Abstract
Transcription elongation factor DSIF/Spt4–Spt5 is capable of promoting and inhibiting RNA polymerase II elongation and is involved in the expression of various genes. While it has been known for many years that DSIF inhibits elongation in collaboration with the negative elongation factor NELF, how DSIF promotes elongation is largely unknown. Here, an activity-based biochemical approach was taken to understand the mechanism of elongation activation by DSIF. We show that the Paf1 complex (Paf1C) and Tat-SF1, two factors implicated previously in elongation control, collaborate with DSIF to facilitate efficient elongation. In human cells, these factors are recruited to the FOS gene in a temporally coordinated manner and contribute to its high-level expression. We also show that elongation activation by these factors depends on P-TEFb-mediated phosphorylation of the Spt5 C-terminal region. A clear conclusion emerging from this study is that a set of elongation factors plays nonredundant, cooperative roles in elongation. This study also shows unambiguously that Paf1C, which is generally thought to have chromatin-related functions, is involve directlyd in elongation control.
Keywords: DSIF, RNA polymerase II, transcription elongation, H2B monoubiquitination
Over the past 30 years, it has become clear that eukaryotic mRNA synthesis is controlled at both the initiation and elongation stages of transcription. Post-initiation transcriptional control is involved in the expression of a diverse array of genes, such as hsp70, MYC, and FOS (Saunders et al. 2006 and references therein). On the Drosophila hsp70 gene, for example, RNA polymerase II (Pol II) pauses at the promoter-proximal region and consequently synthesizes only very short transcripts in an uninduced state, but after heat shock, pausing is suppressed and the full-length mRNA is synthesized. Similar regulation is found in the life cycle of certain viruses (Yamaguchi et al. 2002 and references therein). For example, the switch from latent to productive infection of human immunodeficiency virus (HIV) occurs when the viral protein Tat facilitates transcription from the HIV long terminal repeat by releasing an elongation block. In addition, recent genome-wide chromatin immunoprecipitation (ChIP) analyses have suggested that, in Drosophila and human cells, significant proportions of the genes, including many immediate–early genes and developmental genes, are controlled at a post-initiation step (Price 2008 and references therein). Thus, there is ample evidence that post-initiation control plays an important role in physiological and pathological processes.
More than a dozen transcription elongation factors have been identified in humans. It is generally believed that elongation factors affect Pol II's choice between continuing or stalling RNA synthesis, and thereby affect the overall rate of elongation, by causing conformational changes or chemical modifications to Pol II. For example, TFIIS can reactivate Pol II stalled for a variety of reasons (e.g., nucleotide misincorporation) by inducing cleavage of the nascent transcript at the active site (Fish and Kane 2002). Another factor, known as FACT (for facilitate chromatin transcription), facilitates Pol II passage through nucleosome arrays that are inhibitory to both transcription initiation and elongation by destabilizing nucleosomal structure (Reinberg and Sims 2006). For many of the elongation factors, however, not much is known about their mechanisms of action, and it is also unclear whether they have distinct or redundant functions.
DSIF/Spt4–Spt5 is a heterodimeric elongation factor that is capable of both promoting and inhibiting Pol II elongation (Wada et al. 1998; Yamada et al. 2006). Spt4 and Spt5 are conserved from yeast to humans, and part of Spt5 also shares extensive sequence similarity with NusG, a transcriptional terminator/anti-terminator in bacteria and archaea (Hartzog et al. 1998; Andrulis et al. 2000; Ponting 2002). Although seemingly contradictory to the statement above, DSIF has no discernible effect on transcription in a highly purified in vitro transcription system (Yamaguchi et al. 1999a). It has been known for many years that DSIF inhibits Pol II elongation in collaboration with the negative elongation factor NELF; i.e., the Pol II elongation complex stalls or slows down when bound by DSIF and NELF simultaneously (Yamaguchi et al. 1999a; Cheng and Price 2007). To add complexity, their inhibitory action is regulated by P-TEFb, the positive elongation factor with protein kinase activity. P-TEFb extensively phosphorylates the repetitive C-terminal domain (CTD) of Pol II during elongation, and one of its consequences is the reversal of the elongation block (Yamaguchi et al. 1999a; Cheng and Price 2007). In vivo, such a mechanism is responsible for promoter-proximal pausing, a widespread checkpoint during the transcription cycle (Lee et al. 2008; Price 2008), and plays an important role in expression of Drosophila hsp70, proliferation of HIV, and neuronal differentiation, just to name a few (Andrulis et al. 2000; Guo et al. 2000; Wu et al. 2003; Zhang et al. 2007).
On the other hand, how DSIF simulates elongation is largely unknown. What is known is that Spt5 possesses the C-terminal region (CTR), proline, serine, and threonine-rich repetitive sequence similar to the Pol II CTD, and that P-TEFb-mediated phosphorylation of the Spt5 CTR is required for elongation activation by DSIF (Ivanov et al. 2000; Bourgeois et al. 2002; Yamada et al. 2006). In a previous study, we proposed the “mini-CTD” hypothesis, in which phosphorylated Spt5 CTR functions in a manner analogous to phosphorylated Pol II CTD and serves as an additional “code” for active elongation complexes, possibly as a scaffold for other factors (Phatnani and Greenleaf 2006; Yamada et al. 2006). In this study, we took an activity-based biochemical approach to understand the mechanism of elongation activation by DSIF, leading to the identification of Paf1 complex (Paf1C) and Tat-SF1 as factors that collaborate with DSIF to facilitate productive elongation.
Results
Identification of DSIF coactivator (DC) activity
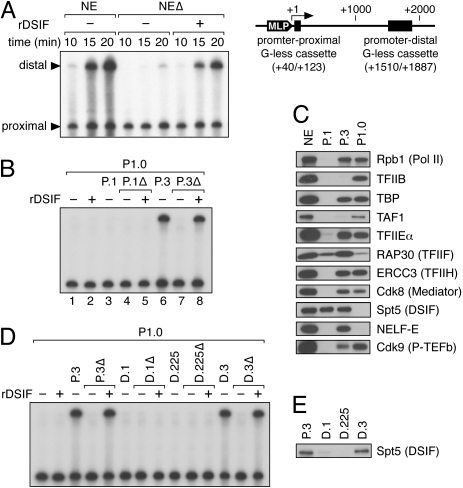
The template pSLG402 (Fig. 1A) contains two G-free cassettes downstream from the adenovirus major late promoter and, hence, short (promoter-proximal) and long (promoter-distal) RNase T1-resistant products are generated. From the resulting data, one can determine the success rate of Pol II elongation; i.e., what percentage of Pol II molecules that have finished transcribing the first G-less sequence (+40/+123) reach the 3′ end of the second G-less sequence (+1887). Thus, the focus of the assay is the elongation block (pause, arrest, or termination) that mature elongation complexes encounter during transcription of the intervening sequence and its suppression by protein factors. Elongation activation by DSIF can be assayed with this template. As illustrated in Figure 1A, transcription of the promoter-distal region is strongly reduced by immunodepletion of DSIF from HeLa nuclear extract (NE) and is restored by add-back of recombinant DSIF (rDSIF), whereas transcription of the promoter-proximal region is only weakly affected by depletion and add-back of DSIF, suggesting that the promoter-proximal product is an appropriate control for elongation stimulation activity of DSIF. The efficiency and specificity of DSIF immunodepletion are demonstrated in Supplemental Figure 1.
Figure 1.
