Abstract
The Floridian marine cyanobacterium Lyngbya confervoides afforded three new cyclodepsipeptides, termed tiglicamides A–C (1–3), along with their previously reported analogues largamides A–C (4–6), all of which possess an unusual tiglic acid moiety. Their structures were deduced by one- and two-dimensional NMR combined with mass spectrometry and the absolute configurations established by chiral HPLC and Marfey’s analysis of the degradation products. Compounds 1–3 moderately inhibited porcine pancreatic elastase in vitro with IC50 values from 2.14 to 7.28 µM. Compounds 1–6 differ from each other by one amino acid residue within the cyclic core structure, suggesting an unusually relaxed substrate specificity of the nonribosomal peptide synthetase that is the putative biosynthetic enzyme responsible for the corresponding amino acid incorporation.
Keywords: Marine cyanobacteria, Lyngbya confervoides, cyclodepsipeptides, nonribosomal peptide synthesis, elastase inhibitors
1. Introduction
Cyanobacteria are a group of microorganisms with the distinction of being the most ancient known organisms on Earth [Schopf, 1996]. The secondary metabolites produced by cyanobacteria are extremely diverse in their structural motifs, with modified polypeptides or peptide–polyketide hybrids being the most commonly encountered. Most of these peptides are thought to be biosynthesized by nonribosomal polypeptide synthetases (NRPS) or the mixed polyketide synthase–NRPS pathways [Tan, 2007]; however, several peptidic natural products were recently shown to be made ribosomally [McIntosh et al., 2009]. The secondary metabolites produced by cyanobacteria exhibit a broad spectrum of biological activities affecting a variety of bacterial, viral, fungal and mammalian targets. Among marine cyanobacteria, the genus Lyngbya is considered to be the most prolific producer of natural products with over 200 compounds reported [Blunt and Munro, 2008].
Here we describe the isolation, structure elucidation and biological evaluation of three new analogues of largamides A–C (4–6) [Plaza and Bewley, 2006], which we named tiglicamides A–C (1–3), from a recollection of the Floridian marine cyanobacterium Lyngbya confervoides that also afforded compounds 4–6 [Matthew et al., 2009]. Our previous chemical investigations of the same species already yielded several structurally unrelated secondary metabolites, including serine protease inhibitors, namely lyngbyastatins 4–6 [Matthew et al., 2007; Taori et al., 2007], pompanopeptin A [Matthew et al., 2008], along with largamides D–H [Plaza and Bewley, 2006]. Due to the structural homology to largamides A–C (4–6), which are moderate inhibitors of porcine pancreatic elastase [Matthew et al., 2009], we tested tiglicamides A–C (1–3) for activity against this enzyme.
