Summary
Dramatic chromosome motion is a characteristic of mid-prophase of meiosis observed across broadly divergent eukaryotic phyla. Although the specific mechanisms of motions vary among organisms studied thus far, the outcome is similar in all cases: vigorous back-and-forth movement (as fast as ~1μm/sec for budding yeast), led by chromosome ends (or near-end regions), and directed by cytoskeletal components through direct association through the nuclear envelope. The exact role(s) of movements remains unknown, although an idea gaining currency is that movement serves as a stringency factor, eliminating unwanted inter-chromosomal associations or entanglements that have arisen as part of the homolog pairing process and, potentially, unwanted associations of chromatin with the nuclear envelope. Turbulent chromosome movements observed during bipolar orientation of chromosomes for segregation could also serve similar roles during mitosis. Recent advances shed light on the contribution of protein complexes involved in the meiotic movements in chromosome dynamics during the mitotic program.
Intriguing back-and-forth chromosomal movements underlie meiotic prophase
Meiosis is the process by which a diploid cell gives rise to the haploid gamete cells required by sexual reproduction. A key feature of this process is the occurrence of two successive rounds of chromosome segregation, first of homologous maternal and paternal chromosomes (‘homologs’) and then of sister chromatids, as during mitosis. The existence and mechanics of the first division require the prior execution of a series of carefully regulated events including recognition, juxtaposition and intimate synapsis of homologs all along their lengths. Part way through this process, chromosomes initiate dramatic dynamic movements (Box 1; supplementary movie 1). Molecular studies are now revealing that these movements involve direct associations of chromosome ends, or near-end regions, with cytoskeletal motion-generating machineries by means of linkages that span the nuclear envelope (NE). Issues of interest raised by these findings include: the role(s) of such movements for the meiotic process, the basis for their temporal regulation, their detailed mechanism(s), and the possibility of functional and mechanistic analogies with general processes common to all types of cells. Also of interest are the chromosome reorganizations that precede these dynamic active mid-prophase events, including changes in the associations of heterochromatic regions and early stages of homolog pairing, the basis for which is still mysterious. Here, we review characteristics of movements accompanying the different phases of prophase and the mechanisms of zygotene/pachytene motion, and the interplay between the different components of the system. Understanding the potential roles of motion, as well as potential mitotic implications, are currently under investigation.
Box 1. Overview of the discovery of meiotic prophase chromosome motion.
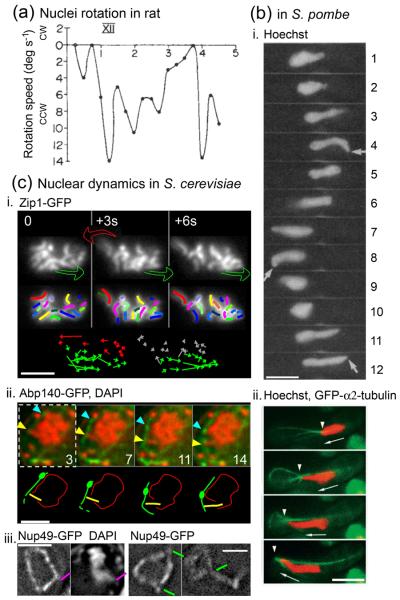
The first evidence that meiotic prophase chromosomes are experiencing rotational movements during the formation of synapsis came from rat spermatocytes [1]. Time-lapse phase-contrast observation of zygotene and pachytene spermatocytes revealed oscillatory movements of both nuclei and chromosomes, the latter also exhibiting more complex tri-dimensional trajectories. The importance and potential widespread existence of similar movements across eukaryotic phyla emerged twenty years later from a very different organism, Schizosaccharomyces pombe. The group of Yasushi Hiraoka described during the zygotene and early pachytene stage of meiotic prophase dynamic and rapid (~5 μm/min) movements along the length of the cell of the telomeres clustered at the SPB (reviewed in [2]; [19]). Such phenomena explained the characteristic “horse-tail” shape of the nucleus previously observed in snapshots of fission yeast meiotic prophase. This observation open the way to a large number of studies deciphering the precise mechanisms of meiotic chromosomes movements, and analyzing the molecular role of motion (see text). Cytoplasmic microtubules play a key role in the S. pombe movements, as they exert a pulling force on the SPB, thus on the clustered telomeres [20; 31]. The force is generated by the minus-end directed motor protein dynein Dhc1 that binds to the cell cortex and whose uneven distribution may account for the back and forth redirection of motion of microtubules along the length of the cell (Figure 1b). In their original paper, Chikashige et al. proposed that motion led by clustered telomeres could be necessary to facilitate pairing by reducing the dimensionality of the search and maybe helping to resolve interlocks [19]. Recent results indeed revealed an role for fission yeast nuclear oscillation in promoting chromosome pairing ([50]; further discussion in text).
Chromosome dynamics during meiotic prophase
Chromosome pairing and synapsis
The central event of meiotic prophase is pairing (and accompanying recombination) of homologs, which involves the stable, close juxtaposition of chromosomes that, in most species, have entered this stage unpaired. The earliest stages of this process are not well understood and likely involve interactions of telomeres/centromeres and between homologous interstitial regions through recombination-independent processes (reviewed in [3-5]; discussed below). Once recombination begins, at the G2/prophase transition, pairing becomes obvious. In many organisms, including plants, animals and filamentous fungi, chromosome axes are first brought close together along their lengths to a separation of ~400 nm, through the actions of axis-associated recombination complexes that then form the links that span the two axes. In a next stage, axes become closely associated at ~100 nm (‘synapsed’) through the synaptonemal complex (SC), a close-packed array of transverse filaments, plus other components [6]. These steps occur during specific stages of meiotic prophase as originally defined by light microscopy: coalignment occurs during leptotene, when chromosomes are ‘threadlike’; SC formation underlies zygotene, when chromosomes are closely and obviously paired in some regions; and pachytene is the stage when SC extends along the full length of all chromosomes (Box 2). These events are all tightly integrated, temporally, spatially and functionally, with a highly regulated series of meiotic recombination events [7-12].