Identification of DC activity. (A) rDSIF promotes transcription elongation in HeLa NE. Transcription reactions contained the template pSLG402 (shown schematically to the right) and either NE or NE devoid of DSIF (NEΔ). Transcription was allowed to proceed for the indicated times. (MLP) Adenovirus major-late promoter. (B) rDSIF does not promote transcription elongation in P1.0. Phosphocellulose fractions (see Fig. 2A) were used instead of NE. “Δ” indicates the fractions that were immunodepleted with anti-Spt5 antibody and were hence devoid of DSIF. (C) Immunoblot analysis of the phosphocellulose fractions. (D) P.3-derived DEAE Sepharose fractions (see Fig. 2A) were assayed for DC activity. (E) Immunoblot analysis of the DEAE Sepharose fractions.
Under different conditions, however, DSIF is unable to stimulate transcription elongation. For example, when P1.0, the 1.0 M KCl phosphocellulose fraction derived from HeLa NE, is used instead of NE, rDSIF has little effect on elongation efficiency (Fig. 1B, lane 2), indicating the requirement for additional factors in DSIF-mediated activation. Here, HeLa NE was fractionated on a phosphocellulose column with increasing concentrations (0.1, 0.3, and 1.0 M) of KCl (Fig. 2A), and these fractions were termed P.1, P.3, and P1.0, respectively. Fractionation of NE is often carried out using 0.1, 0.3, 0.5, and 0.85 M KCl buffer; our P1.0 fraction can be regarded as the combination of 0.5 M and 0.85 M fractions obtained by the conventional method.
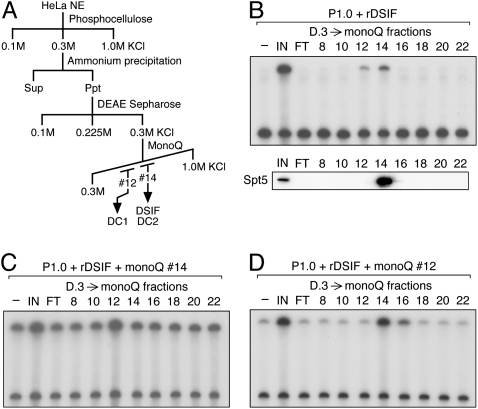
Figure 2.
Separation of DC activity into two components. (A) Scheme for partial purification of DCs. (B–D) D.3-derived mono-Q fractions were assayed for DC activity. In B, P1.0, rDSIF, and one of the column fractions were used as protein source. An anti-Spt5 immunoblot is shown below. In C and D, the same mono-Q fractions were assayed in the presence of an additional mono-Q fraction.
The aim of this study is to identify and characterize such factors, tentatively called DCs. Consistent with the observation that P1.0 supports efficient transcription initiation but does not support efficient elongation (Fig. 1B, lane 1), P1.0 contains Pol II and general transcription factors (GTFs) but is devoid of DSIF (Fig. 1C). We then examined all the phosphocellulose fractions in order to reconstitute the efficient elongation seen with HeLa NE and found that P.3, but not P.1, strongly stimulates elongation in conjunction with P1.0 (Fig. 1B, lanes 3,6). As similar amounts of DSIF were found in P.1 and P.3 (Fig. 1C), something other than DSIF is likely responsible for the differential effects. Moreover, since the stimulatory activity of P.3 was abrogated by depletion of DSIF and was restored by add-back of rDSIF (Fig. 1B, lanes 7,8), the stimulatory activity is DSIF-dependent. Collectively, these results indicate that P.3 contains DCs, factors that collaborate with DSIF to support efficient elongation.
Separation of DC activity into two components
Following concentration by ammonium sulfate precipitation, P.3 was separated on a DEAE Sepharose column (Fig. 2A). Consequently, both the DSIF protein and DC activity were found in the 0.3 M KCl fraction D.3 (Fig. 1D,E). The observation that DC activity cochromatographed with DSIF through these steps suggests that physical interactions may occur between them.
The active fraction was then applied to a mono-Q column. In this and subsequent steps, DC activity was assayed by carrying out transcription in the presence of a constant amount of rDSIF, without prior depletion of endogenous DSIF from each fraction. DSIF was recovered in mono-Q fraction #14, and DC activity also appeared to peak at the same fraction (Fig. 2B). However, it was noticeable that the recovery of DC activity was significantly lower than expected. As a reason for the low yield, we examined the possibility that DC activity was separated into two components: one in fraction #14, and another in other fractions. In search of such secondary activity, all of the mono-Q fractions were assayed in the presence of rDSIF and fraction #14 and, to our surprise, fraction #12 was found to stimulate elongation further (Fig. 2C). We then asked whether the two activities found in fractions #12 and #14 are distinct, by assaying all of the fractions in the presence of rDSIF and fraction #12. Under these conditions, elongation was stimulated by fractions #14 –#16 (Fig. 2D). It is therefore likely that mono-Q fractions #12 and #14 contain distinct DC activities that we hereafter call DC1 and DC2, respectively.
Final purification and identification of DCs
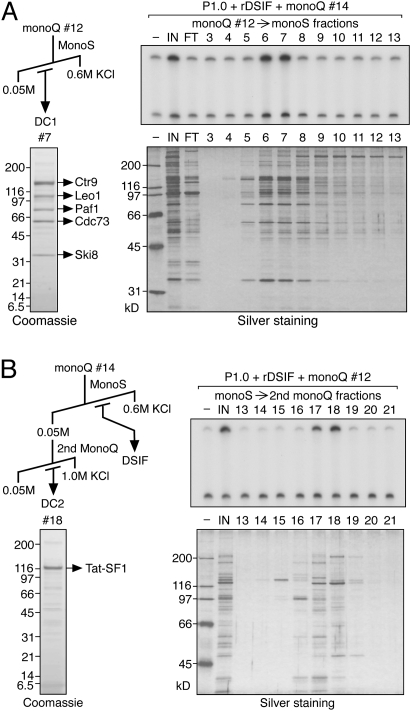
DC1 and DC2 were further purified from the respective mono-Q fractions. Chromatography of the DC1-containing fraction on a mono-S column resulted in the recovery of DC1 activity in fractions 6 and 7, and silver staining revealed several candidate proteins associated with this activity (Fig. 3A). Coomassie staining of the most active fraction revealed the presence of five major polypeptides and, by mass spectrometry, they were identified as Ctr9, Leo1, Paf1, Cdc73, and Ski8, in order of decreasing molecular weight. These proteins are the components of human Paf1C, a conserved factor that was first described in budding yeast as a Pol II-associated complex (Shi et al. 1996). Antibodies were raised against some of the Paf1C components and used to confirm their identities by immunoblotting (Supplemental Fig. 1). These antibodies will be used for ChIP analyses in the subsequent study. Consistent with recent reports that human Paf1C is devoid of Rtf1, an integral subunit of yeast Paf1C (Rozenblatt-Rosen et al. 2005; Yart et al. 2005; Zhu et al. 2005), Rtf1 was not detected in our purified Paf1C fraction (Supplemental Fig. 1).
Figure 3.
Final purification and identification of DCs. (A) Final purification of DC1 was performed according to the scheme shown. Mono-S fractions were assayed for DC1 activity in the presence of P1.0, rDSIF, and crude DC2 (mono-Q #14). The same fractions were analyzed by SDS-PAGE and silver staining. Proteins in an active fraction were also visualized by Coomassie staining and were subjected to LC-MS/MS analysis. (B) Final purification of DC2 was performed according to the scheme shown. Fractions derived from the second mono-Q column were assayed for DC2 activity in the presence of P1.0, rDSIF, and crude DC1 (mono-Q #12). Proteins were analyzed as described in A.
