Abstract
Retroviral integrase enzymes have a nonspecific endonuclease activity that is stimulated by certain compounds, suggesting that integrase could be manipulated to damage viral DNA. To identify integrase stimulator (IS) compounds as potential antiviral agents, we have developed a nonradioactive assay that is suitable for high-throughput screening. The assay uses a 49-mer oligonucleotide that is 5'-labeled with a fluorophore, 3'-tagged with a quencher, and designed to form a hairpin that mimics radioactive double-stranded substrates in gel-based nicking assays. Reactions in 384-well plates are analyzed on a real-time PCR machine after a single heat denaturation and subsequent cooling to a point between the melting temperatures of unnicked substrate and nicked products (no cycling is required). Under these conditions, unnicked DNA reforms the hairpin and quenches fluorescence, whereas completely nicked DNA yields a large signal. The assay was linear with time, stimulator concentration, and amount of integrase, and 20% concentrations of the solvent used for many chemical libraries did not interfere with the assay. The assay had an excellent Z'-factor, and it reliably detected known IS compounds. This assay, which is adaptable to other nonspecific nucleases, will be useful for identifying additional IS compounds to develop the novel antiviral strategy of stimulating integrase to destroy retroviral DNA.
Keywords: Endonuclease, Integrase, Retrovirus, HIV, High-throughput screening
Introduction
Although several drugs are available for human immunodeficiency virus type 1 (HIV-1) infection, the problem of resistance makes it imperative that new agents and strategies be developed. One key target for new antiretrovirals is the integrase enzyme [1], which is present in all infectious retrovirus particles and must be functional for initial virus transmission, continued virus replication, and subsequent development of the acquired immunodeficiency syndrome (AIDS) [2]. Integrase has long been an attractive antiviral target because human cells do not depend on any closely related protein, and the approval of the first integrase inhibitor for clinical use in 2007 validates this enzyme as a fruitful target for drug development [3].
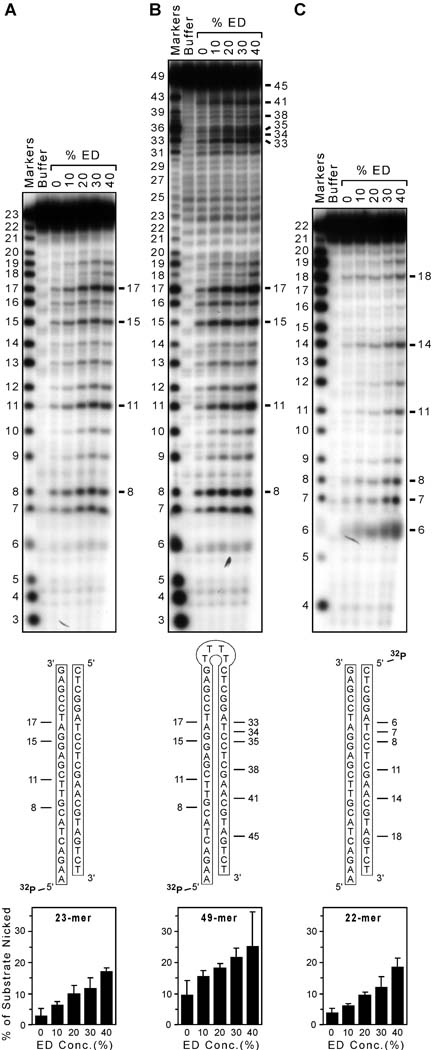
Integrase is a multifunctional enzyme that catalyzes at least two endonuclease reactions that are necessary for virus replication: (i) site-specific nicking to prepare the ends of viral DNA for integration, and (ii) nonspecific insertion of the trimmed viral DNA into cellular DNA [1]. However, integrase also exhibits a nonspecific endonuclease activity that can nick any DNA sequence [4], and this potentially deleterious activity is dramatically stimulated by certain compounds [5]. For example, increasing concentrations of 1,2-ethanediol (ED) stimulate HIV-1 integrase to nick a nonspecific double-stranded oligonucleotide at almost every internal site, with only sites near the DNA ends being spared (Fig. 1A). Although certain sites are preferentially nicked, as is also true for the classic nonspecific nuclease DNase I [4,6], it is remarkable that the 4 sites most frequently chosen by HIV-1 integrase on this particular DNA substrate (the 8, 11, 15, and 17 positions in the autoradiogram in Fig. 1A) follow 3' to C, T, G, and A nucleotides, respectively (as seen in the schematic beneath the autoradiogram in Fig. 1A). This nonspecific nicking activity is exhibited by several retroviral integrases, and under certain conditions, integrase is stimulated to nick more than 50% of nonspecific DNA substrates [5].
FIG. 1. Gel-based nonspecific nicking assay.
(A) Autoradiogram of nicking assays. A nonspecific 23-mer was 5' labeled and annealed to a complementary 22-mer, then incubated with protein buffer as a negative control (the second lane) or HIV-1 integrase with 0, 10, 20, 30, or 40% 1,2-ethanediol (the final 5 lanes). The buffer lane also contained 20% ED, and the first lane shows markers. Nucleotide sizes are indicated at the left, and prominent products are highlighted at the right; no products were evident near the bottom of the gel (not shown). The sequence of the 5' 32P-labeled substrate is depicted beneath the gel, and prominent nicks in the labeled strand are indicated. Two similar experiments were conducted and analyzed as in Materials and methods, and the graph at the bottom shows the percentage of substrate that was nicked as a function of the ED concentration in reactions with integrase (data are mean ± standard error). (B) Similar to panel A but using a 5' 32P-labeled 49-mer designed to form a hairpin as the substrate. (C) Similar to panel A but with the 5' 32P label on the 22-mer strand. (R2 ≥ 0.96 for the regression line for each graph at the bottom, not shown.)
The above facts suggest a novel antiviral strategy in which the nonspecific nicking activity of integrase would be stimulated to damage viral DNA before integration occurs and abort the infection before it becomes established [1,2]. Moreover, any collateral damage to cellular DNA would be limited to cells that had just been entered by HIV, which would also block productive infection and be predicted to have clinical benefit. This idea already has experimental support, including the demonstration that viral DNA in HIV-1 preintegration complexes is susceptible to nucleases [7,8] and the strong antiviral effect that was documented when bacterial nonspecific nucleases were packaged into retroviruses [9,10,11,12]. Further efforts to realize this antiviral strategy will require identification of a potent and relatively nontoxic integrase stimulator (IS) for preclinical testing. To facilitate the identification of such compounds, we have developed a nonradioactive assay that can be used for high-throughput screening (HTS) of libraries of drug-like chemicals to identify IS compounds that induce integrase to nick DNA nonspecifically. This new assay can also be used to study any nonspecific endonuclease.