Box 2. Stages of meiosis.
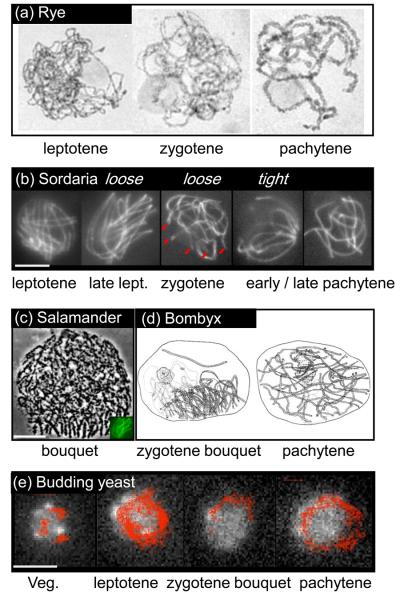
Figure I (a) shows classically fixed and stained chromosomes of rye (Secale cereale) showing leptotene (“threadlike”), zygotene (partial coming-together, i.e. synapsis) and pachytene (full synapsis). (courtesy of D. Zickler). (b) The seven pairs of chromosomes from the filamentous fungus Sordaria macrospora illuminated along their structural axes by Spo76-GFP (courtesy of D. Zickler; [84]; bar = 5μm). From left to right: midleptotene – homolog axes coaligned at ~400 nm and no bouquet; late leptotene – coaligned axes with loose bouquet; zygotene – partial synapsis with loose bouquet (chromosome ends are indicated by red lines); pachytene – full synapsis: early with tight bouquet (left) and late with ends dispersed around the nuclear periphery (right). (c) Bouquet stage in Salamander [adapted with permission from 85], with Sordaria nucleus from (b) shown at lower right for size comparison (bar: 10 μm). (d) Bombyx silkworm oocyte: zygotene nucleus with bouquet (left) and pachytene nucleus with full synapsis and ends dispersed (right) [reproduced with permission from 86; NV nuclear vacuole; N nucleolus]. (e) Telomere positions in meiosis of the budding yeast S. cerevisiae [reproduced from 26; bar ~2μm]. Trajectories of Rap1-GFP telomere foci recorded at a frame rate of 0.5 sec over 7 minutes superimposed over a nucleus of the corresponding stage. Perinuclear telomere foci in vegetative nuclei exhibit relatively constrained movements along the NE. Leptotene: telomeres move around the nucleus; zygotene bouquet: a subset of telomeres appears confined within a subregion of the NE; Pachytene: dynamic dispersal of telomeres around the NE. Note that yeast has 16 pairs of chromosomes, implying that the bouquet contains 64 individual telomeres and 32 pairs of telomeres.
In certain organisms, pairing and recombination are not accompanied by SC formation (e.g. the fission yeast Schizosaccharomyces pombe), whereas in others, such as male Drosophila melanogaster, pairing and stable homolog association occur in the absence of recombination (see for instance [13]). Nonetheless, irrespective of the details, homologous chromosomes must come together in space and generate intimate, stable interactions, and in such a way that they are not topologically entangled with unrelated chromosomes [14].
Dynamic zygotene/pachytene movements
Two related types of chromosome movement are superimposed on the later stages of the pairing process, during the periods when chromosomes are synapsing and synapsed. First, three-dimensional reorganization of chromosome ends, which dynamically cluster into the so-called ‘bouquet configuration’ at zygotene and then decluster back to a dispersed configuration at pachytene (Box 2; [4, 15-18]); and, second, global chromosome motions, which initiate at zygotene and persist into pachytene. The existence of such movements was initially revealed by light-microscopy studies of living meiotic cells from rat spermatocytes [1; Figure 1a]. With the advent of time-lapse fluorescence microscopy, movies of living S. pombe cells revealed dynamic back-and-forth nuclear deformations at the corresponding stage ([2, 19, 20]; Figure 1b). Most recently, analogous motions have been described in budding yeast Saccharomyces cerevisiae (Figure 1c; [21-25]) and in the nematode Caenorhabditis elegans (A. Dernburg cited in [4]). In budding yeast, a bias in the global distribution of telomeres-led chromosomal movements could account for bouquet clustering and declustering [22, 26].
Figure 1.
Nuclear dynamics in rat, the budding yeast S. cerevisiae and the fission yeast S. pombe. (a) Rotating movements of chromosomes in rat spermatocyte nuclei recorded at a time-frame of 2 images per second (adapted with permission from [1]). Rotation speed is in degrees s−1 (Y-axis) and time is in minutes (X-axis). cw stands for clockwise direction above the abscissa and ccw counterclockwise below it; XII stands for the zygotene stage of rat spermatogenesis. (b) Chromosome dynamics in S. pombe (see also Box 1). Scale bars = 5 μm. i) Time-series of chromosomes stained with Hoechst and exhibiting the horsetail movements characteristics of S. pombe meiotic prophase (reproduced with permission from [19]); time indicated in minutes. Arrows indicate the leading edge of the moving nucleus. (ii) Time-series of the microtubule network visualized with GFP-labelled α2-tubulin (in green) and chromosomal mass stained with Hoechst (in red). The arrowheads indicate the positions of the leading edge of the nucleus (images taken every 30 seconds, reproduced with permission from [20]). (c) chromosome and NE dynamics of S. cerevisiae pachytene nuclei (adapted with permission from [22]). Scale bars = 2 μm. i) Top: three frames from a two-dimensional time series of pachytene chromosomes labelled with Zip1-GFP (3 second intervals), with colored arrows indicating dynamic transition between two frames, as defined from the general direction of chromosomes displacement vectors (see below) . Middle: for each frame of the upper lane, chromosomes are outlined to facilitate visual tracking over time. Bottom: the apparent changes in the telomeric position of each chromosome between two frames are represented with colored vectors, whose size and direction reflect the characteristics of the displacement, and whose color reflect the general trend of this vector as compared with its counterparts. ii) Time-series of the actin cable network (labeled with Abp140–GFP, green) and chromosomes (stained with DAPI, red), with the corresponding schematic representations underneath. In the example shown, a chromosome end pointing outward (yellow arrowhead) is moving along the same track and at a pace similar to that of an actin cable carrying a fiduciary mark (turquoise arrowhead). iii) Time-series of pachytene nuclei labeled with Nup49-GFP, with (left) or without (right) concomitant staining of DNA with DAPI. Pink line: concomitant NE deformation and chromosomal protrusion. The typically observed short and long tube-like NE deformations are indicated with green lines.
