Abstract
The natural tropism of most viral vectors, including adeno-associated viral (AAV) vectors, leads to predominant transduction of neurons and epithelia within the central nervous system (CNS) and retina. Despite the clinical relevance of glia for homeostasis in neural tissue, and as causal contributors in genetic disorders such as Alzheimer's and amyotrophic lateral sclerosis, efforts to develop more efficient gene delivery vectors for glia have met with limited success. Recently, viral vector engineering involving high-throughput random diversification and selection has enabled the rapid creation of AAV vectors with valuable new gene delivery properties. We have engineered novel AAV variants capable of efficient glia transduction by employing directed evolution with a panel of four distinct AAV libraries, including a new semi-random peptide replacement strategy. These variants transduced both human and rat astrocytes in vitro up to 15-fold higher than their parent serotypes, and injection into the rat striatum yielded astrocyte transduction levels up to 16% of the total transduced cell population, despite the human astrocyte selection platform. Furthermore, one variant exhibited a substantial shift in tropism toward Müller glia within the retina, further highlighting the general utility of these variants for efficient glia transduction in multiple species within the CNS and retina.
Introduction
Glia play a number of critical roles in the central nervous system (CNS) and peripheral nervous system, including providing structural and nutritional support for neurons, maintaining tissue homeostasis, and participating in signal transmission in neural tissue.1,2 Given these natural roles, glia are an attractive therapeutic target, particularly because many regions of the brain possess significantly more glia than neurons,3 and since it is unlikely that neurons severely afflicted by disease or injury are the optimal source of secreted neuroprotective factors. Furthermore, dysfunction of astrocytes likely plays a significant role in the pathology of genetic diseases, including Alzheimer's4 or amyotrophic lateral sclerosis,5 and reactive astrogliosis has been implicated as detrimental for neuroregeneration. Müller cells, the radial glial cells of vertebrate retina, react similarly to astrocytes in many ocular injuries, including retinal detachment and retinal neovascular disease.
To date, the use of viral vectors including adeno-associated virus (AAV) to transduce glia in vivo has met with limited success,6,7,8,9 even when combined with glial-specific promoters.10 AAV, a nonpathogenic virus in humans, is capable of robust systemic delivery11 and exhibits a strong clinical safety profile.12,13,14 Therefore, we sought to engineer an AAV variant for highly efficient gene delivery to glia.
AAV's single-stranded DNA genome has two open reading frames, including cap, which encodes for the structural proteins VP1-3 that assemble into the icosahedral viral capsid.15 AAV vectors based on the capsids of the natural variants16 mediate efficient gene delivery in a range of tissues.7,11,17 However, within the brain and retina, natural AAV variants predominantly exhibit neuronal tropism,7,9,18,19,20 potentially due to inefficient uptake by astrocytes.21 Some level of astrocyte transduction in vivo has been reported for AAV4 and AAV5,7,8,18,22 though AAV4 transduces astrocytes only within the subventricular zone, and reports of AAV5 transduction of astrocytes have been highly variable, potentially due to differences in vector production and promoter usage.19,23 Recently, AAV9 has been shown to transduce astrocytes via intravenous injection, though expression was observed in other tissues including heart and muscle.24
Engineering the AAV capsid for targeted delivery based on the rational insertion of defined peptide sequences into the capsid has enjoyed some success.25,26,27,28 As a complementary approach, we recently reported that directed evolution can successfully create AAV vectors with novel gene delivery properties,29 and this approach has since been extended to generate highly chimeric AAV variants by family shuffling.30,31,32,33 Here, we successfully merged numerous AAV library strategies to engineer novel AAV vectors for CNS and retinal gene therapy.
A panel of highly diverse (>107 members each) AAV libraries—generated by random mutagenesis, DNA shuffling, AAV peptide display, and a new semi-random loop replacement method—were selected via multiple evolutionary cycles, or genetic diversifications followed by selection on primary human astrocytes. In addition to enhanced properties in vitro, several resulting variants efficiently transduced astrocytes at levels up to 5.5-fold greater than the parental serotypes in the rat striatum, and one vector additionally exhibited enhanced tropism for Müller cells in the retina. These novel vectors further demonstrate the utility of directed evolution for creating novel viruses and may enhance gene therapies for the treatment of numerous neurological disorders.
Results
Barriers in AAV gene delivery to astrocytes
We first determined the relative gene delivery efficiencies of seven AAV serotypes (1, 2, 4–6, 8, and 9) on primary human astrocytes (Figure 1). Iodixanol-purified, high-titer recombinant AAV (rAAV) cytomegalovirus (CMV) –green fluorescent protein (GFP) vectors30 were generated, and we found that the transduction efficiencies of the serotypes varied considerably, over a 500-fold range, with the following order: AAV2 > 1 = 6 = 9 > 5 > 4 > 8. Because high genomic multiplicity of infections (>104) were required to transduce >50% of the astrocytes even for the more efficient natural serotypes, and because all AAV variants are reportedly inefficient on glia in vivo,7,9,18,19,20 we next probed potentially limiting steps in AAV gene delivery to human astrocytes.
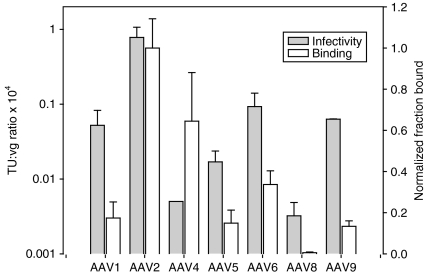
Figure 1.
Characterization of natural AAV serotypes. rAAV CMV-GFP vector transduction efficiencies (gray bars) on primary human astrocytes (gMOI = 103–105) using seven AAV serotypes (1, 4–6, 8, and 9) varied significantly (n = 3, error bars represent SD). Binding levels (white bars) of each serotype to primary human astrocytes matched the relative transduction efficiencies, except for surprisingly high binding levels of AAV4, which transduced primary human astrocytes poorly (n = 3, error bars represent SD). AAV, adeno-associated virus; CMV, cytomegalovirus; GFP, green fluorescent protein; gMOI, genomic multiplicity of infection.
