Abstract
The chemokine stromal cell-derived factor-1 (SDF-1/CXCL12) and its G-protein-coupled receptor (GPCR) CXCR4 play fundamental roles in many physiological processes, and CXCR4 is a drug target for various diseases such as cancer metastasis and human immunodeficiency virus, type 1, infection. However, almost no structural information about the SDF-1-CXCR4 interaction is available, mainly because of the difficulties in expression, purification, and crystallization of CXCR4. In this study, an extensive investigation of the preparation of CXCR4 and optimization of the experimental conditions enables NMR analyses of the interaction between the full-length CXCR4 and SDF-1. We demonstrated that the binding of an extended surface on the SDF-1 β-sheet, 50-s loop, and N-loop to the CXCR4 extracellular region and that of the SDF-1 N terminus to the CXCR4 transmembrane region, which is critical for G-protein signaling, take place independently by methyl-utilizing transferred cross-saturation experiments along with the usage of the CXCR4-selective antagonist AMD3100. Furthermore, based upon the data, we conclude that the highly dynamic SDF-1 N terminus in the 1st step bound state plays a crucial role in efficiently searching the deeply buried binding pocket in the CXCR4 transmembrane region by the “fly-casting” mechanism. This is the first structural analyses of the interaction between a full-length GPCR and its chemokine, and our methodology would be applicable to other GPCR-ligand systems, for which the structural studies are still challenging.
Introduction
Chemokines are a number of small (8–10 kDa) secreted proteins that direct cell migration in immune systems by activating their receptors expressed on the cell surface (1, 2). The chemokine, stromal cell-derived factor-1 (SDF-1,2 also known as CXCL12) (3, 4), and its receptor, CXCR4 (5–7), play many essential physiological roles, such as homeostatic regulation of leukocyte traffic, hematopoiesis, and embryonic development (8–11). The interaction between SDF-1 and CXCR4 also controls cancer metastasis (12, 13), and CXCR4 is a co-receptor for T-tropic strains of human immunodeficiency virus, type 1 (5, 14).
The most abundant splice variant of SDF-1 (SDF-1α) is composed of 68 amino acids, and its NMR (15, 16) and crystal structures (17, 18) demonstrated that SDF-1α assumes a typical chemokine fold as follows: an unstructured N terminus (Lys1–Tyr7) followed by a long flexible loop (N-loop), a three-stranded anti-parallel β-sheet, and an α-helix. The mutational analyses revealed that although the SDF-1α N terminus is critical for the CXCR4-mediated signaling (15), both the N terminus and the N-loop residues are implicated in the receptor binding (15, 18, 19). In addition, recent mutational analysis suggested that the residues on the SDF-1α β-sheet are also important for receptor binding (20).
CXCR4, composed of 352 amino acids, belongs to the class A G-protein-coupled receptor (GPCR) family, with the seven transmembrane (TM) helices. Whereas GPCR activation is mediated by the conformational changes in its TM region (21, 22), the mutational analyses revealed that the CXCR4 N terminus and the extracellular loops (ECLs) are also involved in the ligand binding (23–25). The recently solved NMR structure of a disulfide-bridged dimeric mutant of SDF-1α, complexed with a CXCR4 N-terminal peptide, revealed the N-terminal peptide-binding modes of SDF-1α (20).
Based on these previous studies, a two-step/two-site binding model has been proposed for the SDF-1α-CXCR4 interaction (15). In this model, two independent interactions are hypothesized as follows: the SDF-1α N-loop interacts with the CXCR4 N terminus, and subsequently the SDF-1α N terminus interacts with the CXCR4 TM region to trigger receptor activation.
However, little is known about the structural basis of the recognition mode of SDF-1α with the full-length CXCR4, and there is no direct evidence that the SDF-1α N terminus and the N-loop interact with structurally independent sites on CXCR4. This is mainly because of the difficulties in overexpression, purification, and crystallization of CXCR4. Although the preparative scale heterologous expression of CXCR4, using a baculovirus expression system, has recently been reported (26), the expression level is not sufficient for standard structural analyses, such as crystallography and NMR. In addition, the solubilization of CXCR4 by detergents, which is required for solution NMR measurements, results in a huge molecular mass of over 100 kDa, which also hampers NMR analyses.
We recently established a novel NMR method, transferred cross-saturation (TCS), to identify the molecular interface, the region involved in the molecular interaction, in a large protein complex (27–29) through the cross-saturation phenomenon, which identifies the proximal residues within 5 Å of their binding partner (30). In this method, the molecular weight limitation in NMR measurements is overcome by properly transferring the cross-saturation effects from the receptor-bound ligands to the free ligands. The advantage of the TCS methods is that only substoichiometric amounts of receptors relative to ligands are required. Moreover, the combined usage of TCS and methyl-utilizing cross-saturation (31), which provides much higher sensitivity for the detection of the cross-saturation phenomena, decreases the amount of receptor required.
Here, we report NMR analyses of the interaction between SDF-1α and full-length CXCR4. We applied the methyl-utilizing TCS method to elucidate the CXCR4-binding site of SDF-1α, and we demonstrated that SDF-1α utilizes an extended surface on the multiple region dispersed throughout the protein, consisting of the β-sheet, 50-s loop, and N-loop, in addition to the N terminus, for the receptor binding. We further investigated the effect of AMD3100, which is a selective CXCR4 antagonist that binds to the CXCR4 TM region (32, 33), on the SDF-1α-CXCR4 interaction. Consequently, we found that even in the state with SDF-1α bound to CXCR4, the SDF-1α N terminus is released from CXCR4, upon the addition of AMD3100, whereas the other SDF-1α region still binds to CXCR4. Our data provide the first structural evidence for the existence of two independent interactions between SDF-1α and CXCR4.
EXPERIMENTAL PROCEDURES
Expression and Purification of CXCR4
The cDNA fragment encoding human CXCR4 with the C-terminal 1D4 epitope tag (TTVSKTETSQVAPA) was amplified by PCR and cloned into the pVL1392 vector (Pharmingen) via the XbaI-BamHI sites. Recombinant baculoviruses were generated using a BaculoGold transfection kit (Pharmingen), according to the manufacturer's instructions.
For the large scale expression of CXCR4, 2.8 liters of the expresSF+® cells (Protein Sciences Corp.) in Sf900-II serum-free media (Invitrogen) were grown at 27 °C, in a 3-liter spinner flask (Bellco) equipped with a dissolved oxygen controller (Wakenyaku). The cells, at a density of 1.8–2 × 106 cells/ml, were inoculated with the high titer virus stock (120 ml per 2.8 liters of cells) and were harvested 48 h post infection.