Materials and methods
Reagents
Triton X-100, EDTA, dimethylsulfoxide (DMSO), 1,2-ethandediol, and DNase I were obtained from Sigma Life Science (St. Louis, MO), and Tris and glycerol were from Fisher Scientific (Fair Lawn, NJ). Percentage concentrations of liquid reagents refer to the volume of reagent relative to total volume of solution. Oligodeoxynucleotides (hereafter referred to as oligonucleotides) were purchased from Integrated DNA Technologies, Inc. (Coralville, IA), and melting temperatures were estimated with the formula in the Stratagene (La Jolla, CA) QuikChange Site-Directed Mutagenesis Kit: Tm (in °C) = 81.5 + 0.41(%GC) − 675/N − % mismatches, where %GC is the percentage of G plus C bases, N is the number of bases in the hybrid, and % mismatches was set to zero.
Integrase
The integrase coding region of HIV-1 was cloned into plasmid pQE-30 (Qiagen, Inc., Chatsworth, CA) and the integrase protein was expressed in Escherichia coli M15[pREP4] (Qiagen) and purified by metal affinity chromatography under native conditions, as described previously [13,14], except that the dialysate contained 33 mM Tris-HCl (pH 7.6), 667 mM NaCl, 0.7 mM dithioerythritol, 0.07 mM EDTA, 0.07% Triton X-100, and only 10% glycerol (to limit the amount of glycerol delivered to reactions). Purified protein was diluted to a stock concentration of 4 pmol/µl and stored at −70°C .
Radioactive assay for DNA nicking
Relevant oligonucleotides were gel-purified, then gel-purified again after 5' labeling with 32P [13]. Markers for gel analysis were produced by treatment of the same oligonucleotides with snake venom phosphodiesterase [15]. For reactions in Figs. 1A and 1C, double-stranded substrates were prepared by annealing the labeled oligonucleotide with 2-fold unlabeled complementary oligonucleotide [5]. Integrase assays were conducted for 90 min at 37°C in 10-µl reactions that contained 5 pmol of double-stranded DNA (0.2 pmol radiolabeled), 25 mM Tris-HCl (pH 8.0), 10 mM dithiothreitol (DTT), 10 mM MnCl2, and 1 µl of integrase or protein storage buffer, sometimes supplemented with 10% to 40% 1,2-ethanediol (corresponding to 1.8 to 7.2 M). Reactions were halted by adding loading buffer and heating at 95°C for 5 min, then analyzed on 20% polyacrylamide-7 M urea denaturing gels [5]. Results were quantified from gels in which reaction lanes were separated by empty lanes, and the percentage of nicked substrate was calculated as phosphorimager units for all bands migrating faster than substrate divided by units for the entire lane, with corrections for control reactions that lacked integrase.
Fluorescence assay for DNA nicking
The dual-tagged 49-mer oligonucleotide (Fig. 2) was dissolved in 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA, quantified by spectrophotometry at 260 nm using the extinction coefficient provided by the company, and stored at −70°C. Reactions were assembled in microcentrifuge tubes for time-course studies or directly in 384-well plates, either on ice or using a cooling block if done robotically by an Eppendorf epMotion 5070 Workstation (Mississauga, ONT, Canada). Conditions were identical to the radioactive assay, and reactions were initiated by addition of divalent metal cation and then processed as in Table 1. Fluorescence of completed reactions was measured on an Applied Biosystems (Foster City, CA) 7900HT Real-Time PCR System in the Absolute Quantitation mode for fluorescein (FAM) with the passive reference set to None using the conditions in Table 1.
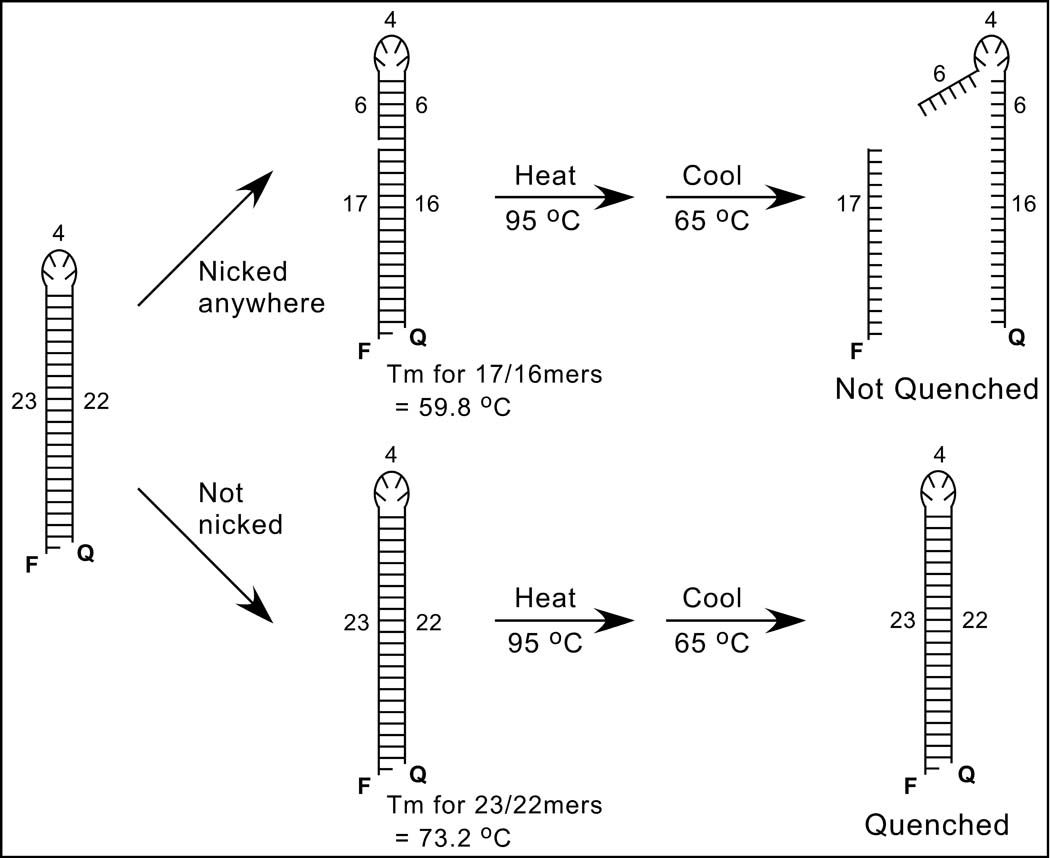
FIG. 2. Nonradioactive nonspecific nicking assay.
A schematic of the solution-based assay is shown. The 49-mer oligonucleotide substrate at the left, which has the sequence shown in Fig. 1B and was designed to form a hairpin, is 5'-labeled with a fluorophore (F) and 3'-tagged with a quencher (Q); numbers refer to the lengths of structural features in nucleotides, and the 5' overhang permits a nontagged version to be 32P-radiolabeled for gel-based assays. The upper scheme shows that nicking on either side of the hairpin (position 17 is used as an example) will separate the F and Q groups, especially after heat-denaturation at 95°C and subsequent cooling to 65°C, which is above the melting temperature (Tm) of potential base-paired sequences (the Tm for the 17-mer and its complementary 16-mer was estimated as 59.8°C). In contrast, the lower scheme shows that unnicked DNA should reanneal and quench because the Tm of the full stem (estimated as 73.2°C) is higher than 65°C.