For DC2, mono-Q fraction #14 containing DSIF and DC2 was applied to a mono-S column. At this step, we were able to separate DC2 from DSIF. DC2 was recovered in the flow-through fraction, whereas DSIF was found in eluate fractions (Supplemental Fig. 2). Subsequent mono-Q column chromatography resulted in the recovery of DC2 activity in fractions 17 and 18, and silver staining revealed an ∼120-kD protein associated with this activity (Fig. 3B). This protein was identified as Tat-SF1 by mass spectrometry and immunoblotting (Supplemental Fig. 1). Tat-SF1 was first described as a factor required for HIV Tat-dependent transcriptional activation in vitro (Zhou and Sharp 1996), and was later shown to play a more general role in transcription elongation (Li and Green 1998).
If Paf1C and Tat-SF1 are responsible for DC activity, these proteins should be present in the phosphocellulose P.3 fraction and absent in the other phosphocellulose fractions. As would be expected from their roles as DCs, Paf1C and Tat-SF1 were found in P.3 almost exclusively (Supplemental Fig. 1).
Cooperative action of DSIF, Paf1C, and Tat-SF1 on elongation
Next, we examined the effects of DSIF, DC1 (Paf1C), and DC2 (Tat-SF1) either individually or in various combinations using DC1 and DC2 each from the final purification step. As we have already seen, elongation is strongly stimulated by combination of the three factors (Fig. 4A). In contrast, they had little effect when used individually or in combinations of two factors (Fig. 4A). The only exception was the combination of DSIF and DC2 (Tat-SF1), in which case transcription elongation was modestly enhanced in a dose-dependent manner.
Figure 4.
Cooperative action of DSIF, Paf1C, and Tat-SF1 on elongation. (A) Transcription assays were carried out in the presence of various combinations of rDSIF and DCs, each from the final purification step. (B) Purified proteins used for in vitro transcription. (C) Transcription assays were carried out in the presence of various combinations of rDSIF, Flag-Paf1C, and rTat-SF1. Below, percent read-through was calculated from relative intensity of the two products by taking into account the number of uridines (labeled nucleotides) in each fragment. (D) Requirement for P-TEFb-mediated phosphorylation of the Spt5 CTR in productive elongation. rDSIF composed of rSpt4 and wild-type rSpt5 or one of its mutants was used in combination with Flag-Paf1C and rTat-SF1. Where indicated, P-TEFb inhibitors such as DRB and Flavopiridol were included. In the top panel, transcription reactions were performed in the presence of [α-32P] UTP, whereas in the bottom panel, an identical set of reactions were performed in the presence of [γ-32P] ATP, followed by immunoprecipitation of Spt5. (E) Pulse-chase transcription assays were carried out as described in the Materials and Methods. The indicated combinations of elongation factors (ECs) were added to reactions 10 min prior to chase.
To determine whether Paf1C and Tat-SF1 are responsible for DC activity, we prepared these proteins as follows. Epitope-tagged Paf1C (Flag-Paf1C) was purified from the NE of HeLa cells stably expressing Flag-Leo1, essentially as described (Zhu et al. 2005), and the integrity of this complex was confirmed by Coomassie staining and immunoblotting (Fig. 4B; Supplemental Fig. 1). Recombinant Tat-SF1 was overexpressed in insect cells and purified from the cell extract by nickel-affinity chromatography (Fig. 4B).
Essentially identical results were obtained with these proteins (Fig. 4C). Thus, the combination of rDSIF, Flag-Paf1C, and rTat-SF1 strongly stimulated elongation, but they had little effect when used individually or in combinations of two factors, except when rDSIF and rTat-SF1 were used together. Elongation efficiency can be calculated from the relative intensity of the two RNA products by taking into account the number of uridines (labeled nucleotides) in each fragment. In our partially reconstituted transcription system, elongation efficiency is almost zero in the absence of DSIF, Paf1C, and Tat-SF1, and is 31% in their presence. These results demonstrate that Paf1C and Tat-SF1 are indeed responsible for DC activity, and that DSIF, Paf1C, and Tat-SF1 stimulate transcription elongation in an interdependent manner.
In a previous study (Yamada et al. 2006), we showed that P-TEFb-mediated phosphorylation of the Spt5 C-terminal repetitive sequence (CTR) is required for elongation activation by DSIF. We therefore examined whether this holds true in our partially reconstituted system by using P-TEFb inhibitors and Spt5 mutants. Note that P-TEFb is supplied from the P1.0 fraction (Fig. 1C). Consequently, P-TEFb kinase inhibitors such as DRB and Flavopiridol strongly inhibited transcription of the promoter-distal region, whereas they had little effect on transcription of the promoter-proximal region (Fig. 4D). Then, rDSIF composed of Spt4 and one of the Spt5 mutants was used in combination with Flag-Paf1C and rTat-SF1. We found that Spt5 TA in which several P-TEFb-targeted threonine residues are replaced by alanines is almost inactive, whereas Spt5 SA in which irrelevant serine residues are replaced by alanines has activity comparable with wild-type Spt5. An identical set of transcription reactions was carried out in the presence of [γ-32P] ATP instead of [α-32P] UTP, and it was found that the level of Spt5 phosphorylation correlates well with the level of transcription (Fig. 4D). These results led us to conclude that P-TEFb-mediated phosphorylation of the Spt5 CTR is required for elongation activation by DSIF, Paf1C, and Tat-SF1.
We also examined a possible role of NELF and TFIIF. The P1.0 fraction is devoid of NELF and contains a limited amount of TFIIF (Fig. 1C). We found that DSIF, Paf1C, and Tat-SF1 facilitate efficient elongation regardless of the presence of excessive NELF or TFIIF (Supplemental Fig. 3).
The above transcription assays have been carried out at low nucleotide triphosphate (NTP) concentrations. This is because we employ a continuous labeling protocol in which the concentration of the labeled nucleotide UTP has to be low enough to allow sensitive detection of products. In order to examine the role of DSIF, Paf1C, and Tat-SF1 at high NTP concentrations, the template pSLG402 was modified so that the first G-less cassette was placed directly downstream from the major-late promoter, and a pulse-chase protocol was employed. Essentially, pulse labeling was performed in the absence of GTP to allow the formation of +83 complexes, elongation factors were added, chase reactions were performed in the presence of 500 μM (each) 4NTPs, and products were analyzed without RNase T1 treatment. As shown in Figure 4E, an approximately twofold increase in the elongation rate is observed in the presence of DSIF, Paf1C, and Tat-SF1, demonstrating that these factors can stimulate elongation even at high NTP concentrations, although to a lesser extent.
We consider the reason why these elongation factors are less efficient in stimulating transcription at high NTP concentrations as follows: Elongation factors are thought to control the elongation process by various mechanisms, some considered to facilitate efficient elongation by suppressing pause and arrest. The observation that DSIF, Paf1C, and Tat-SF1 work more efficiently at low NTP concentrations, where incidence and severity of pause and arrest are higher, indicates that these elongation factors act by suppressing pause and arrest that could occur, for example, when Pol II encounters a DNA sequence intrinsically unfavorable for elongation. We speculate that, in vivo, there are numerous obstacles that necessitate these elongation factors even at physiological NTP concentrations (∼1 mM), because endogenous Pol II genes are typically larger than the DNA template used in vitro.
Physical interactions of DSIF, Paf1C, Tat-SF1, and Pol II
It has been shown that Pol II interacts with DSIF, Paf1C, and Tat-SF1, respectively (Wada et al. 1998; Kim et al. 1999; Zhu et al. 2005), and that DSIF interacts with Tat-SF1 (Kim et al. 1999). In addition, similar chromatographic behavior suggests that DSIF, Paf1C, and Tat-SF1 interact directly with each other. Furthermore, genetic and physical interactions between yeast counterparts of DSIF and Paf1C have been described (Squazzo et al. 2002; Qiu et al. 2006). We investigated their interactions by transient transfection of two constructs and subsequent immunoprecipitation with anti-Flag antibody. Consequently, HA-Spt5 coprecipitated Flag-Paf1, Flag-Leo1, and Flag-Tat-SF1, respectively (Fig. 5A). We then considered the possibility that Spt5 phosphorylation mediates these interactions, as Spt5 phosphorylation is required for Paf1C- and Tat-SF1-dependent productive elongation (Fig. 4D). This possibility was examined by using two phosphorylation-defective Spt5 mutants, TA and ΔCTR. Unexpectedly, these Spt5 mutants also coprecipitated with Flag-Paf1, Flag-Leo1, and Flag-Tat-SF1 at a similar level to wild-type Spt5 (Fig. 5A), suggesting that Spt5 phosphorylation is not required for its interactions with Paf1C and Tat-SF1.