Among the five main classes of proteolytic enzymes (aspartic, serine, cysteine, metallo- and threonine), the serine proteases constitute the most extensively studied enzyme family. Serine proteases are known to regulate important biological processes, which makes them attractive therapeutic targets [Ilies et al., 2002]. Elastase is a serine protease implicated in adult respiratory distress syndrome (ARDS), rheumatoid arthritis, pulmonary emphysema, cystic fibrosis and chronic bronchitis. Despite extensive research efforts, there are relatively few elastase inhibitors in advanced stages of development; however, one of them, sivelestat (ONO-5046), has already been launched in Japan for the treatment of acute lung injury associated with systemic inflammatory response syndrome (SIRS) [Abbenante and Fairlie, 2005]. The investigation of natural products from marine cyanobacteria as a source of novel serine protease inhibitors may eventually aid the development of more promising therapeutic leads.
2. Results and discussion
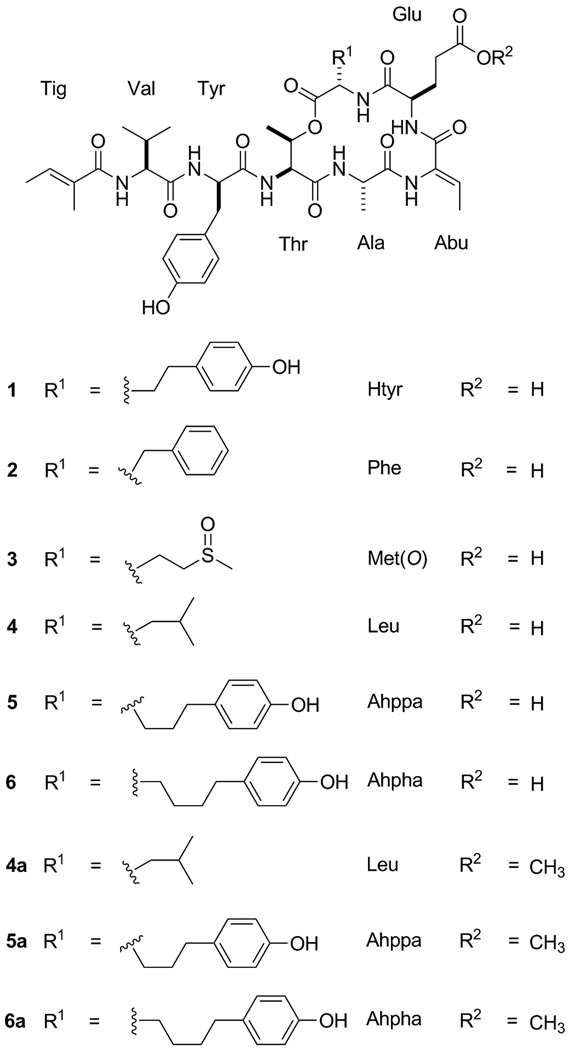
The marine cyanobacterium Lyngbya confervoides collected near Ft. Lauderdale (Florida, USA) was extracted with organic solvents and the organic extract subjected to HP-20 chromatographic fractionation, and several HPLC purifications to yield compounds 1–3 as colorless, amorphous solids. The planar structures of 1–3 (Fig. 1) were determined by a combination of NMR (1H, COSY, TOCSY, ROESY, HSQC, and HMBC) spectroscopic analysis and mass spectrometry. Compound 1 was isolated as a colorless amorphous solid. A pseudomolecular [M + Na]+ ion peak at m/z 928.4032 in the HR-ESI/APCI-MS suggested a molecular formula of C45H59N7O13, which was in agreement with the putative molecular composition based on NMR data. A detailed 2D NMR analysis in DMF-d7 (Table 1) enabled the identification of the five common amino acids alanine (Ala), valine (Val), glutamic acid (Glu), tyrosine (Tyr), threonine (Thr), the two modified amino acid residues homotyrosine (Htyr) and 2-amino-2-butenoic acid (Abu), and a tiglic acid (Tig) unit in 1, thus indicating a striking resemblance to largamides A–C (4–6) (Fig 1). The presence of the tiglic acid unit is corroborated by the appearance of two deshielded methyl groups (δH 1.81 and 1.73) and one olefinic methine proton (δH-3 6.44, br q) in the 1H NMR spectrum; the latter further showed correlations to a quaternary carbon at δ 132.4 (C-2, Tig) and a carbonyl carbon at δ 169.1 (C-1, Tig) in the HMBC spectrum. In addition, the olefinic proton δ 6.44 (H-3) showed a ROESY cross peak to a methyl resonating at δ 1.73 (H3-3) and lacked a correlation to the methyl at δ 1.81 (H3-2), indicating E geometry of the double bond and confirming a tigloyl group in 1 as in 4–6. The geometry of the Abu unit was deduced as Z based on a ROESY cross peak between the Abu NH (δH 10.21) and Abu methyl group (δH 1.78). HMBC analysis supported by ROESY correlations unambiguously established the linear sequence of the amino acid units and tiglic acid moiety (Table 1). The deshielded proton signal at δH 5.39 (Thr) was indicative of a lactone functionality which arises from ester linkage of 1 from the carbonyl of Htyr and the hydroxyl group of Thr. The IR spectrum of 1, displaying absorptions at 1722 and 1652 cm−1 characteristic of amide and ester functionalities, respectively, supported the proposed depsipeptide structure.
Fig. 1.
Structures of tiglicamides A–C (1–3), largamides A–C (4–6) and their corresponding methyl esters 4a–6a.
Table 1.
1H and 13C NMR assignments for tiglicamide A (1) (600 MHz, DMF-d7)
| Unit | C/H no. | δH (J in Hz) | δC, mult. | HMBCa |
|---|---|---|---|---|
| Htyr | 1 | 171.4, qC | ||
| 2 | 4.61, br m | 50.6, CH | 1, 3 | |