Table 1.
Protocol for Processing the Plate-Based Assay for Nonspecific DNA Nicking
| 1. 10-µl reactions are set up in 384-well plates that are kept at 4°C. |
| 2. Plates are sealed, briefly spun, then incubated at 37°C for 90 min. |
| 3. Reactions are stopped by placing in a 65°C oven for 10 min to inactivate integrase. |
| 4. Plates are spun again and placed on the queue for a real-time PCR systema. |
| 5. The machine is programmed for a single denaturation at 95°C for 1 min and renaturation at 65°C for 1 min, followed by 2 min 59 secb at 65°C during which fluorescence is read 21 times in the absolute quantitation mode (no cycling is performed). |
| 6. The data are exported as a Microsoft Excel filec. |
| 7. The data are copied and pasted into a pre-templated Excel filed that automatically averages the final 10 reads for each well, calculates the control data, and displays the results on bar graphs. |
The protocol was developed using an Applied Biosystems 7900HT Real-Time PCR System.
When a data collection phase of precisely 3 min was tried, the machine started another pass across the plate and read one of the outer columns of wells an extra time.
In the absolute quantitation mode with this system, each well generates 31 rows of data or information; thus, the output for a 384-well plate is an Excel file with > 10,000 rows.
Available on request.
Results
A fluorescence assay for nonspecific DNA nicking
Our standard assay for nonspecific DNA nicking uses double-stranded 23-mer oligonucleotides that are 5' 32P-labeled on one strand, and completed reactions are analyzed by denaturing polyacrylamide gel electrophoresis and autoradiography [4]. For example (Fig. 1A, top), HIV-1 integrase nicks the labeled strand of this substrate (shown schematically beneath the autoradiogram) to create shorter products that can be quantified by phosphorimaging (Fig. 1A, bottom). However, radioactive gel-based assays are not appropriate for high-throughput screening. Our initial attempts to convert this assay to a solution-based system using double-stranded 23-mers in which one strand was 5'-labeled with a fluorophore (fluorescein, or FAM) and 3'-tagged with a quencher (Black Hole Quencher-1, or BHQ-1) yielded high background fluorescence even without treatment by a nuclease, likely because the rigidity of double-stranded oligonucleotides separated the 5' and 3' ends [16]. This problem, and the possibility that nicks on the dual-tagged strand might be bridged by the complementary strand, could not be overcome by post-reaction denaturation using alkali or formamide (which caused precipitation or quenched the signal, respectively). However, further efforts led to the successful nonspecific nicking assay depicted in Fig. 2.
We designed a 49-mer oligonucleotide that is 5'-labeled with a fluorophore (FAM, or F in Fig. 2), 3'-tagged with a quencher (BHQ-1, or Q), and can form a hairpin that resembles the 23-base-pair substrate in the radioactive assay (the stem of the 49-mer is missing the final 3' nucleotide to create a 5' overhang for radioactive labeling of a nontagged version). Nicking anywhere in the sequence will unlink the F and Q groups (the upper pathway in Fig. 2, which shows the anticipated prominent nick 17 nucleotides from the 5' end). Separation of the F and Q groups (and unquenching of fluorescence) should occur upon heat denaturation at 95°C and cooling to 65°C, which is above the predicted melting temperature (Tm) for the 17-mer annealed to the 16 complementary nucleotides at the 3' end of the original substrate (Fig. 2). In contrast, in the absence of nicking (the lower pathway in Fig. 2), the stem should reform with zero-order kinetics after heating and subsequent cooling to 65°C, which is below the predicted Tm for the 22 base pairs in the full stem.
The hairpin substrate in Fig. 2 has 3 theoretical advantages compared to linear substrates: (i) the proximity of the F and Q groups should permit efficient quenching and low background fluorescence, (ii) nicking on either side of the stem should be detected, and (iii) a pre-reaction annealing step is unnecessary. The other key aspect of this new assay is the use of a real-time PCR machine to process and read completed reactions without actually cycling. For this purpose, the machine is programmed for a single denaturation and renaturation, followed by a brief incubation at 65°C during which fluorescence is read 21 times in the absolute quantitation mode. Data are exported as a Microsoft Excel file, and the final 10 reads for each well are averaged. To minimize the use of reagents, all reactions are performed in 10-µl volumes in a 384-well plate. The complete protocol is presented in Table 1.
Validation of the fluorescent nicking assay
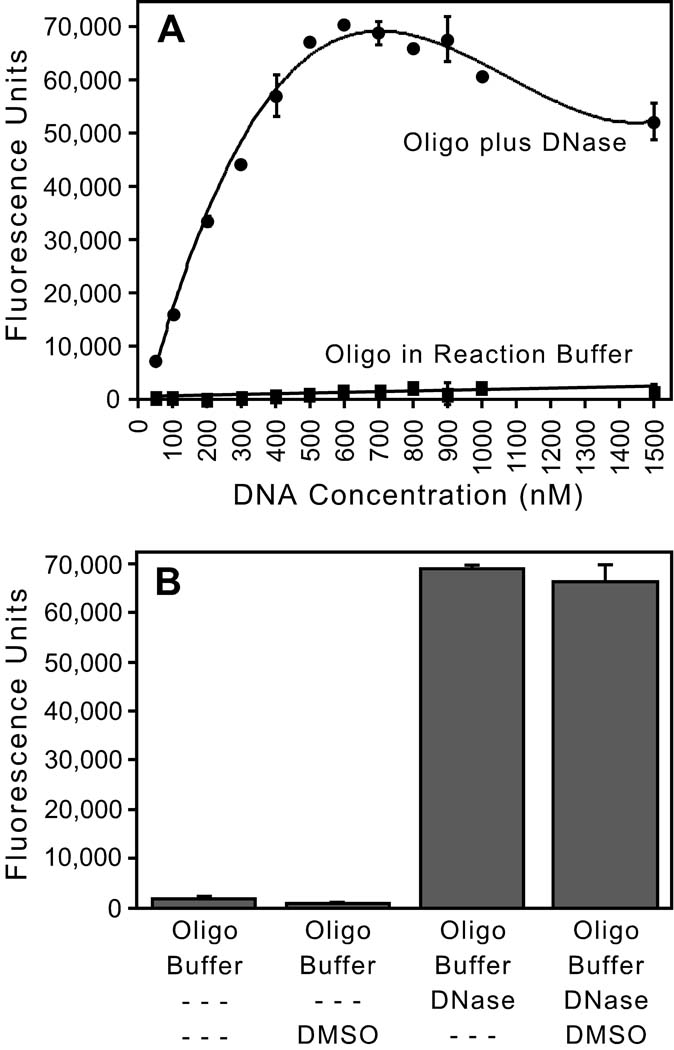
Minimal fluorescence was measured when 0.5 to 15 pmol (50 to 1500 nM) of the F/Q-tagged oligonucleotide was incubated in integrase reaction buffer (Fig. 3A), confirming that the unnicked oligonucleotide is efficiently quenched. In contrast, the signal increased for all DNA concentrations incubated in the same buffer plus an excess of the nonspecific nuclease DNase I (Fig. 3A). The highest readings were ~65,000 fluorescence units, which approximates 216 (or 65,536) and is the theoretical maximum signal from a 16-bit analog-to-digital converter and a charge-coupled device (CCD) detection camera that is saturated. Lower signal at the highest DNA concentrations may be due to the inner filter effect as the fluorophore absorbs some of the input light. A similar curve was obtained used a short, fluorescently-tagged, single-stranded oligonucleotide that lacked a quencher group, confirming the linear response and maximum signal for this system (not shown). We selected 500 nM DNA for subsequent experiments because it is within the linear phase and can yield near-maximum fluorescence (Fig. 3A), providing the largest window to detect stimulation. Although this early experiment used a renaturation temperature of 63°C, similar results were obtained using a cooling temperature of 70°C, which is important because it was reported that interaction of FAM and BHQ-1 can raise the Tm of complementary oligonucleotides by 7°C [17]. Given that FAM fluorescence can decrease 10% at 70°C compared to 63°C [18], we chose an intermediate cooling temperature of 65°C for all experiments. Importantly for the goal of screening chemical libraries, we also found that the presence of 20% dimethylsulfoxide (DMSO) did not unquench the fluorescence of unnicked substrate (Fig. 3B, the first two bars), nor mask the signal from DNA nicked by exposure to DNase (Fig. 3B, the final two bars). Combined with the demonstration that HIV-1 integrase tolerates even higher DMSO concentrations [19], these results indicate that the solvent commonly present in chemical libraries will not interfere with the use of this assay for high-throughput screening for integrase stimulators.