Figure 5.
Physical interactions of DSIF, Paf1C, Tat-SF1, and Pol II. (A) Coimmunoprecipitation of transiently expressed proteins. Following transfection of the indicated constructs into HeLa cells, Flag-tagged proteins were immunopurified from cell extracts using anti-Flag M2 agarose and then immunoblotted with the indicated antibodies. Asterisks denote nonspecific signal. (B) Coimmunoprecipitation was carried out using 100 ng each of rDSIF containing HA-Spt5, rTat-SF1, and rPaf1C containing V5-Paf1 that were individually expressed and purified from Sf-9 cells. (C) NE prepared from HeLa/FH3 cells (Hasegawa et al. 2003) stably expressing the Flag- and His-tagged Rpb3 subunit of Pol II was incubated with anti-Flag M2 agarose to immobilize 1 μg of FH-Pol II to the beads. The Pol II beads were then incubated with rPaf1C (2 μg), rTat-SF1 (2 μg), and rDSIF (2 μg) in various combinations for 2 h at 4°C and washed several times with NP-40 buffer (50 mM Tris at pH 7.9, 150 mM NaCl, 1% NP-40). Bound materials were eluted with Flag peptide and subjected to immunoblotting.
In the above experiments, it cannot be ruled out that the interactions are mediated by an abundant endogenous protein. Hence, we wished to carry out coimmunoprecipitation using purified factors. In order to obtain Paf1C in sufficient quantity, Ctr9, Leo1, V5-Paf1, Cdc73, and Ski8 were coexpressed in insect cells using the baculoviral expression system and were affinity-purified. Purity of rPaf1C and its transcriptional activity are demonstrated in Supplemental Figure 4. Figure 5B unambiguously shows the presence of direct binary interactions among DSIF, Paf1C, and Tat-SF1.
Next, we asked whether these factors interact with Pol II independently, cooperatively, or competitively. Flag-tagged Pol II was immobilized to agarose beads, and purified factors were added in various combinations. DSIF, Paf1C, and Tat-SF1 bound to Pol II when added individually (Fig. 5C, lanes 14–16), but when added together, they bound to Pol II at significantly higher levels (Fig. 5C, lanes 10–13). These results suggest that DSIF, Paf1C, and Tat-SF1 bind to Pol II in a cooperative manner through multiple protein–protein interactions, which could partly explain the mechanism for their cooperative action.
Nonredundant roles of DSIF, Paf1C, and Tat-SF1 in EGF-induced transcription
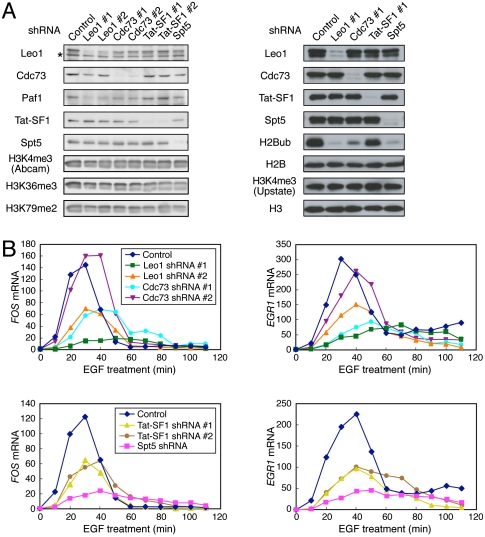
To understand the functional significance of DSIF, Paf1C, and Tat-SF1 in EGF-induced transcription, we knocked down these factors using lentiviral vectors expressing shRNAs. Considering potential off-target effects, two different shRNAs were designed each for Leo1, Cdc73, and Tat-SF1. For Spt5, a well-validated shRNA was used (Yamada et al. 2006). These shRNAs resulted in a significant reduction of each target protein (Fig. 6A). In addition, the Paf1 protein level was partially reduced by Leo1 and Cdc73 shRNAs, suggesting that they contribute to the stability of Paf1C.
Figure 6.
Nonredundant roles of DSIF, Paf1C, and Tat-SF1 in EGF-induced transcription. (A) HeLa cells were transduced with recombinant lentivirus expressing no shRNA (Control) or expressing one of the shRNAs. Four days post-infection, immunoblotting was performed. (B) Four days to 6 d post-infection, the cells were treated with EGF for the indicated times and then harvested for RNA analysis. The expression levels of the FOS and EGR1 mRNAs were quantified by real-time RT–PCR, and the values are expressed as fold changes from the mRNA levels in control virus-transduced, unstimulated cells.
It has been shown that Paf1C plays a critical role in generating transcription-associated histone modifications such as H2B monoubiquitination and H3 methylation at K4, K36, and K79 in yeast, Drosophila, and humans (Krogan et al. 2003; Ng et al. 2003; Zhu et al. 2005; Adelman et al. 2006). In addition, recent studies have shown that the yeast counterpart of DSIF is involved in the recruitment of Paf1C and corresponding histone modifications (Qiu et al. 2006; Zhou et al. 2009; Liu et al. 2009). Under our conditions, knockdown of Leo1 or Cdc73 resulted in a significant reduction in the level of monoubiquitinated H2B (H2Bub) but caused little change in the H3K4me3 (H3K4 trimethylation), H3K36me3, and H3K79me2 (H3K79 dimethylation) levels. Knockdown of Spt5 also selectively eliminated the monoubiquitin mark in HeLa cells. We discuss the seemingly contradictory results in the Discussion section.
We examined functional consequences of the knockdown by real-time RT–PCR analysis. As reported previously (Yamada et al. 2006), knockdown of Spt5 strongly attenuated EGF-induced FOS expression (Fig. 6B). The FOS mRNA level was also reduced by knockdown of Leo1, Cdc73, or Tat-SF1 to varying degrees. EGR1, another EGF-induced gene, was also examined, as this gene is also regulated at a post-initiation step (Wang et al. 2005). Consequently, EGF-induced EGR1 expression was significantly attenuated by the knockdown of Spt5, Tat-SF1, Leo1, or Cdc73 (Fig. 6B). On the other hand, the level of the control GAPDH mRNA was essentially unaffected by the knockdown over the period examined (data not shown). It is noteworthy that the degree of Paf1C knockdown correlates well with the severity of the phenotype. For example, Leo1 shRNA #1 is more potent than shRNA #2 in silencing Leo1 expression and in suppressing EGF induction. Moreover, Leo1 shRNAs are generally more potent than Cdc73 shRNAs in reducing the stability of Paf1C and in suppressing EGF induction. Consistent with these factors being important to the expression of growth-related genes, prolonged culture of Spt5, Leo1, Cdc73, or Tat-SF1 knockdown cells leads to growth arrest (Y Tsugeno and T Yamada, unpubl.). These findings demonstrate that DSIF, Paf1C, and Tat-SF1 play important, nonredundant roles in EGF-induced transcription in living cells.