We first quantified the relative-binding ability of several viral serotypes to astrocytes (Figure 1). Intriguingly, the trend in viral binding (AAV2 > 4 > 6 > 1 = 5 = 9 > 8) closely matched that of viral transduction, except that AAV4 which bound almost as well as AAV2, was extremely poor in transducing astrocytes. The low transduction efficiency observed for AAV4 is thus due to a later step in viral infection, such as internalization or inefficient unpackaging.34 Interestingly, treatment of astrocytes with the proteasome inhibitor MG132 or the genotoxic agent hydroxyurea—which reportedly overcome barriers in vector degradation and genome processing in some systems such as lung epithelium35—resulted in only modest (<1.5-fold) increases in GFP+ cells for all serotypes (data not shown). The data indicate that vector engineering may be required to achieve efficient transduction of astrocytes because even serotypes that bind efficiently to astrocytes (AAV2 and 4) fail to achieve high levels of gene delivery (Figure 1).
Library construction
We conducted directed evolution of the AAV capsid to enhance astrocyte gene delivery using a panel of highly diverse (>107) viral libraries: an AAV2 random mutagenesis library (AAV2-EP),29 a chimeric AAV library (ShH),30 and an AAV2 peptide display library (AAV2 7mer).27,28 In addition, we generated new AAV2-based libraries via a bioinformatics-based surface loop replacement strategy. Previously, peptide-based AAV2 capsid engineering studies have predominantly manipulated the surface loop containing the heparan sulfate proteoglycan (HSPG)–binding site (i.e., ~585RGNR~);25,26,27,28 however, recent studies of the AAV2 capsid have highlighted several other regions that significantly influence the transduction properties of AAV2.30,31,36 Therefore, we chose to engineer four important surface loops37 (Table 1 and Figure 2).
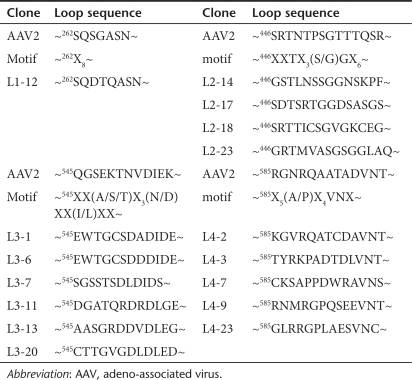
Table 1.
Summary of loop regions within clones selected from AAV random loop libraries after a single round of evolution
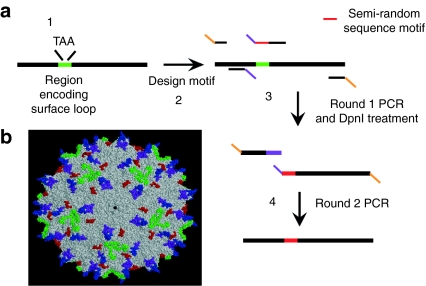
Figure 2.
Design of novel loop replacement library. (a) Library design consisted of introduction of a stop codon within the target loop sequence (1), semi-random motif design (2), an initial PCR round to amplify two overlapping fragments (3), and a final PCR to assemble the full-length cap gene containing the semi-random motif (4). (b) Four separate AAV2 libraries containing semi-random loops [L1, ~262SQSGASN~ (purple); L2, ~446SRTNTPSGTTTQSR~ (blue); L3, ~545QGSEKTNVDIEK~ (red); and L4, ~585RGNRQAATADVNT~ (green)] were constructed, with sequences determined through a bioinformatics analysis of AAV VP1 protein sequences. AAV, adeno-associated virus.
We replaced the wild-type (wt) AAV2 sequence with a semi-random sequence determined by bioinformatics analysis of 130 AAV cap genes obtained from the GenBank database.38 These loops include L1 (~262SQSGASN~, purple in Figure 2b), L2 (~446SRTNTPSGTTTQSR~, blue in Figure 2b), L3 (~545QGSEKTNVDIEK~, red in Figure 2b), and L4 (~585RGNRQAATADVNT~, green in Figure 2b). Specifically, the VP1 protein sequences were aligned to identify positions within these loops highly conserved either in amino acid identity or in chemical properties (e.g., charge, polarity, hydrophobicity, etc.). For each loop, residues that were conserved in >90% of sequences were fixed, whereas amino acids whose identity was limited to one of several residues in >90% of the sequences were encoded with a restricted codon (e.g., RGC for serine and glycine). Finally, all other residues were replaced with a random amino acid, encoded by an NNK codon. The resulting semi-random sequences (Table 1) were introduced into the AAV2 cap gene using spliced overlap extension PCR. This library, which builds upon work conducted with the HSPG-binding site,25,26,27,28 is the first to explore and exploit the modular nature of numerous additional surface loops and to engineer highly diverse libraries guided by bioinformatic analysis of many (>100) serotypes.
Selection and in vitro characterization of successful viral variants
One round of evolution consisted of initial library diversification followed by four or five selection steps. An in vitro selection approach with primary astrocytes from the adult human cerebral cortex was implemented. Briefly, 5 × 105 primary human astrocytes were infected with iodixanol-purified, replication-competent AAV libraries29,30 at a genomic multiplicity of infection of 103, and successful virions were amplified by the later addition of wt adenovirus type 5 to complete one round of selection. After five rounds of selection, preliminary characterization of individual rAAV CMV-GFP generated with capsid clones from the AAV2 EP, AAV2 7mer, and chimeric AAV libraries failed to reveal variants with enhanced gene delivery (Supplementary Table S1, Supplementary Figure S1, and data not shown). Therefore, the cap genes from the AAV2 libraries (EP and 7mer) and the chimeric AAV library were pooled and further diversified using DNA shuffling and random mutagenesis. The resulting library was then subjected to four additional selection steps. Subsequent analysis of the mutants selected in this second round of evolution revealed several clones related to AAV2 and/or AAV6 that exhibited enhanced transduction of primary human astrocytes (two- to fivefold higher than wtAAV2 and tenfold higher than wtAAV6) (Figure 3a, Supplementary Figures S2 and S3). ShH13 was closely related to AAV6, whereas the others were based on AAV2 (Supplementary Figure S2). Some variants (ShH7, ShH14, ShH21, and 2H22) differ from the parent serotype by up to five mutations, whereas several variants (ShH13, ShH18, ShH19, and ShH22) are viral chimeras with capsid regions from several serotypes swapped into either AAV2 (ShH18, ShH19, and ShH22) or AAV6 (ShH13) scaffolds. Interestingly, no variants from the AAV2 7mer were found after the second evolutionary cycle.
