SUMMARY
An important issue for chromatin remodeling complexes is how their bromodomains recognize particular acetylated lysine residues in histones. The Rsc4 subunit of the yeast remodeler RSC contains an essential tandem bromodomain (TBD) that binds acetylated K14 of histone H3 (H3K14ac). We report a series of crystal structures that reveal a compact TBD that binds H3K14ac in the second bromodomain and, remarkably, binds acetylated K25 of Rsc4 itself in the first bromodomain. Endogenous Rsc4 is acetylated only at K25, and Gcn5 is identified as necessary and sufficient for Rsc4 K25 acetylation in vivo and in vitro. Rsc4 K25 acetylation inhibits binding to H3K14ac, and mutation of Rsc4 K25 results in altered growth rates. These data suggest an autoregulatory mechanism in which Gcn5 performs both the activating (H3K14ac) and inhibitory (Rsc4 K25ac) modifications, perhaps to provide temporal regulation. Additional regulatory mechanisms are indicated as H3S10 phosphorylation inhibits Rsc4 binding to H3K14ac peptides.
INTRODUCTION
Chromatin serves a central role in regulating the access of transcription factors to chromosomal loci. The primary repeating unit of chromatin, the nucleosome, helps organize DNA topology by wrapping DNA, a property that can occlude binding sites for regulatory factors and thereby contribute to transcriptional silencing (Kornberg and Lorch, 1999). However, the nucleosome is a dynamic participant in transcriptional activation, because nucleosome remodelers function to reposition nucleosomes to expose the underlying DNA. Furthermore, a large array of covalent modifications occur on the histone components and can serve as binding epitopes for protein domains specialized for their recognition. The principle of histone marking by covalent modification and recognition by specific domains has been termed “the histone code” (Fischle et al., 2003; Strahl and Allis, 2000). These binding domains reside on both chromatin regulators and transcriptional regulators. Thus, most factors are targeted to particular locations in the genome by one of two mechanisms: through interactions with site-specific DNA binding proteins or by using specialized domains to interact with modified histones. The most common posttranslational modification of histones is the acetylation of lysine residues by histone acetyltransferase (HAT) enzymes, which occurs primarily on the flexible N-terminal histone “tails” that emanate from the globular nucleosome core (Kouzarides, 2000). One of the best-studied HAT enzymes is yeast Gcn5, which acetylates lysine 14 of histone H3 (H3K14ac), a modification correlated with transcriptional activation (Brownell et al., 1996; Howe et al., 2001; Lo et al., 2000; Syntichaki et al., 2000; Trievel et al., 1999). Acetylated lysines are typically bound by ~110 amino acid residue structures called bromodomains that also recognize several of the residues flanking the acetyl-lysine, thereby providing acetyl-lysine recognition within a sequence context (Hudson et al., 2000; Mujtaba et al., 2002; Owen et al., 2000). There is considerable interest in determining which bromodomains bind particular histone acetyl-lysines and whether these interactions mediate targeting or some other aspect of regulation.
Complexes that rely on bromodomains for their full function include chromatin remodelers, which use the energy of ATP hydrolysis to move and/or eject nucleosomes to uncover the underlying DNA (Cairns, 2005). Indeed, how remodelers are targeted and regulated is a central question in chromatin biology. Important initial work demonstrated that bromodomains present on the yeast remodeler SWI/SNF are important for the retention of the remodeler on acetylated chromatin templates, consistent with a role for bromodomains in targeting (Hassan et al., 2002, 2006). The paralog of ySWI/SNF is the 15 subunit remodels the structure of chromatin (RSC) complex, which is both abundant and essential in S. cerevisiae (Cairns et al., 1996) and is involved in multiple chromosomal processes including transcriptional regulation, DNA repair, stress response, and chromosome cohesion and segregation (Angus-Hill et al., 2001; Baetz et al., 2004; Cairns et al., 1999; Chai et al., 2005; Chang et al., 2005; Yukawa et al., 1999). Importantly, RSC subunits contain 8 of the 15 bromodomains in S. cerevisiae, indicating that histone acetylation likely plays a central role in recruiting RSC to chromatin and/or in regulating its remodeling activity. Consistent with this notion, acetylation of histones promotes nucleosome remodeling by RSC and the passage of RNA polymerase II through chromatin in vitro (Carey et al., 2006).
The Rsc4 subunit of the RSC complex, which contributes to RSC-activated Pol II transcription in vivo (Kasten et al., 2004; Soutourina et al., 2006), contains a pair of bromodomains, termed BD1 and BD2, that are essential for cell viability (Kasten et al., 2004). BD1 and BD2 are adjacent in the primary protein sequence and together form the Rsc4 tandem bromodomain (TBD, residues 56–304). Our previous studies indicated that the Rsc4 TBD binds H3K14ac (Kasten et al., 2004). For example, the Rsc4 TBD preferentially binds histone H3 N-terminal peptides acetylated at K14, but not at several other positions tested on both the H3 and the H4 tails. Furthermore, conditional rsc4 alleles are lethal in combination with gcn5Δ (Gcn5 acetylates H3K14) or with h3K14 replacements (at 33°C), but not in combination with mutation of other lysine residues in the H3 and H4 tails (Kasten et al., 2004). However, it was unresolved which of the bromodomains (or both) bound H3K14ac, and the clear possibility remained of alternative ligands. Furthermore, it was not known whether other modifications occurring near H3K14ac, such as H3S10 phosphorylation, might affect binding.