Concomitant association of DSIF, Paf1C, Tat-SF1, and Pol II with the FOS gene
To understand the interplay of DSIF, Paf1C, and Tat-SF1 in living cells, we examined their distribution across a gene by ChIP analysis. It has been shown that DSIF and Paf1C are associated with transcriptionally active loci (Andrulis et al. 2000; Zhu et al. 2005); however, direct comparison of their distribution has not been made. On the other hand, Tat-SF1 binding to the genome has not been explored. Here, the FOS gene was analyzed, as expression of this gene is known to be regulated at the elongation step (Saunders et al. 2006). As reported previously (Yamada et al. 2006), significant amounts of Pol II and DSIF were associated with the promoter-proximal region of FOS prior to induction, and their association was extended to the coding and 3′ regions in EGF-stimulated cells (Fig. 7). These results are consistent with the general idea that FOS expression is regulated by promoter-proximal pausing. Then, Tat-SF1 was examined using anti-Flag antibody and HeLa cells stably expressing Flag-Tat-SF1 because anti-Tat-SF1 antibody suitable for ChIP analysis was not available. Tat-SF1 was found associated at least with the promoter-proximal region upon EGF stimulation. Leo1 and Cdc73 were associated with the entire gene in an EGF-dependent manner, but, unlike Pol II, their association appeared to peak in the coding region. Nonetheless, these results confirm that DSIF, Paf1C, and Tat-SF1 are all recruited, along with Pol II, to the FOS gene following EGF treatment.
Figure 7.
Concomitant association of DSIF, Paf1C, Tat-SF1, and Pol II with the FOS gene. ChIP assays were carried out as described in the Materials and Methods. Prior to harvest, HeLa cells were cultured for 18 h in the presence of 0.2% serum and then stimulated for 7.5 min with 0.1 mg/mL EGF or were left untreated. The amplicons used are presented as bars below the diagram of the FOS gene. All of the data are means ± SEM from three independent experiments and are expressed as percent input. Enrichment ratios of histone H3K4me3 to total histone H3 were calculated from respective ChIP data and are expressed in arbitrary units.
Temporal patterns of their association were also examined. Association of Pol II, DSIF, Paf1C, and Tat-SF1 with the promoter-proximal region of FOS started to increase within a few minutes after EGF administration and peaked around 10 min (Supplemental Fig. 5). Taking this and earlier biochemical studies together, these ChIP data suggest that DSIF, Paf1C, and Tat-SF1 are assembled onto the Pol II elongation complex in a temporally coordinated manner, and that this large protein complex is responsible for active transcription.
We then asked whether any histone modifications occur in association with FOS gene induction. Occupancy of histone H3 at the FOS promoter region was significantly reduced after EGF administration, probably due to transcription-coupled histone eviction (Fig. 7; Supplemental Figs. 5, 6). On the other hand, the H3K4me3 level at the FOS promoter region was already high prior to induction, and its relative level was essentially unaffected by EGF treatment. These results suggest that H3K4me3 is not involved in EGF-induced transcriptional change per se.
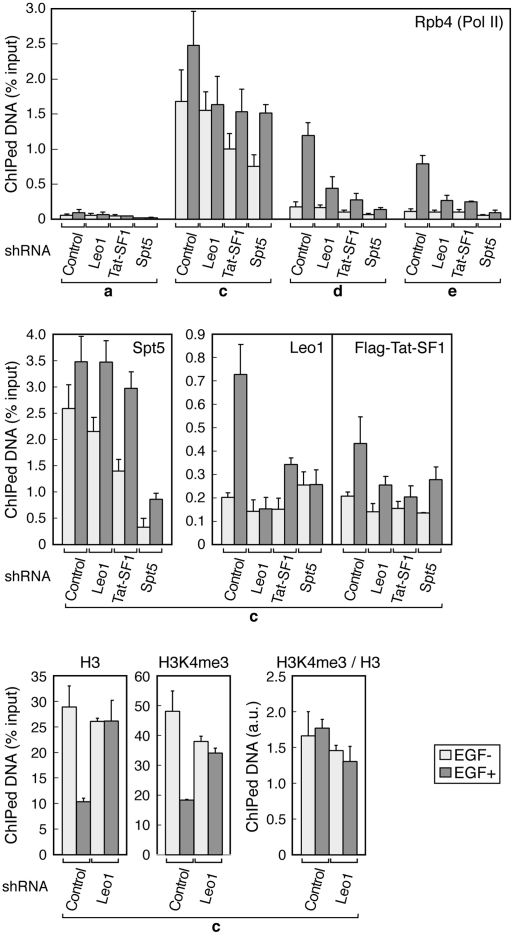
Finally, knockdown ChIP analyses were carried out to examine the role of each elongation factor in greater detail. From the data shown in Figure 8, the following conclusions can be drawn: (1) DSIF, Paf1C, and Tat-SF1 are all important to Pol II elongation on the FOS gene. (2) Leo1 knockdown has no appreciable effect on the H3K4me3 level at the FOS promoter region. (3) DSIF and Tat-SF1 stimulate Paf1C recruitment to the FOS gene following induction. (4) DSIF and Paf1C also contribute to Tat-SF1 recruitment to the FOS gene following induction. (5) DSIF recruitment is relatively unaffected by the knockdown of Paf1C and Tat-SF1. Thus, although for some reason that we do not understand there is a disagreement as to the role of Paf1C and Tat-SF1 in DSIF recruitment to Pol II, these in vitro and in vivo data are largely concordant and together suggest that DSIF, Paf1C, and Tat-SF1 are assembled into the Pol II elongation complex in a cooperative manner. This could partly explain the mechanism of their cooperative action.
Figure 8.
Cooperative assembly of DSIF, Paf1, and Tat-SF1 into the Pol II elongation complex. HeLa cells were transduced with recombinant lentivirus expressing no shRNA (Control), Leo1 shRNA #1, Tat-SF1 shRNA #1, or Spt5 shRNA. Five days to 6 d post-infection, the cells were treated with EGF for 7.5 min or were left untreated and subjected to ChIP. Data are means ± SEM from three independent experiments. Anti-H3K4me3 from Upstate Biotechnologies (07-473) was used where indicated.
Discussion
In this study, we took an activity-based biochemical approach to understand the mechanism of elongation activation by DSIF and found that Paf1C and Tat-SF1, factors implicated previously in elongation control, are required for DSIF-mediated, P-TEFb-dependent elongation activation. Interestingly, DSIF, Paf1C, and Tat-SF1 have no discernible effect on transcription individually but, in combination, strongly enhance Pol II elongation in our partially reconstituted transcription system in vitro. Our data also show that these factors are recruited to the FOS gene in a temporally coordinated manner and contribute to its high-level expression, suggesting that the mechanism found in vitro also occurs in vivo.
Rediscovery of Paf1C as a DC
Paf1C was originally identified in yeast as a Pol II-associated complex by Jaehning and colleagues (Shi et al. 1996), and was subsequently shown to play a critical role in generating transcription-associated histone modifications during elongation (Krogan et al. 2003; Ng et al. 2003). Such chromatin-related functions of Paf1C are conserved in Drosophila and humans (Zhu et al. 2005; Adelman et al. 2006). Our findings are generally consistent with previous studies in yeast showing that Paf1C genetically and physically interacts with the yeast counterpart of DSIF, Spt4–Spt5 (Squazzo et al. 2002; Qiu et al. 2006). More recently, Zhou et al. (2009) and Liu et al. (2009) independently reported that the CTR region of yeast Spt5 is phosphorylated by Bur1/2, one of the yeast counterparts of mammalian P-TEFb, and that the Spt5 CTR is important to Paf1C recruitment and corresponding histone modifications. Their findings are largely consistent with the present study, but are different in that the interaction between human DSIF and Paf1C is not dependent on the presence of the Spt5 CTR.