It would be interesting to know whether these different mechanisms share a same origin and what evolutionary tracks separate them.
Within this general commonality, programmatic diversity among different organisms again occurs. In most organisms, telomeres in the bouquet configuration are mostly colocalized in a general region of the NE surrounding the microtubule organizing center (MTOC). Clustering density can vary from ‘tight’ to loose plus intermediate states. The loose configuration may sometimes precede the tight configuration (e.g. [27]; see Box 2 Fig. Ib). Alternatively, in time-lapse analysis of living S. cerevisiae cells, the two configurations dynamically interconvert: rapid telomere movements lead to transient colocalization and dynamic local clustering/declustering within the vicinity of the spindle-pole body (SPB, the yeast MTOC; [26]; see Box 2 Fig. Ie). However, in S. pombe, telomeres remain tightly clustered to the SPB for the entire length of prophase [reviewed in 2]. Also, in C. elegans, clustering at the NE is mediated by specific subtelomeric regions, rather than telomeres per se as in other organisms [reviewed in 4, 28].
Box 2 Figure I.
Stages of meiotic prophase
Pre-zygotene movement
This review focuses on the programmed motions that initiate during meiotic mid-prophase. It should be noted, however, that interesting dynamic changes also occur prior to this (relatively late) point in the program. Most dramatically, chromosomes exit the preceding mitosis in a ‘Rabl’ configuration, with centromeres clustered together as a consequence of being brought to the corresponding pole of the mitotic spindle and, usually (although not in many fungi and basidiomycetes), free of the nuclear envelope [15, 17, 18]. Thus, bouquet formation is preceded by both loss of the Rabl configuration and association of telomeres to the NE. In addition, centromeres in particular, or heterochromatic regions in general, appear to cluster and decluster dynamically during the early prophase of most organisms, suggesting an important role that remains unclear [reviewed in 29].
During mid-prophase movement, forces generated by the cytoskeleton are transmitted to chromosome ends via bridges that span the NE
The mechanisms responsible for dynamic zygotene/pachytene chromosome movements are now elucidated, or largely so, in a handful of organisms. In all cases described so far, forces appear to be generated via cytoskeleton elements and are exerted on NE-localized chromosome ends via specialized complexes that span the NE (below). Correspondingly, in some cases, force-mediated NE deformations are seen to accompany chromosomal motion. However, the molecular mechanisms by which these effects are implemented are different in different organisms.
In fission yeast, and more recently in budding yeast, the mechanisms responsible for these movements have been partially deciphered. In S. pombe, telomeres cluster together on the NE at the SPB, and these ensembles move back and forth within the cell through direct association with cytoplasmic microtubules that run tangentially to the nuclear periphery and with microtubule-associated dynein motor protein complexes playing a central role [Figure 1bii; 2, 30]. The observed motions can be explained by dynamic redistribution of dynein motors from the ‘leading’ side of the SPB complex to the ‘trailing’ side, as directed by microtubule length and mechanical load [31]. In S. cerevisiae, unpaired and paired telomeres become associated transiently through the NE, to nucleus-hugging actin cables that are continuous with the actin cytoskeleton [see notably 23; see also 21, 26, 22]. In this case, the telomere–actin associations seem to be passive, with actin-tethered chromosome ends moving because of the dynamic tread-milling of the underlying actin cable per se rather than as a cargo being transported along the cable by a motor protein [Figure 1cii; 22]. Interestingly, budding yeast uses an analogous mechanism for retrograde transport of mitochondria from the bud tip to the mother cell tip [32], suggesting that meiotic chromosome movement systems could have evolved from processes used more generally for cytoplasmic transport of vesicles and/or the nucleus itself (see below).
Information for other systems is just beginning to emerge. In C. elegans, microtubule-associated dynein seems to play a role [A. Dernburg cited in 4; 33]. By contrast, in some plants, complete depolymerization of cytoplasmic microtubules by the chemicals vinblastine and amiprophos-methyl does not disrupt telomere clustering in rye anthers, although addition of colchicine at a concentration too low to depolymerize microtubules, does. This result points to a non-cytoplasmic-microtubule mechanism for movement in this organism [16] – for example potentially by means of actin or an alternative cytoskeleton element. Finally, for mammalian meiosis, the nature of the force-generating mechanism is thus far unknown.