To better understand Rsc4 recognition of chromatin, we performed biochemical and genetic studies that showed that only BD2 binds to H3K14ac peptides, and we visualized the acetyl lysine component of this interaction using X-ray crystallography. Serendipitously, crystal structure determination of protein prepared by in vitro acetylation with Gcn5 revealed that an acetylated lysine of Rsc4(K25ac) binds to its own BD1. This interaction was shown to be important in vivo, and the modifying enzyme was identified as Gcn5, the same acetyltransferase that modifies the H3K14 ligand of BD2. Importantly, peptide-binding data showed that binding of Rsc4 K25ac to BD1 impairs the ability of BD2 to bind an H3K14ac peptide, thereby indicating an autoregulation mechanism for recognition of a histone modification.
RESULTS AND DISCUSSION
Structure of the Rsc4 Tandem Bromodomain
Several crystal structures of Rsc4 constructs were determined (Figure 1). The first, Rsc4(36–340), was determined by the SAD method using selenomethionine-substituted protein and refined to a free R value of 21.8% against native data to 1.8 Å resolution. This structure is ordered from residue 36 to residue 320 with no disordered internal loops. Structures of other constructs were subsequently determined by molecular replacement and refined to resolutions of 1.75–2.35 Å and Rfree values of 21.9%–24.8%. While the structures varied significantly in their last ordered residue (313–320), only minor differences were seen for residues 36–312, with root-mean-square deviations (rmsds) of 0.5–1.0 Å following least-squares overlap on 275 pairs of Cα atoms.
Figure 1. Structures of Rsc4.
(A) Domain organization of Rsc4 (top) and crystallized constructs (bottom). Rsc4 structure is the following: bromodomain 1 (cyan), bromodomain 2 (blue), wing insertion (orange), and the binding region for RSC and the RNA polymerases (green) (Kasten et al., 2004; Soutourina et al., 2006). First and last ordered residues in the crystallized constructs are indicated with arrows. Acetylated lysine residues are indicated with a magenta dot (K14 of H3 and K25 of Rsc4).
(B) Orthogonal views of Rsc4(36–340) ribbon diagram. Secondary structures are labeled, and termini are indicated N and C. The asparagine and two tyrosine side chains from each bromodomain that are important for acetyl lysine binding are colored yellow. The wing insertion is colored orange.
(C) Rsc4 amino acid sequence with secondary structures indicated above. Residues not ordered in the Rsc4(36–340) structure are indicated with a dashed line. The red squares indicate interface residues between BD1 (residues 36–162) and BD2 (residues 163–320). The magenta circle indicates the site of acetylation (K25).
The Rsc4 TBD is a compact structure in which each of the individual bromodomains (BD1 and BD2) resembles bromodomains from other proteins (Mujtaba et al., 2002; Owen et al., 2000; Sun et al., 2007). In keeping with standard nomenclature, we name the four bundle helices Z, A, B, and C, with a -1 or -2 suffix to indicate if it is from the first or second bromodomain (Figure 1C). The acetyl-lysine binding pockets are formed primarily by residues within the BC and ZA loops including the short helix Z′. Both BD1 and BD2 conserve two tyrosine residues within Z′ and an asparagine residue within B that are characteristic of bromodomain binding sites. Overlap on Cα atoms with the bromodomain from Gcn5, which shares 19% and 33% sequence identity with Rsc4 BD1 and BD2, respectively, and whose crystal structure has been determined at 1.9 Å resolution (Owen et al., 2000), gives rmsds of 1.8 Å (100 pairs of Cα atoms) for BD1 and 1.6 Å (106 pairs of Cα atoms) for BD2.
The Rsc4 TBD reveals important differences with the previously reported structure of the double bromodomain of TAFII1 (formerly termed TafII250) (Jacobson et al., 2000), the largest subunit of the TBP-associated factors for RNA Pol II transcription. We note that TAFII1 was only crystallized in the absence of ligand. Notably, the relative positions and orientations of the two bromodomains in Rsc4 are very different to those in TAFII1 (see Figure S1 in the Supplemental Data available with this article online). Also, Rsc4 is substantially more compact, with extensive BD1-BD2 interactions (mainly through αC-1 and αB-2) that bury a total of 1621 Å2 of solvent accessible surface area at the bromodomain interface (Figure S2). In comparison, 1122 Å2 are buried at the BD1-BD2 interface in the TAFII1 structure. This supports the impression from genetic data that the Rsc4 TBD functions as a single structural unit (Kasten et al., 2004). In contrast, the two bromodomains of TAFII1 appear to be relatively independent. As a result of these differences, the two acetyl-lysine binding sites of Rsc4 face the same side of the structure in the same relative orientation and are separated by just 20 Å.
Whereas BD1 conforms to the standard bromodomain architecture, BD2 includes an additional “wing” helix (W) inserted between Z-2 and A-2. Because it is adjacent to the presumed binding surface, we hypothesized that W might play an important functional role. We therefore designed a deletion variant that replaced residues 187–206 with a short Ser-Ser-Gly linker, termed rsc4Δ187–206. Although the rsc4Δ187–206 allele does not confer a strong phenotype in isolation, a temperature-sensitive (ts−) phenotype was conferred in combination with gcn5Δ (data not shown), suggesting that the wing might assist BD2 in H3K14ac recognition.