All of the following procedures were carried out at 4 °C. Cells were disrupted by nitrogen cavitation (Parr Bomb) under 600 p.s.i. for 30 min in 250 ml of buffer A (50 mm Tris, pH 8.0, 100 mm NaCl, 1 mm EDTA) with a protease inhibitor mixture (Nacalai Tesque, Inc.), and the lysate was centrifuged at 800 × g for 10 min. The supernatant was centrifuged at 100,000 × g for 60 min, and the resulting membrane pellet was solubilized in 120 ml of buffer B (20 mm Hepes, pH 7.2, 150 mm NaCl, 15% (v/v) glycerol) + 1% n-dodecyl β-d-maltoside (DDM, Dojindo) for 4 h. The solubilized membrane was centrifuged at 75,000 × g for 30 min, and the supernatant was batch-incubated overnight with 2 ml of 1D4-Sepharose beads, in which 3.5–5 mg/ml 1D4 antibodies (University of British Columbia) were coupled to CNBr-activated Sepharose 4B (GE Healthcare). The beads were washed with 60 ml of buffer C (buffer B + 0.1% DDM), and the protein was eluted with 7.5 ml of buffer D (buffer C + 200 μm “TETSQVAPA” peptide (synthesized by Toray Research Center)). The simultaneous concentration and buffer exchange of the eluate were accomplished by a centrifugal filter device (AmiconUltra-15, 30-kDa molecular mass cutoff, Millipore).
Expression and Purification of SDF-1
The cDNA fragment encoding human SDF-1α was amplified by PCR and cloned into the pET-11a vector (Novagen) via the NdeI-BamHI sites to express SDF-1α with an N-terminal methionine extension (MetSDF-1α). All mutants were generated by QuikChange site-directed mutagenesis (Stratagene).
MetSDF-1α was expressed in the Escherichia coli BL21-CodonPlus® (DE3) RP strain (Stratagene). Cells were grown at 37 °C in M9 minimal medium to an A600 of 0.8–1.0, induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and incubated for an additional 6 h. Selective incorporations of 1H and 13C labels into the methyl groups were achieved as described previously (34, 35).
Cells were disrupted by sonication in buffer A. Inclusion bodies were isolated by centrifugation and were solubilized in buffer E (50 mm Tris, pH 8.0, 6 m guanidinium hydrochloride, 10 mm dithiothreitol). The protein was refolded by dialysis against buffer F (buffer A + 0.4 m arginine hydrochloride, 1 mm reduced glutathione, 1 mm oxidized glutathione) and was purified by two cycles of reverse phase high pressure liquid chromatography. The N-terminal methionine extension of SDF-1 did not affect the chemotactic activity (data not shown) as reported previously (36).
Pulldown Analysis
The purified CXCR4 was combined with an excess amount of Cy3–12G5, in which 12G5 antibodies (R & D Systems) coupled to Cy3 mono-reactive dye (GE Healthcare) or SDF-1α, and with 100 μl of 1D4-Sepharose in 500 μl of buffer C. The mixture was incubated at 4 °C for 4 h, and the beads were washed twice with 1 ml of buffer C. The protein was eluted with 500 μl of buffer D. Nonspecific binding of Cy3–12G5 or SDF-1α to 1D4-Sepharose was not detected.
Surface Plasmon Resonance (SPR) Analysis
The FLAG-M5 (Sigma) and 12G5 binding activities of CXCR4 were analyzed by SPR measurements using a BIAcore 2000 instrument (Biacore), as described in the literature (37).
NMR Experiments
NMR spectral assignments were performed using the standard triple resonance experiments (38) with uniformly 13C- and 15N-labeled MetSDF-1α. Assignments of the two diastereotopic methyl groups of leucine and valine residues were achieved as described previously (39). All of the following NMR experiments were carried out at 10 °C in 10 mm sodium phosphate, pH 7.4, 150 mm NaCl, H2O/D2O = 1:99, and 15% (v/v) [2H8]glycerol with a Bruker Avance 800 spectrometer equipped with a cryogenic probe.
In the methyl-utilizing TCS (methyl-TCS) experiments, 100 μm (final concentration) of lyophilized [[U-2H]Ileδ1-,Leu-,Val-13C1H3]MetSDF-1α R8A/R12A was combined with 10 μm CXCR4 or with 10 μm CXCR4 and 1 mm AMD3100 (Sigma). For negative control experiments, 10 μm (final concentration) of lyophilized [U-2H]MetSDF-1α was further added to the sample containing MetSDF-1α R8A/R12A and CXCR4 but not AMD3100. The pulse scheme was as described previously (31) but with the heteronuclear single quantum coherence-type (HSQC-type) sequence with echo/anti-echo gradient coherence selections (40). The irradiation frequency was set at 5.0 ppm, and the maximum radiofrequency amplitude was 0.21 kHz for WURST-20 (the adiabatic factor Q0 = 1) (41). The irradiation time and the additional relaxation times were set to 0.5 and 1.5 s, respectively.
To observe the effect of AMD3100 on the spectra of SDF-1α with an excess amount of CXCR4, 1H-13C heteronuclear multiple quantum coherence (HMQC) spectra (40) were recorded for the following samples: 10 μm (final concentration) of lyophilized [[U-2H]Leu-, Val-13C1H3,12C2H3)]MetSDF-1α combined with the buffer only, with 20 μm CXCR4 in the same buffer, and with 20 μm CXCR4 and 1 mm AMD3100 in the same buffer. All of the recorded spectra were processed by Topspin 2.0 (Bruker) and were analyzed by Sparky (69).
RESULTS
Characterization of the Prepared CXCR4
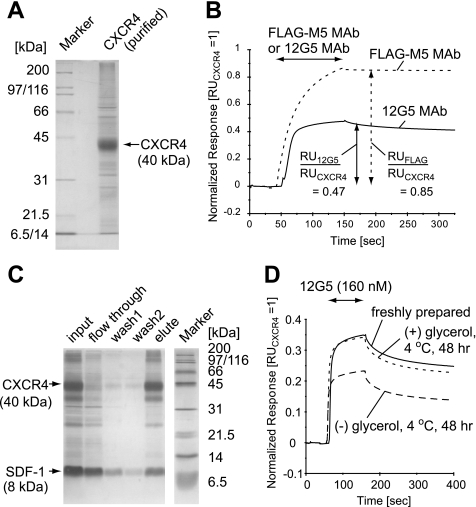
CXCR4 was expressed in insect cells using a baculovirus expression system and was solubilized in 1% DDM. The solubilized CXCR4 with the C-terminal 1D4 epitope tag was purified by 1D4 antibody affinity chromatography. In both SDS-PAGE analysis (Fig. 1A) and Western blotting analysis with 1D4 antibody (supplemental Fig. 1), one major band at an ∼42-kDa band is observed. Considering the molecular weight of CXCR4 (41–43 kDa, including the glycosylation), we conclude that the major band is intact CXCR4. Purity of the intact CXCR4 is >80% as judged from SDS-PAGE (Fig. 1A).
FIGURE 1.
