strength measurement, or on isolated muscle using electrical stimulation. All the results obtained demonstrate an important reduction in muscle strength, indicating that triadin plays an essential role in skeletal muscle function and in skeletal muscle structure. These results indicate that triadin alteration leads to the development of a myopathy, which could be studied using this new animal model.
Introduction
Muscle contraction is activated by Ca2+ release from the sarcoplasmic reticulum in response to plasma membrane depolarization. This process, called excitation-contraction coupling, takes place at the skeletal muscle triad junction, where T-tubules and the sarcoplasmic reticulum terminal cisternae are in close contact (1). Calcium release occurs via the calcium release complex, a macromolecular complex specifically localized in the skeletal muscle triad (2). The main components of this calcium release complex are the ryanodine receptor (RyR)2 and the dihydropyridine receptor (DHPR) (3), both of which are calcium channels. It is now clear that a number of proteins are associated with these two calcium channels to form the calcium release complex, triadin being part of them. Triadin is an integral membrane protein of the sarcoplasmic reticulum, first identified in rabbit skeletal muscle as a 95-kDa glycoprotein specifically located in the triads (4, 5). Because of its co-localization with RyR in the triads, involvement of triadin in excitation-contraction coupling has been postulated (6, 7). Protein interaction studies have shown that the major molecular partners of triadin are RyR (8–10), calsequestrin (CSQ), a protein that traps calcium inside the sarcoplasmic reticulum (11–13), and junctin (14). Functional studies have shown that triadin by itself regulates the activity of the RyR calcium channel in vitro (10, 15).
Triadin is expressed in both cardiac and skeletal muscles. In cardiac muscle, several isoforms of triadin have been identified, the major one being cardiac triadin 1 (16). We have shown that at least four isoforms of triadin are expressed in rat skeletal muscle (17, 18). All these isoforms are issued from the alternative splicing of the same gene (19). We have previously reported that the major skeletal muscle triadin isoform is Trisk 51, both in rat and in human (18, 19). Trisk 95, the “classic” 95-kDa isoform, is expressed in almost equivalent amounts, and Trisk 32, the 32-kDa isoform identical to cardiac triadin 1, represents 10–20% of all triadins (18, 20). Overexpression studies using adenoviruses have shown that Trisk 95 is able to block the depolarization-induced calcium release, but in the same conditions, no effect was observed with Trisk 51 overexpression, and therefore no function could be attributed to this isoform (21). Triadin knockdown has also been realized using small interference RNA in cultured cells (22, 23), and resulted in reduction of the depolarization-induced calcium release. In addition to various functional effects resulting from expression modification, the triadin isoforms have different localizations and partners, and whereas Trisk 95 and Trisk 51 are both located within the triad and are associated with RyR, Trisk 32 is mainly in the longitudinal sarcoplasmic reticulum associated with the inositol trisphosphate receptor (18).
Triadin has mainly been characterized on a molecular point of view, and only a few physiological studies have been performed. To determine functions of triadin in the whole animal, we have engineered a mouse line deleted of all the triadin isoforms. After molecular characterization of the muscles of this mouse, we studied their morphology and function. Muscle function and muscle strength were studied both on whole animal and on isolated muscle, using hanging test or electrical stimulation combined with force measurement and magnetic resonance imaging (MRI). All the results presented in this study confirmed an important reduction in muscle strength in triadin KO animals, and this mouse line could therefore represent a new animal model to study the development of a structural myopathy.
EXPERIMENTAL PROCEDURES
Engineering of the Triadin KO Mouse
The triadin conditional mutant mouse line (triadin flox), as well as the full triadin KO (triadin KO) mouse line were established at the Mouse Clinical Institute/Institut Clinique de la Souris (Illkirch, France). The targeting vector was constructed as follows: a 1.2-kb fragment corresponding to the floxed fragment (comprising the ATG containing exon 1) as well as the 3.7-kb 5′ homolog arm and 3.3-kb 3′ homolog arm were amplified by PCR and subsequently cloned into a Mouse Clinical Institute basic vector. This vector already contained the LoxP sites and the Neomycin resistance cassette, flanked by frt sites, which was then excised after selection using flp. The coding region and the junctions were verified by sequencing. The linearized construct was electroporated in 129S2/SvPas mouse embryonic stem cells. After selection, targeted clones were identified by PCR using external primers and further confirmed by Southern blots with Neo and 3′ external probes. Three positive embryonic stem clones were obtained. One was microinjected into C57BL/6J blastocysts, and derived male chimaeras gave germ line transmission. To generate full triadin KO mice, heterozygote triadin flox mice were crossed with cytomegalovirus Cre+/+ mice (24) expressing the Cre recombinase in the zygote, to generate heterozygous triadin KO mice (triadin+/−). These mice were crossed to generate WT (triadin+/+) and KO (triadin−/−) littermates used in the experiments described here.
Antibodies and Western Blot Analysis
Anti-calsequestrin monoclonal antibody (clone VIIID12) was obtained from Affinity BioReagents. Polyclonal antibody against RyR has been previously described (3), as well as antibodies against Trisk 95 and Trisk 51 (17), and antibodies against the N-terminal end of triadins (25). An antibody against mouse Trisk 32 (C-terminal peptide: RRQQEVQRE) was produced as previously described (18). Rabbit anti-bovine mitochondrial F0/F1-ATPase antibody was previously characterized (26). Rabbit anti-junctin was from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-SERCA, kindly provided by Dr. M. J. Moutin, was produced by rabbit immunization with purified large cytoplasmic loop of the Ca2+-ATPase (27). Secondary antibodies used for Western blot were labeled with horseradish peroxidase (Jackson ImmunoResearch Laboratories).
The presence and the amount of different proteins in samples were assayed by Western blot analysis, using a chemiluminescent reagent (Western lightning Chemiluminescence reagent plus, PerkinElmer Life Sciences) after electrophoretic separation of the protein on a 5–15% acrylamide gel and electrotransfer on Immobilon P (Millipore) (17). The quantification was performed using a ChemiDoc XRS apparatus (Bio-Rad) and the Quantity One Software (Bio-Rad). The signal was normalized to the amount of myosin, measured after staining of the membrane with Coomassie Blue.
Muscle Fibers Preparation and Immunofluorescent Labeling
All procedures using animals were approved by the Institutional ethics committee and followed the guidelines of the National Research Council Guide for the care and use of laboratory animals. Flexor digitorum brevis (FDB) fibers were dissociated according to the procedure described by Pouvreau et al. (28). Intact FDB were excised from the hind paw in Ringer-glucose solution (136 mm NaCl, 10 mm glucose, 5 mm KCl, 2.6 mm CaCl2, 1 mm MgCl2, 10 mm Hepes, pH 7.2) and digested 30–45 min at 37 °C in collagenase (2 mg/ml in Ringer, Sigma-Aldrich). Muscles were washed 10 min at room temperature and kept 2 h at 4 °C in 25 ml of Ringer. Fibers were then dissociated by several flushing steps and seeded on laminin-coated slides 20 min at room temperature. Immunofluorescent labeling was performed using a protocol adapted from Vassilopoulos et al. (18). Fibers were fixed for 15 min with 4% paraformaldehyde in phosphate-buffered saline at room temperature, permeabilized for 5 min with 1% Triton X-100 in phosphate-buffered saline at room temperature, and saturated for 30 min with phosphate-buffered saline supplemented with 0.1% Triton X-100, 0.5% bovine serum albumin, and 2% goat serum. Fibers were incubated 1 h at room temperature with the primary antibodies (dilution, 1:100 to 1:800) in phosphate-buffered saline supplemented with 0.1% Triton X-100, 0.5% bovine serum albumin, and 2% goat serum and then incubated with secondary antibodies for 30 min at room temperature. The nuclei were stained with TO-PRO-3 (Molecular Probes), and the samples were mounted with an anti-fading solution (fluorescent mounting medium, DakoCytomation). For double labeling, the two primary antibodies (from different species) and the two secondary antibodies were added at the same time. The secondary antibodies were labeled with either Alexa-488, Alexa-546 (both from Molecular Probes), or Cy3 (Jackson ImmunoResearch Laboratories). Muscle fibers were analyzed by confocal laser scanning microscopy using a Leica SPE operating system, with sequential excitation of the two fluorophores.