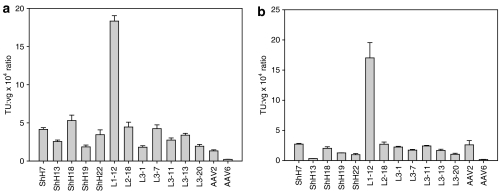
Figure 3.
Enhanced transduction of primary human astrocytes by novel AAV variants. (a) Transduction of primary human astrocytes by rAAV CMV-GFP vectors with capsids selected from the AAV EP, chimera, and random loop libraries demonstrated enhanced gene delivery by numerous viral variants (n = 3, error bars represent SD). (b) Transduction of SHSY-5Y neuroblastoma cells with rAAV CMV-GFP vectors revealed a decrease in transduction for most EP, chimera, and random loop variants, except for L1-12 (n = 3, error bars represent SD). AAV, adeno-associated virus; CMV, cytomegalovirus; GFP, green fluorescent protein.
Clones isolated after a single round of evolution with the loop replacement libraries contained new L1, L2, or L3 regions and were more efficient on astrocytes (2- to 15-fold) relative to wtAAV2 (Figure 3a, Supplementary Figure S3, and Table 1). Furthermore, most of these new functional loops possessed very low sequence similarity to wtAAV2, demonstrating that these regions of the AAV2 capsid are highly modular. Intriguingly, some variants (L2-17, L3-6, L4-7, L4-9, and L4-23) exhibited considerable reductions in viral infectivity (more than tenfold), potentially due to the loss of important complementary interactions between some residues within these loops and structurally adjacent regions of the capsid.
To characterize the gene delivery properties of the selected variants further, we analyzed their relative transduction efficiencies on SHSY-5Y (a human neuroblastoma cell line). Interestingly, efficiencies for most of the viral variants were significantly less (two- to fivefold) than that of wtAAV2 (Figure 3b), perhaps implying potential astrocytic selectivity compared to AAV2's strong neuronal tropism.7,21 Gene delivery efficiencies of these novel variants varied on other glial cells, including rat astrocytes (where ShH19 exhibited highest transduction levels) and U87MG glioblastoma cells (Supplementary Figure S4). Importantly, all of the AAV2-based variants exhibited HSPG dependencies similar to wtAAV2, except for two (L1-12 and L3-7) that were more dependent on HSPG than wtAAV2 (Supplementary Figure S5). Therefore, the enhanced transduction efficiencies of these AAV2-like mutants are likely due to improvements in either secondary receptor interactions or later stages of viral infection.
In vivo gene delivery of selected viral mutants
To further demonstrate the potential of these novel variants for in vivo gene delivery to astrocytes, we generated double-stranded (ds) AAV vectors expressing GFP under the control of the hybrid CAG (CMV early enhancer/chicken β-actin) promoter, which has been shown to mediate more robust gene expression within the CNS, and in particular within astrocytes, than the CMV promoter.10,18,19 Capsids from the most efficient human astrocyte variant (L1-12), the most efficient rat astrocyte variant (ShH19, Supplementary Figure S4), the AAV6-like variant (ShH13), and the corresponding control parent serotypes AAV2 and AAV6 were analyzed. High-titer rAAV dsCAG-GFP vectors were purified via the same method (iodixanol gradient ultracentrifugation followed by heparin affinity chromatography), to avoid differences in vector purity that have been implicated in altering astrocyte transduction in vivo.19
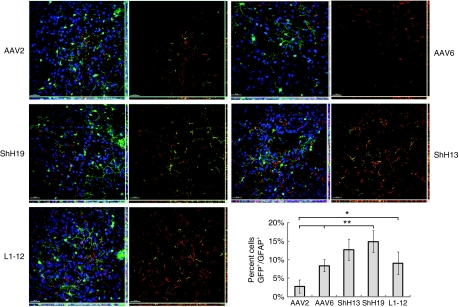
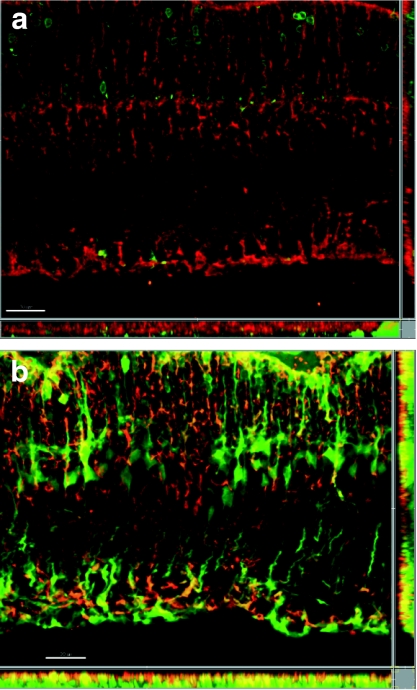
A volume of 3 µl DNase-resistant genomic particles (5 × 108 vg) were stereotactically injected into striatum of female rats, and all AAV variants and control serotypes exhibited robust GFP expression 3 weeks after injection (Figure 4). For all variants and serotypes, >99.5% of the GFP+ cells were either NeuN+ or GFAP+, with minimal transduction of other cell types, such as microglia (data not shown). Similar to previous reports utilizing the CAG promoter,18,23 wtAAV2 transduced astrocytes infrequently (2.7 ± 1.8%), whereas wtAAV6 mediated somewhat higher gene delivery to astrocytes (8.4 ± 1.6%). ShH13 transduction of astrocytes was not significantly higher than wtAAV6. In contrast, the AAV2-like variants ShH19 and L1-12 transduced 5.5-fold (14.9 ± 3.0%) and 3.3-fold (9.0 ± 3.0%) greater numbers of astrocytes, respectively, compared to wtAAV2. Furthermore, these enhanced transduction levels may further increase when utilized in a human, as the selections were performed with primary human cells. To explore further the utility of these variants in glia in vivo, we injected recombinant variants into the subretinal space in rat eyes to observe their efficiency in infecting a glial subtype within the retina, Müller cells. Three weeks after injection, observation of GFP expression revealed that ShH13 exhibited the most pronounced shift in tropism toward glia, with expression localized predominantly to Müller cells and retinal pigment epithelium, while its parent vector AAV6 showed much lower GFP expression and primarily within photoreceptors and the retinal pigment epithelium (Figure 5).
Figure 4.