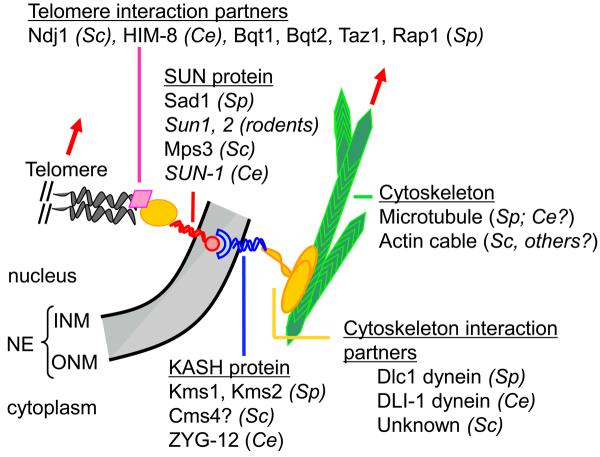
The NE–cytoskeleton associations involved in these dynamic chromosomal movements involve the same types of proteins used by non-meiotic cells to link the nuclear envelope directly to the global cytoskeleton, where the resulting associations anchor, move and position the nucleus within the cell [reviewed in 34-37]. Typically, such processes involve proteins of the SUN (for: S. pombe Sad1 and Caenorhabditis elegans Unc-84 proteins [38]) and KASH (for: Klarsicht/ANC-1/Syne-1 homology [39]) domain families. These two types of proteins locate within the inner and outer nuclear membrane of the NE, respectively, and interact physically with each other in the lumen, thereby directly connecting the involved NE regions with the cytoskeleton networks [35, 37]. The precise structures of the interaction and the stoichiometries of the proteins involved remain unknown [37]. Notably: how do the relatively small (<35 aa) C-terminal domain of the KASH protein interact with the SUN protein in the perinuclear space, and also whether both KASH and SUN proteins are needed so that the telomere/NE attachment appears mature? These molecules provide not only molecular connections but structural reinforcement such that the forces generated by movement of the nucleus do not (usually) rupture the association sites.
During meiosis, the SUN domain protein Mps3 in S. cerevisiae [23, 40], Sun1 and Sun2 in rodents [41, 42], Sun-1 in C. elegans [33] and Sad1 in S. pombe [43] are all implicated in meiotic telomere–NE attachment. In S. pombe, additional components of telomere–NE–cytoskeleton complexes are now characterized, with Bqt1 and Bqt2 connecting telomeres to Sad1, which itself connects to the KASH domain protein Kms1, which in turn interacts with the dynein light chain Dlc1 [43-46]. In S. cerevisiae, no KASH domain protein has been identified thus far as playing a role with Mps3, but the putative tail-anchored membrane protein Csm4 is a good candidate for playing a similar role [22-25]. Furthermore, at the chromosomal level, the meiosis-specific telomere-binding protein Ndj1 has been shown to tether telomeres to the NE during meiosis by means of interactions with Mps3 [40, 23; Figure 2]. In addition, Mps3 is also involved in the Sir4–Esc1 pathway of telomere recruitment to the nuclear membrane during vegetative growth [47]. In mammals, while the motion-generating system has not been identified, Sun1 and Sun2 are inner nuclear membrane (INM) components of meiosis-specific telomere–NE attachments and presumably are thus, by analogy, involved [41, 42]. In C. elegans, the KASH domain protein Zyg-1 interacts with Sun-1 to link chromosomes to microtubules [4].
Figure 2.
Schematic representation of meiotic chromosome telomere–NE attachment. The names of the identified protein members of these complexes are indicated for the nematode C. elegans (Ce), the budding yeast S. cerevisiae (Sc), the fission yeast S. pombe (Sp) and rodents. NE, nuclear envelope, INM, inner nuclear membrane; ONM, outer nuclear membrane. Telomeres (grey helix) connect to the NE through telomere interaction partners (schematized with a pink diamond and a yellow oval), and SUN-domain protein spanning the INM (in red). SUN-domain proteins can interact in the nuclear membrane lumen (in grey) with KASH domain proteins that span the ONM (in blue). Molecular motors or yet-to-be-identified partners (in yellow) then connect the telomere–NE complexes to the cytoskeleton (in green). Displacement of either the cytoskeleton network or the molecular motors induces the displacement of the underlying telomere and chromatin (red arrows).
Two aspects of the mid-prophase motion phenomenon remain poorly understood in all systems. First, SUN-domain proteins relocate during meiotic prophase, from other locations (principally the vicinity of the SPB in yeast mitotic cells, the centrosome in C. elegans and a more homogeneous rim in mammalian somatic cells) to chromosome extremities [33, 42, 48]. The temporal and spatial signals for this relocalization remain unclear [42]. Second, what are the signals that activate the sequential events and the motion itself? Importantly, in budding yeast, this signal must act after telomere–NE association, which is complete prior to onset of zygotene motion. An obvious possibility is regulatory activation of the association between telomere–NE ensembles and actin cables, either directly or through effects on SUN-domain protein localization, similar to the anchoring of dynein to the cortex by a meiosis-specific gene product [49].
Role(s) of chromosome motion for mid-prophase of meiosis
Pairing? Maybe not
Since the first description of the bouquet stage in the early 20th century [see 17], the primary rationale for its existence was thought to be to promote the coming-together of homologous chromosomes. Indeed, association and alignments of homologs in the bouquet prior to the onset of pairing could reduce the dimensionality and distances involved in homology searching. Modern studies have shown that homolog pairing can be aberrant (although never completely abrogated) in the presence of mutations impairing chromosome movement [e.g. 50, 51] or in mutants likely defective in such movement [33] and, conversely, ectopic recombination frequencies increase in correlation with closeness to telomeres [52, 53]. Nonetheless, significant challenges to this original idea are beginning to emerge, pointing to alternative or additional roles. Most importantly, in several cases, bouquet formation appears to occur too late in the pairing process to be the primary mediator of homology searching [reviewed in 17; see Box 2b]. Furthermore, in budding yeast, dynamic movement begins exactly at zygotene, i.e. concomitant with SC formation and thus well into the recombination process, when stable strand exchange intermediates are already formed between homologous interacting DNAs [7, 21-23]. Additionally, in both fission yeast and budding yeast, mutations conferring disruption of telomere motion have only relatively modest effects on either recombination or pairing [24, 25, 46, 54]. Finally, in the three organisms studied, the movements of mid-prophase chromosomes have a back-and-forth character (rotationally within rat spermatocytes nuclei, longitudinally in S. pombe, and circum-nuclear in S. cerevisiae), which does not have the effect of ‘stirring’ chromosomes (Figure 1). Indeed, different chromosomes tend to move coordinately either because their telomeres are stably colocalized (S. pombe) or because of other types of connections (S. cerevisiae). For these reasons, we want to focus more on the possibility that the crucial functional effect is chromosome movement, including but not limited to movement into and out of the bouquet configuration, rather than the existence of a bouquet configuration per se.