H3K14ac Binds Preferentially to Rsc4 BD2
We took both in vivo and in vitro approaches to determine which of the two Rsc4 bromodomains binds H3K14ac. Our in vivo approach involved isolating mutations specifically impaired at individual bromodomains and testing them in combination with gcn5Δ and h3K14 mutations. First, we made mutations in BD1 or BD2 of two tyrosine residues that are highly conserved in all bromodomains (Figure S3) and are important for recognizing the acetyl moiety in acetyl-lysine. Replacement of these tyrosines with alanine resulted in the rsc4 alleles rsc4 Y92A Y93A (BD1 mutant), rsc4 Y225A Y226A (BD2 mutant), and the combined rsc4 Y92A Y93A Y225A Y226A (BD1&2 mutant) alleles. Each encoded a stable derivative that was fully capable of assembly into the RSC complex (data not shown). The isolated BD mutant alleles lack clear plate phenotypes, likely due to the fact that the normal interaction of bromodomains with their substrates involves an interaction with the acetyl moiety (which is compromised) and also the peptide sequence (which is retained). However, the combined BD1&2 mutations conferred lethality (even in WT GCN5 background, Figure 2A). This demonstrates that the two bromodomains are partially redundant; compromising the acetyl-binding pocket of both bromodomains is needed to confer inviability. We note that redundancy can result from the two bromodomains binding to different ligands; compromising one bromodomain makes Rsc4 reliant on the alternative bromodomain and its ligand(s) to conduct an essential function.
Figure 2. Rsc4 BD2 Interacts Genetically and Biochemically with Histone H3K14ac.
(A) Complementation of rsc4 BD mutants in WT (YBC627 and YBC2499), gcn5Δ (YBC2352), and histone h3K14G (YBC2501) strains. Patches were incubated for 2 days at 30°C. The RSC4 construct location in the groups of five is provided by the key at top: vector, pRS314; RSC4, pRS314.RSC4 (p1060); BD1 mutant, pRS314.rsc4 Y92A Y93A (p1462); BD2 mutant, pRS314.rsc4 Y225A Y226A (p1463); BD1&BD2 mutant, pRS314.rsc4 Y92A Y93A Y225A Y226A (p1471). Strains were grown on selective media with or without 5-FOA, as 5-FOA enforces the loss of the pRS316.RSC4 plasmid (right panels).
(B) Rsc4 BD2 is required for acetyl-specific binding to the H3K14 peptide. The binding of purified Rsc4(46–334) derivatives to biotinylated histone H3 tail peptides conjugated to streptavidin beads was examined by western blot. The H3 peptides extend from amino acid 1 to 39 of the H3 sequence and are either unmodified or acetylated at K14 as indicated. Binding avidity was further examined by increasing salt stringency in the wash buffers, as indicated. Western blots were probed with anti-Rsc4 polyclonal antibodies.
(C) Rsc4 BD2 is sufficient to bind to the H3K14ac peptide. Western blot analysis of purified Rsc4(36–321) TBD and Rsc4(157–321) BD2, as in (B).
(D) Binding by Rsc4 TBD and BD2 to H3K14ac is inhibited by H3S10ph. Western blot analysis of purified Rsc4 TBD bound to the H3 tail peptides, as in (B).
(E) Acetyl-lysine bound within the BD2 pocket of Rsc4(36–340) following soaking with H3(6–18) K14ac peptide. Difference density (Fo − Fc), contoured at 3.0 × rmsd, was phased on protein model refined prior to inclusion of the ligand. The closely overlapping acetyl lysine side chains from a Gcn5 bromodomain:peptide complex structure (Owen et al., 2000) are shown colored white following superposition of the protein structures.
In support of the two bromodomains binding different ligands, we found that rsc4 Y92A Y93A gcn5Δ combinations were lethal, whereas rsc4 Y225A Y226A gcn5Δ combinations grew well (Figure 2A). As above, lethality could result from compromising both BD1 and BD2; BD1 is compromised through mutation of the tyrosine residues and BD2 through the lack of the acetyl group on the H3K14 substrate in the gcn5Δ background. Consistent with this notion, synthetic lethality was also observed when the BD1 mutant was combined with the h3K14G mutant (Figure 2A). These data implicate BD2 in binding H3K14ac, because mutations in BD2 that prevent the recognition of the acetyl moiety (Y225A, Y226A) are not expected to be exacerbated by the absence of the acetyl group on the substrate (from gcn5Δ). In contrast, the synthetic lethality observed by combining a BD1 mutation with gcn5Δ implicates an important function for BD1 in binding a ligand(s) distinct from H3K14ac.