| 3 | 2.12, m; 1.82, m | 33.9, CH2 | 2, 3 | |
| 4 | 2.47, m (2H) | 30.8, CH2 | 2, 3, 5, 6/10 | |
| 5 | 132.2, qC | |||
| 6/10 | 7.04, d (8.0) | 129.8, CH | 4 | |
| 7/9 | 6.71, d (8.0) | 115.1, CH | 4, 5 | |
| 8 | 156.3, qC | |||
| OH | 9.31, s | 7/9 | ||
| NH | 7.72, d (9.4) | 1 (Glu) | ||
| Glu | 1 | 171.2, qC | ||
| 2 | 4.55, br m | 52.9, CH | ||
| 3 | 2.50, m; 2.19, m | 26.7, CH2 | 2, 4, 5 | |
| 4 | 2.56, m; 2.47, m | 30.7, CH2 | 2, 3, 5 | |
| 5 | 174.9, qC | |||
| OH | not observed | |||
| NH | 7.55, d (8.6) | 1 (Abu) | ||
| Abu | 1 | 163.8, qC | ||
| 2 | 129.5, qC | |||
| 3 | 6.57, br q (6.8) | 128.7, CH | 1, 4 | |
| 4 | 1.78, d (7.0) | 12.4, CH3 | 1, 2, 3 | |
| NH | 10.21, s | 1 (Ala) | ||
| Ala | 1 | 175.5, qC | ||
| 2 | 4.36, br q (6.7) | 50.3, CH | 3 | |
| 3 | 1.40, d (6.7) | 16.4, CH3 | 1, 2 | |
| NH | 8.86, br s | 1 (Thr) | ||
| Thr | 1 | 170.2, qC | ||
| 2 | 4.78, br m | 55.5, CH | 1 | |
| 3 | 5.39, br q | 73.1, CH | 1 | |
| 4 | 1.19, d (5.9) | 15.9, CH3 | 2, 3 | |
| NH | 7.89, d (8.2) | 1 (Tyr) | ||
| Tyr | 1 | 172.4, qC | ||
| 2 | 4.77, br m | 55.4, CH | 1, 1 (Val) | |
| 3 | 3.08, dd (−13.2, 3.9) | 37.8, CH2 | 2, 4, 5/9 | |
| 2.84, dd (−13.2, 9.6) | ||||
| 4 | 128.2, qC | |||
| 5/9 | 7.12, d (7.8) | 130.5, CH | 3, 6/8, 7 | |
| 6/8 | 6.75, d (7.8) | 115.1, CH | 5/9, 7 | |
| 7 | 156.7, qC | |||
| OH | 9.35, s | 6/8, 7 | ||
| NH | 8.08, br d (7.5) | |||
| Val | 1 | 171.8, qC | ||
| 2 | 4.30, br dd | 58.8, CH | 1, 3, 4, 5 | |
| 3 | 2.04, m | 31.3, CH | 1, 2, 4, 5 | |
| 4 | 0.76, d (6.3) | 19.3, CH3 | 2, 3, 5 | |
| 5 | 0.73, d (6.3) | 17.9, CH3 | 2, 3, 4 | |
| NH | 7.29, d (8.8) | 1 (Tig) | ||
| Tig | 1 | 169.1, qC | ||
| 2 | 132.4, qC | |||
| 3 | 6.44, br q (6.3) | 130.1, CH | 1, 4 | |
| 4 | 1.73, br d (6.3) | 13.4, CH3 | 2, 3 | |
| 5 | 1.81, br s | 12.2, CH3 | 1, 2, 3 |
Protons showing long-range correlation with indicated carbon.
An [M + Na]+ peak at m/z 898.3935 in the HR-ESI/APCI-MS of 2 in conjunction with 2D NMR data suggested a molecular formula of C44H57N7O12 for compound 2. The 1H NMR spectrum indicated that compound 2 is closely related to 1. 2D NMR analysis (1H, COSY, HSQC, HMBC, ROESY) provided further evidence for the presence of Ala, Val, Thr, Glu, and two aromatic amino acid residues, only one of which was para-substituted (Tyr) in 2. The Htyr unit in 1 was replaced by a Phe residue in 2 (Table 2), which accounted for the 30 mass unit difference (CH2O) compared with 1. The presence of the Phe residue in the cyclic core was confirmed based on an HMBC correlation of the Phe NH proton to C-1 of the Glu residue (Table 2).
Table 2.
NMR data for tiglicamides B (2) and C (3) in DMF-d7 (600 MHz)
| Tiglicamide B (2) | Tiglicamide C (3) | ||||||
|---|---|---|---|---|---|---|---|
| Unit | C/H no. | δH (J in Hz) | δC, mult. | HMBCa | δH (J in Hz) | δC, mult. | HMBCa |
| Pheb / Met(O)c | 1 | 170.9, qC | 170.7, qC | ||||
| 2 | 4.77, br m | 52.9, CH | 1, 3 | 4.80, br m | 50.0, CH | 1 | |
| 3 | 3.29, dd (−13.6, 4.9) | 37.6, CH2 | 2, 4, 5/8 | 2.31/2.26d, m | 24.85, 24.90d, | 2, 4 | |
| 2.76, dd (−13.6, 10.3) | 1.96/1.86d, m | CH2 | |||||
| 4 | 138.5, qC | 2.81/2.75d, m | 50.56, 50.60d, | 3 | |||
| 2.71/ 2.63d , m | CH2 | ||||||
| 5/9 | 7.29, m | 129.7, CH | 3, 6/9, 7 | – | – | ||
| 6/8 | 7.27, m | 128.3, CH | 3, 5/8, 7 | – | – | ||