Electron Microscopy
Extensor digitorum longus (EDL) and soleus (Sol) muscles of 5- to 6-month-old mice were excised and cut into small pieces. After three washes in cacodylate buffer 0.1 m, muscles were post-fixed in 2.5% glutaraldehyde, cacodylate buffer (0.1 m) for 2 h at 4 °C. After three washes in cacodylate buffer, T-tubules were stained in 1% OsO4, cacodylate buffer (0.1 m) for 1 h on ice. Muscles were washed 5 min in cacodylate buffer (0.1 m) and four times in H2O and then stained in 1% uranyl acetate, pH 4, 4 °C overnight. After dehydration with a graded series of alcohols and infiltrated in epoxy resin (Fluka), fibers were horizontally embedded in flat molds. For T-tubule staining (adaptation from Galbiati et al. (31)), 75 mm CaCl2 was added in the post-fixation solution, 0.4% K3Fe(CN)6 was added in the OsO4 solution, and the mixture was incubated for 2 h at room temperature. 70 nm sections were cut from the blocks with the ultramicrotome Leica Ultracut S and post-stained for 10 min at room temperature with 5% uranyl acetate, pH 4, washed four times in water, incubated 5 min at room temperature in 4% lead citrate, and washed four times in water. The sections were observed with a JEOL JEM 1200EX II at 60 kV.
Muscle Homogenates Preparation
Two types of skeletal muscle, i.e. EDL and Sol, were collected from young (10 weeks) or adult (6–9 months) mouse hind legs. Crude homogenates were prepared from each muscle by homogenization in 200 mm sucrose, 20 mm HEPES (pH 7.4), 0.4 mm CaCl2, 2 mm leupeptin, 100 mm phenylmethylsulfonyl fluoride, 1 mm diisopropyl fluorophosphate. The amount of total protein was estimated using a modified Folin assay in presence of SDS.
Cell Culture and Intracellular Calcium Measurements
Primary cultures of neonatal mouse skeletal muscle (WT and KO) were obtained as described previously (17). Changes in intracellular calcium were measured on WT or KO 3–4 days primary myotubes, using the calcium-dependent fluorescent dye Fluo 4-AM (Molecular Probes), as described previously (21). Calcium imaging was performed in Krebs buffer (136 mm NaCl, 5 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 10 mm HEPES, pH 7.4). To obtain a calcium-free Krebs solution, CaCl2 was left out, while 1 mm EGTA, 10 μm La3+, and 50 μm Cd2+ were added. Thapsigargin (Molecular Probes) was diluted at 1 μm in this medium and applied simultaneously with caffeine (40 mm) as described previously (29, 30). Fluorescence was measured by confocal laser scanning microscopy, using a Leica TCS-SPE operating system, in the xyt mode. Data are given as mean ± S.E., and n represents the number of myotubes in each condition.
Hang Tests
For the first test, animals (6-month-old females) were placed on wires stretched between two poles 50 cm apart and were allowed to freely move along the wire. Each animal was tested four or five times during a 7-day period, and all the tests were used for the statistical analysis. For the second test, animals (10 weeks and 6 months old) were positioned on a cross-wired surface where they could hold on with all four paws. The grid was then turned up side down. We measured the time during which animals were able to stay on wires before falling. The test was repeated three times successively for each animal, and only the best score was used for the statistical analysis. After 5-min hanging (300 s), the test was stopped.
Isolated Muscle Contraction Measurements
Fast (EDL) or slow (Sol) twitch muscles, isolated from 6-month-old females, were placed horizontally in an experimental chamber continuously superfused (10 ml·min−1) with Krebs' solution (135 mm NaCl, 5 mm KCl, 2.5 mm CaCl2, 1 mm MgSO4, 10 mm glucose, 10 mm NaHCO3, 10 mm Hepes, pH 7.2, at room temperature) equilibrated with 95% O2 plus 5% CO2.
One end of the muscle was attached to a rod while the other was attached to a capacitive force transducer. Two platinum electrodes placed adjacent to the muscle were used to deliver short (1 ms), supramaximal pulses to elicit single twitches. Force responses were digitized at 2 kHz by using a Digidata 1200 A/D card and stored with Axotape software (Axon Instruments, Foster City, CA). Muscles were then stretched by adjusting the position of the transducer to a length that produced the maximal force response and allowed to equilibrate for 60 min.
Single pulses (1 Hz) were used to elicit single twitches. At least 10 twitches were measured under these conditions from each animal, and the averaged amplitude of all transients was used to characterize the given animal. Tetanic contraction was obtained by stimulation at 200 Hz during 200 ms for EDL and at 100 Hz during 500 ms for Sol.
Fatigue Test on Isolated Muscle
To test muscle fatigability a series of tetanic contractions (altogether 150) at a frequency of 0.5 Hz was evoked (32). The amplitude of each tetanus was then normalized to that of the largest in the series (usually the first or the second) and plotted as a function of the position within the series.
Animal Preparation for NMR Studies
Nine WT and nine triadin KO 5-month-old males were used for these experiments. For each animal, MRI functional investigation was performed in the left gastrocnemius muscles. Mice were initially anesthetized in an induction chamber (Equipement Vétérinaire Minerve, France) using 4% isoflurane (Forene, Abbot France, Rungis, France) in 33% O2 and 66% N2O. Once both lower hind limbs were shaved, the anesthetized animal was placed supine in a customized cradle especially designed for a strictly non-invasive muscle investigation. This cradle integrates two rod-shaped transcutaneous electrodes (located above the knee and under the heel, respectively) connected to an electrical stimulator (Stimulator I series, Hugo Sachs Elektronik, Harvard Apparatus, Holliston, MA) and an ergometer consisting of a 9 × 24 mm foot pedal coupled to a force transducer. The transducer was constructed by sticking a strain gauge (1-LY11-6/120A, HBM GmbH, Darmstadt, Germany) according to the manufacturer specifications on a 9 × 32 mm Bakelite slat (0.4-mm thickness) in a Wheatstone bridge design. The foot was positioned on the ergometer pedal, and the hind limb was immobilized in the cradle. In that position, the hind limb was centered inside a 20-mm-diameter 1H Helmholtz imaging coil, and the belly of the gastrocnemius muscle was located above an elliptic 31P-MRS surface coil (8 × 12 mm). The pedal position was adjusted to modify the angle between the foot and the lower hind limb so that the gastrocnemius muscle was passively stretched at rest and produced a maximum isometric twitch tension in response to supramaximal square wave pulses (1.5-ms duration). Supramaximal current (usually 2–4 mA) was determined by inducing contractions with increasing current. Throughout the experiment, anesthesia was maintained by gas inhalation with a facemask continuously supplied with 2.5% isoflurane in 33% O2 (0.4 liter/min) and 66% N2O (0.8 liter/min) and connected to an open-circuit gas anesthesia machine (Isotec 3, Ohmeda Medical, Herts, UK). Animal body temperature was maintained through a feedback loop, including an electrical heating blanket (Prang & Partner AG, Pfungen, Switzerland), a temperature control unit (ref 507137, Harvard Apparatus) and a customized rectal thermometer constructed with an NTC thermistor SMC series (NCP18XW220J03RB-2.2K, Murata, Kyoto, Japan).