FIG. 3. Validation of the fluorescence-based assay.
(A) Effect of DNA concentration. Increasing concentrations of the F/Q-tagged oligonucleotide depicted in Fig. 2 were incubated in integrase reaction buffer or in the same buffer plus 50 µg DNase I, and reactions were conducted and analyzed as in Table 1 (except the cooling temperature in this early experiment was 63°C). The means ± SD of duplicate reactions are shown. (B) Effect of DMSO. 500 nM of the F/Q-tagged 49-mer was incubated in reaction buffer without or with 20% DMSO (the first 2 bars, respectively) or in reaction buffer plus 1 µg DNase I without or with 20% DMSO (the last 2 bars, respectively), and analyzed as in Table 1; the means ± SD of quadruplicate reactions are shown. Figs. A and B are representative of at least two experiments for each condition.
To assess how HIV-1 integrase nicks the hairpin DNA, an untagged version of the 49-mer was 5'-labeled with 32P and tested with integrase in the presence of increasing concentrations of 1,2-ethanediol (Fig. 1B). The hairpin substrate was compared to two linear substrates that mimic its stem and were 5'-radiolabeled either on the 23-mer strand (Fig. 1A) or the complementary 22-mer strand (Fig. 1C). The sequences of all 3 substrates and the positions of radiolabels are shown schematically in the middle of Fig. 1 beneath the gels. The autoradiogram at the top of Fig. 1B shows that integrase created many nicks along the 49-mer. Remarkably, the preferred nicks at positions 8, 11, 15, and 17 precisely matched those made on the labeled 23-mer strand (Fig. 1A). Additional prominent nicks in the 49-mer were noted at positions 33, 34, 35, 38, 41, and 45 near the top of the gel in Fig. 1B (sizes were confirmed by longer runs on other gels). These nicks correspond precisely to the preferred nicks at the 6, 7, 8, 11, 14, and 18 positions on the 22-mer strand (Fig. 1C; that these are the same positions in best seen schematically in the middle of Fig. 1). Thus, nicking of the 49-mer hairpin faithfully reflects how integrase nicks both strands of the linear substrates.
Fig. 1B also shows that few nicks were created near the loop in the middle of the 49-mer, as indicated by the lack of prominent bands between positions 17 and 33. That all of the preferential nicks in the 49-mer are within 17 nucleotides of one of the ends (best seen in the middle of Fig. 1B) strongly supports the rationale of the fluorescence assay, because it indicates that the nick at the 17-mer position (the example used in Fig. 2) gives an upper limit of melting temperatures for nicked DNA, i.e., few molecules nicked by integrase should reanneal at 65°C.
The autoradiograms in Fig. 1 also show that integrase was stimulated to nick each of the radiolabeled substrates by increasing amounts of 1,2-ethanediol, confirming that ED is an IS compound even with the 10-fold increased concentrations of substrate DNA (compared to prior reports [4,5]) that were used in Fig. 1 to match conditions in the fluorescence assay. The small amount of nicking in the absence of ED is due to baseline nicking by integrase plus any stimulation by the 1% glycerol (another IS compound [13]) provided by the integrase storage buffer. Quantitation of two different experiments indicated that the 49-mer was nicked to approximately twice the extent of either linear substrate (the graphs at the bottom of Fig. 1), consistent with the idea that Fig. 1B monitors nicking of either side of the hairpin. Thus, as predicted, the 49-mer in the fluorescence assay has the potential to reveal greater nicking than linear substrates in the radioactive assay. The fluorescence assay also detected stimulation by increasing amounts of glycerol (data not shown), and other experiments confirmed that an active-site mutant of HIV-1 integrase was inactive in this assay (not shown).