Yeast Paf1C also appears to play roles other than histone modification, such as 3′-end processing of mRNAs and small nucleolar RNAs (snoRNAs) (Penheiter et al. 2005; Sheldon et al. 2005). A chromatin-independent role of yeast Paf1C in transcription elongation was also described by Rondón et al. (2004). In this pioneering work, Rondón et al. (2004) carried out in vitro transcription assays using a naked DNA template and whole-cell extracts prepared from yeast strains lacking individual components of Paf1C. Consequently, cell extracts from Δpaf1 and Δcdc73 showed reduced elongation efficiency, whereas those from Δleo1 and Δrtf1 showed a wild-type level of efficiency (Rondón et al. 2004). Thus, these results generally support the idea that Paf1C stimulates transcription elongation on a naked DNA template, but the underlying cause of the observed transcriptional defect remains uncertain. Here, we provided compelling biochemical evidence that human Paf1C has bona fide elongation stimulation activity on a naked DNA template and, surprisingly, that this activity is dependent on other elongation factors; i.e., DSIF and Tat-SF1. This Paf1C activity can be distinguished from its chromatin-dependent elongation stimulation activity described by Pavri et al. (2006), as their reconstituted transcription system is devoid of DSIF and Tat-SF1, whereas ours is devoid of chromatin structure.
While DSIF, Paf1C, and Tat-SF1 are clearly capable of stimulating Pol II elongation independently of chromatin in vitro, some of the knockdown effect on gene expression that we observed in HeLa cells may be caused by the removal of the monoubiquitin mark. Particularly interesting in this regard is the finding by Pavri et al. (2006) that H2Bub functions cooperatively with FACT to facilitate chromatin transcription independently of H3K4 methylation. We envisage that DSIF, Paf1C, and Tat-SF1 on the one hand directly regulate Pol II elongation and, on the other hand, act through H2B monoubiquitination, and thereby activate transcription of target genes in vivo.
Why did knockdown of Leo1 and Cdc73 not lead to the reduction of the H3K4me3 level? In fact, to our knowledge, there are no inconsistencies between our data and published data in a strict sense. In yeast, whereas paf1 or rtf1 deletion causes a significant reduction of H3K4 and H3K79 methylation, leo1 deletion does not affect these histone modifications significantly (Tenney et al. 2006), and there has been no report on the role of Ctr9p and Cdc73p in histone modifications. In Drosophila and human cells, knockdown of Rtf1, Ctr9, and Ski8 affects the H3K4me3 level (Zhu et al. 2005; Adelman et al. 2006), but the role of the other Paf1C components in histone modifications has been unclear. The apparently variable results may be because different Paf1C components have different levels of importance to its function as a complex. Another related question is why knockdown of Leo1, Cdc73, and Spt5 did not affect the H3K4me3 level despite the strong reduction in the H2Bub level. Possibly, after knockdown of Leo1 or Cdc73, residual Paf1C activity (and hence residual H2Bub) may be sufficient for maintaining the H3K4me3 level. Alternatively, while in yeast H2B monoubiquitination is a critical determinant for H3K4me3 (Nakanishi et al. 2009), this may not be true in humans. Whereas Set1 is the sole enzyme responsible for H3K4 methylation in yeast, at least 10 known or predicted H3K4 methyltransferases exist in humans (Ruthenburg et al. 2007). Enzymes that act independently of Paf1C and H2B monoubiquitination may be partly responsible for H3K4me3 in humans.
Rediscovery of Tat-SF1 as a DC
Tat-SF1 was described originally as a coactivator of HIV Tat by Sharp and colleagues (Zhou and Sharp 1996), and was later shown to have general elongation stimulation activity (Li and Green 1998). However, its precise mechanisms of action have not been elucidated. Here, we established that Tat-SF1 function is dependent on DSIF and Paf1C. This is, to our knowledge, the first demonstration that Tat-SF1 and Paf1C interact physically and functionally with each other. On the other hand, the connection between Tat-SF1 and DSIF is not unprecedented. Parada and Roeder (1999) described Tat-SF—a large multiprotein complex that is not fully defined but is thought to contain Tat-SF1, DSIF, Nucleolin, and P-TEFb—as a factor responsible for Tat-dependent transcriptional activation. Kim et al. (1999) independently reported copurification of Tat-SF1 with DSIF and RAP30, the small subunit of TFIIF. The relevance of these findings to our study is unclear, because other components of the previously described complexes, such as Nucleolin, RAP30, and Pol II, are absent in our purified coactivator fractions. However, it is quite possible that DSIF, Paf1C, and Tat-SF1 are responsible for the stimulatory activity detected and sought after in the previous studies.
Tat-SF1 is highly conserved from yeast to humans. Although Tat-SF1 function in species other than humans is largely unknown, its yeast counterpart, Cus2p, was shown to interact genetically and physically with the splicing machinery (Yan et al. 1998). Strong sequence conservation of DSIF, Paf1C, and Tat-SF1 supports the idea that the mechanism found in this study is conserved throughout evolution. Moreover, considering the accumulating evidence that DSIF, Paf1C, and Tat-SF1 are involved in mRNA processing (Yan et al. 1998; Wen and Shatkin 1999; Penheiter et al. 2005; Sheldon et al. 2005), it is tempting to speculate that their interplay occurs not only during elongation control but also during control of mRNA processing.
Nonredundant roles of transcription elongation factors
We found that P-TEFb-mediated phosphorylation of the Spt5 CTR is required for elongation stimulation by DSIF, Paf1C, and Tat-SF1, but is not required for their interactions (Figs. 4, 5). What, then, is the actual function of Spt5 phosphorylation in productive elongation? Our data do not exclude the possibility that Spt5 phosphorylation is not essential but enhances the stability of the DSIF–Paf1C–Tat-SF1 interactions. Another interesting possibility is that phosphorylation of the Spt5 CTR contributes to productive elongation by triggering recruitment of other unidentified factors. Since our partially reconstituted transcription system contains the crude P1.0 fraction, such factors may have been supplied from this fraction.
Although this study identified Paf1C, Tat-SF1, and P-TEFb as necessary for DSIF-mediated elongation activation, it remains to be determined whether these factors are sufficient for the activation. Transcription assays using an oligo(dC)-tailed template are useful for studying elongation mechanisms, as this template circumvents the requirement for numerous factors involved in promoter-specific transcription initiation. Despite many attempts, we were unable to reproduce elongation activation by DSIF, Paf1C, Tat-SF1, and P-TEFb using this template and purified Pol II (Y Chen, Y Yamaguchi, and H Handa, unpubl.). Although still inconclusive, these preliminary data suggest that other factors, possibly factors interacting with the Spt5 CTR in a phosphorylation-dependent manner, may also be required for DSIF-dependent elongation activation. This role might be played instead by TFIIF, because TFIIF has been strongly implicated in DSIF-, NELF-, and P-TEFb-dependent elongation control (Cheng and Price 2007).
Of a dozen elongation factors identified so far, many have been thought to possess similar biochemical activity to enhance Pol II elongation. In fact, however, only a few, such as Elongin and ELL, have been shown to stimulate elongation in a highly purified transcription system (Aso et al. 1995; Shilatifard et al. 1996), and the others either have little effect in a highly purified system (e.g., DSIF) or remain to be examined in this respect (e.g., Tat-SF1). In addition, there have been many unanswered questions regarding possible interplay among different elongation factors, such as whether elongation factors with apparently similar activities have distinct or redundant functions. This study revealed a surprising interplay of three (or four, if P-TEFb is taken into account) elongation factors, and established that these factors play nonredundant, cooperative roles in productive elongation. The requirement for a set of elongation factors in transcription elongation is reminiscent of the requirement for a set of GTFs in transcription initiation, although it should be noted that Pol II elongation generally occurs without any additional factors, whereas GTFs are absolutely required for correct initiation of transcription. Future experiments will focus on establishment of a minimal set of factors required for productive elongation and elucidation of the molecular mechanism involved.