Whole Animal Stimulation Protocol and Force Measurements
The transcutaneous stimulation protocol consisted in 6 min of repeated isometric contractions at a frequency of 1.7 Hz. Analog electrical signal coming out from the force transducer was amplified with a customized amplifier (Operational amplifier AD620, Analog Devices, Norwood, MA; gain = 70 dB; bandwidth, 0–5 kHz) converted to a digital signal (PCI-6220, National Instrument, Austin, TX) and recorded on a personal computer using the WinATS software version 6.5 (Sysma, Aix-en-Provence, France). Isometric force production per twitch (in newtons) was averaged every 30 s and normalized to the MRI-calculated muscle volume.
MRI Data Acquisition and Processing
Investigations were performed in a 4.7-Tesla horizontal magnet (47/30 Biospec Avance, Bruker, Karlsruhe, Germany) equipped with a Bruker 120-mm BGA12SL (200 mT/m) gradient insert. Ten non-contiguous axial scout slices (1 mm thickness, spaced 0.5 mm) covering the region from the knee to the ankle were selected across the lower hind limb. Multiecho images of these slices (8 echoes, 10.587-ms spaced, 1000-ms repetition time, 42 × 42 mm2 field-of-view, 256 × 256 acquisition matrix, and 3.2-min acquisition time) were recorded at rest. 31P-MRS spectra (32 scans, 8-kHz sweep width, and 2048 data points) from the gastrocnemius region were continuously acquired in 60-s blocks throughout a standardized experimental protocol consisting of 6-min rest, 6-min stimulation, and 16-min recovery. MRI data acquisition was gated to muscle stimulation to reduce potential motion artifacts due to contraction. MRI data were processed using a customized image analysis program developed on the IDL platform (Interactive Data Language, Research System, Inc., Boulder, CO). For each MRI slice, a gastrocnemius region was manually outlined to measure the corresponding cross-sectional areas (in mm2). The gastrocnemius muscle volume (mm3) was calculated as the sum of volumes included between consecutive slices. PCr, inorganic phosphate (Pi), and ATP peaks were identified by a time-domain fitting routine using the AMARES-MRUI Fortran code and using appropriate prior knowledge (33). Signal areas were corrected for magnetic saturation effects using fully relaxed spectra collected at rest with a repetition time of 20 s. Intracellular pH (pHi) was calculated from the chemical shift of the Pi signal relative to PCr (34). Time points for the time course of pHi and phosphorylated metabolite concentrations were assigned to the midpoints of the acquisition intervals.
Statistical Analysis
Unless otherwise stated, data were pooled over the animals within the same group and are presented as means ± S.E. Significance of the differences between WT and KO animals was assessed using Student's t test, assuming significance at p < 0.05.
NMR variables evolving with respect to time during the stimulation period (isometric force production, metabolite concentrations, and pHi) were analyzed with one-way repeated-measurements analysis of variance followed by post-hoc Tukey's tests using JMP software (SAS Institute Inc., Cary, NC).
RESULTS
Mouse Model Development
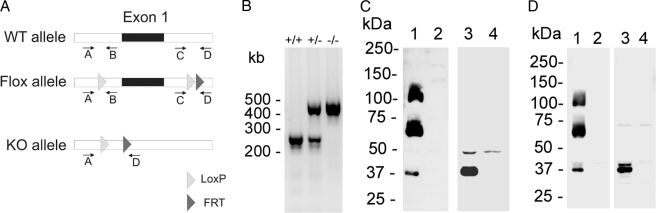
To delete triadin from skeletal and cardiac muscles, we have engineered a triadin mouse in which the first exon has been flanked by two lox-P sites (Fig. 1A). After mating these “floxed” mice with mice expressing the Cre recombinase in the whole animal, we produced a triadin null mouse line, in which triadin expression has been abolished in all tissues. KO (−/−) and WT (+/+) animals were obtained by mating heterozygote mice (+/−).The genotype of each animal was double-checked by PCR, using specific primers (Fig. 1B). At first sight, KO animals were indiscernible from WT animals, and the KO mice grew up and reproduced normally. Nevertheless, the adult KO animals seemed quieter in their cage, which could reflect cardiac or skeletal muscles problems. Before performing the full physiological characterization of the animals, we performed molecular controls, to evaluate the extent of protein modifications induced by deletion of triadin gene. We first checked by Western blot the absence of all triadin isoforms in skeletal as well as in cardiac muscles. We used different anti-triadin antibodies, because some react with all triadins and some are specific for single isoforms. No specific signal was detected with an antibody directed against the common N-terminal end of triadin, either in skeletal muscle or in cardiac muscle (Fig. 1C). To confirm the absence of all triadin, and particularly the expression of proteins truncated in the N-terminal end (possibly due to an in-frame initiation with another ATG), the reactivity with specific C-terminal antibodies was also assayed (Fig. 1D). Using a mixture of anti-Trisk 95, anti-Trisk 51, and anti-Trisk 32, a similar result was observed: no triadin was detected in the cardiac and the skeletal muscles of the KO animals. These experiments demonstrate that the deletion of the first exon in triadin gene has indeed led to the production of a full triadin KO animal.
FIGURE 1.
Molecular characterization of the triadin KO mouse. A, schematic representation of the wild-type, floxed, and KO allele, and positioning of the primers used for the PCR amplification. B, PCR amplification of genomic DNA was performed with a mix containing primers A, C, and D. This gives rise to a band of 240 bp on WT animals (+/+, primers C and D), a band of 477 bp on KO animals (−/−, primers A and D), and both on heterozygotes animals (+/−). The DNA fragment between primers A and D is too large to be amplified on WT animals. C, Western blot analysis using an anti-N-terminal antibody on WT skeletal muscle (lane 1), KO skeletal muscle (lane 2), WT heart (lane 3), and KO heart (lane 4). The signals corresponding to Trisk 95 (100 kDa), Trisk 51 (60 kDa), and Trisk 32 (37 kDa) was absent in KO muscles (skeletal and cardiac). D, Western blot analysis using a mix of specific antibodies against Trisk 95, Trisk 51, and Trisk 32, on WT skeletal muscle (lane 1), KO skeletal muscle (lane 2), WT heart (lane 3), and KO heart (lane 4). The signals corresponding to Trisk 95 (100 kDa), Trisk 51 (60 kDa), and Trisk 32 (37 kDa) was absent in KO muscles (skeletal and cardiac).