Optimization of reaction parameters and performance of the assay
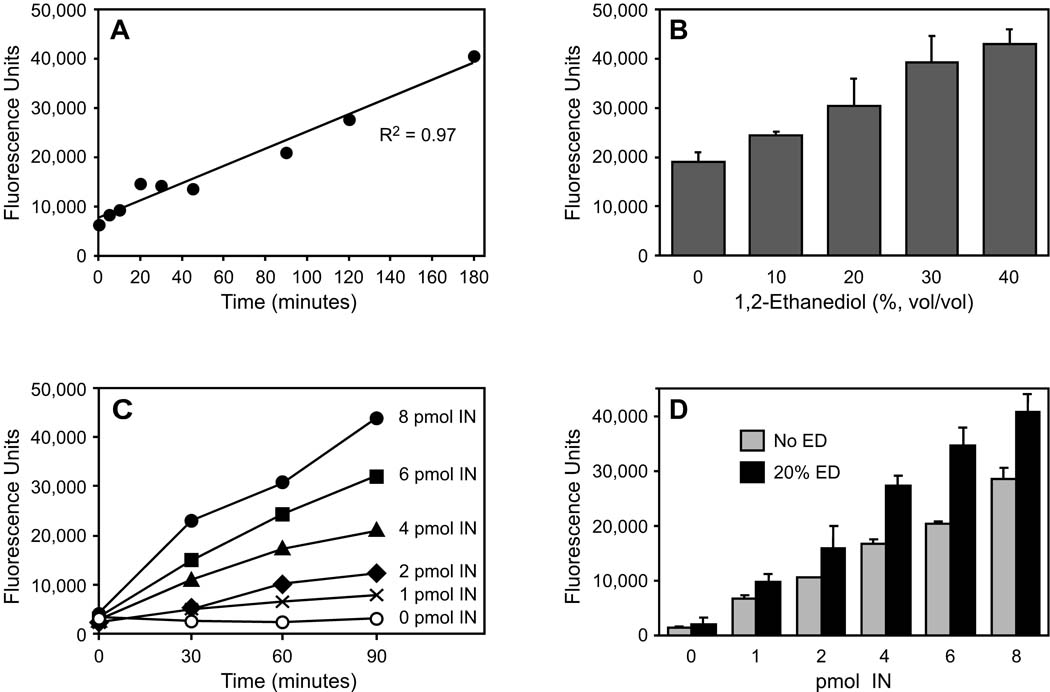
Our standard 10-µl radioactive assays are incubated for 90 minutes and use 0.5 pmol (50 nM) substrate DNA with 4 pmol (400 nM) of viral enzyme. To optimize reaction parameters for the nonradioactive assay using 500 nM DNA, we first performed time course experiments. In reactions containing integrase with 20% ED, fluorescence increased linearly for 3 hours (Fig. 4A); in particular, 90 minutes was in the linear phase of the reaction. We next performed 90- minute reactions and found that 10% to 40% ED stimulated nicking in a dose-dependent manner, with 20% ED in the linear region of the dose response (Fig. 4B). We also found that with 20% ED, the signal increased over time for a range of integrase concentrations and that the signal increased with the amount of integrase (Fig. 4C). In fact, the slopes of the regression lines for the data in Fig. 4C (not shown) increased linearly with the concentration of integrase (R2 = 0.998). Thus, 4 pmol (400 nM) of integrase is in the linear portion of the enzyme response. Additionally, 20% ED stimulated nicking at all concentrations of integrase tested in triplicate 90-minute reactions (Fig. 4D; as in the radioactive assay, nicking by integrase in the absence of ED is due to baseline nicking plus any effect from glycerol in the protein buffer). Although the amount of stimulation by ED tended to increase at higher integrase concentrations, so did the amount of unstimulated nicking (Fig. 4D). In particular, 4 pmol of integrase nicely balances the needs of minimizing baseline nicking and having the potential to readily reveal stimulation. Similar results to those shown in Figs. 4A to 4D were obtained with at least two preparations of integrase.
FIG. 4. Parameters of the fluorescence assay.
Except as noted, all reactions were conducted and analyzed as in Materials and methods. (A) Time course of integrase-mediated nicking. 500 nM of the F/Q-tagged 49-mer was incubated with 4 pmol HIV-1 integrase and 20% ED in replicate microcentrifuge tubes, and at various times individual reactions were heated at 65°C to inactivate integrase and then transferred to a 384-well plate for analysis. (B) Dose-dependant stimulation by 1,2-ethanediol. 500 nM of the F/Q-tagged 49-mer was incubated for 90 minutes with 4 pmol HIV-1 integrase and the indicated concentrations of ED; the means ± SD of triplicate reactions are shown (R2 = 0.99 for the regression line, not shown). (C) Effect of integrase concentration. 500 nM of the F/Q-tagged 49-mer was incubated with increasing amounts of HIV-1 integrase in the presence of 20% ED in microcentrifuge tubes, and at various times aliquots were removed, heated at 65°C, then transferred to a 384-well plate for analysis. (D) Stimulation of varying integrase concentrations by ED. 500 nM of the F/Q-tagged 49-mer was incubated for 90 minutes with increasing amounts of HIV-1 integrase with or without 20% ED present; the means ± SD of triplicate reactions are shown (R2 = 0.98 for each regression line, not shown; the p-values comparing the bars were 0.04 for 1 pmol integrase, 0.08 for 2 pmol integrase, < 0.002 for 4 or 6 pmol integrase, and < 0.006 for 8 pmol integrase). The number of pmols indicated in Figs. C and D is per 10-ul reaction.
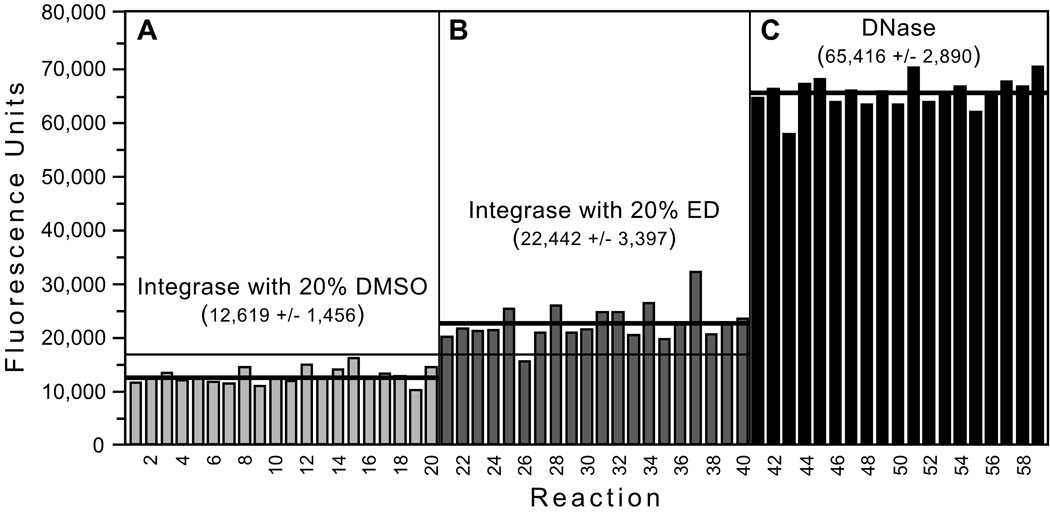
Using the optimized conditions (500 nM DNA, 90 minutes incubation, 4 pmol integrase), 3 sets of replicate reactions were performed with (i) integrase plus DMSO as negative controls for baseline nicking (Fig. 5A), (ii) integrase plus ED as an example of a known stimulator (Fig. 5B), or (iii) DNase I as positive controls for maximum nicking (Fig. 5C). The coefficients of variation (calculated as the standard deviation [SD] divided by the mean) were ≤ 15% for each condition. The dynamic range (the difference between the means of the positive and negative controls) was ~53,000; the signal-to-background (the ratio of the means of the positive to negative controls) was 5.2; and the signal-to-noise (the ratio of the dynamic range to the SD of the negative controls) was 36 [20]. Moreover, none of the negative controls (Fig. 5A), but 19 of 20 reactions with ED (Fig. 5B, all but reaction 26), yielded higher fluorescence than the mean + 3 SD for the negative controls. Thus, the assay detected stimulation of integrase-mediated nicking by a known IS compound with a sensitivity of 95%. A subsequent experiment using a robotic workstation similarly identified ED as a stimulator 39 of 40 times (not shown). We also calculated the Z'-factor, which is a function of the dynamic range of an assay and the variation (i.e., the SD) of the controls; values greater than 0.5 are considered optimal for high-throughput screening assays [20]. Importantly, the Z'-factor from the data in Fig. 5 was excellent at 0.75 (range of 0.74 to 0.83 in similar experiments), and it averaged 0.68 in experiments done robotically.