An NMR titration experiment indicated that the Rsc4-H3K14ac peptide dissociation constant is approximately 1–2 mM (data not shown). This low inherent binding is not unusual for bromodomains (Hudson et al., 2000; Shen et al., 2007; Sun et al., 2007) and presumably reflects the involvement of additional contacts with the nucleosome in the context of the intact RSC complex. It does, however, limit the choice of binding assay, and we therefore estimated relative binding affinities using biotinylated H3 tail peptides (residues 1–39) that were either acetylated at K14 or unmodified, and were immobilized on streptavidin beads. This analysis revealed that Rsc4(46–334) binds H3K14ac peptides in preference to unmodified peptides (Figure 2B). Consistent with the genetic experiments, the Y92F variant (intact BD2) binds H3K14ac peptides preferentially in the same manner as the wild-type (WT) sequence, whereas preferential binding is abolished with the equivalent mutation in BD2 (Y225F) (Figure 2B). Moreover, a Rsc4(157–321) BD2 derivative that lacked BD1 entirely still demonstrated a preferential interaction with H3K14ac peptides compared to unmodified peptides (Figure 2C). Not surprisingly, given the extensive interactions between BD1 and BD2, peptide binding to the isolated BD2 structure was weaker than to the TBD, and the isolated BD1 was insoluble (data not shown). The binding assay was further used to map the Rsc4 interaction to residues 6–21 of the H3K14ac peptide, with further truncation from the N terminus preventing binding to the intact Rsc4(36–321) (data not shown). Thus, these data indicate that Rsc4 BD2 preferentially binds the H3 tail acetylated at K14 with other interactions also contributing to binding affinity. We also examined whether or not modifications near H3K14 affect H3K14ac recognition. Previous work suggested that H3K9 acetylation does not affect Rsc4 binding (Kasten et al., 2004). However, S10 phosphorylation has previously been linked to H3K14 acetylation (Lo et al., 2000) and had not been examined in our prior studies. We find that S10 phophorylation antagonizes selective binding of Rsc4(36–321) to H3K14ac peptides, raising the possibility that this modification may restrict RSC binding in vivo (Figure 2D).
Structure of Rsc4 BD2 Bound to H3K14ac
To visualize the H3-Rsc4 interaction, we determined the structure of a peptide complex by soaking Rsc4(36–340) crystals in high concentrations (~40 mM) of H3(6–18)K14ac (Table 1). Density for the peptide backbone is not apparent in this 1.7 Å resolution structure but is clearly defined for an acetylated lysine side chain in the BD2 binding site in a conformation that overlaps closely with the structure of a Gcn5 bromodomain ligand complex (Figure 2E). Unfortunately, density was not reliably interpretable beyond the side chain, and it was not possible to determine the path of the bound peptide backbone. Numerous data sets were obtained and structures determined at 1.7–2.3 Å resolution for Rsc4(36–340) cocrystallized or soaked with H3 peptide, but ligand density was always limited to the Kac side chain. The BD1 and BD2 pockets are open in the crystal lattice, although they are each within 6 Å of significant crystal contacts. Because in vitro binding appears normal in the 6–21 H3K14ac peptide but is abolished in a 10–21 peptide (data not shown), it seems likely that lattice contacts prevent secondary interactions that are important for full binding affinity and block the peptide from adopting the bound conformation outside of the primary binding pocket. Nevertheless, this structure supports the genetic and biochemical studies by showing that H3K14ac can bind Rsc4 BD2.
Table 1.
Data Collection and Refinement Statistics
| Rsc4(36–340) |
Rsc4(36–340) |
Rsc4(1–321) |
Rsc4(1–340) |
|
|---|---|---|---|---|
| Histone (6–18, K14ac) Peptide Soak | Histone Chimera, Acetylated | Acetylated | ||
| Data Collection | ||||
| Space group | R32 | R32 | C2221 | C21 |
| Cell dimensions (Å) | a = b = 95.9, c = 233.5 | a = b = 95.0, c = 233.1 | a = 86.1, b = 91.0, c = 263.2 | a = 123.3, b = 83.2, c = 127.1, β = 109.33 |
| Resolution (Å) | 20–1.80 | 50–1.75 | 50–2.20 | 50–2.35 |
| Resolution Outer Shell (Å) | 1.86–1.80 | 1.81–1.75 | 2.28–2.20 | 2.43–2.35 |
| No. reflections | 514,412 | 335,001 | 548,136 | 906,673 |
| Unique reflections | 37,132 | 39,000 | 49,197 | 50,527 |
| Rsym (%) | 5.5 (36.9) | 6.8 (59.7) | 7.8 (49.4) | 7.6 (50.3) |
| I/σ(I) | 25.5 (1.6) | 33.7 (2.2) | 32.0 (2.8) | 26.9 (3.2) |
| Completeness (%) | 96.2 (67.4) | 94.6 (72.4) | 95.3 (83.9) | 99.5 (99.7) |
|
| ||||
| Refinement | ||||
| Rwork/Rfree (%) | 17.6/21.8 | 18.4/21.9 | 20.1/24.8 | 18.0/22.3 |
|
| ||||
| Number of Atoms | ||||
| Protein | 2744 | 2669 | 5342 | 7790 |