| 7 | 7.19, m | 126.4, CH | 5/8, 6/9 | – | – | ||
| S-Me | – | – | – | 2.55, 2.52d, s | 38.7, 37.6d, CH3 | 4 | |
| NH | 7.84, d (9.4) | 2, 1 (Glu) | 7.71, d (9.4) | 1(Glu) | |||
| Glu | 1 | 171.1, qC | 171.7, qC | ||||
| 2 | 4.42, br m | 52.4, CH | 3, 4 | 4.48, br m | 53.6, CH | 3, 4 | |
| 3 | 2.44, m ; 2.10, m | 27.0, CH2 | 2, 4, 5 | 2.47, m ; 2.18, m | 26.9, CH2 | 1, 2, 4, 5 | |
| 4 | 2.50, m ; 2.41, m | 30.6, CH2 | 2, 3, 5 | 2.60, m ; 2.48, m | 30.9, CH2 | 2, 3, 5 | |
| 5 | 174.9, qC | 175.0, qC | |||||
| OH | not observed | not observed | |||||
| NH | 7.60, d (9.0) | 2, 1 (Abu) | 7.49, d (8.6), 7.48, d | 1 (Abu) | |||
| (8.6)e | |||||||
| Abu | 1 | 163.9, qC | 164.9, qC | ||||
| 2 | 130.9, qC | 130.6, qC | |||||
| 3 | 6.65, q (7.0) | 129.0, CH | 1, 4 | 6.54, qd (7.0, 1.3)/6.53, | 129.3, CH | 1, 4 | |
| qd (7.0, 1.3)e | |||||||
| 4 | 1.80, d (7.0) | 12.6, CH3 | 1, 2, 3 | 1.77, dq (7.1, 1.3) | 12.6, CH3 | 1, 2, 3 | |
| NH | 10.23, s | 1, 1 (Ala) | 10.25, s | 1, 1 (Ala) | |||
| Ala | 1 | 175.7, qC | 176.1, qC | ||||
| 2 | 4.39, br m | 50.2, CH | 1, 3 | 4.34, br m | 50.6, CH | 1, 3 | |
| 3 | 1.42, d (6.8) | 16.4, CH3 | 1, 2 | 1.40, d (7.0) | 16.7, CH3 | 1, 2 | |
| NH | 8.89, br s | 1, 2, 3 | 8.90, d (3.0)/8.89, d (3.2)e | 1, 3 | |||
| Thr | 1 | 170.4, qC | 170.3, qC | ||||
| 2 | 4.77, br m | 55.2, CH | 1 | 4.80, br m | 55.5, CH | 1 | |
| 3 | 5.45, br q | 72.9, CH | 5.429, qd (6.4, | 73.9, CH | 1, 1 (Met(O)) | ||
| 2.9)/5.427, qd (6.4, 2.9)e | |||||||
| 4 | 1.18, d (5.0) | 15.7, CH3 | 2, 3 | 1.20, d (6.4) | 16.1, CH3 | 2, 3 | |
| NH | 7.92, d (8.2) | 2, 1 (Tyr) | 7.95, d (8.1) | 2, 1 (Tyr) | |||
| Tyr | 1 | 172.7, qC | 172.8, qC | ||||
| 2 | 4.77, br m | 55.3, CH | 3 | 4.79, br m | 55.8, CH | 1, 3, 1 (Val) | |
| 3 | 3.08, dd (−13.7, 4.5) | 37.7, CH2 | 2, 4, 5/9 | 3.06, dd (−14.0, 4.8) | 38.1, CH2 | 1, 2, 4, 5/9 | |
| 2.83, dd (−13.7, 10.1) | 2.83, dd (−14.0, 10.0) | ||||||
| 4 | 128.5, qC | 128.4, qC | |||||
| 5/9 | 7.13, d (7.8) | 130.5, CH | 3, 6/8, 7 | 7.12, d (8.5) | 131.1, CH | 3, 6/8, 7 | |
| 6/8 | 6.73, d (7.8) | 115.2, CH | 5/9, 7 | 6.71, d (8.5) | 115.6, CH | 5/9, 7 | |
| 7 | 157.0, qC | 157.3, qC | |||||
| OH | 9.37, br s | 6/8, 7 | 9.35, s | 6/8, 7 | |||
| NH | 8.09, d (8.1) | 1 (Val) | 8.12, d (8.5) | 2, 1 (Val) | |||
| Val | 1 | 171.8, qC | 172.0, qC | ||||
| 2 | 4.28, br t (7.3) | 58.8, CH | 1, 3, 4, 5, 1 (Tig) | 4.29, dd (6.8, 2.7)/4.28, | 59.2, CH | 1, 3, 4, 5, 1 (Tig) | |
| dd (6.8, 2.7)e | |||||||
| 3 | 2.02, m | 31.4, CH | 1, 2, 4, 5 | 2.02, m | 31.5, CH | 1, 2, 4, 5 | |
| 4 | 0.74, d (6.8) | 19.4, CH3 | 2, 3, 5 | 0.75, d (6.8) | 19.7, CH3 | 2, 3, 5 | |
| 5 | 0.72, d (6.8) | 18.0, CH3 | 2, 3, 4 | 0.72, d (6.8) | 18.3, CH3 | 2, 3, 4 | |
| NH | 7.26, d (7.3) | 2, 1 (Tig) | 7.27, d (8.6) | 1, 2, 1 (Tig) | |||
| Tig | 1 | 169.2, qC | 169.3, qC | ||||
| 2 | 132.5, qC | 132.7, qC | |||||
| 3 | 6.43, br q (6.8) | 130.3, CH | 1, 4, 5 | 6.43, qq (7.0, 1.5) | 130.6, CH | 1, 4, 5 | |
| 4 | 1.70, br d (6.8) | 13.5, CH3 | 2, 3 | 1.72, dq (7.0, 1.5) | 13.7, CH3 | 2, 3 | |
| 5 | 1.79, br s | 12.3, CH3 | 1, 2, 3 | 1.80, br s | 12.4, CH3 | 1, 2, 3 | |
Protons showing long-range correlation with indicated carbon.
Refers to compound 2.
Refers to compound 3.
Signal doubling because of diastereomers at chiral S*.
Signal doubling observed.