A stringency factor for eliminating unwanted connections?
One proposal suggests that that motion would resolve topological entanglements among chromosomes that impede completion of recombination and juxtapositioning of homologs [8, 22, 24]. Topological ‘interlockings’ are seen at the zygotene stage and undergo resolution during the pachytene stage in several organisms, with strong indications that there is a specific mechanism for their resolution during those stages [55]. Furthermore, a back-and-forth motion could be highly appropriate to ‘get the kinks out’ of a set of chromosomes that are in the final stages of pairing and synapsis. In accord with such a possibility, a maize mutation that disrupts formation of the bouquet configuration, and thus potentially chromosome movement, results in accumulation of prophase interlocks [pam1; 56].
Movements could also help to free chromosomes from interconnections of other kinds. In budding yeast, at pachytene, motion led by one chromosome end causes other, unrelated, chromosomes to move coordinately [Figure 1ci], implying the existence either of direct nonspecific interchromosomal links and/or parallel independent association of multiple ends to a particular patch of NE substructure [22]. Such connections must ultimately be eliminated if homologs are to segregate cleanly from one another, independent of other chromosomes at meiosis I; mid-prophase movement could help to accomplish this task. Indeed, colocalization of telomeres (or, in C. elegans, potentially analogous ‘pairing centers’; [4]) in a polarized configuration might specifically promote inappropriate connections that must be resolved by the dynamic motions of telomere redistribution during bouquet exit. Similarly, it has been suggested that chromosome movement might eliminate ectopic recombination in which a sequence at one position interacts with an homologous position located at a non-corresponding position on another chromosome [16, 22, 23, 52, 53, 57, 58]. In accord with this possibility, abrogation of motion confers a slight increase in ectopic recombination in budding yeast [for discussion, see 23]; however, in fission yeast, while abrogation of telomere clustering significantly increases ectopic recombination, mutation induced abrogation of motion does not [57, 58]. Similarly, motion might help to disrupt nonhomologous associations between centromeres that often precede their homologous association (reviewed in [2]). Another potential role for motion could be to abrogate the local associations that occur between chromatin and the NE or nuclear pore complexes that have been recently revealed to modulate transcription [59-61] or DNA repair [62-64]. Overall, dynamic programmed movements could be a way to free chromosomes from all inappropriate or leftover interconnections and associations they might have been acquired. In summary: dynamic chromosome movements could well serve as a ‘stringency factor’ to eliminate unwanted (and thus presumably weaker) connections so that chromosomes can be fully and intimately synapsed along their lengths by the end of pachytene. Cytological studies (such as screening for interlocks by visualizing axis components) and genetics and chromosomal dynamics analysis of mutants of the members involved in the telomere–NE–cytoskeleton complexes should provide further insights on the actual role of motion.
Evolutionary note
It would be interesting to know whether telomere- (or centromere- or pairing-center-) directed homolog pairing comprises a ‘primordial’ mechanism that preceded evolution of recombination-mediated pairing or whether recombinational mechanisms that had evolved for mitotic double-strand break repair were the original pairing process. The need for a mechanical stringency factor would be a central requirement of both scenarios.
Other models
Situations where telomere dynamics are abrogated by mutation or by drug(s) are characterized by variable delays and/or defects in recombination, assayed genetically or physically and/or in recombination-mediated pairing, manifested as juxtaposition of fluorescently tagged loci at corresponding positions on homologs [24, 25, 56-58]. The fact that these delays and defects do not impair totally the progression of meiosis has led to the suggestion that motion could be required for completion of interhomolog interactions. One specific hypothesis is that telomere clustering plays a supporting role in accelerating the rate of recombinational progression [reviewed in 16]. A second hypothesis is that motion is required to mechanically promote steps of recombination [25]. Since resolution of the recombination intermediates single end invasion and double Holiday junction does not require motion [22], these steps would be early post-double strand breaks recombination steps. However, mutation-induced delays could be indirect regulatory checkpoints effects that delay recombination progression. For example, mutations of the NE/telomeric ensembles could disorganize those complexes thus generating a checkpoint response. By this model, regulatory delays would reflect defects in basic telomere biology irrespective of any effects on chromosome motion [24]. Alternatively, or in addition, these delays could be triggered by primary defects in eliminating unwanted connections. For example, interlocking of chromosomes will tend to preclude completion of recombinational interactions in the affected region, and the presence of such, even one or a few, stalled recombinational interactions could trigger a checkpoint surveillance response, which in turn would confer broader defects and delays on a nucleus-wide basis.
Generalizations for chromosomes beyond meiotic prophase
The notion that vigorous movements serve to eliminate unwanted connections can be extended also to the prometaphase/metaphase stages, in mitotic as well as meiotic cells. During these periods, chromosomes undergo dramatic back-and-forth movements directed by kinetochore–microtubule associations as part of the process by which sisters (or homologs) become oriented to opposite poles [65]. Indeed, nonspecific physical connections are frequently observed for late-stage mitotic chromosomes, particularly but not exclusively at their ends [66]; conversely, the somatic pairing that is robust throughout most of the cell cycle in Drosophila is partially disrupted during the segregation process in euchromatic regions [67].