Characterization of the purified CXCR4. A, purified CXCR4 was loaded on a 12% SDS-polyacrylamide gel. The gel was stained with Coomassie Brilliant Blue. B, normalized SPR responses for FLAG-M5 binding to N-terminally FLAG-tagged CXCR4 (dotted line) and 12G5 binding to CXCR4 (solid line). C, SDF-1 binding activity of the purified CXCR4 analyzed by a pulldown assay. Each fraction was loaded on a 15% SDS-polyacrylamide gel, which was silver-stained. D, normalized SPR responses for 12G5 binding to CXCR4, incubated under various conditions. B and D, the entire sensorgrams were divided by the CXCR4 capture levels (RUCXCR4). RU, resonance units; MAb, monoclonal antibody.
To examine the conformational integrity of the obtained CXCR4, the binding activity with the anti-CXCR4 antibody 12G5, which recognizes ECL2 of CXCR4 in its native conformation (42), was analyzed by SPR experiments (37). The normalized response of 12G5 for CXCR4 immobilized on a sensor chip was about half that of a FLAG-M5 antibody for N-terminally FLAG-tagged CXCR4 (Fig. 1B), suggesting that ∼50% of the obtained CXCR4 was correctly folded. A pulldown assay using fluorescently labeled 12G5 revealed that 50–100 μg of the correctly folded CXCR4 was obtained from 1 liter of insect cell culture. We also examined the SDF-1α binding activity of the obtained CXCR4 by pulldown assays. Almost stoichiometric amounts of SDF-1α, relative to the correctly folded CXCR4 (∼50% of the ∼42-kDa band), were co-precipitated (Fig. 1C), suggesting that the obtained CXCR4 is able to bind to SDF-1α.
It is well known that detergent-solubilized GPCRs are extraordinarily labile. Because we also found that all of the DDM-solubilized CXCR4 aggregated after 48 h of incubation at room temperature (data not shown), we examined the stability of the solubilized CXCR4 under various conditions. Although an incubation of the sample solution at a low temperature prevented the aggregation of CXCR4 (data not shown), the normalized response of 12G5 binding to CXCR4 by the SPR analyses was reduced to ∼60% after 48 h of incubation, even at 4 °C (Fig. 1D). Further addition of glycerol to the sample solution markedly prevented the reduction of the response to 12G5 (Fig. 1D). Therefore, we performed the subsequent NMR analyses at a low temperature, in the presence of glycerol, and within 48 h after purification.
Methyl-utilizing TCS Experiments
To identify the SDF-1α residues in close proximity to CXCR4, methyl-TCS experiments were carried out. Fig. 2A shows an outline of the methyl-TCS experiment performed in this study. We prepared highly deuterated SDF-1α, with protons selectively incorporated into the 13C-labeled valine, leucine, and isoleucine (δ1 only) methyl groups (34). 1H NMR spectrum of the labeled SDF-1α showed that the deuteration level was sufficiently high (>98%, data not shown) for the methyl-TCS experiments (31). The methyl-TCS experiments were carried out under conditions with an excess amount (10-fold) of the labeled SDF-1α relative to the CXCR4. In the methyl-TCS experiments, irradiation with a frequency corresponding to the methylene, methine, and aromatic protons (2.5–7.5 ppm) was applied to the mixture of the nonlabeled CXCR4 and the labeled SDF-1α. The saturation caused by the irradiation was not kept within the CXCR4 molecule but was transferred to the SDF-1α residues in close proximity to the CXCR4, through the cross-saturation phenomena (30). If the complex has an exchange rate between the free and bound states that is faster than the longitudinal relaxation rates of the methyl protons of SDF-1α (1–2 s−1), then the saturation in the proximal residues was sufficiently transferred to the free state of SDF-1α. As a result, the proximal residues can be identified by the selective intensity reductions of the methyl resonances in the 1H-13C HSQC spectra of unbound SDF-1α.
FIGURE 2.
Scheme and the spectra of the methyl-TCS experiments. A, schematic representation of methyl-TCS experiments. The saturation of the nonlabeled CXCR4 caused by the irradiation is transferred to the free state of the methyl-protonated SDF-1α mutant. B, schematic representation of the negative control experiments for methyl-TCS. The addition of equal amounts of deuterated wild-type SDF-1α to CXCR4 inhibits the specific interaction between CXCR4 and the methyl-protonated SDF-1α mutant. C and D, lower panel shows the two-dimensional 1H-13C HSQC spectra of 100 μm [[U-2H[Ileδ1-, Leu-, Val-13C1H3)]SDF-1α R8A/R12A with 10 μm nonlabeled CXCR4. D, 10 μm [U-2H]SDF-1α was further added (negative control experiments). Only the regions of the leucine methyl signals are shown (the full-size spectrum and assignments of each resonances are shown in supplemental Fig. 2). In the upper panels, the black lines show the 1H one-dimensional slices through the Leu55δ1 and Leu62δ2 signals (the corresponding 13C frequencies are indicated with cyan lines in the two-dimensional spectra). The red lines show the same 1H one-dimensional slices with selective irradiations of CXCR4.
To achieve efficient exchange between the free and bound states of SDF-1α, we utilized the SDF-1α mutant R8A/R12A for the subsequent TCS experiments, because the introduction of mutations into Arg8 and Arg12 reportedly reduces the receptor affinity but does not affect the chemotactic efficacy (18, 19).
Despite the substantial improvement of the sample preparation procedure, the obtained CXCR4 samples significantly contained denatured CXCR4 and impurities. In addition, DDM micelles were also included in the samples. Therefore, to selectively observe the specific interaction between correctly folded CXCR4 and SDF-1α, we subtracted the nonspecific binding effects from the TCS results by the control experiments, in which the specific binding surface on the correctly folded CXCR4 was selectively blocked by the addition of the stoichiometric amount of perdeuterated wild-type SDF-1α (Fig. 2B). The difference in reduction ratio (ΔRR), which represents the specific interaction between SDF-1α and CXCR4, was calculated for each resonance by subtracting the intensity reduction ratio in the control experiment from that in the TCS experiment.
Fig. 2, C and D, shows the 1H-13C HSQC spectra of SDF-1α R8A/R12A observed in the TCS and control experiments, respectively. In the TCS experiment, several resonances, including Leu55δ2, showed significantly higher intensity reductions upon irradiation than in the control experiment, whereas other resonances, including Leu62δ1, exhibited similar intensity reductions in the two experiments. The ΔRR was calculated for each resonance (Fig. 3, A and B), and the residues were colored according to their ΔRR values on the SDF-1α structure in the free state (Fig. 3C). The residues were categorized into three classes, based on their ΔRR values as follows: high ΔRR values (>0.125), Val23γ1, Leu26δ1, Ile28, Leu29, Val39, Val49γ2, and Leu55; moderate ΔRR values (0.10–0.125), Val3, Leu5, Val18γ1, Leu26δ2, Ile51, and Ile58; and low ΔRR values (<0.10), Val23γ2, Leu36, Ile38, Leu42, Leu62, and Leu66. Because Val18γ1, Val23γ2, Leu26δ2, Ile51, and Ile58 methyl groups are completely buried within the SDF-1α molecule (supplemental Fig. 3), these resonances were excluded from the following considerations.