FIG. 5. Performance of the assay.
Using the optimized conditions, 3 sets of 20 reactions were set up with: (A) 4 pmol integrase plus 20% DMSO as negative controls for baseline nicking, (B) 4 pmol integrase plus 20% ED as an example of a known integrase stimulator, and (C) 10 µg of DNase I as positive controls for maximum nicking (one reaction from the third set is not shown and was excluded from analysis because it had minimal fluorescence and apparently did not receive DNase). Reactions were conducted and analyzed as in Table 1. The mean ± SD is shown for each set of reactions, thick lines are drawn at each mean value, and the thin horizontal line indicates the mean + 3 SD for the negative controls.
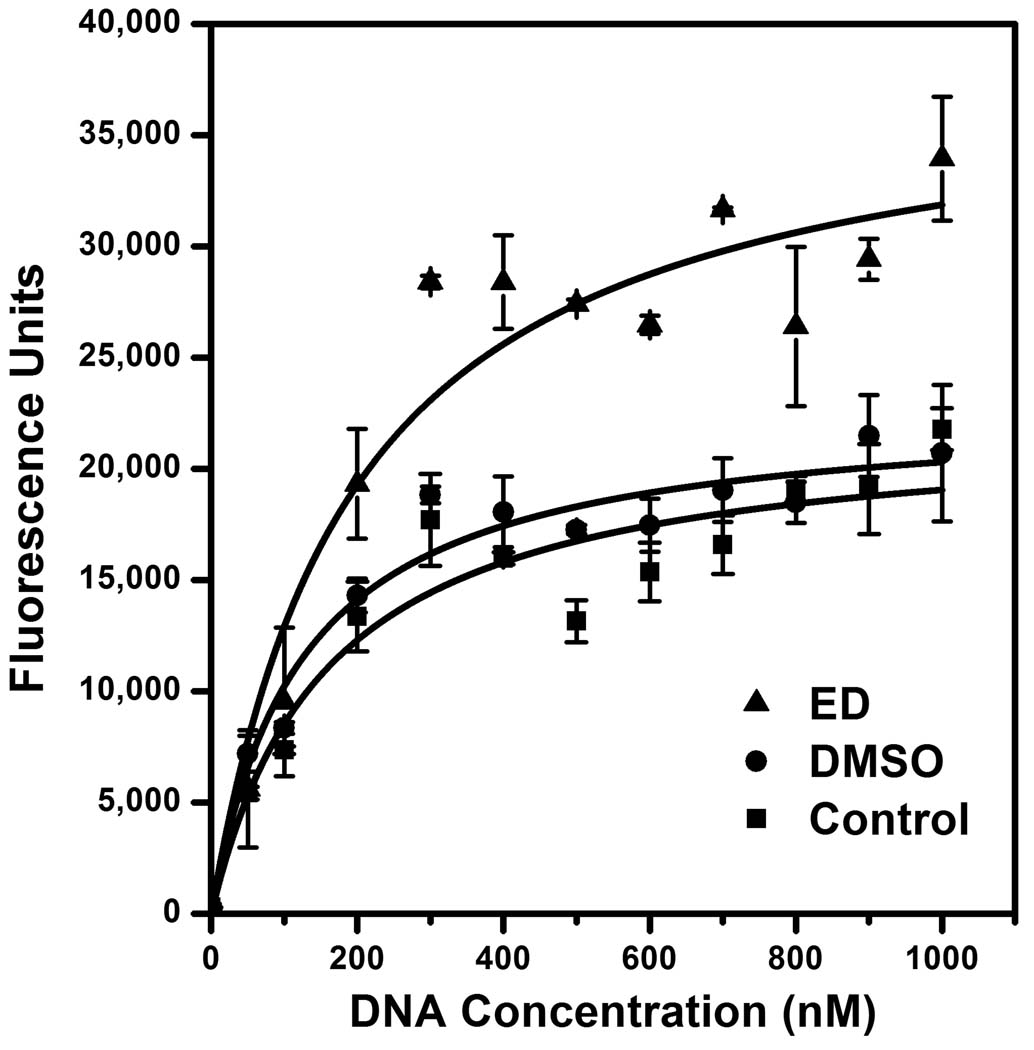
To investigate the mechanism of stimulation by 1,2-ethanediol, we performed kinetic studies by varying the concentration of the F/Q-tagged DNA substrate. The results from 3 independent experiments (Fig. 6) show that ED increased the Vmax of integrase from 22,300 ± 2,600 units/time (mean ± SD) with water as the control to 37,300 ± 2,200 units/time in the presence of 20% ED (p < 0.02 by the paired t-test). In contrast, ED did not significantly affect the Km of integrase in these experiments (182 ± 48 nM DNA for the water control vs. 204 ± 75 nM DNA for 20% ED). Thus, ED acts as an integrase stimulator by speeding up catalysis. Importantly, 20% DMSO did not significantly affect the Vmax (23,000 ± 2,800 units/time) or Km (138 ± 56 nM DNA), validating it as an appropriate negative control for use in high-throughput screening. The data from Fig. 6 also show that the optimized DNA concentration of 500 nM DNA (Fig. 3A) exceeds the Km by approximately 3-fold, further validating the final reaction conditions.
FIG. 6. Kinetic analysis.
Reactions were set up and analyzed as in Materials and methods using optimized conditions (90 minutes and 4 pmol integrase) but the amount of substrate DNA was varied. Three reaction series (containing 20% ED, 20% DMSO, or extra water as a control) were compared. The experiment was repeated on 3 separate days, and the means ± standard errors are shown. The Vmax and Km data were calculated and the curves were fit using the OriginPro 8 program (OriginLab Corporation, Northampton, MA).
Discussion
Retroviral integrase catalyzes the insertion of viral DNA into cellular DNA, which is required for retroviral replication and pathogenesis. Thus, many HTS assays for inhibitors of purified integrase have been described [21,22,23,24,25,26,27,28,29,30]. However, the discovery that integrase also has a nonspecific endonuclease activity that can be stimulated to achieve high levels of DNA nicking immediately suggested that integrase has the potential to damage viral DNA [4]. Thus, integrase possesses an activity that could be targeted for an antiviral strategy not involving inhibition, as previously suggested by others [2]. At least 4 chemicals have been identified as stimulators of integrase-mediated nonspecific nicking: glycerol; 1,2-ethanediol; 1,2-propanediol; and 1,3-propanediol [4,5]. These agents dramatically stimulate integrase to nick DNA, albeit only at high concentrations. Although these compounds participate in integrase-catalyzed alcoholysis as the attacking nucleophilic molecule that joins to DNA at the site of nicking, this characteristic would not be required for all stimulators given that integrase can use water as the nucleophile for nonspecific hydrolysis [4]. Thus, the limiting factor in developing this novel antiviral strategy is identification of potent integrase stimulators. The recent report of successful screening for stimulators of another enzyme, glucokinase [31], provides strong support for this idea.