Roles and effects beyond the chromosomes
Roles for motion other than for chromosome dynamics have also been suggested. In higher plants, nuclear pores cluster to the same regions as telomeres during the bouquet stage [reviewed in 17], leading to the suggestion that this configuration might promote direct loading of newly synthesized proteins onto chromosome ends [D. Zickler, pers. commun.]. Also, stretching forces transmitted through the cytoskeleton to the nuclear membrane of mouse fibroblasts appear to activate stretch-sensitive channels within the membrane that lead to transient elevation of nuclear [Ca2+], presumably through its release from the endoplasmic reticulum [68]. Perhaps the NE deformations that accompany meiotic chromosome movement play an analogous role at that stage.
An integrated mechanical system
All of the components involved in meiotic chromosome movements are in direct mechanical linkage. In this regard, it is interesting that budding yeast pachytene chromosomes are semi-flexible objects having flexural persistence lengths (i.e. the distance over which the correlation between the direction of two vectors tangent to the polymer is lost) comparable to those of actin filaments [22]. Also in budding yeast [22], visualization of a fluorescently tagged nuclear pore component reveals that the NE undergoes a cone-like deformation at the point of end-directed chromosome movement, implying that pulling encounters resistance from the envelope substructure and proteins of the inner and outer membranes [Figure 1ciii, pink line]. Occasionally small thin projections are also observed, implying that this substructure occasionally ruptures, leading to formation of a membrane tubes [Figure 1ciii, green line]. Additionally, the chromosomes are not simply free in solution; instead they sit in a crowded environment and are likely engaged in non-specific interconnections with other objects from the nucleoplasm that, in addition, might be connected to one another (see above). Thus, chromosomes are expected to resist telomere-led movement, and there are hints to this effect. For example, in budding yeast, force exerted on the end of one chromosome results in coordinate movements of other nearby chromosomes, perhaps through inter-chromosomal linkages; however, occasional membrane tubes also contain a chromosome, and, when that occurs, the orphaned chromosome is always small. As small chromosomes are less likely to be involved in linkages with other chromosomes, this finding points to a constraining role of such linkages.
The zygotene/pachytene period of chromosome movement could therefore provide an interesting system not only for studying the mechanical capacities of the motion-generating system but, concomitantly, the properties of the nucleus and the partitioning of mechanical load between the NE and the underlying chromosomes. For a whole mammalian nucleus, the primary force-bearing component can be the chromatin (for instance in human stem cells) and/or the lamina of the NE (for instance in human epithelial cells) [69, 70]. During meiosis, chromatin is more compact during zygotene (when motion is biased toward the bouquet tendency) and more expanded during pachytene (when chromosome ends disperse throughout the nuclear periphery). We have suggested that this change in chromatin state also produces an overall change in whole chromosome stiffness (persistence length) that could be important for driving the transition from the zygotene telomere-clustering state to the pachytene state where telomeres are dispersed. It would be interesting to know whether changes in chromatin state confer changes in the deformability of the NE [e.g. 69] that are also seen during this transition. Similarly, it would be interesting to know whether the force required to move a chromosome varies over time in relation to these effects.
Emerging roles for the members of NE bridging complexes in regulating DNA metabolism
Two recent findings highlight the potential of bridging members from nuclear envelope proteins to play new and unsuspected roles in chromatin metabolism and organization in non-meiotic cells. First, the SUN–KASH complex couples the microtubule network and heterochromatin/centromeric regions in interphase S. pombe cells [71]. Similarly, in S. cerevisiae, the N-terminal domain of the SUN-protein Mps3 is required for telomere tethering to the nuclear periphery in vegetative cycling cells [47]. Second, in budding yeast, a DNA double-strand break whose repair is impaired or delayed is recruited to the nuclear periphery, where it associates with Mps3 [64; see also 72, 73]. The involvement of Mps3, and the consequences of what could be a secondary response to persistent unrepaired breaks remain, however, poorly understood.
It should also be recalled that disruption of actin polymerization is required for movement of an interphase chromosomal site activated for transcription away from the NE [74]. More generally, actin plays important roles in transcription-related phenomena. While the present consensus suggests that a pool of nuclear actin monomers plays important roles within this compartment [74-78], it is conceivable that future work will reveal a role for cytoplasmic actin, as well as nuclear actin, in some of the described phenomena or others. In particular, identification of SUN–KASH linkages between cytoplasm and NE, plus appreciation that certain diseases, such as muscular dystrophy or cardiomyopathy [79], are attributable to defects in lamin structure, have triggered multiple lines of investigation into whether/how mechanical forces, potentially even transduced directly from the cell surface, could influence the NE and its linked chromosomal regions [e.g. 69, 80-83].
Clearly, fully understanding of the important role(s) that dynamic motor-mediated chromosome movements play in meiosis is an outstanding goal (Box 3). Understanding these molecular mechanisms will also shed light on the dynamic phenomena observed during the basic mitotic cell cycle.
Box3. Future issues and questions of special interests.
- Deciphering the precise nature of the KASH / SUN domain protein complexes, including i) the stoichiometry and ii) the physical interactions among the proteins involved, and iii) additional partners of the complex.
- Deducing the structure of telomeres/NE complexes, from individual chromatid telomeres through KASH proteins.
- Understanding the mechanical coupling among involved components in response to force.
- Relating dynamic telomere-led motions to bouquet dynamics.
- What are the force generating mechanisms in species other than S. pombe?
- Do changes in chromatin state during meiotic prophase drive changes in the nature of chromosome motion at zygotene vs. pachytene?
- Does motion disentangle interlocked chromosomes and/or resolve ectopic chromosomal interactions?
Supplementary Material
movie of S. cerevisiae Zip1-GFP chromosome, 1 sec intervals [22; see also 21, 23 for more examples]. Colored arrows indicate pronounced telomereled movements.