HR-ESI/APCI-MS analysis for compound 3 provided an [M + Na]+ peak at m/z 898.3591, suggesting a molecular formula of C40H57N7O13S. The 1H NMR of 3 displayed signals characteristic for 1 and 2 with the exception of the fewer aromatic signals and two additional methyl singlets at δH 2.55/2.52 (1:1), which integrated together for one methyl group (S-Me). A detailed 2D NMR analysis (COSY, TOCSY, HSQC, HMBC, ROESY) revealed that compound 3 contained only one aromatic amino acid residue and differs from 1 or 2 only by the presence of a methionine sulfoxide (Met(O)) unit instead of Htyr or Phe, respectively (Table 2). The Met(O) residue was verified by 2D NMR analysis, where the two methyl singlets at δH 2.55/2.52 showed HSQC correlations to the doubled signals at δC 38.7/37.6 and HMBC correlations to δC 50.56/50.60. This doubling of signals in the ratio of 1:1 suggested the occurrence of epimeric R and S sulfoxides. The IR absorption at 1037 cm−1 also supported the sulfoxide assignment. The sulfoxide is most likely an isolation artifact formed by oxidation of the methionine-containing natural product [Matthew et al., 2008; Gunasekera et al., 2008; Harrigan et al., 1999].
The absolute configuration of all amino acid units in compounds 1–3 was deduced by chiral HPLC of the acid hydrolysis products, revealing d onfiguration for glutamic acid and tyrosine and l onfiguration for all other amino acids. However, none of the chiral HPLC conditions employed were successful in resolving the d/l-isomeric peaks for Phe. Hence the acid hydrolyzate of 2 was subjected to Marfey’s analysis [Fujii et al.,1997], establishing l onfiguration for Phe in 2.
Compounds 1–3 were tested for serine protease-inhibitory activity. They showed moderate activity against porcine pancreatic elastase in vitro with IC50 values ranging from 2.14 to 7.28 µM (Table 3). While the described compounds are two to three orders of magnitudes less potent against the same enzyme or other mammalian elastases than lyngbyastatins 4–7 [Matthew et al., 2007; Taori et al., 2007] or ONO-5046 [Kawabata et al., 1991], preliminary data suggested some selectivity towards elastase. The activities of two other serine proteases tested (chymotrypsin, trypsin) were not compromised by compounds 1–3 at concentrations up to 50 µM. These results are consistent with those previously reported for their analogues (4–6) [Matthew et al., 2009]. Since we also isolated the corresponding largamide methyl esters 4a–6a (presumably isolation artifacts), we were able to probe the effect of methylation at that position. Compounds 4a–6a retained low-micromolar inhibitory activity (Table 3), indicating that the carboxylic acid residue is not a requisite element for elastase-inhibitory activity [Matthew et al., 2009].
Table 3.
Inhibition of porcine pancreatic elastase
3. Conclusion
The Lyngbya confervoides that yielded tiglicamides A–C (1–3) is a particularly prolific source of secondary metabolites [Matthew et al., 2007, 2008, 2009; Taori et al., 2007], with already 13 previously reported structures belonging to six different structural families [Sharp et al., 2009]. The similarity to the largamides A–C (4–6) and, specifically, the variability of the last amino acid position (N→C) suggests relaxed substrate specificity of the corresponding putative biosynthetic NRPS enzyme, allowing the incorporation of at least six different amino acids, viz. Htyr (1), Phe (2), Met (3), Leu (4), 2-amino-5-(4’-hydroxy-phenyl)pentanoic acid (Ahppa) (5), and 2-amino-5-(4’-hydroxy-phenyl)hexanoic acid (Ahpha) (6) at that position. Largamides A–C (4–6) were major metabolites in this cyanobacterium, while tiglicamides A–C (1–3) were only minor metabolites. It is unclear if the differing yields are reflective of the relative efficiencies of substrate activation or due to genetic heterogeneity of the cyanobacterial samples and consequently biosynthetic enzymes, although the 16S rDNA sequence was identical for at least three distinct collections at different times [Sharp et al., 2009]. Detailed genetic studies of this intriguing cyanobacterium will provide novel insights into the biosynthesis of compounds 1–6 and other co-produced L. confervoides metabolites.
4. Experimental
4.1. General experimental procedures
Optical rotation was measured on a Perkin Elmer 341 polarimeter. UV spectra were recorded using a SpectraMax M5 (Molecular Devices). 1H and 2D NMR spectra for 1 and 2 were recorded in DMF-d7 on a Bruker 600 MHz spectrometer equipped with a 1-mm high-temperature superconducting cryogenic probe and 3 was recorded in 5 mm cryogenic probe operating at 600 MHz and 150 MHz using residual solvent signals as the internal standard (δH 8.02, δC 162.7). HSQC experiments were optimized for 1JCH = 145 Hz, and HMBC experiments were optimized for nJC,H = 7 Hz for 1 and 2 and 10 Hz for 3. HRMS data were obtained using an Agilent LC-TOF mass spectrometer equipped with an APCI/ESI multimode ion source detector (UCR Mass Spectrometry Facility, University of California at Riverside), and low resolution mass spectra were obtained on a A3200 Q TRAP LC/MS/MS (hybrid triple quadrupole linear ion trap mass spectrometer, Applied Biosystems, USA) with an electrospray ionization (ESI) interface operated in positive mode. HPLC-based compound isolation was performed on a Shimadzu LC-20AT prominence LC with peak detection by a Shimadzu SPD-M20A prominence diode array detector.