Materials and methods
Purification and identification of DCs
HeLa NE and its phosphocellulose column fractions (P.1, P.3, and P1.0) were prepared as described (Wada et al. 1998). The P.3 fraction (∼100 mg) from 100 mL of HeLa NE was concentrated 10-fold by precipitation with saturated ammonium sulfate. After centrifugation, the precipitates were resuspended and dialyzed against HGE.1 (20 mM HEPES [at H 7.9, 20% glycerol, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.1 M KCl). The number following HGE denotes the molar concentration of KCl. Concentrated P.3 was applied to a 20-mL DEAE Sepharose column equilibrated with HGE.1, and bound proteins were step-eluted with HGE.225, HGE.3, and HGE1.0. DC activity was found in the 0.3 M KCl fraction (D.3). Then, D.3 was applied directly (in 1-mg aliquots) to a mono-Q PC 1.6/5 column equilibrated with HGE.3, and bound proteins were eluted with a 1.5-mL linear gradient from 0.3 to 1.0 M KCl. DC activity was separated at this step, and the activities peaking at fractions #12 and #14 were termed DC1 and DC2, respectively. For purification of DC1, the mono-Q fraction #12 was dialyzed against HGE.05 and subjected to a mono-S PC 1.6/5 column. Bound proteins were eluted with a 1.5-mL linear gradient from 0.05 to 0.6 M KCl. At this step, five polypeptides were found associated with DC1 activity and, therefore, these polypeptides were excised from an SDS gel and subjected to LC-MS/MS analysis. For purification of DC2, the mono-Q fraction #14 was dialyzed against HGE.05 and subjected to a mono-S PC 1.6/5 column. Since DC2 activity was found in the flow-through fraction, this fraction was applied directly to the second mono-Q column, and bound proteins were eluted with a 1.5-mL linear gradient from 0.05 to 1.0 M KCl. At this step, a single polypeptide was found associated with DC2 activity and, therefore, this polypeptide was excised from an SDS gel and subjected to LC-MS/MS analysis.
Continuous-labeling transcription assays were performed using pSLG402 (Lee and Greenleaf 1997) essentially as described (Guo et al. 2000), except that 6 μL of P1.0 were used instead of NE to support transcription initiation. Where indicated by “+” or “1×,” the following proteins were also included: rDSIF, 0.6 pmol each of rSpt5 and rSpt4; native Paf1C, 0.4 pmol; native Tat-SF1, 0.5 pmol; Flag-Paf1C, 0.4 pmol; rTat-SF1, 2.5 pmol. Following preincubation of 20-μL reactions for 45 min, 5 μL of 4NTPs (final concentrations of 30 μM ATP, 300 μM CTP, 30 μM GTP, 2.5 μM [α-32P] UTP) were added, and initiation/elongation was allowed to proceed for 20 min unless stated otherwise.
Pulse-chase transcription assays were performed using pML-DGL1, a pSLG402 derivative in which the first G-less cassette of 83 base pairs (bp) in length was placed directly downstream from the major-late promoter. Following 45 min of preincubation, 3NTPs (final concentrations of 500 μM ATP, 500 μM CTP, 0.5 μM [α-32P] UTP) were added and incubated for 20 min to allow the formation of +83 complexes. Then, chase reactions were carried out in the presence of 500 μM (each) 4NTPs for 30–60 sec.
Recombinant plasmids, recombinant proteins, and cell lines
pFlag-Leo1 and pFlag-Paf1 were gifts from D. Reinberg. pFastBac-HT-myc-Tat-SF1 was a gift from K. Nagata. For mammalian expression of Tat-SF1, sequence encoding Tat-SF1 and the C-terminal Flag tag (a gift from Q. Zhou) was subcloned into pcDNA3. For mammalian expression of Spt5, sequence encoding the N-terminal HA tag and Spt5 was subcloned into pcDNA3. Spt5 TA and SA were described previously (Yamada et al. 2006). Spt5ΔCTR lacks amino acids 758–936 and is identical to Spt5Δ repeat described previously (Yamaguchi et al. 1999b).
To establish a HeLa cell line stably expressing Flag-Leo1, HeLa cells were transfected with pFlag-Leo1 and selected for 2 wk in the presence of 500 μg/mL G418. A number of clonal cell populations were isolated, and a cell line expressing Flag-Leo1 at a moderate level was selected. A HeLa cell line stably expressing Flag-Tat-SF1 was similarly established using pcDNA3-Flag-Tat-SF1.
Preparation of rSpt5 and rSpt4 was detailed in Yamaguchi et al. (1999b). Flag-tagged Paf1C was prepared essentially as described (Zhu et al. 2005) with minor modifications. Briefly, NE was prepared from HeLa/Flag-Leo1 cells and was loaded onto an anti-Flag affinity column. After stringent wash with HGE.5, bound proteins were eluted with Flag peptide and further subjected to mono-Q column chromatography. For preparation of rTat-SF1, recombinant baculovirus was produced with pFastBac-myc-Tat-SF1 and Bac-to-Bac Baculovirus Expression System (Invitrogen). His- and myc-tagged Tat-SF1 was overexpressed in baculovirus-infected Sf9 cells and purified by nickel-affinity chromatography.
For baculoviral expression of Paf1C, Cdc73/HRPT2 cDNA was obtained from Addgene (Rozenblatt-Rosen et al. 2005); Paf1 and Leo1 cDNAs were provided by D. Reinberg (Zhu et al. 2005); Ctr9/p150TSP cDNA was provided by S. Desiderio (Malek et al. 1996); and Ski8 cDNA was cloned by RT–PCR from HeLa total RNA. These cDNAs, except Paf1, were subcloned into pFastBac 1. Paf1 cDNA was subcloned into a pFastBac HT-B derivative that was engineered to produce an N-terminal His- and V5-tagged protein. Five recombinant baculoviruses generated from these constructs were coinfected into Sf9 cells and, 48 h post infection, cell lysates were prepared and subjected to anti-V5-affinity chromatography.
shRNA-mediated knockdown and RNA analysis
Double-stranded oligonucleotides encoding shRNAs were cloned into pBS-U6 as described (Yamada et al. 2006). The following 21-nucleotide (nt) sequences were used as shRNA targets: Spt5 shRNA, 5′-AAGAACUGGGCGAGUAUUACA-3′; Tat-SF1 shRNA #1, 5′-AAGAAUUGUACGGAGACGGCA-3′; Tat-SF1 shRNA #2, 5′-AAGAUGAAAUUAGAGGCUACA-3′; Leo1 shRNA #1, 5′-AAGUUUUACGAAUGAAACGCA-3′; Leo1 shRNA #2, 5′-AAGAUAAUAGUGGAACCAUGG-3′; Cdc73 shRNA #1, 5′-AAGUAUAGACAGAAGCGCUCC-3′; Cdc73 shRNA #2, 5′-AAGAAACCACGAAUUGAGGAU-3′. After functional validation, cassettes including a mouse U6 promoter were excised and subcloned into pLenti6 (Invitrogen). Recombinant lentiviruses were produced and concentrated prior to use according to standard procedures. In knockdown experiments, HeLa cells were infected with recombinant lentivirus expressing no shRNA or expressing one of the shRNAs and were selected in the presence of 4 μg/mL Blasticidin. Four days to 6 d post-infection, the cells were either lysed with high-salt buffer (50 mM Tris-HCl at pH 7.9, 500 mM NaCl, 1% NP-40) for immunoblotting or harvested for RNA analysis. Spliced mRNAs for FOS, EGR1, and GAPDH were quantified by one-step RT–PCR using QuantiTect SYBR Green RT–PCR kit (Qiagen) and an ABI 7300 real-time PCR system (Applied Biosystems).