FIGURE 3.
Determination of the CXCR4 interface on SDF-1α by the methyl-TCS experiments. A, ratio of signal intensity reductions originating from the isoleucine, leucine, and valine methyl resonances, with and without irradiation. The purple bars represent the ratios from the methyl-TCS experiment (Fig. 2, A and C), and the green bars represent those from the negative control experiment (Fig. 2, B and D). The ratios for the Val18γ1 and Val49γ2 signals were not determined (N.D.), because these two signals were severely overlapped. The error bars represent the experimental errors, calculated from the root sum square of (noise level/signal intensity) in the two spectra, with and without irradiation. B, differences in the signal reduction ratios (ΔRR values) between the methyl-TCS experiment (purple bars in A) and the control experiment (green bars in A). The error bars represent the experimental errors, calculated from the root sum square of errors in two experiments. Red, light red, orange, and cyan bars represent the signals with ΔRR > 0.150, 0.125, 0.100, and <0.100, respectively. Methyl protons completely buried in the structures are shaded in gray. C, mapping of the affected residues on the SDF-1α structure (Protein Data Bank code 1VMC). The SDF-1α structure is shown in a CPK representation. Isoleucine, leucine, and valine residues are colored according to the ΔRR values of their methyl resonances, as in B. For the leucine and valine residues, the larger ΔRR of their two methyl resonances was utilized.
Our relaxation matrix calculations (31) suggested that the ligand methyl protons within 5 Å of the receptor protons exhibited the intensity reductions of more than 0.1. In addition, when the ligand methyl protons are close to each other, the ligand methyl protons more than 5 Å away from the receptor protons also exhibit some intensity reductions, because of the spin diffusion effects within the ligand molecule. However, because of the fast internal motions of methyl protons, the spin diffusion effects should be efficiently suppressed (31). Therefore, the spin diffusion effect should provide only minor effects relative to the cross-saturation between CXCR4 and SDF-1α.
The deuteration levels of SDF-1α were sufficiently high, and thus most of the effects of the residual protons in SDF-1α have been eliminated from the TCS results. Although the residual proton effect is enhanced in the high molecular weight systems, the enhanced effect was usually <0.1, even in the case of the ∼150-kDa the complex between fragment B of protein A and IgG (31). Consequently, the high ΔRR values (>0.1) should represent the cross-saturation effect originating from CXCR4.
The cross-saturation effect is relatively insensitive to the irradiation bandwidth, due to the effective spin diffusion within the CXCR4 molecule, whereas the residual proton effect is sensitive to the irradiation bandwidth. Therefore, we carried out the TCS experiments with reduced irradiation bandwidth (4.5–7.5 ppm). Although the residues with low ΔRR values are remarkably affected by the irradiation bandwidth, the residues with high ΔRR values (>0.1), such as Leu29δ2, showed little dependence of ΔRR on the irradiation bandwidth (data not shown), suggesting that the cross-saturation is dominant in residues with high ΔRR values (>0.1).
The methyl resonances with high ΔRR values (>0.125) formed a contiguous surface on the SDF-1α structure as follows: starting from the β1-strand (Val23, Leu26, and Ile28), extending through one side of the β-sheet (Leu29, Val39, and Val49), and ending at the 50-s loop (Leu55). In addition, the resonances from the SDF-1α N terminus (Val3 and Leu5) also exhibited moderate ΔRR values, which should represent the close proximity of these protons to CXCR4, because the effect of spin diffusion would be negligible for these isolated spins. Consequently, we conclude that Val3, Leu5, Val23, Leu26, Ile28, Leu29, Val39, Val49, and Leu55 of SDF-1α are in close proximity to CXCR4 in the SDF-1α-CXCR4 complex.
Effect of AMD3100 on the SDF-1α-CXCR4 Interaction
To elucidate the CXCR4-binding mode of SDF-1α in more detail, the SDF-1α-CXCR4 interaction was further investigated under conditions where the CXCR4 TM region was blocked by the CXCR4-selective antagonist AMD3100 (32, 33).
We first carried out TCS experiments in the presence of AMD3100 to examine its effect on the interaction between SDF-1α and CXCR4. The calculated ΔRR values are shown in Fig. 4A, and the residues are colored according to their ΔRR values on the SDF-1α structure in the free state (Fig. 4B). As a result, the resonances from the N-terminal residues (Val3 and Leu5) showed almost no ΔRR values in the presence of AMD3100, whereas significant ΔRR values were observed for these residues in the absence of AMD3100 (Fig. 4, B and C). Except for the N terminus, the residues of the SDF-1α with significant ΔRR values (>0.10) in the presence of AMD3100 included Val23γ1, Leu26δ1, Ile28, Leu29δ2, Val49γ2, and Leu55 and are almost identical to those in the absence of AMD3100.
FIGURE 4.
Results of the methyl-TCS experiments performed in the presence of AMD3100. A, ΔRR was calculated for each resonance, as in Fig. 3B. Red, light red, orange, and cyan bars represent the signals with ΔRR > 0.125, 0.100, 0.075, and <0.075, respectively. Methyl protons completely buried in the structures are shaded in gray. B, mapping of the affected residues on the SDF-1α structure (Protein Data Bank code 1VMC), as in Fig. 3C. Colors are the same as in A.
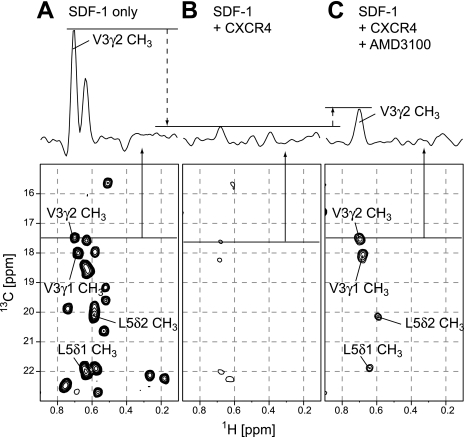
We next observed the effect of AMD3100 on the spectra of wild-type SDF-1α with an excess amount of CXCR4. Fig. 5A shows the 1H-13C HMQC spectrum of wild-type SDF-1α, with protons selectively incorporated into the 13C-labeled leucine and valine methyl groups (35). All 26 methyl resonances from the eight leucine and five valine residues were observed. Upon the addition of an excess amount of CXCR4, the signal intensities were remarkably decreased (Fig. 5B), indicating a significant increase in the molecular weight of SDF-1α. Upon the further addition of AMD3100, the intensities of several signals, with chemical shifts almost identical to those of the methyl resonances in the N-terminal residues (Val3 and Leu5) in the free state, were recovered (Fig. 5C).
FIGURE 5.
Spectra of SDF-1α with an excess amount of CXCR4 and CXCR4-AMD3100. A–C, lower panels show the two-dimensional 1H-13C HMQC spectra of 10 μm [[U-2H]Leu-,Val-13C1H3,12C2H3)]SDF-1α in the free state (A), with an excess amount (20 μm) of CXCR4 (B), and with an excess amount (20 μm) of CXCR4 and 1 mm AMD3100 (C). The upper panels show the 1H one-dimensional slice in each 1H-13C HMQC spectrum at the corresponding 13C frequencies indicated with lines in the two-dimensional spectra.