Enhanced in vivo astrocyte tropism of evolved AAV variants. Representative images of coronal brain sections 3 weeks after injection with dsCAG-GFP vectors using capsids from AAV2, AAV6, ShH13, ShH19, and L1-12 show robust levels of GFP expression (green). Representative images (original magnification ×25) of cell types reveal enhanced astrocyte transduction levels by ShH19 (P < 0.01 compared to AAV2/6), and L1-12 (P < 0.02 compared to AAV2) compared to the parent AAV2 and AAV6 vectors: GFP+ cells (green), NeuN+ neurons (blue), and GFAP+ astrocytes (red). For each variant, the image on the right indicates areas of colocalization between GFP and GFAP (yellow). The corresponding x-z and y-z planes are shown on the bottom and on the right side, respectively, for each image. Bar = 50 µm. Quantified percentage of astrocytes transduced (GFP+/GFAP+) within the total GFP+ cell population (n = 4, error bars represent SD). For each vector, over 950 GFP+ cells were counted and analyzed for costaining with either NeuN (neuron) or GFAP (astrocytes). AAV, adeno-associated virus; GFP, green fluorescent protein.
Figure 5.
Enhanced transduction of retinal Müller glia in vivo by ShH13. Representative images (original magnification ×25) of subretinal injections with dsCAG-GFP vectors with capsids from ShH13 and AAV6 reveal a pronounced shift in viral tropism toward Müller glia in the ShH13 variant compared to the parent AAV6 vector: GFP expression (green), overlay with GS+ Müller glia (red), isolation of areas of colocalization (yellow). The corresponding x-z and y-z planes are shown on the bottom and on the right side, respectively, for each image. Bar = 30 µm. (a) AAV6 expression is predominantly seen in photoreceptors and RPE (not shown), whereas (b) ShH13 shows robust expression in Müller glia and RPE (not shown). AAV, adeno-assocaited virus; RPE, retinal pigment epithelium.
Discussion
Glia are a potentially important therapeutic target within the CNS and retina, either to counteract disease pathology directly involving these cells or to harness them to secrete therapeutic proteins and thereby protect neighboring neurons from injury or disease. Most current AAV serotypes fail to efficiently transduce astrocytes or Müller glia in vivo, and even serotypes such as AAV4 and AAV5 that can infect astrocytes either transduce only a subset within a specific brain region (the subventricular zone for AAV4)8 or exhibit variable efficiencies (AAV5).7,18 Additionally, recent work in the retina utilizing AAV vectors that are more resistant to proteasome-mediated degradation resulted in greatly improved efficiencies, yet still relatively low gene delivery to Müller glia.20 Accordingly, we have employed directed evolution to engineer a novel AAV vector capable of efficient gene delivery to astrocytes and potentially other glia cells. Furthermore, while the selections were performed on human cell in vitro, the resulting variants exhibited enhanced infection of astrocytes upon injection into the rat striatum (ShH19 and L1-12) (Figure 4) or Müller glia within the rat retina (ShH13) (Figure 5), making them attractive candidates for future studies in disease models. These results suggest that directed evolution coupled with stringent selections can engineer novel capsid variants that can effectively transduce related cells across multiple species.
Barriers to efficient AAV gene delivery—such as poor binding, proteasome degradation, and genome processing—vary greatly among cell types.25,27,28,35 Our results indicate that while efficient cell binding is likely one such barrier present within astrocytes, downstream barriers may also play a significant role as evidenced by the binding results for AAV4 (Figure 1) and the improved efficiencies of numerous AAV2- and AAV6-based variants with similar HSPG dependencies as their parents (Supplementary Figure S5). Of particular note, a previous study using fluorescently labeled AAV2 failed to detect any significant viral uptake in astrocytes,21 reflecting poor internalization of AAV2, potentially due to inefficient interactions with secondary protein receptors that facilitate receptor-mediated endocytosis.36 Importantly, while a lack of detailed mechanistic knowledge of specific gene delivery barriers in this and other cases can preclude rational design strategies to improve vector properties, a broader “black box” approach to engineer efficient AAV vectors through random diversification and high-throughput selection can still succeed.
Recent efforts in AAV capsid engineering30,31,39 have suggested that certain loop regions may be highly modular and may therefore enable functional loop swapping methods among various serotypes. Based on these observations, we developed a new semi-random loop replacement strategy for the engineering of novel vectors. Strikingly, after four rounds of selection, many of these variants, which contained new loop sequences of up to 14 amino acids in length, demonstrated enhanced levels of gene delivery (Figure 3a). For example, replacement of loop 2 (L1, ~262SQSGASN~) of wtAAV2 (shown in purple in Figure 2b) with an octamer sequence, analogous to the size of wtAAV6, yielded clones with improved gene delivery efficiencies on a panel of cell types (Figure 3, Supplementary Figures S4 and S5). This loop—whose length in natural serotypes varies from four (AAV4, AAV11, and AAV12) to eight (AAV1, AAV5, AAV6, and AAV9) amino acids—modulates the tropism of AAV vector, potentially via interactions with secondary protein receptors.30,36
Our loop-engineering strategy focused on identifying residues that are conserved among the serotypes. However, it does not detect amino acid positions that may have coevolved with other positions within a given serotype, due for example to physical contact between a loop and another region in the tertiary or quaternary structure of the capsid, but whose identity may not be strictly conserved from serotype to serotype. For instance, this approach initially considered amino acid 553 within L3 as random; however, all six selected clones retained the aspartic acid of wtAAV2 at this location. Within the AAV2 capsid structure,37 D553 (within L3) forms an ionic interaction with R459 (within L2), which may help stabilize these two capsid loops and ensure proper capsid assembly. Similar analysis of the selected L2 sequences revealed the retention of a hydrogen bond acceptor (serine) or basic residue (arginine) at the randomized amino acid position 447 (R for wtAAV2), which interacts with both E499 and N551. Future refinement in this approach may incorporate information regarding key structural contacts37,40,41 to aid the engineering of novel AAV vectors with customized gene delivery properties.