| Solvent | 351 | 253 | 369 | 506 |
| Average Isotropic B factor(Å)2 | 30.6 | 34.2 | 51.4 | 40.7 |
|
| ||||
| Ramachandran Plot, Nonglycine Residue in | ||||
| Most favorable region (%) | 92.1 | 91.7 | 91.4 | 91.7 |
| Allowed region (%) | 7.9 | 8.3 | 8.2 | 8.2 |
|
| ||||
| Rmsd | ||||
| Bond lengths (Å) | 0.016 | 0.016 | 0.013 | 0.016 |
| Bond angles (°) | 1.241 | 1.406 | 1.416 | 1.211 |
Rsc4 K25ac Binds BD1
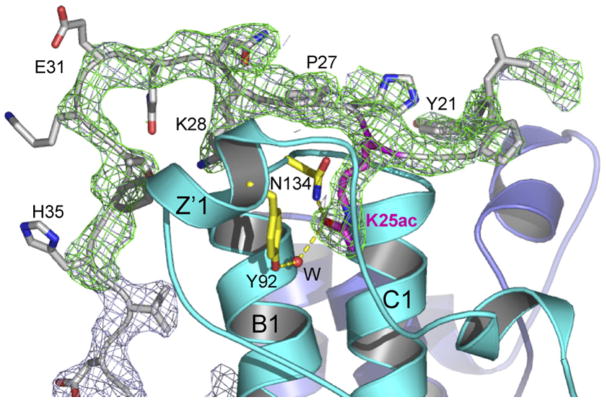
In an effort to overcome the problem of low-affinity binding and to visualize a bound H3 peptide better, we took a chimera approach in which a very high local concentration of H3 peptide might be achieved by expressing the H3 tail as a fusion with the Rsc4 N terminus. The fusion included H3 residues 6–18, followed by eight residues containing a thrombin cleavage site, followed by Rsc4 residues 1–321 (Figure 1A). This construct contained the entire N-terminal sequence of Rsc4, whereas our initial crystal structures lacked the first 35 residues. The purified chimeric protein was acetylated at H3K14 using purified recombinant Gcn5, with progress of the reaction monitored by migration of the H3 peptide (following treatment and release with thrombin) using acid-urea gel electrophoresis. Acetylation was also monitored by western analysis, which revealed high levels of H3K14ac and very low levels of H3K9ac (data not shown). To more definitively identify the acetylated lysines, we subjected tryptic fragments to mass spectrometric analysis. This revealed nearly complete acetylation at H3K14 and, surprisingly, nearly complete acetylation at Rsc4 K25. As described below, the modification of K25 was unexpected but highly fortuitous. The acetylated chimeric H3-Rsc4 protein was crystallized and the structure determined. Counter to the design goal, H3 residues were not ordered, and the BD2 pocket appeared to be empty. This was disappointing, but given the lack of information about likely binding orientation, the failure was not surprising. Remarkably, however, Rsc4 K25ac was present in the BD1 pocket, and the surrounding residues were well ordered, starting at residue 19 (Figure 3). An essentially identical arrangement was revealed in the subsequently determined crystal structure of Rsc4(1–340) bearing K25ac. This derivative bore its natural N terminus and lacked the H3 peptide, and quantitative acetylation at K25 (by Gcn5) was verified by mass spectrometry (data not shown). This binding could be recapitulated in trans, as a peptide containing Rsc4 residues 18–34 acetylated at K25 interacted with the TBD protein in vitro, though with low affinity (Figure S4).
Figure 3. Binding of Rsc4 K25ac in BD1.
Acetylated K25 binds BD1 and orders flanking residues. The Fo − Fc map (3.0 × rmsd, green) and 2Fo − Fc map (1.2 × rmsd, gray) were phased from the protein model refined in the absence of residues 19–35. Residues involved in K25ac recognition are shown in yellow.
Rsc4 K25 Is Acetylated by Gcn5 In Vivo
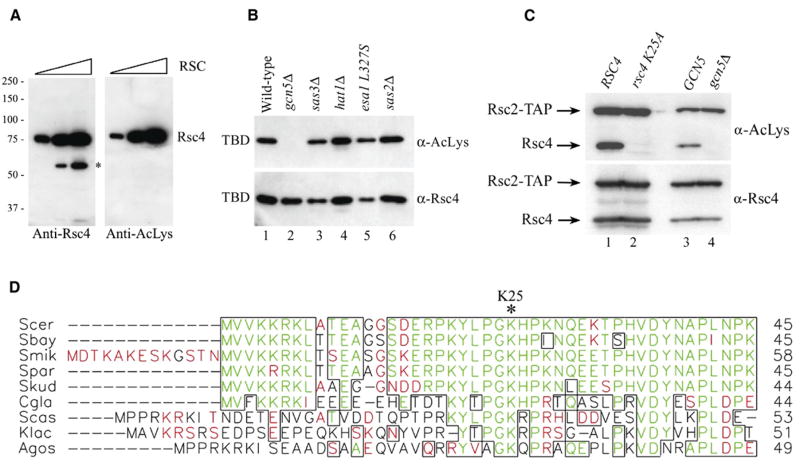
Motivated by the surprising crystallographic observation that Rsc4 K25ac binds BD1, we sought to determine if this modification occurs in vivo. Western blot analysis of purified RSC complex using an antibody against acetyl-lysine revealed a single dominant band that comigrated with Rsc4 (Figure 4A). To identify the acetyltransferase that modifies Rsc4 in vivo, we expressed in yeast a Rsc4(1–340) construct that also included a nuclear targeting sequence and a 10 × HIS tag. We then purified this protein with nickel chelating chromatography and examined its acetylation state. Western analysis revealed clear acetylation in the WT strain and in a wide variety of strains lacking specific acetyltransferases. However, Rsc4 K25 acetylation was abolished in a strain lacking Gcn5 (Figure 4B). As a definitive test for the presence of Gcn5-dependent Rsc4 K25ac in the RSC complex in vivo, we made a Rsc4 derivative with a single amino acid replacement (K25A) as the sole source of Rsc4. This derivative or the WT Rsc4 was expressed in WT cells and purified as a component of the RSC complex using a TAP tag on the Rsc2 subunit (Figure 4C, lanes 1 and 2). Importantly, substitution of K25 abolished the acetylation of Rsc4 (Figure 4C, lanes 1 and 2). Furthermore, acetylation of WT Rsc4 present in the RSC complex was abolished when Rsc4 was isolated from a strain lacking Gcn5 (Figure 4C, lanes 3 and 4). The functional importance of K25 is supported by sequence alignment, which reveals that this lysine is highly conserved within yeast Rsc4 orthologs (Figure 4D). Moreover, G24 and P27 are also highly conserved, and the GKXP sequence matches the Gcn5 recognition motif determined from structural analysis of a Gcn5 HAT-histone H3 complex (Rojas et al., 1999).