DISCUSSION
CXCR4-binding Site on SDF-1α
The methyl-TCS experiments utilize isoleucine, leucine, and valine methyl groups as probes to identify the molecular interface. Our previous data base analyses demonstrate that the probability for the methyl protons to be located within 3 Å of their binding partner in a protein-protein complex is comparable or even larger than that of amide protons (31). In addition, the TCS experiments identify the residues located within 5 Å of the protons of their binding partners (30). Whereas methyl groups with small solvent exposure (accessible surface area ∼0.1) cannot make van der Waals contact, they can be within 5 Å of the protons of their binding partners, as in the case of the complex between fragment B of protein A and IgG (31). Therefore, the methyl groups detected in the methyl-TCS experiments would demonstrate the regions constituting the binding interfaces.
Our TCS experiments revealed that the SDF-1α N terminus (Val3 and Leu5), the β1-strand (Val23, Leu26, and Ile28), one side of the β-sheet (Leu29, Val39, and Val49), and the 50-s loop (Leu55) are in close proximity to CXCR4 in the SDF-1α-CXCR4 complex (Fig. 3). It should be noted that the remarkable intensity reduction was observed for Val49γ2, which is close to the N-loop. Therefore, we conclude that SDF-1α utilizes an extended surface on the multiple region dispersed throughout the protein, consisting of the β-sheet, 50-s loop, and N-loop, in addition to the N terminus, for the receptor binding.
These results are in good agreement with the previous mutational analyses (15, 18–20), where the deletion of the N terminus (Lys1–Val3), and the substitutions of Arg8, Arg12, and Arg47, which are close to the Leu29, Val39, and Val49 methyl groups, significantly affected the receptor binding affinities (supplemental Fig. 4), whereas Val18, which is in N-loop together with Arg12, is not considered in our TCS experiments, because Val18γ1 is completely buried in the molecule. In addition, the CXCR4-binding site determined in the TCS experiment contains a more extensive surface than that proposed in previous mutational studies, suggesting that a broad area of CXCR4 is responsible for the SDF-1α binding.
The extensive surface may be composed of the region interacting with the CXCR4 N terminus and that interacting with the CXCR4 extracellular loop. The CXCR4 N terminus was suggested to bind to the SDF-1α N-loop (15). This is consistent with both the membrane proximal region of the CXCR4 N terminus and the latter half of the SDF-1α N-loop that contain acidic and basic residues. The SDF-1α β-sheet and 50-s loop would bind to the CXCR4 region, including the extracellular loops. Further investigations are necessary to determine the precise configuration between SDF-1α and CXCR4.
In contrast to most of the chemokine-chemokine receptor interactions (2), the SDF-1α-CXCR4 interaction exhibits relatively stringent specificity; SDF-1α binds to only CXCR4 and CXCR7, and CXCR4 exclusively binds to SDF-1α. Our TCS and mutational results, together with the previous mutational results, provide detailed information about the specific SDF-1α recognition mechanism of CXCR4. CXCR4 recognizes an extensive surface on the SDF-1α β-sheet, 50-s loop, and N-loop, which contains a variety of basic (such as Arg12 and Arg47), acidic (such as Glu15 and Asp52), and hydrophobic residues (such as Leu29, Val49, and Leu55), with their side chains exposed on the molecular surface. This combination of different types of residues in the extensive binding site identified in this study would substantially contribute to the specific interaction.
Most chemokines, including SDF-1α, tend to dimerize at high concentrations (16, 36, 43–45). The interactions between chemokines and glycosaminoglycans promote the chemokine dimerizations (43, 46), and the recent NMR analyses revealed that the interaction between SDF-1α and CXCR4 N-terminal peptides also promote the SDF-1α dimerizations (20, 44). On the other hand, the monomeric SDF-1α is relevant to the signal transduction, because the disulfide-bridged dimeric mutant of SDF-1α exhibits slightly larger Ca2+ influx EC50 values and the chemotactic antagonism (20). In addition, it has recently been reported that the SDF-1α monomer and not the dimeric mutant of SDF-1α is relevant in its cardioprotective effect in vivo (47). In this study, almost stoichiometric amounts of SDF-1α, relative to the correctly folded CXCR4, were co-precipitated in the pulldown assay, suggesting that the stoichiometry between CXCR4 and SDF-1α is 1:1 (Fig. 1C). Therefore, we performed the TCS experiments under the conditions of 100 μm SDF-1α, where SDF-1α assumes the monomeric form. In our TCS experiments, such a 1:1 binding mode should be selectively observed by using the negative control experiments, in which the 1:1 binding mode was blocked by the addition of the stoichiometric amount of wild-type SDF-1α relative to the folded CXCR4. Interestingly, the CXCR4-binding site of SDF-1α determined in this study partially overlapped the SDF-1α dimer interface (Leu26δ1 and Ile28δ1) (Fig. 3), suggesting that the SDF-1α monomer would serve as a more effective ligand for CXCR4. Our results did not provide any information about the function of SDF-1α dimer, which might be more effective in the presence of glycosaminoglycans, and further investigations are necessary to address the physiological relevance of the SDF-1α monomer-dimer equilibrium.
There is increasing evidence that the GPCRs, including chemokine receptors, exist as homodimers or heterodimers on the cell surface (48–51). However, the functional consequences of the receptor dimerizations are still under debate, because of the difficulties in the detection and the control of the receptor oligomerization states (52–54). In this study, we did not control the oligomerization of CXCR4, and further studies are necessary to investigate whether the CXCR4 dimerizations affect the ligand binding. For elucidating the relevance of GPCR dimerization, TCS experiments using monomeric or dimeric GPCR trapped by mutations (55) or cross-linkings (56) would be helpful.
It is well established that CXCR4 is post-translationally modified by sulfation of its tyrosines at the N terminus. Although it has been reported that CXCR4 expressed in Hi5 insect cells was tyrosine-sulfated (26), the sulfated residues and the extent of sulfation have not yet been determined. Although the tyrosine sulfation is reportedly important for the high affinity ligand binding of both the CXCR4 (57) and CXCR4 N-terminal peptides (20, 44), CXCR4 lacking the tyrosine sulfation site can also bind to SDF-1α with substantial affinity (57) and has signal transduction activity (24, 25). In addition, the complex structures of SDF-1α and CXCR4 N-terminal peptides have recently been solved, and the structures with and without tyrosine sulfations are almost identical (20). Therefore, we conclude that the tyrosine sulfation does not change the binding mode of the SDF-1α-CXCR4 interaction and that our data represent the SDF-1α-CXCR4 interactions in their native forms.