Analysis of the novel AAV2 and AAV6 variants yields several interesting insights. Within the AAV2-like clones, mutations N496H, located at the apex of the threefold axis of symmetry, and Q598L, located within the crater at the threefold axis, may play a role in the enhanced transduction (Figure 3, Supplementary Figures S2 and S3). These sites may thus represent new targets for vector engineering. Interestingly, none of the mutations in the AAV6-based mutant ShH13 is predicted to be surface-exposed, and these changes may thus indirectly alter receptor binding via modulating the surface structure of the viral capsid, or alter later infection steps such as intracellular trafficking or vector unpackaging. For example, one such interior mutation (F129L) lies within the VP1-unique region but does not overlap with previously identified functional domains within VP1.42 In a recent directed evolution study, we demonstrated that changes within this region can significantly enhance the gene delivery properties of the evolved variant, potentially due to changes in endosomal escape of the virus.33 Finally, the most efficient AAV variants in vivo (ShH19 and L1-12) acquired very similar loop 2 sequences (SQDTQASN and SASTGASN for L1-12 and ShH19, respectively, compared to SQSGASN for wtAAV2, shown in purple in Figure 2b) through distinct methods (i.e., shuffling for ShH19 and loop replacement for L1-12).30,36 Together, these results highlight how exploring complementary yet distinct libraries can reveal key common features (i.e., capsid regions) that may be exploited in future directed evolution experiments.30
The ability of ShH13 to transduce two glia cell types, astrocytes and Müller cells, suggests a common mechanism for the improved gene delivery. Müller glia possess apical-basal polarity, and many astrocytes also retain some features of this polarity.43 In epithelial cells, cell polarity has previously been shown to result in inefficient trafficking of AAV to the nucleus,35 and an analogous nonproductive infection may occur within some glia. ShH13, which contains only mutations on the interior of the capsid and within the VP1-unique region, may exhibit enhanced transport through the cytoplasm through altered PLA2 activity.33,42 Alternatively, the variant could utilize a receptor more highly expressed in astrocytes and Müller cells relative to the surrounding neuronal cells, such as epidermal growth factor receptor or fibroblast growth factor receptor 3.44,45
In summary, by employing directed evolution with a diverse array of novel AAV libraries including a new peptide loop replacement library, we engineered novel AAV vectors capable of highly efficient delivery to astrocytes in vitro and importantly to astrocytes and Müller glia in vivo. These AAV capsids in conjunction with cell-specific promoters, such as GFAP, and other recently identified capsid mutations20 may yield further enhancements in the targeted gene delivery to astrocytes and Müller glia to treat a variety of genetic diseases. For example, modulating mutant superoxide dismutase-1 expression within astrocytes may reduce neuronal cell death in amyotrophic lateral sclerosis,5 or therapeutic gene delivery may prevent the accumulation of amyloid β-plaques in Alzheimer's disease.4 Furthermore, because Müller cells span the entire retina and surround every class of neuron present within this tissue, transduction of Müller cells would permit spread of neurotrophic factors throughout all layers of this tissue to significantly augment existing therapies for retinitis pigmentosa, age-related macular degeneration and neovascularization.12,13,14,46 These novel capsids may thus endow AAV vectors with the capacity to treat a variety of neurodegenerative disorders in disease models and ultimately in the clinic.
Materials and Methods
Cell lines and viral production. Cell lines were cultured at 37 °C and 5% CO2, and unless otherwise mentioned were obtained from the American type culture collection (Manassas, VA). Primary human astrocytes from the cerebral cortex (ScienCell, San Diego, CA) were cultured according to the manufacturer's instructions. AAV293 cells (Stratagene, La Jolla, CA), rat astrocytes (primary cells derived from adult rat brain), and the human glioblastoma cell line U87MG were cultured in Dulbecco's modified Eagle's medium. SHSY-5Y, a human neuroblastoma cell line, was cultured in minimum essential medium, alpha modification (αMEM) (Sigma-Aldrich, St Louis, MO). All media were supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and 1% penicillin/streptomycin (Invitrogen).
Library generation and viral production. Random mutagenesis libraries were generated by subjecting cap genes from AAV2 to error-prone PCR using CAP For and CAP Rev as forward and reverse primers, respectively, as previously described.29 All primer sequences are provided in Supplementary Table S2. Peptide display libraries were generated similar to previous reports.27,28 Briefly, a unique AvrII was introduced into pSub2Cap2 between amino acid 587 and 588 by PCR mutagenesis. A random 21 nucleotide insert, 7mer For, was used to synthesize dsDNA inserts, along with the antisense primer 7mer Rev. The resulting dsDNA inserts were cloned into the AvrII site of pSub2Cap2 after digestion with NheI. A previously generated AAV library constructed by DNA shuffling of cap genes from AAV1, 2, 4–6, 8, and 9 was also used.30
The novel random loop replacement libraries were constructed as follows (Figure 2). First, sequences of 130 AAV cap genes were obtained from the GenBank database38 and aligned with BioEdit (www.mbio.ncsu.edu/BioEdit/bioedit.html). Positions within each wtAAV2 loop (Table 1) were then analyzed for several characteristics: amino acid identity, charge, polarity, and hydrophobicity. Any feature contained within >90% of the sequences was then encoded into the new AAV2 loop sequence using defined or restricted codons. All other positions were then replaced by a random codon (NNK). Next, site-directed mutagenesis was used to introduce a stop codon (TAA) into each of the loops of interest within the AAV2 cap gene to generate four template plasmids and prevent carryover of wtAAV2 sequences into the final viral library. Spliced overlap extension was performed using two partial cap products: P1, which contains fully wtAAV2 sequence (using flanking forward and loop reverse primers), and P2, which contains a wtAAV2 anchor sequence to anneal with the 3′-end of P1 followed by the semi-randomized loop sequence and the remainder of the wtAAV2 sequence (using loop forward and flanking reverse primers). The resulting products were gel purified, and various molar ratios were used as templates for a second PCR using the flanking primers. Primers used for the mutagenesis are provided in Supplementary Table S2. Finally, the resulting cap genes were cloned into pSub2 for replication competent AAV production.29
The rcAAV libraries and rAAV vectors expressing GFP under a CMV promoter were packaged as previously described.29,30 DNase-resistant genomic titers were obtained via quantitative PCR, and transduction titers were obtained by flow cytometry.29,30
Library selection and evolution. Primary human astrocytes (5 × 105) were infected with each viral library at a genomic multiplicity of infection of 103. Twelve hours after infection, the media were changed, and cells were infected with wt adenovirus to amplify successful AAV variants. Forty-eight hours later, cells were harvested and lysed to yield the viral pool for the next selection round. After four such rounds of selection, successful cap genes were recovered by PCR. The resulting pool of cap genes was subjected to DNA shuffling and EP PCR to generate a new library for the next selection rounds.29,30 In total, the AAV2 EP/7mer library and chimeric AAV library were subjected to two evolutionary cycles (i.e., mutagenesis plus selection steps), and the random loop libraries were subjected to one evolutionary cycle.