Acknowledgements
We thank D. Zickler for allowing reproduction of unpublished pictures in Box 2 Fig. I, panels a and b, and Y. Hiraoka, M. Parvinen, and H. Scherthan for allowing reproduction of their work. This work was supported by a grant to N.K. (NIH RO1 GM-044794 and GM 025326).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parvinen M, Soderstrom KO. Chromosome rotation and formation of synapsis. Nature. 1976;260:534–535. doi: 10.1038/260534a0. [DOI] [PubMed] [Google Scholar]
- 2.Chikashige Y, et al. Another way to move chromosomes. Chromosoma. 2007;116:497–505. doi: 10.1007/s00412-007-0114-8. [DOI] [PubMed] [Google Scholar]
- 3.Bozza CG, Pawlowski WP. The cytogenetics of homologous chromosome pairing in meiosis in plants. Cytogenet Genome Res. 2008;120:313–319. doi: 10.1159/000121080. [DOI] [PubMed] [Google Scholar]
- 4.Bhalla N, Dernburg AF. Prelude to a division. Annu Rev Cell Dev Biol. 2008;24:397–424. doi: 10.1146/annurev.cellbio.23.090506.123245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerton JL, Hawley RS. Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat Rev Genet. 2005;6:477–487. doi: 10.1038/nrg1614. [DOI] [PubMed] [Google Scholar]
- 6.Tesse S, et al. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci U S A. 2003;100:12865–12870. doi: 10.1073/pnas.2034282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 8.Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma. 2006;115:158–174. doi: 10.1007/s00412-006-0048-6. [DOI] [PubMed] [Google Scholar]
- 9.Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–194. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- 10.Borner GV. Balancing the checks: surveillance of chromosomal exchange during meiosis. Biochem Soc Trans. 2006;34:554–556. doi: 10.1042/BST0340554. [DOI] [PubMed] [Google Scholar]
- 11.Blat Y, et al. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 12.Padmore R, et al. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- 13.Vazquez J, et al. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr Biol. 2002;12:1473–1483. doi: 10.1016/s0960-9822(02)01090-4. [DOI] [PubMed] [Google Scholar]
- 14.Kleckner N, Weiner BM. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harb Symp Quant Biol. 1993;58:553–565. doi: 10.1101/sqb.1993.058.01.062. [DOI] [PubMed] [Google Scholar]
- 15.Scherthan H. A bouquet makes ends meet. Nat Rev Mol Cell Biol. 2001;2:621–627. doi: 10.1038/35085086. [DOI] [PubMed] [Google Scholar]
- 16.Harper L, et al. A bouquet of chromosomes. J Cell Sci. 2004;117:4025–4032. doi: 10.1242/jcs.01363. [DOI] [PubMed] [Google Scholar]
- 17.Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 18.Bass HW. Telomere dynamics unique to meiotic prophase: formation and significance of the bouquet. Cell Mol Life Sci. 2003;60:2319–2324. doi: 10.1007/s00018-003-3312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chikashige Y, et al. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- 20.Ding DQ, et al. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci. 1998;111(Pt 6):701–712. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- 21.Scherthan H. Telomere attachment and clustering during meiosis. Cell Mol Life Sci. 2007;64:117–124. doi: 10.1007/s00018-006-6463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koszul R, et al. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008;133:1188–1201. doi: 10.1016/j.cell.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conrad MN, et al. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 2008;133:1175–1187. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- 24.Wanat JJ, et al. Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet. 2008;4:e1000188. doi: 10.1371/journal.pgen.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosaka H, et al. Csm4-dependent telomere movement on nuclear envelope promotes meiotic recombination. PLoS Genet. 2008;4:e1000196. doi: 10.1371/journal.pgen.1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trelles-Sticken E, et al. Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J Cell Biol. 2005;170:213–223. doi: 10.1083/jcb.200501042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storlazzi A, et al. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 2003;17:2675–2687. doi: 10.1101/gad.275203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips CM, Dernburg AF. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev Cell. 2006;11:817–829. doi: 10.1016/j.devcel.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 29.Stewart MN, Dawson DS. Changing partners: moving from nonhomologous to homologous centromere pairing in meiosis. Trends Genet. 2008;24:564–573. doi: 10.1016/j.tig.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto A, Hiraoka Y. Cytoplasmic dynein in fungi: insights from nuclear migration. J Cell Sci. 2003;116:4501–4512. doi: 10.1242/jcs.00835. [DOI] [PubMed] [Google Scholar]
- 31.Vogel SK, et al. Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol. 2009;7:e1000087. doi: 10.1371/journal.pbio.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boldogh IR, Pon LA. Mitochondria on the move. Trends Cell Biol. 2007;17:502–510. doi: 10.1016/j.tcb.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Penkner A, et al. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev Cell. 2007;12:873–885. doi: 10.1016/j.devcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Starr DA, Fischer JA. KASH 'n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays. 2005;27:1136–1146. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- 35.Tzur YB, et al. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat Rev Mol Cell Biol. 2006;7:782–788. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- 36.Wilhelmsen K, et al. KASH-domain proteins in nuclear migration, anchorage and other processes. J Cell Sci. 2006;119:5021–5029. doi: 10.1242/jcs.03295. [DOI] [PubMed] [Google Scholar]
- 37.Starr DA. A nuclear-envelope bridge positions nuclei and moves chromosomes. J Cell Sci. 2009;122:577–586. doi: 10.1242/jcs.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malone CJ, et al. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- 39.Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- 40.Conrad MN, et al. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104:8863–8868. doi: 10.1073/pnas.0606165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding X, et al. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell. 2007;12:863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 42.Schmitt J, et al. Transmembrane protein Sun2 is involved in tethering mammalian meiotic telomeres to the nuclear envelope. Proc Natl Acad Sci U S A. 2007;104:7426–7431. doi: 10.1073/pnas.0609198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chikashige Y, et al. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 44.Tang X, et al. Bqt2p is essential for initiating telomere clustering upon pheromone sensing in fission yeast. J Cell Biol. 2006;173:845–851. doi: 10.1083/jcb.200602152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miki F, et al. Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol Genet Genomics. 2004;270:449–461. doi: 10.1007/s00438-003-0938-8. [DOI] [PubMed] [Google Scholar]