Two-step Binding Model for the SDF-1α-CXCR4 Interaction
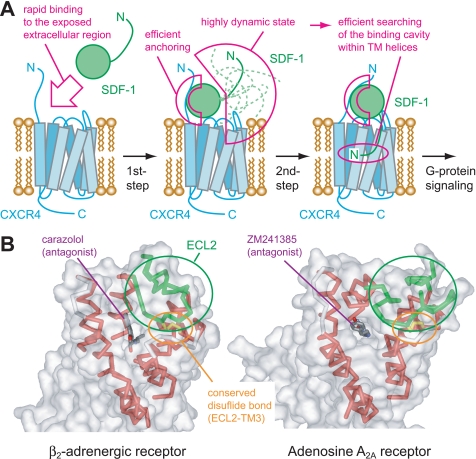
In the methyl-TCS experiment with AMD3100 (Fig. 4), the loss of the ΔRR values on the N-terminal residues revealed that the SDF-1α N terminus is not responsible for CXCR4 binding in the presence of AMD3100, whereas the similar ΔRR values on the SDF-1α β-sheet and 50-s loop demonstrated that the binding of the SDF-1α β-sheet and 50-s loop to CXCR4 is unaffected by AMD3100. In the NMR observations of SDF-1α with an excess amount of CXCR4 (Fig. 5), the reappearance of the N-terminal signals upon the addition of AMD3100 indicated the highly dynamic nature of the SDF-1α N terminus, whereas the absence of the NMR signals from the other SDF-1α region, even in the presence of AMD3100, suggested that the stable interaction between the SDF-1α region without N terminus and the CXCR4-AMD3100 complex still exists. Considering that AMD3100 binds to the TM region of CXCR4 (32), these NMR experiments demonstrated that the SDF-1α N terminus should bind to the CXCR4 TM region, whereas the other SDF-1α region should bind to the CXCR4 extracellular regions. These two interactions should occur independently, because AMD3100 could block only the interaction between the SDF-1α N terminus and the CXCR4 TM region. The SDF-1α N-loop residues are suggested to bind to the CXCR4 N terminus (15). Considering that Val49γ2, which is close to the N-loop, showed significant ΔRR even in the presence of AMD3100, the interaction between the SDF-1α N-loop and the CXCR4 N terminus would be also independent of the interaction between the SDF-1α N terminus and the CXCR4 TM region. Consequently, we can provide novel structural evidence that strongly supports the two-step binding model, as illustrated in Fig. 6A, with the characteristic features of the two independent interactions. The interaction between SDF-1α and CXCR4 in the presence of AMD3100 should mimic the 1st step of this two-step binding model. Although the direct observation of this 1st step intermediate state is difficult because of its low population, we successfully trapped this state by adding AMD3100, which inhibits the formation of the final 2nd step complex.
FIGURE 6.
Physiological role of the two-step mechanism for the SDF-1α-CXCR4 interaction. A, schematic diagram of the two-step mechanism for the SDF-1α-CXCR4 interaction. The 1st step interaction between the SDF-1α β-sheet, 50-s loop, and N-loop and the CXCR4 extracellular region facilitates the rapid binding and efficient anchoring of SDF-1α on the extracellular side of CXCR4. The SDF-1α N terminus is highly dynamic even in this state, which is used to search for the binding cavities buried within the TM helices. Consequently, the 2nd step interaction between the SDF-1α N terminus and the CXCR4 TM region is formed, and the SDF-1α N terminus triggers the conformational changes in the CXCR4 TM to induce G-protein signaling. B, crystal structures of two GPCRs, β2-adrenergic receptor (Protein Data Bank code 2RH1), and adenosine A2A receptor (Protein Data Bank code 3EML). Small molecule antagonists (shown as sticks) are buried in the deep cavities within the TM helices (red ribbons). The entrances of these cavities are restricted by the long ECL2s (green ribbons) and the conserved disulfide bonds between ECL2 and TM3 (yellow sticks). Solvent-accessible surfaces of the receptors are also shown (transparent gray surfaces).
The recently solved structures of β2-adrenergic receptor (58, 59) and adenosine A2A receptor (60) indicated that the binding pocket within the TM helices is partially covered by ECL2 (Fig. 6B). Considering the fact that CXCR4 also possesses the ECL2 with the similar length and the conserved disulfide bond between ECL2 and TM3, it is difficult for SDF-1α to bind the buried pocket, because SDF-1α is much larger than most of the other GPCR ligands. Therefore, it is likely that the rate-limiting step for the receptor binding of SDF-1α would be the opening of ECL2 to provide the space for inserting the critical residues into the TM region.
The two-step interaction, described above, would be important for the efficient binding between SDF-1α and CXCR4 (Fig. 6A). The independent interaction between the SDF-1α β-sheet, 50-s loop, and N-loop and the CXCR4 extracellular region should facilitate the rapid and efficient anchoring of SDF-1α near the CXCR4 TM region. Even in this 1st step bound state, the SDF-1α N terminus should exhibit a highly dynamic nature, as shown in the spectrum of SDF-1α with CXCR4 and AMD3100 (Fig. 5C), in the limited space. Because an unstructured protein has a large capture radius, the dynamics of the SDF-1α N terminus would increase the efficiency for searching the space for its binding site, through the well known “fly-casting” mechanism of intrinsically disordered proteins (61). This fly-casting effect, together with the trapping mechanism of SDF1-α to glycosaminoglycans (46), which elevates the local concentration of SDF1-α, would enhance the rate of the complex formation. Consequently, the SDF-1α N terminus can bind to the buried cavities within the CXCR4 TM helices to trigger the conformational changes in the CXCR4 TM region, which lead to the G-protein signaling.
Because all known chemokines have flexible N-terminal regions that are critical for signaling through chemokine receptors, the two-step mechanism should be generalized across the chemokine families. Such a mechanism may also be present in other GPCRs with peptide ligands, including the class B GPCRs, with relatively large extracellular domains for anchoring part of their ligands.
Applicability of Methyl-TCS to GPCRs
GPCRs constitute the largest family of cell surface signaling receptors, with >800 members. GPCRs play crucial roles in many physiological processes, through signal transduction across cell membranes in response to a variety of stimuli (62). Although GPCRs include more than 30% of modern drug targets (63), the tertiary structures of only four GPCR structures have been solved to date (58–60, 64, 65). Despite the recent advances in protein chemistry and x-ray crystallography, structural determinations of GPCRs are still challenging, and laborious optimizations of crystallization conditions for each GPCR are necessary, which hampers the analyses of multiple GPCR-ligand interactions. Although solution NMR provides powerful tools for investigations of the protein-protein interactions, NMR analyses with the full-length GPCRs are very limited to date (66–68).
One of the problems in solving GPCR structures and investigating the GPCR-ligand interactions is their low expression levels in heterologous expression systems, and another problem is their extreme instability in detergent micelles, which often yield nonfunctional receptors during solubilization and purification. Therefore, the experimental conditions of expression, solubilization, and purification should be extensively optimized. In addition, the percentages of correctly folded receptors should be estimated, and the effect of denatured receptors on the structural analyses should be considered.