Figure 4. Rsc4 K25 Is Acetylated In Vivo by Gcn5.
(A) Rsc4 is acetylated in vivo. Increasing amounts of purified RSC complex (87.5, 350, and 1400 ng) were analyzed by western blot with anti-Rsc4 or anti-acetyl-lysine antibodies. An asterisk denotes a proteolytic fragment of Rsc4 that is evident when RSC is overloaded.
(B) Rsc4 is acetylated in vivo by Gcn5. Western blot analysis of partially purified (via nickel, NTA-Agarose) Rsc4(1–340) TBD (p2243) following expression in WT or various HAT mutant strains. Strains were the following: WT, (YBC1895); gcn5Δ, (YBC1662); sas3Δ, (YBC1911); hat1Δ, (YBC2493); esa1 L327S, (LPY3430); and sas2Δ, (YBC1857).
(C) Rsc4 resident in RSC complex is acetylated at K25 by Gcn5 in vivo. Western blot analysis of partially purified RSC isolated via the Rsc2-TAP tag from strains bearing either RSC4 or the rsc4 K25A mutant or GCN5 or gcn5Δ. The protein A portion of the TAP tag (not cleaved in this procedure) serves as an internal control, as it is recognized by all antibodies, including anti-acetyl-lysine or anti-Rsc4. Strains included the following: RSC4, (YBC2814); rsc4 K25A, (YBC2815); GCN5, (YBC2825); gcn5Δ, (YBC2822).
(D) Alignment of the amino termini of Rsc4 orthologs from various yeast species (Clustal W). Identical amino acids are shown in green, and similar amino acid residues are shown in red, with regions of high similarity blocked together (PrettyPlot). Rsc4 K25, asterisk at top. Species include the following: S. cerevisiae (Scer), S. bayanus (Sbay), S. mikatae (Smik), S. paradoxus (Spar), S. kudrievzevii (Skud), C. glabrata (Cgla), S. castellii (Scas), K. lactis (Klac), and A. gossypii (Agos).
Rsc4 K25 Is Important for Fitness and Gene Expression
To examine the in vivo consequences of Rsc4 K25 acetylation, we made a K25A mutation and assessed phenotypes. The Rsc4 K25A mutation in isolation (and also K25R and K25Q) conferred only weak plate phenotypes, such as slightly slower growth on minimal medium under moderate heat stress (Figure S4). However, combining K25A with a conditional rsc4 allele (rsc4-2, which has a point mutation in each bromodomain [Kasten et al., 2004]) greatly enhanced temperature sensitivity (Figure S4). This supports the proposal that acetylation of Rsc4 K25 contributes to RSC function in vivo. Subtle phenotypic effects can confer a fitness advantage that might be more clearly revealed in a growth competition assay. We therefore measured the fitness of the rsc4 K25A mutant in direct competition with an isogenic WT strain and quantified gene fitness by the selection coefficient (s), which was obtained by comparing growth of the rsc4 K25A mutant and the WT RSC4 strain in coculture (Table 2). In this analysis, positive values of s indicate a selective disadvantage for the K25A mutant, while negative values of s indicate a greater selective advantage compared to WT. Surprisingly, under rich growth conditions (YPD, 30°C) the K25A mutant has a greater selective advantage (s = −0.018), indicating a 1.8% increase in the mutant K25A allele per generation in the coculture population under these conditions. In contrast, growth in minimal medium conditions (SD, 30°C) provides a selective advantage for the WT (s = 0.021), and this advantage is further increased (s = 0.087) when grown in minimal medium at elevated temperature (SD, 37°C). The loss of fitness for the K25A mutation when grown in minimal media conditions is highly significant (p < 0.01) and corresponds to an approximate 2.1% (30°C) or 8.7% (37°C) decrease in the K25A allele every generation.
Table 2.
Competition of the rsc4 K25A Strain against Wild-Type
| Media | s | Impacta | n | Number of Cultures | p |
|---|---|---|---|---|---|
| YPD 30°C | −0.018 ± 0.0036 | 1.8% | 6 | 7 | <0.01 |
| SD 30°C | 0.021 ± 0.0069 | −2.1% | 6 | 8 | <0.01 |
| SD 37°C | 0.087 ± 0.0059 | −8.7% | 6 | 4 | <0.01 |
Media are described in the Experimental Procedures section. Selection coefficients (s) ± the standard deviation for rsc4 K25A relative to the WT strain were measured as described in the Experimental Procedures section. A positive value for s indicates a greater selective advantage for the WT strain, and a negative value indicates that the mutant has a greater selective advantage. Indicated are the number of time points (n) at which genotype frequencies were measured and the number of cultures analyzed for each growth condition. The statistical significance (p) for a test of the null hypothesis that the selection coefficient is not different from zero was determined.
Impact indicates the approximate percent change in the mutant allele relative to the WT allele per generation in coculture.