Immunological analyses
Anti-Rpb4, anti-Spt5, anti-Leo1, and anti-Cdc73 antibodies were produced in-house in rabbits. Anti-Rpb4 was raised against histidine-tagged full-length Rpb4. Anti-Spt5 was raised against the C-terminal fragment (amino acids 937–1087) of Spt5 fused to glutathione-S-transferase. Anti-Leo1 was raised against the N-terminal fragment (amino acids 90–320) of Leo1 fused to glutathione-S-transferase. Anti-Cdc73 was raised against histidine-tagged full-length Cdc73. Anti-Paf1 was a gift from D. Reinberg. The following commercial antibodies were also used: anti-H3 (Abcam, ab1791), anti-H3K4me3 (Abcam, ab8580), anti-H3K4me3 (Upstate Biotechnologies, 07-473), anti-H3K36me3 (Abcam, ab9050), anti-H3K79me2 (Abcam, ab3594), anti-H2B (Abcam, ab1790), anti-H2Bub (Upstate Biotechnologies, 05-1312).
ChIP assays were performed according to standard procedures (Aida et al. 2006). Of the five primer sets used, c, d, and e were described previously (Yamada et al. 2006). The remaining primer sets are as follows: a, 5′-TGCAGTCACCTCCTCTGGGA-3′ and 5′-GGCTGGTGCTCACTGTAATG-3′; f, 5′-ACTACTCAGGAGGCTGAGGC-3′ and 5′-TGATTGAGGTCCCCAGGAGC-3′. Absolute quantification of target sequences was performed by real-time PCR.
Immunoprecipitations and immunodepletions were carried out essentially as described (Yamaguchi et al. 1999a; Narita et al. 2007).
Acknowledgments
We thank P. Sharp, J. Kim, Q. Zhou, D. Reinberg, B. Zhu, K. Nagata, and S. Desiderio for plasmids and antibodies. We also thank S. Chanarat for his contribution during the initial stage of this project, and J. Kato and S. Okamoto for technical assistance. This work was supported by Special Coordination Funds for Promoting Science and Technology from the Japan Science and Technology Agency (to H.H.) and by a Grant-in-Aid for Scientific Research on Priority Areas from MEXT (to Y.Y.). This work was also supported in part by a Grant of the Global COE Program from MEXT.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1834709.
Supplemental material is available at http://www.genesdev.org.
References
- Adelman K, Wei W, Ardehali MB, Werner J, Zhu B, Reinberg D, Lis JT. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol Cell Biol. 2006;26:250–260. doi: 10.1128/MCB.26.1.250-260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Chen Y, Nakajima K, Yamaguchi Y, Wada T, Handa H. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol Cell Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: Roles in promoter proximal pausing and transcription elongation. Genes & Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso T, Lane WS, Conaway JW, Conaway RC. Elongin (SIII): A multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- Bourgeois CF, Kim YK, Churcher MJ, West MJ, Karn J. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol Cell Biol. 2002;22:1079–1093. doi: 10.1128/MCB.22.4.1079-1093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- Fish RN, Kane CM. Promoting elongation with transcript cleavage stimulatory factors. Biochim Biophys Acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- Guo S, Yamaguchi Y, Schilbach S, Wada T, Lee J, Goddard A, French D, Handa H, Rosenthal A. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature. 2000;408:366–369. doi: 10.1038/35042590. [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes & Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa J, Endou M, Narita T, Yamada T, Yamaguchi Y, Wada T, Handa H. A rapid purification method for human RNA polymerase II by two-step affinity chromatography. J Biochem. 2003;133:133–138. doi: 10.1093/jb/mvg013. [DOI] [PubMed] [Google Scholar]
- Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Yamaguchi Y, Wada T, Handa H, Sharp PA. Tat-SF1 protein associates with RAP30 and human SPT5 proteins. Mol Cell Biol. 1999;19:5960–5968. doi: 10.1128/mcb.19.9.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Lee JM, Greenleaf AL. Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J Biol Chem. 1997;272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Green MR. The HIV-1 Tat cellular coactivator Tat-SF1 is a general transcription elongation factor. Genes & Dev. 1998;12:2992–2996. doi: 10.1101/gad.12.19.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol. 2009;29:4852–4863. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek SN, Yang CH, Earnshaw WC, Kozak CA, Desiderio S. p150TSP, a conserved nuclear phosphoprotein that contains multiple tetratricopeptide repeats and binds specifically to SH2 domains. J Biol Chem. 1996;271:6952–6962. doi: 10.1074/jbc.271.12.6952. [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Lee JS, Gardner KE, Gardner JM, Takahashi YH, Chandrasekharan MB, Sun ZW, Osley MA, Strahl BD, Jaspersen SL, et al. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J Cell Biol. 2009;186:371–377. doi: 10.1083/jcb.200906005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yung TMC, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. NELF interacts with CBC and participates in 3′-end processing of replication-dependent histone mRNAs. Mol Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Parada CA, Roeder RG. A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 1999;18:3688–3701. doi: 10.1093/emboj/18.13.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. A posttranscriptional role for the yeast Paf1–RNA polymerase II complex is revealed by identification of primary targets. Mol Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes & Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Ponting CP. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 2002;30:3643–3652. doi: 10.1093/nar/gkf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH. Poised polymerases: On your mark…get set…go! Mol Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Qiu H, Hu C, Wong CM, Hinnebusch AG. The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol Cell Biol. 2006;6:3135–3148. doi: 10.1128/MCB.26.8.3135-3148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D, Sims RJ., 3rd de FACTo nucleosome dynamics. J Biol Chem. 2006;281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- Rondón AG, Gallardo M, García-Rubio M, Aguilera A. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 2004;5:47–53. doi: 10.1038/sj.embor.7400045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, Resau JH, Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Sheldon KE, Mauger DM, Arndt KM. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Finkelstein A, Wolf AJ, Wade PA, Burton ZF, Jaehning JA. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol Cell Biol. 1996;16:669–676. doi: 10.1128/mcb.16.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney K, Gerber M, Ilvarsonn A, Schneider J, Gause M, Dorsett D, Eissenberg JC, Shilatifard A. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc Natl Acad Sci. 2006;103:11970–11974. doi: 10.1073/pnas.0603620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk A. Mediator requirement for both recruitment and post-recruitment steps in transcription initiation. Mol Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes & Dev. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-H, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinski J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour DS. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes & Dev. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Yamaguchi Y, Inukai N, Kamijo S, Mura T, Handa H. P-TEFb-mediated phosphorylation of the hSpt5 C-terminal repeats is critical to processive transcription elongation. Mol Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999a;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J Biol Chem. 1999b;274:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Deléhouzée S, Handa H. HIV and HDV: Evolution takes different paths to relieve blocks in transcriptional elongation. Microbes Infect. 2002;4:1169–1175. doi: 10.1016/s1286-4579(02)01641-6. [DOI] [PubMed] [Google Scholar]
- Yan D, Perriman R, Igel H, Howe KJ, Neville M, Ares M., Jr CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol. 1998;18:5000–5009. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yart A, Gstaiger M, Wirbelauer C, Pecnik M, Anastasiou D, Hess D, Krek W. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol Cell Biol. 2005;25:5052–5060. doi: 10.1128/MCB.25.12.5052-5060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Klatt A, Gilmour DS, Henderson AJ. Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J Biol Chem. 2007;282:16981–16988. doi: 10.1074/jbc.M610688200. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Sharp PA. Tat-SF1: Cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science. 1996;274:605–610. doi: 10.1126/science.274.5287.605. [DOI] [PubMed] [Google Scholar]
- Zhou K, Kuo WH, Fillingham J, Greenblatt JF. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci. 2009;106:6956–6961. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes & Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]