In vitro transduction and cell-binding analysis. Transduction studies using rAAV CMV-GFP were performed with 5 × 104 cells (primary human astrocytes, rat astrocytes, U87MG, CHO, pgsD, and SHSY-5Y) in 12-well plates. Cells were transduced with rAAV GFP vectors at a genomic multiplicity of infection of 103–105 (n = 3), and the percentage of GFP-expressing cells was determined by flow cytometry 48 hours after infection. Viral-binding assays were performed as previously described.33
In vivo characterization. ds rAAV vectors expressing GFP under a CAG promoter—composed of the CMV enhancer, chicken β-actin promoter, and rabbit β-globin intron10—were packaged with capsids from AAV2, AAV6, ShH13, ShH19, and L1-12 and purified using iodixanol gradient ultracentrifugation followed by heparin column purification as previously described.29 rAAV vectors were stereotaxically injected into the striatum of the rat brain (AP, +0.2; ML, ±3.5; DV, −4.5 from skull) of adult female Fischer 344 rats (100 g, 6 weeks). The animals were anesthetized with ketamine (90 mg/kg animal) and xylazine (10 mg/kg animal) prior to injection, and 3 µl of high-titer AAV vectors (5 × 108 vg/µl) were injected using Hamilton syringe as described.47 After 3 weeks, animals (n = 4 per vector) were transcardially perfused with 4% paraformaldehyde in phosphate- buffered saline, and the retrieved brains were postfixed by immersing in 4% paraformaldehyde overnight at 4 °C, with subsequently stored in 30% sucrose for cryoprotection before sectioning.
Coronal sections (40 µm) were cut using a freezing, sliding microtome. Cell phenotype was identified by the primary antibodies mouse-anti NeuN (1:200; Chemicon, Billerica, MA) and guinea pig-anti GFAP (1:1,000; Advanced ImmunoChemical, Long Beach, CA), and GFP expression was amplified using primary rabbit anti-GFP (1:2,000; Invitrogen). Corresponding secondary antibodies (labeled with AlexaFluor 488, 546, and 633) were used for detection. For nucleus staining, some sections were counterstained using TO-PRO-3 (1:2,000; Invitrogen). Animal protocols were approved by the UCB Animal Care and Use Committee and conducted in accordance with National Institutes of Health guidelines.
rAAV6 and ShH13 were also injected subretinally into adult wt Sprague Dawley rats. Before vector administration, rats were anesthetized with ketamine (72 mg/kg) and xylazine (64 mg/kg) by intraperitoneal injection. An ultrafine 30½-gauge disposable needle was inserted through the sclera to create an entryway for a Hamilton syringe for subsequent injection of 3 µl of high-titer AAV vector (1× 1012 vg/ml) between the retinal pigment epithelium and photoreceptor layer. Three weeks after vector injection, the eyes were enucleated, a hole was made in the cornea, and eyes were fixed in 10% neutral buffered formalin for 2–3 hours. The cornea and lens were removed, and the resulting eyecups were stored in 30% sucrose overnight. Eyes were then embedded in optimal cutting temperature embedding compound (OCT; Miles Diagnostics, Elkhart, IN) and oriented for making 5–10-µm thick transverse retinal sections using a freezing, sliding microtome. Müller glia were visualized using the primary antibody rabbit anti-glutamine synthetase (1:2,500; Sigma, St Louis, MO), and the corresponding Cy3-conjugated secondary antibody was used for detection. All procedures were handled according to the ARVO Statement for the Use of Animals and approved by the UCB Animal Care and Use Committee.
SUPPLEMENTARY MATERIALFigure S1. Transduction efficiencies of AAV2 variants from the random heptamer library after a single round of evolution. Transduction of primary human astrocytes with the rAAV CMV-GFP vectors with capsids selected from the AAV2 random heptamer library demonstrated only modest (<1.5-fold) first round improvements in viral transduction (n = 3, error bars represent SD), highlighting the need in this case for multiple rounds of evolution.Figure S2. Primary amino acid sequence alignment of AAV variants from chimeric AAV and AAV EP libraries along with parent serotypes. Distinct colors were used to shade each amino acid.Figure S3. Transduction efficiencies of other AAV variants on primary human astrocytes (n = 3, error bars represent SD).Figure S4. Transduction efficiencies of AAV variants on other glial cell lines. Tranduction of rat astrocytes with rAAV CMV-GFP clones from the both the chimeric (A) and random loop libraries (B) revealed significantly different trends compared to transduction of primary human astrocytes (n = 3, error bars represent SD). Tranduction of U87MG glioblastoma cells with rAAV CMV-GFP clones from the both the chimeric (C) and random loop libraries (D) revealed significantly different trends compared to transduction of primary human astrocytes (n = 3 and error bars represent SD).Figure S5. Heparan sulfate dependence of AAV variants. Transduction of parent CHO cell line and a CHO mutant deficient in heparan sulfate proteoglycan (HSPG) (psgD) demonstrated HSPG dependence of all AAV2-like variants from the chimeric (A) and random loop libraries (B) (n = 3, error bars represent SD).Table S1. Summary of peptides within clones selected from AAV2 7mer library after a single round of evolution.Table S2. Oligonucleotide sequences for construction of viral libraries.
Supplementary Material
Transduction efficiencies of AAV2 variants from the random heptamer library after a single round of evolution. Transduction of primary human astrocytes with the rAAV CMV-GFP vectors with capsids selected from the AAV2 random heptamer library demonstrated only modest (<1.5-fold) first round improvements in viral transduction (n = 3, error bars represent SD), highlighting the need in this case for multiple rounds of evolution.
Primary amino acid sequence alignment of AAV variants from chimeric AAV and AAV EP libraries along with parent serotypes. Distinct colors were used to shade each amino acid.
Transduction efficiencies of other AAV variants on primary human astrocytes (n = 3, error bars represent SD).
Transduction efficiencies of AAV variants on other glial cell lines. Tranduction of rat astrocytes with rAAV CMV-GFP clones from the both the chimeric (A) and random loop libraries (B) revealed significantly different trends compared to transduction of primary human astrocytes (n = 3, error bars represent SD). Tranduction of U87MG glioblastoma cells with rAAV CMV-GFP clones from the both the chimeric (C) and random loop libraries (D) revealed significantly different trends compared to transduction of primary human astrocytes (n = 3 and error bars represent SD).