Expression Levels of the Proteins Involved in Calcium Homeostasis
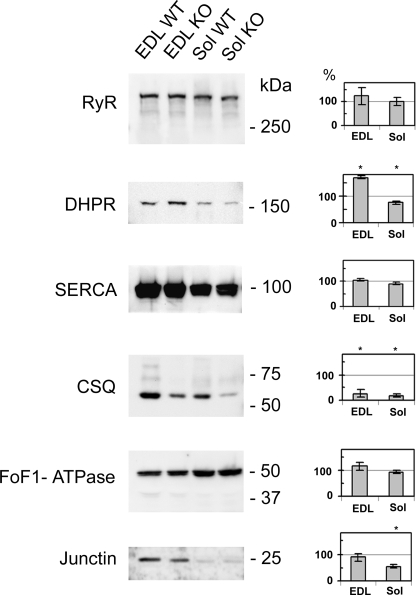
We used quantitative Western blotting to evaluate the modification of expression induced by triadin suppression in the different proteins involved in calcium homeostasis. The expression levels of RyR, DHPR, SERCA, CSQ, Junctin, and the mitochondrial F0/F1-ATP synthase were evaluated on EDL and Sol from three different WT or KO animals of 6–9 months age (Fig. 2). Compared with the expression level in WT muscles, set to 100% as reference, no modification was observed for RyR (125% ± 35%, p = 0.166 in EDL and 102% ± 17%, p = 0.295 in Sol), for SERCA (104% ± 5%, p = 0.49 in EDL and 90% ± 6%, p = 0.23 in Sol) and for F0/F1-ATPase (116% ± 6%, p = 0.099 in EDL and 94% ± 8%, p = 0.823 in Sol). Junctin was not modified in EDL (89%± 12%, p = 0.48) and decreased in Sol (55% ± 12%, p = 0.023). An important modification was observed for the α1 subunit of DHPR, which was increased to 170% ± 14%, p = 0.004 in EDL, and reduced in Sol (75% ± 7%, p = 0.018). The major modification concerns CSQ, with a reduction to 27% ± 4% (p < 0.001) in EDL and 19% ± 4% (p < 0.001) in Sol. Similar results were obtained on younger animals (10 weeks, data not shown). DHPR expression was significantly increased in EDL (157% ± 18%) and reduced in Sol (81% ± 5%). CSQ expression was reduced in both muscle (56% ± 5% in EDL and 20% ± 2% in Sol). The other proteins were not significantly modified at 10 weeks.
FIGURE 2.
Western blot analyzes of EDL and Soleus. 40–80 μg of muscle homogenates was loaded onto 5–15% polyacrylamide gel. The homogenates were prepared using EDL and soleus (Sol) isolated either from wild type (WT) or triadin KO (KO) animal. After electrophoresis separation and Western blot analysis using the indicated antibody (RyR, DHPR, SERCA, CSQ, FoF1-ATPase, and junctin), the membranes were stained with Coomassie Blue to evaluate the amount of myosin in each lane, and the signal in Western blot was normalized to the amount of myosin. The corresponding quantification is presented on the histogram on the right of each blot, the control in each WT condition being set at 100%.
Influence of Triadin Deletion on the Localization of Its Partners
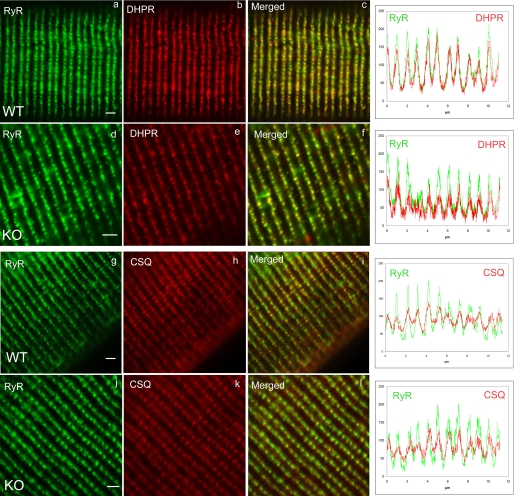
Triadin has been shown to interact with RyR and CSQ and was proposed to anchor the luminal CSQ at the junctional part of sarcoplasmic reticulum, in the triad region (12). Therefore, triadin deletion could result in CSQ delocalization, whereas no effect is expected a priori on DHPR or RyR localization. To test this hypothesis, double immunofluorescent labeling was performed on dissociated FDB fibers of WT or KO animals, using RyR/DHPR antibodies or RyR/CSQ antibodies (Fig. 3). As expected, no modification was observed in DHPR and RyR localization (Fig. 3, a–f, and associated quantification graphs), which are perfectly co-localized and display the typical labeling of the triads in double rows of dots. But, CSQ was also perfectly co-localized with RyR, in contrast to our initial hypothesis (Fig. 3, g–l, and associated graphs).
FIGURE 3.
Double immunofluorescent labeling on isolated muscle fibers. Muscle fibers were isolated from wild type (WT) or triadin KO (KO) FDB and used to perform double immunofluorescent labeling, with antibodies directed against RyR and DHPR (panels a–f) or RyR and CSQ (panels g–l). The fluorescence intensity as a function of distance on the fiber was plotted for six successive triads, and was reported on the adjacent graph for each labeling. The color code is preserved, and the green curve corresponds to the green immunolabeling, whereas the red curve corresponds to the red labeling. Scale bar, 1 μm.
Morphological Modifications of KO Muscle
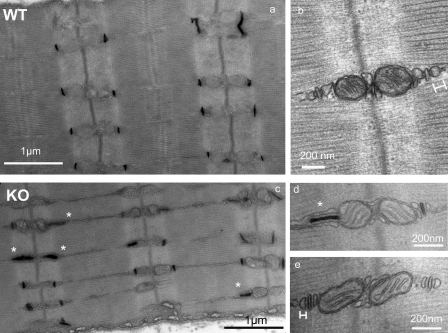
To evaluate more precisely the influence of triadin deletion on muscle ultrastructure, KO and WT muscles were analyzed by electron microscopy. No modification was observed in soleus (data not shown), whereas in EDL, numbers of triads were in abnormal orientation (Fig. 4), being longitudinal or oblique instead of transversal. The abnormal orientation was better visualized after T-tubules labeling (Fig. 4, a and c), and the number of triads with longitudinal or oblique orientation was evaluated to 25% (n = 722) in KO instead of 3% (n = 680) in WT. It is noteworthy that, when the triads are longitudinally orientated, they usually lose the associated mitochondria. At last we observed a flattening of the terminal cisternae, which are significantly reduced from 68 ± 3 nm (n = 34) to 26 ± 1 nm (n = 55), as illustrated on Fig. 4 (b and e).
FIGURE 4.
Electron microscopy analysis of EDL muscle. EDL were isolated from WT or KO animals and examined using electron microscopy at different magnifications, with (a, c, and d) or without (b and e) tubule labeling. A number of triads are in abnormal orientation in KO muscle (white stars) and are presented mainly in a longitudinal orientation (c and d). The width of the terminal cisternae was measured as shown in b and e, respectively, on WT and KO muscles.
Effect of Triadin Deletion on Calcium Fluxes
Calcium imaging studies were performed to assess the ability of triadin KO myotubes to release calcium after stimulation and to evaluate the amount of calcium releasable upon stimulation: the free SR calcium directly released after stimulation (the amount of which is reflected in the amplitude of the peak), and the total amount of calcium stored (free and CSQ-bound). The data are presented on Fig. 5. Membrane depolarization induced by addition of 40 mm KCl, in the presence of extracellular calcium, results in a massive calcium release. The amplitude of the peak is slightly but not significantly (p = 0.14) reduced in triadin KO myotubes (Fig 5A). The return to basal level is significantly slower (p < 0.05) in KO compared with WT (Fig. 5A) reflecting either a reduction in the reuptake process or an additional intracellular influx.