In this study, 100–200 μg of CXCR4, about half of which were correctly folded, were obtained from 1 liter of insect cell culture, and it retained the native conformation for 48 h (Fig. 1). The yield was far below the practical amounts needed for conventional NMR and crystallographic studies. In addition, the low temperature and the addition of glycerol, which were required for the stabilization of CXCR4, induced broadening of the NMR signals. However, the methyl-TCS methods, which are highly sensitive methods, were applicable even in such cases, because only ∼10 μm of receptor is sufficient for the determinations of the receptor-binding sites in ligand molecules with this method. In addition, the effects of denatured CXCR4 could be properly subtracted by the negative control experiment (Fig. 2B).
In this study, we have provided the first structural view of the two-step interaction between a chemokine and its full-length G-protein-coupled receptor. Our strategy with the methyl-TCS experiments used in this study would be generally applicable to GPCR-ligand interactions and hence would provide a favorable alternative for the elucidation of the ligand recognition mechanisms of GPCRs. In addition, we have shown that the drug-induced modulation of the GPCR-ligand interaction could be clearly detected by NMR experiments. Such experiments should be useful to elucidate whether and how the drug functions and thus would support drug development targeted to GPCRs.
This work was supported by a grant from the Japan New Energy and Industrial Technology Development Organization.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and additional references.
- SDF-1
- stromal cell-derived factor-1
- GPCR
- G-protein-coupled receptor
- TM
- transmembrane
- ECL
- extracellular loop
- TCS
- transferred cross-saturation
- methyl-TCS
- methyl-utilizing transferred cross-saturation
- SPR
- surface plasmon resonance
- HSQC
- heteronuclear single quantum coherence
- HMQC
- heteronuclear multiple quantum coherence
- DDM
- n-dodecyl β-d-maltoside.
REFERENCES
- 1.Baggiolini M. (1998) Nature 392, 565–568 [DOI] [PubMed] [Google Scholar]
- 2.Murphy P. M., Baggiolini M., Charo I. F., Hébert C. A., Horuk R., Matsushima K., Miller L. H., Oppenheim J. J., Power C. A. (2000) Pharmacol. Rev. 52, 145–176 [PubMed] [Google Scholar]
- 3.Tashiro K., Tada H., Heilker R., Shirozu M., Nakano T., Honjo T. (1993) Science 261, 600–603 [DOI] [PubMed] [Google Scholar]
- 4.Nagasawa T., Kikutani H., Kishimoto T. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 2305–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Y., Broder C. C., Kennedy P. E., Berger E. A. (1996) Science 272, 872–877 [DOI] [PubMed] [Google Scholar]
- 6.Bleul C. C., Farzan M., Choe H., Parolin C., Clark-Lewis I., Sodroski J., Springer T. A. (1996) Nature 382, 829–833 [DOI] [PubMed] [Google Scholar]
- 7.Oberlin E., Amara A., Bachelerie F., Bessia C., Virelizier J. L., Arenzana-Seisdedos F., Schwartz O., Heard J. M., Clark-Lewis I., Legler D. F., Loetscher M., Baggiolini M., Moser B. (1996) Nature 382, 833–835 [DOI] [PubMed] [Google Scholar]
- 8.Nagasawa T., Hirota S., Tachibana K., Takakura N., Nishikawa S., Kitamura Y., Yoshida N., Kikutani H., Kishimoto T. (1996) Nature 382, 635–638 [DOI] [PubMed] [Google Scholar]
- 9.Tachibana K., Hirota S., Iizasa H., Yoshida H., Kawabata K., Kataoka Y., Kitamura Y., Matsushima K., Yoshida N., Nishikawa S., Kishimoto T., Nagasawa T. (1998) Nature 393, 591–594 [DOI] [PubMed] [Google Scholar]
- 10.Zou Y. R., Kottmann A. H., Kuroda M., Taniuchi I., Littman D. R. (1998) Nature 393, 595–599 [DOI] [PubMed] [Google Scholar]
- 11.Nagasawa T., Tachibana K., Kawabata K. (1999) Adv. Immunol. 71, 211–228 [DOI] [PubMed] [Google Scholar]
- 12.Müller A., Homey B., Soto H., Ge N., Catron D., Buchanan M. E., McClanahan T., Murphy E., Yuan W., Wagner S. N., Barrera J. L., Mohar A., Verástegui E., Zlotnik A. (2001) Nature 410, 50–56 [DOI] [PubMed] [Google Scholar]
- 13.Balkwill F. (2004) Nat. Rev. Cancer 4, 540–550 [DOI] [PubMed] [Google Scholar]
- 14.Princen K., Schols D. (2005) Cytokine Growth Factor Rev. 16, 659–677 [DOI] [PubMed] [Google Scholar]
- 15.Crump M. P., Gong J. H., Loetscher P., Rajarathnam K., Amara A., Arenzana-Seisdedos F., Virelizier J. L., Baggiolini M., Sykes B. D., Clark-Lewis I. (1997) EMBO J. 16, 6996–7007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gozansky E. K., Louis J. M., Caffrey M., Clore G. M. (2005) J. Mol. Biol. 345, 651–658 [DOI] [PubMed] [Google Scholar]
- 17.Dealwis C., Fernandez E. J., Thompson D. A., Simon R. J., Siani M. A., Lolis E. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6941–6946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohnishi Y., Senda T., Nandhagopal N., Sugimoto K., Shioda T., Nagal Y., Mitsui Y. (2000) J. Interferon Cytokine Res. 20, 691–700 [DOI] [PubMed] [Google Scholar]
- 19.Murphy J. W., Cho Y., Sachpatzidis A., Fan C., Hodsdon M. E., Lolis E. (2007) J. Biol. Chem. 282, 10018–10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veldkamp C. T., Seibert C., Peterson F. C., De la Cruz N. B., Haugner J. C., 3rd, Basnet H., Sakmar T. P., Volkman B. F. (2008) Sci. Signal. 1, ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobilka B. K. (2007) Biochim. Biophys. Acta 1768, 794–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weis W. I., Kobilka B. K. (2008) Curr. Opin. Struct. Biol. 18, 734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doranz B. J., Orsini M. J., Turner J. D., Hoffman T. L., Berson J. F., Hoxie J. A., Peiper S. C., Brass L. F., Doms R. W. (1999) J. Virol. 73, 2752–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brelot A., Heveker N., Montes M., Alizon M. (2000) J. Biol. Chem. 275, 23736–23744 [DOI] [PubMed] [Google Scholar]