Heparan sulfate dependence of AAV variants. Transduction of parent CHO cell line and a CHO mutant deficient in heparan sulfate proteoglycan (HSPG) (psgD) demonstrated HSPG dependence of all AAV2-like variants from the chimeric (A) and random loop libraries (B) (n = 3, error bars represent SD).
Summary of peptides within clones selected from AAV2 7mer library after a single round of evolution.
Oligonucleotide sequences for construction of viral libraries.
Acknowledgments
We thank Smita Agrawal for providing the rat astrocytes and Karen Guerin and Kate Kolstad for technical assistance with the retinal injections. This work was supported by Alzheimer's Association award NIRG-05-13529, NIH R21EY016994, NIH T32 GM08352, and a NSF graduate fellowship (to J.T.K.).
REFERENCES
- Volterra A., and , Meldolesi J. Astrocytes, from brain glue to communication elements: the revolution continues. Nat Rev Neurosci. 2005;6:626–640. doi: 10.1038/nrn1722. [DOI] [PubMed] [Google Scholar]
- Nedergaard M, Ransom B., and , Goldman SA. New roles for astrocytes: redefining the functional architecture of the brain. Trends Neurosci. 2003;26:523–530. doi: 10.1016/j.tins.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Sherwood CC, Stimpson CD, Raghanti MA, Wildman DE, Uddin M, Grossman LI, et al. Evolution of increased glia-neuron ratios in the human frontal cortex. Proc Natl Acad Sci USA. 2006;103:13606–13611. doi: 10.1073/pnas.0605843103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagele RG, Wegiel J, Venkataraman V, Imaki H., and , Wang KC. Contribution of glial cells to the development of amyloid plaques in Alzheimer's disease. Neurobiol Aging. 2004;25:663–674. doi: 10.1016/j.neurobiolaging.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw B, Su M, ter Horst M, Iwata S, Rodijk M, Hoeben RC, et al. Increased glia-specific transgene expression with glial fibrillary acidic protein promoters containing multiple enhancer elements. J Neurosci Res. 2006;83:744–753. doi: 10.1002/jnr.20776. [DOI] [PubMed] [Google Scholar]
- Davidson BL, Stein CS, Heth JA, Martins I, Kotin RM, Derksen TA, et al. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc Natl Acad Sci USA. 2000;97:3428–3432. doi: 10.1073/pnas.050581197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Martins IH, Chiorini JA., and , Davidson BL. Adeno-associated virus type 4 (AAV4) targets ependyma and astrocytes in the subventricular zone and RMS. Gene Ther. 2005;12:1503–1508. doi: 10.1038/sj.gt.3302554. [DOI] [PubMed] [Google Scholar]
- Allocca M, Mussolino C, Garcia-Hoyos M, Sanges D, Iodice C, Petrillo M, et al. Novel adeno-associated virus serotypes efficiently transduce murine photoreceptors. J Virol. 2007;81:11372–11380. doi: 10.1128/JVI.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimons HL, Bland RJ., and , During MJ. Promoters and regulatory elements that improve adeno-associated virus transgene expression in the brain. Methods. 2002;28:227–236. doi: 10.1016/s1046-2023(02)00227-x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhu T, Qiao C, Zhou L, Wang B, Zhang J, et al. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat Biotechnol. 2005;23:321–328. doi: 10.1038/nbt1073. [DOI] [PubMed] [Google Scholar]
- Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- Cideciyan AV, Aleman TS, Boye SL, Schwartz SB, Kaushal S, Roman AJ, et al. Human gene therapy for RPE65 isomerase deficiency activates the retinoid cycle of vision but with slow rod kinetics. Proc Natl Acad Sci USA. 2008;105:15112–15117. doi: 10.1073/pnas.0807027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire AM, Simonelli F, Pierce EA, Pugh EN Jr, Mingozzi F, Bennicelli J, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Lusby EW., and , Berns KI. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, et al. Clades of adeno-associated viruses are widely disseminated in human tissues. J Virol. 2004;78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplitt MG, Leone P, Samulski RJ, Xiao X, Pfaff DW, O'Malley KL, et al. Long-term gene expression and phenotypic correction using adeno-associated virus vectors in the mammalian brain. Nat Genet. 1994;8:148–154. doi: 10.1038/ng1094-148. [DOI] [PubMed] [Google Scholar]
- Harding TC, Dickinson PJ, Roberts BN, Yendluri S, Gonzalez-Edick M, Lecouteur RA, et al. Enhanced gene transfer efficiency in the murine striatum and an orthotopic glioblastoma tumor model, using AAV-7- and AAV-8-pseudotyped vectors. Hum Gene Ther. 2006;17:807–820. doi: 10.1089/hum.2006.17.807. [DOI] [PubMed] [Google Scholar]
- Klein RL, Dayton RD, Tatom JB, Henderson KM., and , Henning PP. AAV8, 9, Rh10, Rh43 vector gene transfer in the rat brain: effects of serotype, promoter and purification method. Mol Ther. 2008;16:89–96. doi: 10.1038/sj.mt.6300331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrs-Silva H, Dinculescu A, Li Q, Min SH, Chiodo V, Pang JJ, et al. High-efficiency transduction of the mouse retina by tyrosine-mutant AAV serotype vectors. Mol Ther. 2009;17:463–471. doi: 10.1038/mt.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett JS, Samulski RJ., and , McCown TJ. Selective and rapid uptake of adeno-associated virus type 2 in brain. Hum Gene Ther. 1998;9:1181–1186. doi: 10.1089/hum.1998.9.8-1181. [DOI] [PubMed] [Google Scholar]
- Taymans JM, Vandenberghe LH, Haute CV, Thiry I, Deroose CM, Mortelmans L, et al. Comparative analysis of adeno-associated viral vector serotypes 1, 2, 5, 7, and 8 in mouse brain. Hum Gene Ther. 2007;18:195–206. doi: 10.1089/hum.2006.178. [DOI] [PubMed] [Google Scholar]
- Peel AL., and , Klein RL. Adeno-associated virus vectors: activity and applications in the CNS. J Neurosci Methods. 2000;98:95–104. doi: 10.1016/s0165-0270(00)00183-7. [DOI] [PubMed] [Google Scholar]
- Foust KD, Nurre E, Montgomery CL, Hernandez A, Chan CM., and , Kaspar BK. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod A, Ried M, Wobus C, Lahm H, Leike K, Kleinschmidt J, et al. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat Med. 1999;5:1052–1056. doi: 10.1038/12491. [DOI] [PubMed] [Google Scholar]