4.2. Marine cyanobacterial samples
Samples of Lyngbya confervoides were collected at approximately 15 m depth from reefs near the Port Everglades Inlet, Fort Lauderdale, Florida, USA (26°05.9902’N, 80°05.0184’W) in August 2004 and May and August 2005. S. Golubic identified the cyanobacterium [Paul et al., 2005] and its 16S rDNA gene sequence has been reported [Paul et al., 2005; Sharp et al., 2009].
4.3. Extraction and isolation
The freeze-dried organisms collected through 2004–2005 (~2700 g dry weight) were extracted with EtOAc–MeOH (1:1) to afford a crude extract (~400 g) which was suspended in water (500 mL) then defatted with hexanes (500 mL × 3; ~2 g). The concentrated aqueous layer enriched with salt was further partitioned between n-BuOH (250 mL × 3) and H2O. The combined n-BuOH extract (12 g) was applied on a diaion HP-20 (Supelco) resin (120 g), and subsequently fractionated with H2O and increasing concentrations of MeOH, and then with MeCN and finally with CH2Cl2 to yield 8 fractions [Fr. 1: H2O (100%, 2 L, ~6.8 g); Fr. 2: H2O:MeOH (80:20, 1 L, 854 mg); Fr. 3: H2O:MeOH (50:50, 1 L, 272 mg); Fr. 4: H2O:MeOH (50:50 to 25:75, 1 L, 400 mg); Fr. 5: H2O:MeOH (25:75 to 0:100, 1 L, 430 mg); Fr. 6: MeOH (100%, 1 L, 950 mg); Fr. 7: MeCN (100%, 1 L, 490 mg), Fr. 8: CH2Cl2 (100%, 1 L, 457 mg)]. Fractions 5 and 6 were subjected to reversed-phase preparative HPLC (Phenomenex Luna-C18 10 μ, 100 × 21.20 mm, 5.0 mL/min; PDA detection at 200–400 nm) using a MeOH–0.05 % aqueous TFA linear gradient (40–100% over 30 min and then 100% MeOH for 15 min). The largamide- and tiglicamide-rich fractions eluting between tR 15–25 min were collected and subjected to repeated semipreparative reversed-phase HPLC (Phenomenex Synergi 4u Hydro-RP, 250 ×10 mm, 2.0 mL/min; PDA detection at 200–400 nm) using two sequential linear gradients of MeOH in 0.05% aqueous TFA (60–90% over 25 min, 90–100% over 10 min) to give semi-pure compounds 1–6 eluting between tR 16–21 min. The final purification of the compounds was achieved by means of a Phenomenex Luna Phenyl-hexyl column 10 × 250 mm, using the same HPLC conditions as described above to afford tiglicamide A (1), tR 16.0 min (1.2 mg), tiglicamide B (2), tR 21.8 min (0.8 mg), and tiglicamide C (3), tR 13.1 min (0.4 mg) together with known compounds largamide A (4), tR 20.3 min (8.0 mg), largamide B (5), tR 18.2 min (8.0 mg), largamide C (6), tR 20.0 min (12.0 mg) and methyl esters of largamides A–C (4a–6a) [4a, tR 23.3 min (2.5 mg); 5a, tR 21.1 min (6.6 mg); 6a, tR 22.8 min (3.7 mg)]. These methyl esters were presumably isolation artifacts generated from 4–6, respectively.
4.4. Absolute configuration of amino acid units in compounds 1–3 by chiral HPLC analysis
Compounds 1, 2, and 3 (0.1 mg each) were treated with 6 N HCl (0.3 mL) and heated at 116 °C for 24 h. A portion of each hydrolyzate was evaporated to dryness and the residues were resuspended in H2O (100 µL) and then subjected to chiral HPLC analysis (Phenomenex, Chirex 3126 N,S-dioctyl-(d)-penicillamine, 250 mm × 4.60 mm, 5 µm; solvent, 2 mM CuSO4–MeCN; UV detection 254 nm; flow rate 1.0 mL/min). The absolute configuration of the common amino acid units in 1–3 (tR, min) were established as l-Ala (10.4), l-Thr (11.2) (solvent 2 mM CuSO4), l-Val (17.9), d-Glu (58.0), and d-Tyr (61.0) (solvent 2 mM CuSO4–MeCN, 95:5) by comparison with those of authentic standards. The retention times for the standards were as follows (tR, min) d-Ala (14.6), l-allo-Thr (15.5), d-Thr (13.5), d-allo-Thr (17.4) (solvent 2 mM CuSO4), d-Val (22.8), l-Glu (53.5), and l-Tyr (54.0) (solvent 2 mM CuSO4–MeCN, 95:5). Acid hydrolyzates of 1–3 also yielded peaks for l-Htyr (39.5 min; solvent 2 mM CuSO4–MeCN, 90:10), l/d-Phe (49.5/49.5 min; solvent 2 mM CuSO4–MeCN, 85:15), and l-Met(O) (13.6, 14.7; solvent 2 mM CuSO4), indicating the presence of l-Htyr, l/d-Phe, and l-Met(O) units in 1, 2, and 3, respectively. The standard for d-Htyr eluted at tR 58.5 min (solvent 2 mM CuSO4–MeCN, 90:10) and for d-Met(O) at tR 16.5, 18.5 min (solvent 2 mM CuSO4). However the peaks for l/d-somers of Phe did not resolve under the above mentioned chiral HPLC conditions; hence the hydrolyzate of 3 was further subjected to Marfey’s analysis.