- 46.Miki F, et al. The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol Biol Cell. 2002;13:930–946. doi: 10.1091/mbc.01-11-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bupp JM, et al. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J Cell Biol. 2007;179:845–854. doi: 10.1083/jcb.200706040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaspersen SL, et al. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J Cell Biol. 2002;159:945–956. doi: 10.1083/jcb.200208169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamashita A, Yamamoto M. Fission yeast Num1p is a cortical factor anchoring dynein and is essential for the horse-tail nuclear movement during meiotic prophase. Genetics. 2006;173:1187–1196. doi: 10.1534/genetics.105.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding DQ, et al. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev Cell. 2004;6:329–341. doi: 10.1016/s1534-5807(04)00059-0. [DOI] [PubMed] [Google Scholar]
- 51.Trelles-Sticken E, et al. Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J Cell Biol. 2000;151:95–106. doi: 10.1083/jcb.151.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldman AS, Lichten M. Restriction of ectopic recombination by interhomolog interactions during Saccharomyces cerevisiae meiosis. Proc Natl Acad Sci U S A. 2000;97:9537–9542. doi: 10.1073/pnas.97.17.9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlecht HB, et al. Compartmentalization of the yeast meiotic nucleus revealed by analysis of ectopic recombination. Genetics. 2004;168:1189–1203. doi: 10.1534/genetics.104.029157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wells JL, et al. Homologous chromosome pairing in Schizosaccharomyces pombe. Yeast. 2006;23:977–989. doi: 10.1002/yea.1403. [DOI] [PubMed] [Google Scholar]
- 55.von Wettstein D, et al. The synaptonemal complex in genetic segregation. Ann. Rev. Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]
- 56.Golubovskaya IN, et al. The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays L.) Genetics. 2002;162:1979–1993. doi: 10.1093/genetics/162.4.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niwa O, et al. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. Embo J. 2000;19:3831–3840. doi: 10.1093/emboj/19.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis L, Smith GR. The meiotic bouquet promotes homolog interactions and restricts ectopic recombination in Schizosaccharomyces pombe. Genetics. 2006;174:167–177. doi: 10.1534/genetics.106.059733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taddei A, et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 60.Cabal GG, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 61.Schmid M, et al. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 62.Nagai S, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Therizols P, et al. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J Cell Biol. 2006;172:189–199. doi: 10.1083/jcb.200505159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oza P, et al. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- 66.Poirier MG, Marko JF. Micromechanical studies of mitotic chromosomes. Curr Top Dev Biol. 2003;55:75–141. doi: 10.1016/s0070-2153(03)01002-0. [DOI] [PubMed] [Google Scholar]
- 67.Dernburg AF, et al. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- 68.Itano N, et al. Cell spreading controls endoplasmic and nuclear calcium: a physical gene regulation pathway from the cell surface to the nucleus. Proc Natl Acad Sci U S A. 2003;100:5181–5186. doi: 10.1073/pnas.0531397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dahl KN, et al. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys J. 2005;89:2855–2864. doi: 10.1529/biophysj.105.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pajerowski JD, et al. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci U S A. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.King MC, et al. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–438. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schober H, et al. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009;23:928–938. doi: 10.1101/gad.1787509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kalocsay M, et al. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 74.Chuang CH, et al. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 75.Ye J, et al. Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes Dev. 2008;22:322–330. doi: 10.1101/gad.455908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dundr M, et al. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McDonald D, et al. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol. 2006;172:541–552. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gieni RS, Hendzel MJ. Actin dynamics and functions in the interphase nucleus: moving toward an understanding of nuclear polymeric actin. Biochem Cell Biol. 2009;87:283–306. doi: 10.1139/O08-133. [DOI] [PubMed] [Google Scholar]
- 79.Gruenbaum Y, et al. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 80.Wang N, et al. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 81.Cohen TV, Stewart CL. Fraying at the edge mouse models of diseases resulting from defects at the nuclear periphery. Curr Top Dev Biol. 2008;84:351–384. doi: 10.1016/S0070-2153(08)00607-8. [DOI] [PubMed] [Google Scholar]
- 82.Jaalouk DE, Lammerding J. Mechanotransduction gone awry. Nat Rev Mol Cell Biol. 2009;10:63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Janmey PA, McCulloch CA. Cell mechanics: integrating cell responses to mechanical stimuli. Annu Rev Biomed Eng. 2007;9:1–34. doi: 10.1146/annurev.bioeng.9.060906.151927. [DOI] [PubMed] [Google Scholar]
- 84.van Heemst D, et al. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell. 1999;98:261–271. doi: 10.1016/s0092-8674(00)81020-x. [DOI] [PubMed] [Google Scholar]
- 85.Kezer J, et al. The meiotic structure and behavior of the strongly heteromorphic X/Y sex chromosomes of neotropical plethodontid salamanders of the genus Oedipina. Chromosoma. 1989;98:433–442. [Google Scholar]
- 86.Rasmussen SW. The meiotic prophase in Bombyx mori females analyzed by three dimensional reconstructions of synaptonemal complexes. Chromosoma. 1976;54:245–293. doi: 10.1007/BF00293453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
movie of S. cerevisiae Zip1-GFP chromosome, 1 sec intervals [22; see also 21, 23 for more examples]. Colored arrows indicate pronounced telomereled movements.