- Shi W, Arnold GS., and , Bartlett JS. Insertional mutagenesis of the adeno-associated virus type 2 (AAV2) capsid gene and generation of AAV2 vectors targeted to alternative cell-surface receptors. Hum Gene Ther. 2001;12:1697–1711. doi: 10.1089/104303401750476212. [DOI] [PubMed] [Google Scholar]
- Perabo L, Büning H, Kofler DM, Ried MU, Girod A, Wendtner CM, et al. In vitro selection of viral vectors with modified tropism: the adeno-associated virus display. Mol Ther. 2003;8:151–157. doi: 10.1016/s1525-0016(03)00123-0. [DOI] [PubMed] [Google Scholar]
- Müller OJ, Kaul F, Weitzman MD, Pasqualini R, Arap W, Kleinschmidt JA, et al. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat Biotechnol. 2003;21:1040–1046. doi: 10.1038/nbt856. [DOI] [PubMed] [Google Scholar]
- Maheshri N, Koerber JT, Kaspar BK., and , Schaffer DV. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- Koerber JT, Jang JH., and , Schaffer DV. DNA shuffling of adeno-associated virus yields functionally diverse viral progeny. Mol Ther. 2008;16:1703–1709. doi: 10.1038/mt.2008.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Asokan A, Wu Z, Van Dyke T, DiPrimio N, Johnson JS, et al. Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Mol Ther. 2008;16:1252–1260. doi: 10.1038/mt.2008.100. [DOI] [PubMed] [Google Scholar]
- Grimm D, Lee JS, Wang L, Desai T, Akache B, Storm TA, et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffon KJ, Koerber JT, Dickey DD, Murtha M, Keshavjee S, Kaspar BK, et al. Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc Natl Acad Sci USA. 2009;106:3865–3870. doi: 10.1073/pnas.0813365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zincarelli C, Soltys S, Rengo G., and , Rabinowitz JE. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- Duan D, Yue Y, Yan Z, Yang J., and , Engelhardt JF. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J Clin Invest. 2000;105:1573–1587. doi: 10.1172/JCI8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochrie MA, Tatsuno GP, Christie B, McDonnell JW, Zhou S, Surosky R, et al. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J Virol. 2006;80:821–834. doi: 10.1128/JVI.80.2.821-834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, et al. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci USA. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J., and , Wheeler DL.2008GenBank Nucleic Acids Res 36database issue): D25–D30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Storm T., and , Kay MA. Characterization of the relationship of AAV capsid domain swapping to liver transduction efficiency. Mol Ther. 2007;15:1955–1962. doi: 10.1038/sj.mt.6300293. [DOI] [PubMed] [Google Scholar]
- Nam HJ, Lane MD, Padron E, Gurda B, McKenna R, Kohlbrenner E, et al. Structure of adeno-associated virus serotype 8, a gene therapy vector. J Virol. 2007;81:12260–12271. doi: 10.1128/JVI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindasamy L, Padron E, McKenna R, Muzyczka N, Kaludov N, Chiorini JA, et al. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J Virol. 2006;80:11556–11570. doi: 10.1128/JVI.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod A, Wobus CE, Zádori Z, Ried M, Leike K, Tijssen P, et al. 2002The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity J Gen Virol 83Pt 5): 973–978. [DOI] [PubMed] [Google Scholar]
- Abbott NJ. Blood–Brain Barriers: From Ontogeny to Artificial Interfaces. Wiley-VCH: Weinheim; 2006. The bipolar astrocyte: polarized features of astrocytic glia underlying physiology, with particular reference to the blood-brain barrier. In: Dermietzel, EA (ed.) pp. 189–208. [Google Scholar]
- Cinaroglu A, Ozmen Y, Ozdemir A, Ozcan F, Ergorul C, Cayirlioglu P, et al. Expression and possible function of fibroblast growth factor 9 (FGF9) and its cognate receptors FGFR2 and FGFR3 in postnatal and adult retina. J Neurosci Res. 2005;79:329–339. doi: 10.1002/jnr.20363. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS.et al. (2008A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function J Neurosci 28264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg KP, Geller SF, Schaffer DV., and , Flannery JG. Targeted transgene expression in muller glia of normal and diseased retinas using lentiviral vectors. Invest Ophthalmol Vis Sci. 2007;48:1844–1852. doi: 10.1167/iovs.05-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K, Kaspar BK, Gage FH., and , Schaffer DV. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nature Neuroscience. 2003;6:21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transduction efficiencies of AAV2 variants from the random heptamer library after a single round of evolution. Transduction of primary human astrocytes with the rAAV CMV-GFP vectors with capsids selected from the AAV2 random heptamer library demonstrated only modest (<1.5-fold) first round improvements in viral transduction (n = 3, error bars represent SD), highlighting the need in this case for multiple rounds of evolution.
Primary amino acid sequence alignment of AAV variants from chimeric AAV and AAV EP libraries along with parent serotypes. Distinct colors were used to shade each amino acid.
Transduction efficiencies of other AAV variants on primary human astrocytes (n = 3, error bars represent SD).
Transduction efficiencies of AAV variants on other glial cell lines. Tranduction of rat astrocytes with rAAV CMV-GFP clones from the both the chimeric (A) and random loop libraries (B) revealed significantly different trends compared to transduction of primary human astrocytes (n = 3, error bars represent SD). Tranduction of U87MG glioblastoma cells with rAAV CMV-GFP clones from the both the chimeric (C) and random loop libraries (D) revealed significantly different trends compared to transduction of primary human astrocytes (n = 3 and error bars represent SD).
Heparan sulfate dependence of AAV variants. Transduction of parent CHO cell line and a CHO mutant deficient in heparan sulfate proteoglycan (HSPG) (psgD) demonstrated HSPG dependence of all AAV2-like variants from the chimeric (A) and random loop libraries (B) (n = 3, error bars represent SD).
Summary of peptides within clones selected from AAV2 7mer library after a single round of evolution.
Oligonucleotide sequences for construction of viral libraries.