To facilitate identification of IS compounds, a reliable HTS assay is needed. Although a nonradioactive assay for nonspecific nicking by DNase I has been described [32], that assay used a DNA substrate that was 5'-digoxigenin-labeled and 3'-biotin-tagged, required enzyme-linked antibodies to develop the signal, and measured nicking as decreased digoxigenin remaining bound to wells. It would be preferable to have a solution-based assay that does not require a solid support or multiple post-reaction processing steps and measures nicking as increased signal. Others have used DNA substrates tagged with both a fluorophore and a quencher so that nicking separates the tags and increases the fluorescence signal. Variations of this idea have been described for nonspecific nicking of single-stranded DNA [33], site-specific nicking of both strands of double-stranded DNA by restriction endonucleases [34,35,36], or specific nicking at the ends of viral DNA by integrase [24,30]. However, these reports are not directly adaptable to nonspecific nicking of double-stranded DNA that might occur far from the fluorescent and quencher groups. We therefore developed the assay shown in Fig. 2.
The first key aspect of this new assay was the use of a 49-mer oligonucleotide that can fold back on itself to form a hairpin that mimics the duplex substrates in radioactive gel-based assays for nonspecific nicking. Indeed, integrase nicked a radiolabeled version of this substrate with the identical site-preferences it exhibited on both strands of the linear substrate (Fig. 1). The F/Q-tagged version of this DNA (Fig. 2) is reminiscent of molecular beacons used as hybridization probes for real-time PCR analyses [37]. However, the stem in typical beacons is much shorter, and the signal in those systems does not result from nicking the dual-tagged probe but from disrupting the stem-loop structure as annealing occurs to the accumulating amplified sequence. In fact, the second key aspect of this new assay is the use of a real-time PCR machine, but not for amplification of DNA. Rather, the thermal cycler is used to process completed reactions by controlling a single heat denaturation and subsequent renaturation, then measuring the fluorescence of each well at a relatively high temperature. Selection of a renaturation setting between the melting temperatures of the unnicked substrate and potential nicked products was critical for this purpose, and the use of a PCR machine to evaluate reactions distinguishes this assay from others that used hairpin substrates but assessed DNA nicking close to the fluorescent and quencher tags at temperatures ≤ 37°C [30,35,36].
This assay also facilitated kinetic analyses. Although various estimates of kinetic parameters for HIV-1 integrase under diverse reaction conditions have been described, the Km of 182 nM and Vmax of 1.9 nM/min from this assay under nonstimulating conditions (Fig. 6, using a conversion factor of 65,000 fluorescence units = 500 nM DNA for the Vmax) are similar to the apparent Km of 145 nM and Vmax of 0.7 nM/min reported with an assay that used fluorescence resonance energy transfer to assess nicking of viral DNA [38].
The simple protocol in Table 1 has many attributes that make this assay ideal for high-throughput screening. In particular, each plate requires less than 10 minutes on a PCR machine, and 20% concentrations of the solvent used for many chemical libraries did not interfere with the assay (Fig. 3B and Fig. 6). The assay yielded low background in the absence of nicking and high fluorescence after maximal nicking (Fig. 3A), and it was linear with respect to time (Fig. 4A), stimulator concentration (Fig. 4B), and amount of integrase (Fig. 4C). Moreover, stimulation of HIV-1 integrase-mediated nicking by known IS compounds was readily detected (e.g., Fig. 5B). The Z'-factor for the assay was also excellent, whether reactions were done manually (Fig. 5) or were automated and done robotically (not shown). Finally, hits can be rapidly recognized because the output is a customized bar graph (Table 1 and Fig. 5). Of course, application of this assay to screening will require validation of any hits in secondary assays to exclude artifacts, such as autofluorescence or compound-induced dissociation of the hairpin. The current report, however, demonstrates that the goal of developing a nonradioactive plate-based assay that reliably detects nonspecific nicking by HIV-1 integrase has been achieved. Thus, this assay is now available for screening large collections of drug-like chemicals for agents that induce integrase to damage DNA. Although the assay was designed to identify stimulation of nicking, it can also detect inhibition of nicking. Thus, this fluorescence assay should be useful for identifying stimulators or inhibitors of any nonspecific endonuclease.
Acknowledgements
We thank the Drug Discovery, Development, and Delivery Core of the Dept. of Pharmacology and Penn State Hershey Cancer Institute; Rob Brucklacher and Georgina Bixler of the Functional Genomics Core Facility; Amy Harper for early efforts at developing a nonradioactive assay; Ira Ropson; Yue Sheng (Applied Biosystems); and Fred Krebs (Drexel University). This work was supported by Public Health Service grant R21AI075929 from the Microbicide Innovation Program of the Office of Women's Health and National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Craigie R. HIV integrase, a brief overview from chemistry to therapeutics. J. Biol. Chem. 2001;276:23213–23216. doi: 10.1074/jbc.R100027200. [DOI] [PubMed] [Google Scholar]
- 2.Brown PO. Integration. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1997. pp. 161–203. [Google Scholar]
- 3.Havlir DV. HIV integrase inhibitors--out of the pipeline and into the clinic. N. Engl. J. Med. 2008;359:416–418. doi: 10.1056/NEJMe0804289. [DOI] [PubMed] [Google Scholar]
- 4.Katzman M, Sudol M. Nonspecific alcoholysis, a novel endonuclease activity of human immunodeficiency virus type 1 and other retroviral integrases. J. Virol. 1996;70:2598–2604. doi: 10.1128/jvi.70.4.2598-2604.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skinner LM, Sudol M, Harper AL, Katzman M. Nucleophile selection for the endonuclease activities of human, ovine, and avian retroviral integrases. J. Biol. Chem. 2001;276:114–124. doi: 10.1074/jbc.M007032200. [DOI] [PubMed] [Google Scholar]
- 6.Matsuda M, Ogoshi H. Specificity of DNase I. J. Biochem. (Tokyo) 1966;59:230–235. [PubMed] [Google Scholar]