We also examined the rsc4 K25A mutant for changes in gene expression by performing transcriptional profiling from cells grown in minimal medium at slightly elevated temperature (33°C). In the K25A mutant, 80 genes were upregulated 2-fold or greater, and 61 genes were downregulated 2-fold or greater (Tables S1 and S2). Among the upregulated class, we did not observe a greater affect on genes of a particular inherent transcriptional frequency. Common classes of upregulated genes include those involved in cell wall integrity, those involved in the response to cell stress, and those encoding proteins that reside in the cell membrane. Misregulation of genes involved in cell wall integrity has been observed with other rsc mutants, including conditional rsc4 alleles (Angus-Hill et al., 2001; Kasten et al., 2004). Taken together, our genetic analysis, fitness determination, and gene profiling analysis demonstrate a significant role for Rsc4 K25 in RSC function in vivo.
Rsc4 K25ac Binding to BD1 Inhibits H3K14ac Binding to BD2
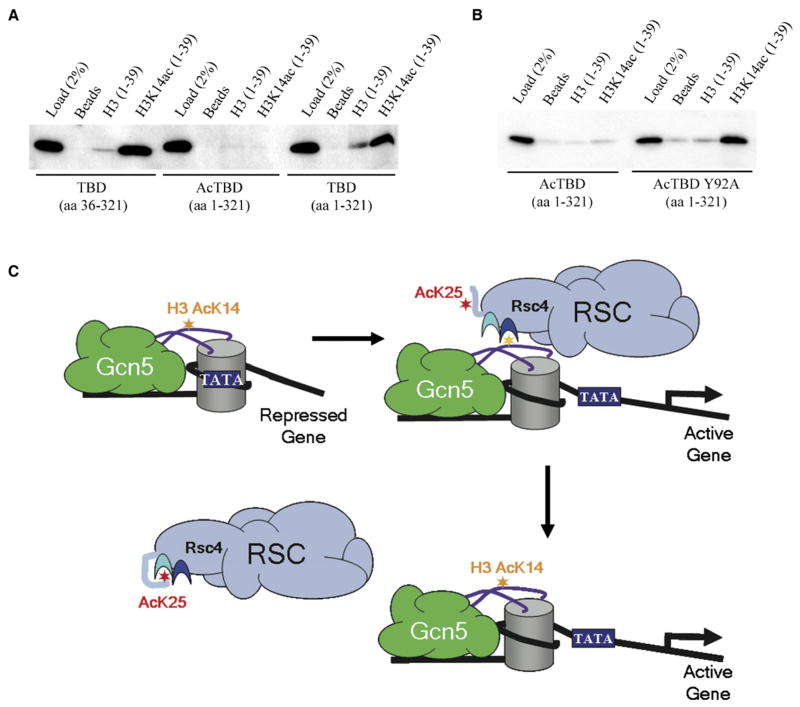
Having demonstrated that Rsc4 K25 is acetylated by Gcn5 both in vitro and in vivo, we investigated possible mechanistic roles for this modification. In principle, the binding of K25ac in BD1 could have a positive, neutral, or negative impact on H3K14ac binding to BD2. To test this, we applied the histone tail peptide binding assay utilized above and prepared both K25-acetylated and nonacetylated versions of Rsc4(1–321). The results show that K25 acetylation of Rsc4(1–321) significantly inhibited binding of H3K14ac peptides to BD2 (Figure 5A). The importance of K25ac binding to BD1 for this inhibition was tested more precisely by using Rsc4(1–321) Y92A protein, which is crippled for binding to BD1. Importantly, binding to H3K14ac peptides is largely restored in the Y92A derivative, thereby indicating that binding of K25ac to BD1 is coupled with inhibition of H3K14ac binding to BD2 (Figure 5B). The mechanism for this inhibition is not immediately obvious, because BD1 and BD2 are separated by 20 Å. Our preferred model is that N-terminal residues that become ordered upon binding of K25ac to BD1 overlap the site of H3 residues that bind cooperatively with the K14ac - BD2 interaction, although an allosteric propagation of a subtle conformational change is also possible.
Figure 5. Inhibition of H3K14ac Binding Involves Rsc4 K25ac Binding to BD1.
(A) Rsc4 K25 acetylation inhibits H3K14ac binding. Binding of purified nonacetylated and acetylated TBD to biotinylated histone H3 tail peptides conjugated to streptavidin beads was examined by western analysis. Rsc4(36–321) TBD was expressed from p1617 and Rsc4(1–321) TBD was expressed from p1616. Rsc4(1–321) TBD was acetylated by Gcn5 where indicated, and acetylation at K25 was confirmed by mass spectrometry analysis (data not shown). Binding and wash buffers contained 100 mM NaCl.
(B) Western blot analysis of purified WT and mutant acetylated Rsc4(1–321) TBD bound to H3 peptides. Bead bindings and washes were conducted at 150 mM NaCl. Rsc4 K25ac(1–321) TBD was expressed from p1617 and Rsc4 K25Ac(1–321) Y92A TBD was expressed from p2296. Proteins were acetylated by Gcn5, and acetylation was confirmed by mass spectrometry analysis (data not shown).
(C) Model for Rsc4 autoregulation. Gcn5 acetylation of H3K14 facilitates Rsc4 interaction with H3K14ac and also the subsequent release of Rsc4 from chromatin by acetylating Rsc4 K25, which binds in BD1 and antagonizes Rsc4 binding to H3K14ac.