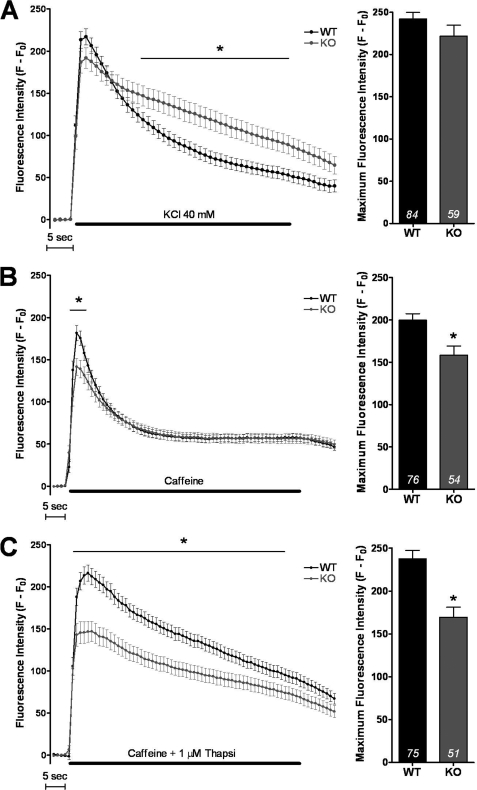
FIGURE 5.
Calcium imaging in WT or KO cultured myotubes. A, KCl depolarization (40 mm) was induced in presence of 2 mm external calcium. The left curves represent the fluorescence variation in WT (black curve) or KO myotubes (gray curve). The histogram on the right presents the maximum value ± S.E. of fluorescence in control myotubes and in KO myotubes with the number of myotubes tested indicated in each histogram. The asterisk indicates statistically significant difference (p < 0.05, Student t test). B, caffeine (40 mm) was used to directly stimulate RyR in the presence of 2 mm external calcium. The curve and the histogram present, respectively, the fluorescence variation and the maximum fluorescence in WT and KO myotubes. C, the same caffeine stimulation was performed in the presence of 1 μm thapsigargin, 0.5 mm Cd2+, 0.1 mm La3+, in WT and KO myotubes, to evaluate the amplitude of the calcium store.
Direct stimulation of RyR by caffeine was performed, either in the presence of extracellular calcium (Fig. 5B) or in the presence of Cd2+ or La3+ to block extracellular calcium entry and thapsigargin to block SERCA and SR calcium reuptake (Fig. 5C). In the presence of extracellular calcium, the maximal amplitude of the calcium release is significantly reduced in KO myotubes (Fig. 5B, p = 0.002), but the return to basal level is identical. This indicates a reduction in the free SR calcium rapidly releasable upon stimulation, but no modification in the cytosolic calcium uptake process. Therefore the effect observed in Fig. 5A (more slowly return to basal level) is most probably due to an increased extracellular calcium influx via DHPR in correlation with an increased expression of the protein.
When the same experiment was performed while preventing Ca2+ influx from extracellular medium as well as SR re-uptake using caffeine in presence of Cd2+, La3+, and thapsigargin (Fig. 5C), the amplitude of the peak was significantly reduced in KO (p < 0.001), definitively pointing to a reduction in the amplitude of the calcium store.
Physiological Studies
To test the overall muscle performance of triadin KO mice, the animals were subjected to different strength tests, at various ages. They were either allowed to freely walk on a wire stretched between two rods or hung gripping a cross-wired surface with all four paws. KO mice performed significantly worse than their WT littermates as assessed by the time until they fell. The time spent walking the wire was 50 ± 13 s for the WT (n = 5 females, 6 months old) as compared with 23 ± 5 s (n = 6 females, 6 months old) for the KO mice (p < 0.05). Similar results were obtained with the wire grid test, with a hanging time of 290 ± 10 s (n = 15 animals both sex, 10 weeks old) and 45 ± 12 s (n = 11 animals both sex, 10 weeks old, p < 0.001) for the WT and KO mice, respectively. The weight of the animals had been followed during 2 months, and no difference was noticed between WT and KO (data not shown). The same wire grid test was also performed at 6 months and gave similar results (data not shown). These data suggested that KO mice might be weaker than WT mice.
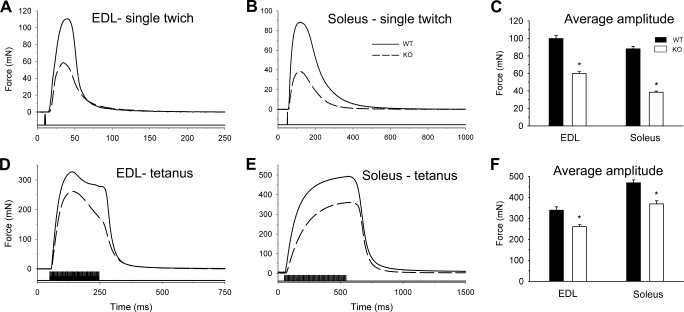
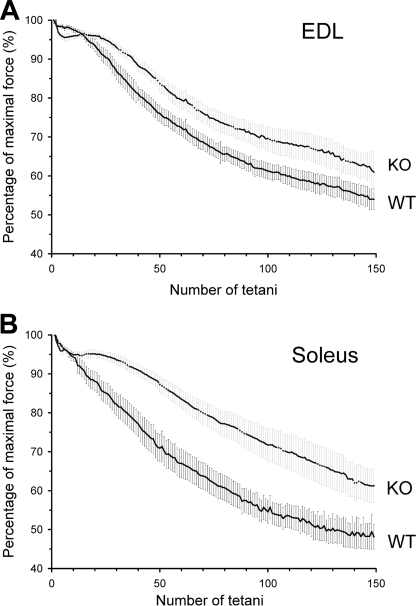
To assay the muscle strength independently of any other parameter (equilibrium, walking ability, etc.), muscle performance was measured on isolated muscles. Fast and slow twitch muscles were dissected from the animals, and twitch and tetanic force were recorded. Fig. 6 presents individual traces of twitches (Fig. 6, A and B) and tetani (Fig. 6, D and E) from both EDL and Sol of WT and KO animals. As depicted in Fig. 6 both single twitches and tetani were smaller in amplitude for KO than for WT mice. Pooled data for the amplitudes of twitches and tetani are presented in Fig. 6, C and F, respectively. Averages present a slightly greater reduction in twitch force for Sol as compared with EDL muscles in the KO mice. This difference was, however, not present for the tetani. Overall, both twitch and tetanic forces were significantly reduced in fast and slow twitch muscles of KO animals. To test how muscles from WT and KO animals endure fatigue, series of tetani were applied. Fig. 7 illustrates the evolution of the maximal amplitude of the tetani as normalized to the maximal amplitude in a given series. In these experiments, the absolute force was always greater in WT than in KO mice. The decline in tetanic force was slower for both muscle types in KO than in WT animals suggesting a slower evolution of fatigue. At the end of the test, the maximal force of EDL muscle was 54% ± 3% of the initial value for WT and 61% ± 5% in KO mice, and in Sol, it was 48% ± 3% in WT and 61% ± 4% in KO mice. Nevertheless, even at the end of the fatigue run the absolute force was not greater in KO than in WT mice (not illustrated). Interestingly, the reduction in peak tetanic force underwent a slight but sudden drop followed by a plateau-like phase early in the run giving rise to a shoulder-like part in the data measured on KO mice. Such a phenomenon was never observed on either EDL or soleus from control mice, and could point to a possible modification in muscle metabolism.
FIGURE 6.
Force transients in isolated muscles of wild-type and triadin knock-out mice. A and B, single twitches elicited by short (1 ms), supramaximal (5 V) stimulation on EDL (A) and Sol (B) from WT (continuous line) and KO (dashed line) mice. D and E, tetani elicited by a train of supramaximal stimulation at 200 Hz for 200 ms on EDL (D) and at 100 Hz for 500 ms on Sol (E) from WT (continuous line) and KO (dashed line) mice. C and F, data were pooled from 5 WT and 6 KO mice, after averaging the given parameter over each individual record (usually 10) measured on an individual muscle. Average amplitudes of the twitches (C) and of the tetani (F) are shown. Asterisks denote the significant difference between WT and KO mice (p < 0.05).