- 7.Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Wei SQ, Engelman A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J. Biol. Chem. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- 9.Natsoulis G, Seshaiah P, Federspiel MJ, Rein A, Hughes SH, Boeke JD. Targeting of a nuclease to murine leukemia virus capsids inhibits viral multiplication. Proc. Natl. Acad. Sci. U. S. A. 1995;93:364–368. doi: 10.1073/pnas.92.2.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schumann G, Qin L, Rein A, Natsoulis G, Boeke JD. Therapeutic effect of Gag-nuclease fusion protein on retrovirus-infected cell cultures. J. Virol. 1996;70:4329–4337. doi: 10.1128/jvi.70.7.4329-4337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.VanBrocklin M, Federspiel MJ. Capsid-targeted viral inactivation can eliminate the production of infectious murine leukemia virus in vitro. Virology. 2000;267:111–123. doi: 10.1006/viro.1999.0113. [DOI] [PubMed] [Google Scholar]
- 12.Schumann G, Hermankova M, Cannon K, Mankowski JL, Boeke JD. Therapeutic effect of a Gag-nuclease fusion protein against retroviral infection in vivo. J. Virol. 2001;75:7030–7041. doi: 10.1128/JVI.75.15.7030-7041.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katzman M, Sudol M. Mapping domains of retroviral integrase responsible for viral DNA specificity and target site selection by analysis of chimeras between human immunodeficiency virus type 1 and visna virus integrases. J. Virol. 1995;69:5687–5696. doi: 10.1128/jvi.69.9.5687-5696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harper AL, Sudol M, Katzman M. An amino acid in the central catalytic domain of three retroviral integrases that affects target site selection in nonviral DNA. J. Virol. 2003;77:3838–3845. doi: 10.1128/JVI.77.6.3838-3845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katzman M, Katz RA, Skalka AM, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J. Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 17.Marras SA, Kramer FR, Tyagi S. Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Res. 2002;30:e122. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marras SA. Selection of fluorophore and quencher pairs for fluorescent nucleic acid hybridization probes. Methods Mol. Biol. 2006;335:3–16. doi: 10.1385/1-59745-069-3:3. [DOI] [PubMed] [Google Scholar]
- 19.Morgan AL, Katzman M. Subterminal viral DNA nucleotides as specific recognition signals for human immunodeficiency virus type 1 and visna virus integrases under magnesium-dependent conditions. J. Gen. Virol. 2000;81:839–849. doi: 10.1099/0022-1317-81-3-839. [DOI] [PubMed] [Google Scholar]
- 20.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 21.Craigie R, Mizuuchi K, Bushman FD, Engelman A. A rapid in vitro assay for HIV DNA integration. Nucleic Acids Res. 1991;19:2729–2734. doi: 10.1093/nar/19.10.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hazuda DJ, Hastings JC, Wolfe AL, Emini EA. A novel assay for the DNA strand-transfer reaction of HIV-1 integrase. Nucleic Acids Res. 1994;22:1121–1122. doi: 10.1093/nar/22.6.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vink C, Banks M, Bethell R, Plasterk RHA. A high-throughput, non-radioactive microtiter plate assay for activity of the human immunodeficiency virus integrase protein. Nucleic Acids Res. 1994;22:2176–2177. doi: 10.1093/nar/22.11.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SP, Censullo ML, Kim HG, Knutson JR, Han MK. Characterization of endonucleolytic activity of HIV-1 integrase using a fluorogenic substrate. Anal.Biochem. 1995;227:295–301. doi: 10.1006/abio.1995.1284. [DOI] [PubMed] [Google Scholar]
- 25.Chang YC, Ching TT, Syu WJ. Assaying the activity of HIV-1 integrase with DNA-coated plates. J. Virol. Methods. 1996;59:135–140. doi: 10.1016/0166-0934(96)02033-2. [DOI] [PubMed] [Google Scholar]
- 26.Hwang Y, Rhodes D, Bushman F. Rapid microtiter assays for poxvirus topoisomerase, mammalian type IB topoisomerase and HIV-1 integrase: application to inhibitor isolation. Nucleic Acids Res. 2000;28:4884–4892. doi: 10.1093/nar/28.24.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.David CA, Middleton T, Montgomery D, Lim HB, Kati W, Molla A, Xuei X, Warrior U, Kofron JL, Burns DJ. Microarray compound screening (micro ARCS) to identify inhibitors of HIV integrase. J Biomol Screen. 2002;7:259–266. doi: 10.1177/108705710200700309. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Klock H, Yin H, Wolff K, Bieza K, Niswonger K, Matzen J, Gunderson D, Hale J, Lesley S, Kuhen K, Caldwell J, Brinker A. Homogeneous high-throughput screening assays for HIV-1 integrase 3'-processing and strand transfer activities. J Biomol Screen. 2005;10:456–462. doi: 10.1177/1087057105275212. [DOI] [PubMed] [Google Scholar]
- 29.John S, Fletcher TM, 3rd, Jonsson CB. Development and application of a high-throughput screening assay for HIV-1 integrase enzyme activities. J Biomol Screen. 2005;10:606–614. doi: 10.1177/1087057105276318. [DOI] [PubMed] [Google Scholar]
- 30.He HQ, Ma XH, Liu B, Zhang XY, Chen WZ, Wang CX, Cheng SH. High-throughput real-time assay based on molecular beacons for HIV-1 integrase 3'-processing reaction. Acta Pharmacol Sin. 2007;28:811–817. doi: 10.1111/j.1745-7254.2007.00561.x. [DOI] [PubMed] [Google Scholar]
- 31.Grimsby J, Sarabu R, Corbett WL, Haynes NE, Bizzarro FT, Coffey JW, Guertin KR, Hilliard DW, Kester RF, Mahaney PE, Marcus L, Qi L, Spence CL, Tengi J, Magnuson MA, Chu CA, Dvorozniak MT, Matschinsky FM, Grippo JF. Allosteric activators of glucokinase: potential role in diabetes therapy. Science. 2003;301:370–373. doi: 10.1126/science.1084073. [DOI] [PubMed] [Google Scholar]
- 32.Mouratou B, Rouyre S, Pauillac S, Guesdon JL. Development of non-radioactive microtiter plate assays for nuclease activity. Anal. Biochem. 2002;309:40–47. doi: 10.1016/s0003-2697(02)00272-5. [DOI] [PubMed] [Google Scholar]
- 33.Li JJ, Geyer R, Tan W. Using molecular beacons as a sensitive fluorescence assay for enzymatic cleavage of single-stranded DNA. Nucleic Acids Res. 2000;28:E52. doi: 10.1093/nar/28.11.e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh SS, Eis PS, Blumeyer K, Fearon K, Millar DP. Real time kinetics of restriction endonuclease cleavage monitored by fluorescence resonance energy transfer. Nucleic Acids Res. 1994;22:3155–3159. doi: 10.1093/nar/22.15.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biggins JB, Prudent JR, Marshall DJ, Ruppen M, Thorson JS. A continuous assay for DNA cleavage: the application of "break lights" to enediynes, iron-dependent agents, and nucleases. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13537–13542. doi: 10.1073/pnas.240460997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang CJ, Li JJ, Tan W. Using molecular beacons for sensitive fluorescence assays of the enzymatic cleavage of nucleic acids. Methods Mol. Biol. 2006;335:71–81. doi: 10.1385/1-59745-069-3:71. [DOI] [PubMed] [Google Scholar]
- 37.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 38.Lee SP, Kin HG, Censullo ML, Han MK. Characterization of Mg2+-dependent 3'-processing activity for human immunodeficiency virus type 1 integrase in vitro: real-time kinetic studies using fluorescence resonance energy transfer. Biochemistry. 1995;34:10205–10214. doi: 10.1021/bi00032a014. [DOI] [PubMed] [Google Scholar]