4.5. Absolute configuration of Phe in 2 by Marfey’s Analysis
A portion of the acid hydrolyzate of compound 2 was treated with 1 M NaHCO3 (5 µL) and 1% v/v solution of 1-fluoro-2,4-dinitrophenyl-5-l-leucinamide (l-FDLA) in acetone and heated at 40 °C for 60 min. The reaction mixture was then cooled, acidified with 2 N HCl (10 µL), dried and dissolved in H2O–MeCN (1:1, 250 µL). Aliquots were subjected to reversed-phase HPLC (Alltech Alltima HP C18 HL 5μ, 250 × 4.6 mm, PDA detection at 200–500 nm; flow rate 0.8 mL/min) using a linear gradient of MeOH in 0.1% (v/v) aqueous TFA (50–100% MeOH over 40 min). The retention times (tR, min) of the derivatized amino acids in the hydrolyzate of compound 2 matched with those of l-Phe (25.8 min) and not of d-Phe (33.0 min).
4.6. Tiglicamide A (1)
Colorless, amorphous solid; [α]20 D −50 (c 0.07, MeOH); UV (MeOH) λmax (log ε) 220 (4.28), 280 (3.34) nm; IR (film) νmax 3330 br, 2926, 1722, 1666, 1615 br, 1430, 1124 cm−1; 1H NMR, 13C NMR, and HMBC data, see Table 1; HR-ESI/APCI-MS m/z [M + Na]+ 928.4032 (calcd for C45H59N7O13Na, 928.4069).
4.7. Tiglicamide B (2)
Colorless, amorphous solid; [α]20 D −45 (c 0.04, MeOH); UV (MeOH) λmax (log ε) 220 (4.26), 280 (3.15) nm; IR (film) νmax 3330 br, 2930, 1722, 1652, 1610, 1514, 1444 cm−1; 1H NMR, 13C NMR, and HMBC data, see Table 2; HR-ESI/APCI-MS m/z [M + Na]+ 898.3935 (calcd for C44H57N7O12Na, 898.3963).
4.8. Tiglicamide C (3)
Colorless, amorphous solid; [α]20 D −56 (c 0.04, MeOH); UV (MeOH) λmax (log ε) 220 (4.27), 280 (3.18) nm; IR (film) νmax 2945 br, 1730, 1670, 1559, 1458, 1055, 1037 cm−1; 1H NMR, 13C NMR, and HMBC data, see Table 2; HR-ESI/APCI-MS m/z [M + Na]+ 898.3591 (calcd for C40H57N7O13SNa, 898.3633).
4.9. Largamide A methyl ester (4a)
Colorless, amorphous solid; [α]20 D −53 (c 0.03, MeOH); UV (MeOH) λmax (log ε) 220 (4.30), 280 (3.24) nm; 1H NMR data, see Table S1; HR-ESI/APCI-MS m/z [M + Na]+ 878.4277 (calcd for C42H61N7NaO12, 878.4276).
4.10. Largamide B methyl ester (5a)
Colorless, amorphous solid; [α]20 D −68 (c 0.17, MeOH); UV (MeOH) λmax (log ε) 220 (4.29), 280 (3.49) nm; 1H NMR data, see Table S1; HR-ESI/APCI-MS m/z [M + Na]+ 956.4353 (calcd for C47H63N7NaO13, 956.4382).
4.11. Largamide C methyl ester (6a)
Colorless, amorphous solid; [α]20 D −60 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 220 (4.43), 280 (3.45) nm; 1H NMR data, see Table S1; HR-ESI/APCI-MS m/z [M + Na]+ 970.4533 (calcd for C48H65N7NaO13, 970.4538).
4.12. Serine protease inhibition assays
Compounds 1–3 were tested for serine protease-inhibitory activity against porcine pancreatic elastase, α-chymotrypsin from bovine pancreas, or trypsin from porcine pancreas, in the presence of chromogenic substrates (N-succinyl-Ala-Ala-Ala-p-nitroanilide for elastase, N-succinyl-Gly-Gly-Phe-p-nitroanilide for chymotrypsin, N-α-benzoyl-dl-arginine 4-nitroanilide hydrochloride for trypsin). Activity was determined by measuring the increase in absorbance at 405 nm over 30 min in a microplate reader using phenylmethylsulfonyl fluoride (PMSF) and lyngbyastatin 4 as positive controls [Matthew et al., 2007]. All assays were performed in quadruplicate (1–3) or triplicate (4a–6a) and IC50 values were expressed as mean ± S.D.