FIGURE 7.
Fatigability of wild-type and knock-out mice. Evolution of the amplitude of tetani on EDL (A) and Sol (B). Altogether 150 tetani (200 and 500 ms long stimulation at 200 and 100 Hz for EDL and Sol, respectively), repeated every 2 s, were evoked on both muscle types, and their amplitudes were normalized to the maximal value in the given train. Measurements were pooled from all mice of the given batch to yield the data points presented in the figure.
Gastrocnemius Muscle Volume and Mechanical Measurements
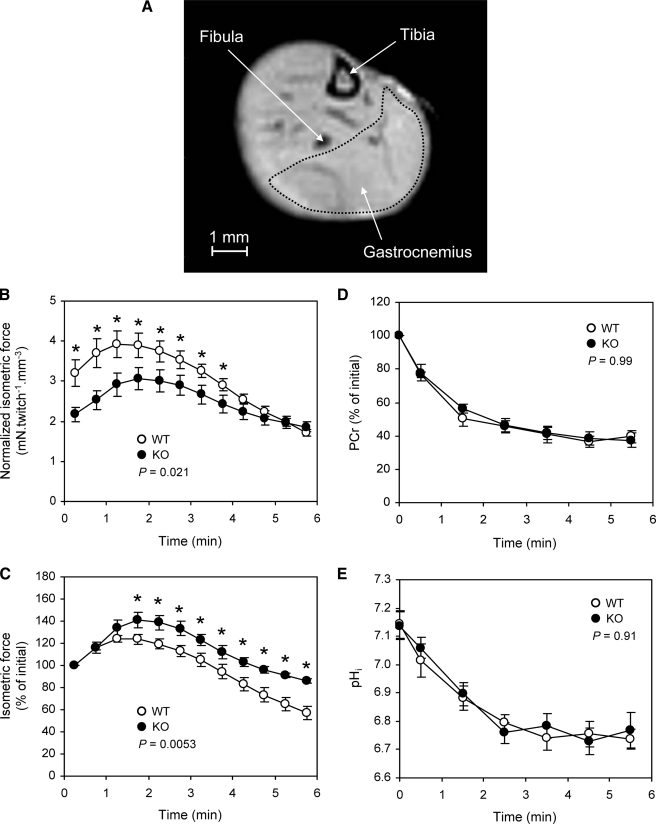
To confirm the results observed in isolated muscle, analyses of muscle morphology, function, and metabolism were performed using NMR combined with muscle contraction. The gastrocnemius muscle was stimulated on an anesthetized animal, to induce its contraction, while 1H and 31P NMR spectra were registered. Muscle volume measured using MRI (Fig. 8A) was significantly larger in WT mice (109.4 ± 3.3 mm3, n = 9) than in KO mice (92.6 ± 4.3 mm3, n = 9, p < 0.05), but no difference in the animal weight was measured (28.6 ± 1.4 g for WT versus 26.8 ± 0.9 g for KO).
FIGURE 8.
NMR analysis. A, NMR image of a hind mouse leg showing delimitation of gastrocnemius. B, isometric force normalized to muscle volume and (C) relative isometric force during 6 min of repeated isometric contractions induced electrically at 1.7 Hz in gastrocnemius. Values are shown as means ± S.E. The p value is the result of a one-way repeated-measures analysis of variance. *, significant difference between both groups for corresponding time points (p < 0.05). D, changes in gastrocnemius PCr level and (E) intracellular pH during 6 min of repeated isometric contractions induced electrically at 1.7 Hz. Values are shown as means ± S.E. The p value is the result of a one-way repeated-measures analysis of variance. The first time point (t = 0) indicates the resting value.
Isometric force production was lower in KO mice throughout the whole 6-min stimulation protocol (Fig. 8B). For each group, force transiently increased in the early stage of the stimulation period, to reach a maximal value (Fig. 8B). This increase was significantly larger in the KO group (+40.7 ± 7.0%) when compared with control animals (+24.0 ± 3.3%) (Fig. 8C). Afterward, force decreased progressively until the end of the stimulation period (Fig. 8, B and C). The corresponding decrease, measured at the end of the stimulation period and illustrating muscle fatigue, was smaller in the KO group (85.9 ± 2.4% of the initial value) as compared with controls (57.1 ± 6.0% of the initial value) (Fig. 8C).
Energy Metabolism
At rest, the PCr/ATP ratio did not differ between both groups (Table 1). Basal pHi was similar in WT (7.14 ± 0.05) and KO (7.14 ± 0.04) (Fig. 8D). During the stimulation period, the time courses of PCr (Fig. 8D) and pHi (Fig. 8E) were similar in both groups. PCr was rapidly consumed at the onset of the stimulation period, and the corresponding initial rates of PCr consumption were identical (Table 1). After 2 min of stimulation, PCr reached a steady state that was maintained until the end of the stimulation period. At that time, PCr levels were 39.7 ± 3.6% and 37.4 ± 4.5% of their basal values in WT and KO groups, respectively. At the onset of the post-stimulation period, the rate of PCr resynthesis did not differ between groups (Table 1).
TABLE 1.
Gastrocnemius energy metabolism in WT and KO mice
Values are means ± S.E. VPCrstim is the rate of PCr degradation at the start of the stimulation period; VPCrrec is the rate of PCr resynthesis at the start of the post-stimulation period.
| WT | KO | |
|---|---|---|
| Basal PCr/ATP ratio | 2.52 ± 0.08 | 2.48 ± 0.23 |
| VPCrstim (%.min−1) | 67.0 ± 7.9 | 51.4 ± 7.2 |
| VPCrrec (%.min−1) | 34.6 ± 3.9 | 30.6 ± 6.1 |
In both groups, pHi, which reflects the anaerobic metabolism, fell rapidly in the early stages of the stimulation period to reach a steady-state after 3 min of stimulation (Fig. 8E). This steady-state remained fairly constant until the end of the stimulation period, when pHi was 6.74 ± 0.04 in WT and 6.77 ± 0.06 in KO animals, respectively.
DISCUSSION
Genetic ablation of a protein has been one of the most powerful technique used to identify the precise spectrum of its functions. It has been successfully applied to a few members of the skeletal calcium release complex (RyR1 (35), DHPR (36–38), and CSQ1 (39)), to identify which precise function could be associated or not with each protein. Whereas deletion of RyR results in perinatal death, as deletion of the α1 subunit of DHPR, deletion of CSQ1, that was considered as essential, has surprisingly no effect on mouse survival or on muscle function (39). Triadin precise function is still unknown, but as the triadin gene is expressed in heart, it was initially imagined that its deletion would lead to embryonic death, as for RyR2 (40). Surprisingly, these mice are viable, as observed in a similar triadin KO mouse model recently developed (30), and contrary to CSQ KO mice, presented important muscle impairment, which has been characterized in the present work.
Triadin gene undergoes an important alternative splicing, and some additional triadin isoforms could possibly be still unidentified. To delete all the triadin isoforms, which share at least the eight first exons, we suppressed the first exon from the triadin gene. As expected, both in cardiac and skeletal muscles of these mice, all known triadin isoforms were deleted. Nevertheless, triadin suppression affects neither the mouse survival nor its reproduction, as observed in the similar model recently reported (30).