- 25.Zhou N., Luo Z., Luo J., Liu D., Hall J. W., Pomerantz R. J., Huang Z. (2001) J. Biol. Chem. 276, 42826–42833 [DOI] [PubMed] [Google Scholar]
- 26.Dukkipati A., Vaclavikova J., Waghray D., Garcia K. C. (2006) Protein Expr. Purif. 50, 203–214 [DOI] [PubMed] [Google Scholar]
- 27.Nakanishi T., Miyazawa M., Sakakura M., Terasawa H., Takahashi H., Shimada I. (2002) J. Mol. Biol. 318, 245–249 [DOI] [PubMed] [Google Scholar]
- 28.Shimada I. (2005) Methods Enzymol. 394, 483–506 [DOI] [PubMed] [Google Scholar]
- 29.Shimada I., Ueda T., Matsumoto M., Sakakura M., Osawa M., Takeuchi K., Nishida N., Takahashi H. (2009) Prog. Nuclear Magn. Reson. Spectrosc. 54, 123–140 [Google Scholar]
- 30.Takahashi H., Nakanishi T., Kami K., Arata Y., Shimada I. (2000) Nat. Struct. Biol. 7, 220–223 [DOI] [PubMed] [Google Scholar]
- 31.Takahashi H., Miyazawa M., Ina Y., Fukunishi Y., Mizukoshi Y., Nakamura H., Shimada I. (2006) J. Biomol. NMR 34, 167–177 [DOI] [PubMed] [Google Scholar]
- 32.Schols D., Struyf S., Van Damme J., Esté J. A., Henson G., De Clercq E. (1997) J. Exp. Med. 186, 1383–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerlach L. O., Skerlj R. T., Bridger G. J., Schwartz T. W. (2001) J. Biol. Chem. 276, 14153–14160 [DOI] [PubMed] [Google Scholar]
- 34.Goto N. K., Gardner K. H., Mueller G. A., Willis R. C., Kay L. E. (1999) J. Biomol. NMR 13, 369–374 [DOI] [PubMed] [Google Scholar]
- 35.Tugarinov V., Kay L. E. (2004) J. Biomol. NMR 28, 165–172 [DOI] [PubMed] [Google Scholar]
- 36.Holmes W. D., Consler T. G., Dallas W. S., Rocque W. J., Willard D. H. (2001) Protein Expr. Purif. 21, 367–377 [DOI] [PubMed] [Google Scholar]
- 37.Navratilova I., Sodroski J., Myszka D. G. (2005) Anal. Biochem. 339, 271–281 [DOI] [PubMed] [Google Scholar]
- 38.Clore G. M., Gronenborn A. M. (1994) Methods Enzymol. 239, 349–363 [DOI] [PubMed] [Google Scholar]
- 39.Senn H., Werner B., Messerle B. A., Weber C., Traber R., Wüthrich K. (1989) FEBS Lett. 249, 113–118 [Google Scholar]
- 40.Davis A. L., Keeler J., Laue E. D., Moskau D. (1992) J. Magn. Reson. 98, 207–216 [Google Scholar]
- 41.Kupce E., Wagner G. (1995) J. Magn. Reson. B. 109, 329–333 [Google Scholar]
- 42.Baribaud F., Edwards T. G., Sharron M., Brelot A., Heveker N., Price K., Mortari F., Alizon M., Tsang M., Doms R. W. (2001) J. Virol. 75, 8957–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veldkamp C. T., Peterson F. C., Pelzek A. J., Volkman B. F. (2005) Protein Sci. 14, 1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veldkamp C. T., Seibert C., Peterson F. C., Sakmar T. P., Volkman B. F. (2006) J. Mol. Biol. 359, 1400–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baryshnikova O. K., Sykes B. D. (2006) Protein Sci. 15, 2568–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson Z., Proudfoot A. E., Handel T. M. (2005) Cytokine Growth Factor Rev. 16, 625–636 [DOI] [PubMed] [Google Scholar]
- 47.Veldkamp C. T., Ziarek J. J., Su J., Basnet H., Lennertz R., Weiner J. J., Peterson F. C., Baker J. E., Volkman B. F. (2009) Protein Sci. 18, 1359–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Babcock G. J., Farzan M., Sodroski J. (2003) J. Biol. Chem. 278, 3378–3385 [DOI] [PubMed] [Google Scholar]
- 49.Percherancier Y., Berchiche Y. A., Slight I., Volkmer-Engert R., Tamamura H., Fujii N., Bouvier M., Heveker N. (2005) J. Biol. Chem. 280, 9895–9903 [DOI] [PubMed] [Google Scholar]
- 50.Springael J. Y., Urizar E., Parmentier M. (2005) Cytokine Growth Factor Rev. 16, 611–623 [DOI] [PubMed] [Google Scholar]
- 51.Wang J., Norcross M. (2008) Drug Discov. Today 13, 625–632 [DOI] [PubMed] [Google Scholar]
- 52.Bouvier M., Heveker N., Jockers R., Marullo S., Milligan G. (2007) Nat. Methods 4, 3–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gurevich V. V., Gurevich E. V. (2008) Trends Neurosci. 31, 74–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chabre M., le Maire M. (2005) Biochemistry 44, 9395–9403 [DOI] [PubMed] [Google Scholar]
- 55.Hernanz-Falcón P., Rodríguez-Frade J. M., Serrano A., Juan D., del Sol A., Soriano S. F., Roncal F., Gómez L., Valencia A., Martínez-A C., Mellado M. (2004) Nat. Immunol. 5, 216–223 [DOI] [PubMed] [Google Scholar]
- 56.Jastrzebska B., Maeda T., Zhu L., Fotiadis D., Filipek S., Engel A., Stenkamp R. E., Palczewski K. (2004) J. Biol. Chem. 279, 54663–54675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Farzan M., Babcock G. J., Vasilieva N., Wright P. L., Kiprilov E., Mirzabekov T., Choe H. (2002) J. Biol. Chem. 277, 29484–29489 [DOI] [PubMed] [Google Scholar]
- 58.Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosenbaum D. M., Cherezov V., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Yao X. J., Weis W. I., Stevens R. C., Kobilka B. K. (2007) Science 318, 1266–1273 [DOI] [PubMed] [Google Scholar]
- 60.Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wright P. E., Dyson H. J. (2009) Curr. Opin. Struct. Biol. 19, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lagerström M. C., Schiöth H. B. (2008) Nat. Rev. Drug Discov. 7, 339–357 [DOI] [PubMed] [Google Scholar]
- 63.Schlyer S., Horuk R. (2006) Drug Discov. Today 11, 481–493 [DOI] [PubMed] [Google Scholar]
- 64.Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 66.Inooka H., Ohtaki T., Kitahara O., Ikegami T., Endo S., Kitada C., Ogi K., Onda H., Fujino M., Shirakawa M. (2001) Nat. Struct. Biol. 8, 161–165 [DOI] [PubMed] [Google Scholar]
- 67.Kisselev O. G., Kao J., Ponder J. W., Fann Y. C., Gautam N., Marshall G. R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4270–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koenig B. W., Kontaxis G., Mitchell D. C., Louis J. M., Litman B. J., Bax A. (2002) J. Mol. Biol. 322, 441–461 [DOI] [PubMed] [Google Scholar]
- 69.Goddard T. D., Kneller D. G. (2006) SPARKY 3, University of California, San Francisco [Google Scholar]