Supplementary Material
Acknowledgments
This work was funded by NIGMS grant P41GM086210. We thank L. Fisher and K. Banks (Broward County Environmental Protection Department) and C. Ross, A. Capper, S. Reed and R. Ritson-Williams (Smithsonian Marine Station) for assistance with collections and C. Ross for assistance with extractions of the cyanobacterium. The 600 MHz 1-mm triple-resonance HTS cryogenic probe was developed through collaboration between the University of Florida, NHMFL, and Bruker Biospin. This is contribution #794 from the Smithsonian Marine Station at Fort Pierce.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Information
Supplementary Information associated with this article include Table S1 with 1H NMR data for 4a–6a, 1D and 2D NMR spectra for 1–3 and 1H NMR spectra for 4–6 and 4a–6a.
References
- Abbenante G, Fairlie DP. Protease inhibitors in the clinic. Med. Chem. 2005;1:71–104. doi: 10.2174/1573406053402569. [DOI] [PubMed] [Google Scholar]
- Blunt JW, Munro MH. Dictionary of Marine Natural Products with CD-ROM. Boca Raton: Chapman & Hall CRC; 2008. [Google Scholar]
- Fujii K, Ikai Y, Mayumi T, Oka H, Suzuki M, Harada KI. A nonempirical method using LC/MS for determination of the absolute configuration of constituent amino acids in a peptide: combination of Marfey’s method with mass spectrometry and its practical application. Anal. Chem. 1997;69:5146–5151. [Google Scholar]
- Gunasekera SP, Williams RR, Paul VJ. Carriebowmide, a new cyclodepsipeptide from the marine cyanobacterium Lyngbya polychroa. J. Nat. Prod. 2008;71:2060–2063. doi: 10.1021/np800453t. [DOI] [PubMed] [Google Scholar]
- Harrigan GG, Luesch H, Yoshida WY, Moore RE, Nagle DG, Paul VJ. Symplostatin 2: a dolastatin 13 analogue from the marine cyanobacterium Symploca hydnoides. J. Nat. Prod. 1999;62:655–658. doi: 10.1021/np980321c. [DOI] [PubMed] [Google Scholar]
- Ilies MA, Supuran CT, Scozzafava A. Therapeutic applications of serine protease inhibitors. Expert Opinion on Therapeutic Patents. 2002;12:1181–1214. [Google Scholar]
- Kawabata K, Suzuki M, Sugitani M, Imaki K, Toda M, Miyamoto T. ONO-5046, a novel inhibitor of human neutrophil elastase. Biochem. Biophys. Res. Commun. 1991;177:814–820. doi: 10.1016/0006-291x(91)91862-7. [DOI] [PubMed] [Google Scholar]
- Matthew S, Ross C, Rocca JR, Paul VJ, Luesch H. Lyngbyastatin 4, a dolastatin 13 analogue with elastase and chymotrypsin inhibitory activity from the marine cyanobacterium Lyngbya confervoides. J. Nat. Prod. 2007;70:124–127. doi: 10.1021/np060471k. [DOI] [PubMed] [Google Scholar]
- Matthew S, Ross C, Paul VJ, Luesch H. Pompanopeptins A and B, new cyclic peptides from the marine cyanobacterium Lyngbya confervoides. Tetrahedron. 2008;64:4081–4089. [Google Scholar]
- Matthew S, Paul VJ, Luesch H. Largamides A–C, tiglic acid-containing cyclodepsipeptides with elastase-inhibitory activity from the marine cyanobacterium Lyngbya confervoides. Planta Med. 2009;75:528–533. doi: 10.1055/s-0029-1185332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JA, Donia MS, Schmidt EW. Ribosomal peptide natural products: bridging the ribosomal and nonribosomal worlds. Nat. Prod. Rep. 2009;26:537–559. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul VJ, Thacker RW, Banks K, Golubic S. Benthic cyanobacterial bloom impacts the reefs of South Florida (Broward County, USA) Coral Reefs. 2005;24:693–697. [Google Scholar]
- Plaza A, Bewley CA. Largamides A–H, Unusual cyclic peptides from the marine cyanobacterium Oscillatoria sp. J. Org. Chem. 2006;71:6898–6907. doi: 10.1021/jo061044e. [DOI] [PubMed] [Google Scholar]
- Schopf JW. Cyanobacteria: Pioneers of early earth. Nova Hedwigia. 1996;112:12–32. [Google Scholar]
- Sharp K, Arthur KE, Gu L, Ross C, Harrison G, Gunasekera SP, Meickle T, Matthew S, Luesch H, Thacker RW, Sherman DH, Paul VJ. Phylogenetic and chemical diversity of three chemotypes of bloom-forming Lyngbya species (cyanobacteria: Oscillatoriales) from reefs of southeastern Florida. Appl. Environ. Microbiol. 2009;75:2879–2888. doi: 10.1128/AEM.02656-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan LT. Bioactive natural products from marine cyanobacteria for drug discovery. Phytochem. 2007;68:954–979. doi: 10.1016/j.phytochem.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Taori K, Matthew S, Rocca JR, Paul VJ, Luesch H. Lyngbyastatins 5–7, potent elastase inhibitors from Floridian marine cyanobacteria, Lyngbya spp. J. Nat. Prod. 2007;70:1593–1600. doi: 10.1021/np0702436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.