As an adaptative mechanism, the expression levels of some potential triadin partners have been modified in our triadin KO model. Calsequestrin expression level undergoes the major modification, confirming the strong relationship between triadin and CSQ. The present reduction in the CSQ expression level is larger than that observed in the triadin KO mouse model described previously (4-fold reduction in all muscle types in the present work, whereas a 1.5-fold reduction was shown by Shen et al. (30)). It is noteworthy that triadin suppression did not induce any modification in CSQ triad localization. Therefore, if triadin is indeed responsible of anchoring CSQ at the terminal cisternae of sarcoplasmic reticulum, it is not the only protein involved in this function, and most probably junctin, which is not modified in this triadin KO mouse line, is a good candidate to fulfill this function (41). The important reduction in CSQ expression could indicate either that triadin is able to regulate CSQ gene expression or that a defect in CSQ anchoring leads to its degradation, because no mislocalization was observed. The second protein for which expression has been surprisingly modified in the present mouse model is the α1 subunit of DHPR, which is overexpressed 1.7-fold in fast (EDL) muscle, but not in slow (soleus) muscle. This is consistent with the absence of defects observed in soleus, and it could be hypothesized that longitudinal or oblique T-tubules allow the presence of higher amounts of DHPR. The modification of these two proteins is observed at different ages (10 weeks and 6 months), but it is more marked at 6 months, pointing to a possible accentuation of the phenotype with aging. This could explain why, in the model described by Shen et al. (30), the modifications of the different proteins expression were less pronounced, and actually, there are no major differences in the expression levels of the proteins assessed between the two models. The mechanisms leading to DHPR overexpression are poorly understood, because no direct relationship has been shown between DHPR and triadin, and most probably reflect an adaptation of the muscle to improve muscle function in absence of triadin.
In agreement with previous results (30), ultrastructural analysis on our KO mice revealed that 25% of triads have an abnormal orientation. Considering the similar results reported in both studies, this strongly supports an involvement of triadin in the setting up or in the maintenance of triad. As for CSQ anchoring, it is mostly probable that triadin is not the only protein involved in this function. One hypothesis could be that triadin would strengthen the triad structure and allow it to be maintained during contraction. Further experiments will be needed to clarify this issue.
One of our first observations is a muscle weakness, in contrast with what has been published on the first triadin KO mouse (30), with an obvious defect in the time the mice spent on the grid of their cage. This prompted us to search for objective evaluation of muscle strength. Considering that the reduction in the time on the grid could induce reduction in the food intake, and in animal development, one could expect a corresponding weight reduction. No modification has been observed for the animal weight; therefore, the muscle weakness observed in these mice does not modify their growth and reproduction. A number of tests was performed to evaluate muscle strength at the whole animal level. All these tests clearly demonstrated a significant muscle strength reduction, at all ages assessed, in males as well as in females, and this result was further confirmed in isolated muscle. Interestingly, this muscle weakness was accompanied by an improved resistance to fatigue, in all muscles tested (EDL, soleus, and gastrocnemius).
As CSQ expression is reduced in these muscles, could the observed effects be attributed to this decrease? It is probably not the case, or only partially so, because none of the defects observed here were similar to defects seen in the CSQ KO mouse (39), except for the reduction in terminal cisternae thickness, which is most probably directly related to CSQ reduction. In contrast with the CSQ1 KO model, the triadin KO mouse showed no increase either in RyR or mitochondria amount, and no duplication or triplication of triad. In addition, no muscle weakness was observed in CSQ1 KO mouse, contrary to triadin KO mouse. Therefore, the physiological effects observed in this KO mouse are clearly different from CSQ deletion-induced defects and can clearly be attributed to triadin deletion.
The muscle weakness observed in triadin KO mouse is probably due to reduction of releasable calcium observed in primary myotubes upon RyR stimulation (either by DHPR or by caffeine). This effect was not observed in CSQ1 KO mouse, which also presented a reduction in the calcium store, perhaps because of a compensatory overexpression of RyR and mitochondria in this model (39). The muscle weakness could also be attributed to an increase in intracellular calcium concentration measured in the other triadin KO mouse model (30), but this is less probable as in human primary culture, a similar intracellular calcium increase has been measured associated (Central Core Disease patients) or not (Malignant hyperthermia susceptible patients) with muscle weakness (42). Therefore, in human muscle culture, the increase in intracellular calcium concentration has not been correlated to muscle weakness.
One major common effect observed in both CSQ KO and triadin KO is improved resistance to fatigue. Fatigue resistance has been shown in a number of mice models, and different explanations have been proposed. In CSQ1 KO, this resistance has been related to an increased amount of mitochondria (39), which is not observed in our model. In a mouse model KO for sarcalumenin (43), another SR calcium-binding protein of skeletal and cardiac muscle, muscle resistance to fatigue has been observed and clearly attributed to increased store-operated calcium entry. This hypothesis could most probably be raised for our model, as we have previously demonstrated that Trisk 95 overexpression in rat myotubes resulted in decreased store-operated calcium entry (44). Therefore, it could be hypothesized that triadin deficiency induces a reduction in the SR-releasable calcium, resulting in two compensatory effects: increased DHPR expression and an increase in store-operated calcium entry to compensate for the defect in SR calcium release by extracellular calcium influx.
Which function of triadin can then be postulated from this model? Triadin is clearly not essential for triad formation, because triads are still present. Nevertheless, it could be involved in maintaining triads during contraction, and in anchoring mitochondria close to triads. A precise stoichiometry for triadin in the complex is also clearly associated with a correct function of the calcium release complex. We have previously shown that triadin Trisk 95 overexpression results in blocking of excitation-contraction coupling (21), and here we show that triadin ablation induced muscle function impairment and modifications in depolarization induced calcium release. All these results confirm that triadin is an essential link within the calcium release complex. The triadin KO mouse presents objective muscle dysfunctions and is, therefore, undoubtedly suffering from myopathy as identical energy consumption produced a reduced strength. This myopathy is a structural myopathy; the observed defect is a modification in the structure of the muscle fiber. Mutation in the triadin gene in human could probably lead to a myopathy, and this gene should be screened in patients presenting with a myopathy of unknown origin. This mouse line could therefore be a model for studying the development of a structural myopathy and its evolution during aging and/or exercise.
Acknowledgments
The mouse mutant lines were established at the Mouse Clinical Institute (Institut Clinique de la Souris, Mouse Clinical Institute/Institut Clinique de la Souris) in the Targeted Mutagenesis and Transgenesis Dept. with funds from the GIS-Maladies Rares. We thank Dr. D. Metzger for his help in the design of triadin KO mouse construct.
This work was supported by grants from Association Française contre les Myopathies, the Agence Nationale de la Recherche-Maladies Rares, and the Hungarian Research Found (Grant OTKA K75604).
- RyR
- ryanodine receptor
- CSQ
- calsequestrin
- DHPR
- dihydropyridine receptor
- EDL
- extensor digitorum longus
- FDB
- flexor digitorum brevis
- KO
- knock out
- PCr
- phosphocreatine
- SERCA
- sarco-endoplasmic reticulum Ca2+ ATPase
- Sol
- soleus
- SR
- sarcoplasmic reticulum
- WT
- wild type
- MRI
- magnetic resonance imaging.
REFERENCES
- 1.Flucher B. E. (1992) Dev. Biol. 154, 245–260 [DOI] [PubMed] [Google Scholar]
- 2.Flucher B. E., Franzini-Armstrong C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8101–8106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marty I., Robert M., Villaz M., Lai Y., De Jongh K., Catterall W. A., Ronjat M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 2270–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandt N. R., Caswell A. H., Wen S. R., Talvenheimo J. A. (1990) J. Membr. Biol. 113, 237–251 [DOI] [PubMed] [Google Scholar]
- 5.Kim K. C., Caswell A. H., Talvenheimo J. A., Brandt N. R. (1990) Biochemistry 29, 9281–9289 [DOI] [PubMed] [Google Scholar]
- 6.Caswell A. H., Brandt N. R., Brunschwig J. P., Purkerson S. (1991) Biochemistry 30, 7507–7513 [DOI] [PubMed] [Google Scholar]
- 7.Knudson C. M., Stang K. K., Moomaw C. R., Slaughter C. A., Campbell K. P. (1993) J. Biol. Chem. 268, 12646–12654 [PubMed] [Google Scholar]
- 8.Liu G., Pessah I. N. (1994) J. Biol. Chem. 269, 33028–33034 [PubMed] [Google Scholar]
- 9.Caswell A. H., Motoike H. K., Fan H., Brandt N. R. (1999) Biochemistry 38, 90–97 [DOI] [PubMed] [Google Scholar]
- 10.Groh S., Marty I., Ottolia M., Prestipino G., Chapel A., Villaz M., Ronjat M. (1999) J. Biol. Chem. 274, 12278–12283 [DOI] [PubMed] [Google Scholar]
- 11.Guo W., Campbell K. P. (1995) J. Biol. Chem. 270, 9027–9030 [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi Y. M., Alseikhan B. A., Jones L. R. (2000) J. Biol. Chem. 275, 17639–17646 [DOI] [PubMed] [Google Scholar]
- 13.Shin D. W., Ma J., Kim D. H. (2000) FEBS Lett. 486, 178–182 [DOI] [PubMed] [Google Scholar]
- 14.Zhang L., Kelley J., Schmeisser G., Kobayashi Y. M., Jones L. R. (1997) J. Biol. Chem. 272, 23389–23397 [DOI] [PubMed] [Google Scholar]
- 15.Ohkura M., Furukawa K., Fujimori H., Kuruma A., Kawano S., Hiraoka M., Kuniyasu A., Nakayama H., Ohizumi Y. (1998) Biochemistry 37, 12987–12993 [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi Y. M., Jones L. R. (1999) J. Biol. Chem. 274, 28660–28668 [DOI] [PubMed] [Google Scholar]
- 17.Marty I., Thevenon D., Scotto C., Groh S., Sainnier S., Robert M., Grunwald D., Villaz M. (2000) J. Biol. Chem. 275, 8206–8212 [DOI] [PubMed] [Google Scholar]
- 18.Vassilopoulos S., Thevenon D., Rezgui S. S., Brocard J., Chapel A., Lacampagne A., Lunardi J., Dewaard M., Marty I. (2005) J. Biol. Chem. 280, 28601–28609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thevenon D., Smida-Rezgui S., Chevessier F., Groh S., Henry-Berger J., Beatriz, Romero N., Villaz V., DeWaard M., Marty I. (2003) Biochem. Biophys. Res. Commun. 303, 669–675 [DOI] [PubMed] [Google Scholar]
- 20.Marty I., Fauré J., Fourest-Lieuvin A., Vassilopoulos S., Oddoux S., Brocard J. (2009) J. Physiol. 587, 3117–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smida Rezgui S. S., Vassilopoulos S., Brocard J., Platel J. C., Bouron A., Arnoult C., Oddoux S., Garcia L., De Waard M., Marty I. (2005) J. Biol. Chem. 280, 39302–39308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Li X., Duan H., Fulton T. R., Eu J. P., Meissner G. (2009) Cell Calcium 45, 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fodor J., Gönczi M., Sztretye M., Dienes B., Oláh T., Szabó L., Csoma E., Szentesi P., Szigeti G. P., Marty I., Csernoch L. (2008) J. Physiol. 586, 5803–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupé V., Davenne M., Brocard J., Dollé P., Mark M., Dierich A., Chambon P., Rijli F. M. (1997) Development 124, 399–410 [DOI] [PubMed] [Google Scholar]
- 25.Marty I., Robert M., Ronjat M., Bally I., Arlaud G., Villaz M. (1995) Biochem. J. 307, 769–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lunardi J., Dupuis A., Frobert Y., Grassi J., Vignais P. V. (1989) FEBS Lett. 245, 223–228 [DOI] [PubMed] [Google Scholar]
- 27.Moutin M. J., Cuillel M., Rapin C., Miras R., Anger M., Lompré A. M., Dupont Y. (1994) J. Biol. Chem. 269, 11147–11154 [PubMed] [Google Scholar]
- 28.Pouvreau S., Royer L., Yi J., Brum G., Meissner G., Ríos E., Zhou J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5235–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee E. H., Cherednichenko G., Pessah I. N., Allen P. D. (2006) J. Biol. Chem. 281, 10042–10048 [DOI] [PubMed] [Google Scholar]
- 30.Shen X., Franzini-Armstrong C., Lopez J. R., Jones L. R., Kobayashi Y. M., Wang Y., Kerrick W. G., Caswell A. H., Potter J. D., Miller T., Allen P. D., Perez C. F. (2007) J. Biol. Chem. 282, 7864–7874 [DOI] [PubMed] [Google Scholar]
- 31.Galbiati F., Engelman J. A., Volonte D., Zhang X. L., Minetti C., Li M., Hou H., Jr., Kneitz B., Edelmann W., Lisanti M. P. (2001) J. Biol. Chem. 276, 21425–21433 [DOI] [PubMed] [Google Scholar]
- 32.Westerblad H., Dahlstedt A. J., Lännergren J. (1998) J. Physiol. 510, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanhamme L., van den Boogaart A., Van Huffel S. (1997) J. Magn. Reson. 129, 35–43 [DOI] [PubMed] [Google Scholar]
- 34.Arnold D. L., Matthews P. M., Radda G. K. (1984) Magn. Reson. Med. 1, 307–315 [DOI] [PubMed] [Google Scholar]
- 35.Takeshima H., Iino M., Takekura H., Nishi M., Kuno J., Minowa O., Takano H., Noda T. (1994) Nature 369, 556–559 [DOI] [PubMed] [Google Scholar]
- 36.Knudson C. M., Chaudhari N., Sharp A. H., Powell J. A., Beam K. G., Campbell K. P. (1989) J. Biol. Chem. 264, 1345–1348 [PubMed] [Google Scholar]
- 37.Gregg R. G., Messing A., Strube C., Beurg M., Moss R., Behan M., Sukhareva M., Haynes S., Powell J. A., Coronado R., Powers P. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13961–13966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahern C. A., Powers P. A., Biddlecome G. H., Roethe L., Vallejo P., Mortenson L., Strube C., Campbell K. P., Coronado R., Gregg R. G. (2001) BMC Physiol. 1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paolini C., Quarta M., Nori A., Boncompagni S., Canato M., Volpe P., Allen P. D., Reggiani C., Protasi F. (2007) J. Physiol. 583, 767–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeshima H., Komazaki S., Hirose K., Nishi M., Noda T., Iino M. (1998) EMBO J. 17, 3309–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tijskens P., Jones L. R., Franzini-Armstrong C. (2003) J. Mol. Cell Cardiol. 35, 961–974 [DOI] [PubMed] [Google Scholar]
- 42.Ducreux S., Zorzato F., Müller C., Sewry C., Muntoni F., Quinlivan R., Restagno G., Girard T., Treves S. (2004) J. Biol. Chem. 279, 43838–43846 [DOI] [PubMed] [Google Scholar]
- 43.Zhao X., Yoshida M., Brotto L., Takeshima H., Weisleder N., Hirata Y., Nosek T. M., Ma J., Brotto M. (2005) Physiol. Genomics 23, 72–78 [DOI] [PubMed] [Google Scholar]
- 44.Vassilopoulos S., Brocard J., Garcia L., Marty I., Bouron A. (2007) Cell Calcium 41, 179–185 [DOI] [PubMed] [Google Scholar]