Models for Biological Mechanism
Our structural, biochemical, and genetic data have established that H3K14ac binds Rsc4 BD2 and that additional interactions with flanking residues in the H3 tail (not resolved in the structure) also contribute to binding affinity. The interaction is inherently weak, but it is specific and biologically important, and binding in the biological context is presumably enhanced by cooperative effects involving the additional DNA- and nucleosome-interacting domains, such as the SWIRM, SANT, and additional bromodomains of RSC subunits (Boyer et al., 2002; Da et al., 2006; Yu et al., 2003). Weak interactions are appropriate components of such regulatory switches, because they are reversible and avoid saturation with incorrect binding partners, such as other acetylated lysine residues. Our surprising finding that Rsc4 K25 is acetylated by Gcn5 both in vitro and in vivo reveals that yeast Gcn5 is both a HAT and a protein acetyltransferase. This is consistent with the roles of related enzymes such as PCAF and p300 in vertebrates (Gu and Roeder, 1997; Mujtaba et al., 2002). Also surprising was our discovery that acetylated Rsc4 K25 bound to BD1. The biological relevance of the interaction is indicated by our growth competition and transcriptional profiling data. The magnitude of the selection coefficients obtained shows that prolonged stress conditions such as those examined would cause the loss of the mutant population (8.7% loss per generation). The likely mechanistic basis for the biological loss of fitness is our finding that this interaction inhibits binding of H3K14ac peptides to BD2. Another direction that merits future study is that BD1 and/or BD2 likely has additional binding partners; these bromodomains are essential for viability, whereas their currently identified ligands are not. To find them, we have tried many other histone tail peptide substrates in our in vitro binding assays, including H3 K9ac, H3 K23ac, H3 K9ac K18ac, and H3 K18ac K27ac in addition to the series tested previously (Kasten et al., 2004). However, none of these provided acetyl-enhanced binding. In principle, the binding of Rsc4 TBDs to alternative ligands might be regulated by the mechanisms revealed here. For example, the binding of Rsc4 K25ac to BD1 might enhance the binding of another ligand to BD2. Alternatively, BD1 may bind an alternative ligand on a histone tail when K25 is not acetylated, leaving the BD1 pocket available and perhaps enabling cooperativity with H3K14ac bound in BD2.
Gcn5 acetylates ligands for both BD1 and BD2, and these ligands compete for binding to Rsc4. This raises the possibility that Gcn5 activity serves as a switch; Gcn5 acetylation of H3K14 would favor RSC-nucleosome binding, and this interaction would be countered by acetylation of Rsc4 K25. One attractive possibility is that this mechanism regulates the residence time of RSC to sites of remodeling (Figure 5C). In this model, activators recruit Gcn5 to promoter regions where it acetylates H3K14. This recruits RSC to promote nucleosome sliding and enhance promoter accessibility and also places Rsc4 in the vicinity of Gcn5, which then acetylates K25 and triggers release of RSC from the now-remodeled nucleosome. Finally, auto-regulatory mechanisms, in which a binding protein or enzyme adopts a repressed conformation in response to intramolecular binding of a posttranslationally attached group, have been extensively characterized for kinases (reviewed in Kuriyan and Cowburn, 1997) and recently described for ubiquitylation (Hoeller et al., 2006). Our demonstration that RSC function is optimized by an analogous approach raises the possibility that mechanisms of this type might be widespread for acetylation and other modifications associated with chromatin dynamics.
EXPERIMENTAL PROCEDURES
See the Supplemental Data for details of media, strain lists, plasmid lists, protein expression and purification, in vitro acetylation, and crystallographic methods.
Histone Tail Binding Assay
Biotinylated histone tail peptides were bound to streptavidin beads (Invitrogen) as previously described (Kasten et al., 2004) and resuspended in a 50% slurry. Binding assays were conducted by rotating 15 μl of the peptide/bead slurry (20 nmol peptide/100 μl bed volume beads) with 500 ng purified Rsc4 protein in peptide binding buffer (PBB) (20 mM Tris [pH 7.5], 150 mM NaCl, 5% glycerol, 0.05% Tween-20, 1 mM EDTA, 1 mM β-mercaptoethanol, protease inhibitors) at 4°C for 3 hr. Typically, the beads were washed twice with PBB and twice with PBB containing 250 mM NaCl or the alternative NaCl concentrations indicated, followed by elution with 4 × SDS sample buffer.
RSC Purification, Extract Preparation, Immunoprecipitation, and Antibodies
RSC was purified as previously described (Saha et al., 2002). Whole-cell extracts were prepared as previously described (Cairns et al., 1999). Partially purified RSC was analyzed for Rsc4 acetylation in the gcn5Δ and rsc4 K25A mutant and was derived from whole-cell extracts lysed in 3 × lysis buffer (60 mM HEPES [pH 7.6], 30% glycerol, 750 mM NaCl, 0.3% Tween-20, 30 mM EDTA, 1.5 mM DTT, protease inhibitors), which were then bound to IgG beads, washed three times with IPP150 (20 mM HEPES [pH 7.6], 10% glycerol, 150 mM NaCl, 0.1% Tween-20, 10 mM EDTA, 0.5 mM DTT, protease inhibitors), and eluted by boiling in 4 × SDS sample buffer prior to western blot analysis. The anti-Rsc4 antibody was made to the full-length protein and was previously described (Kasten et al., 2004). The anti-acetyl lysine antibody was from Cell Signaling.
Affinity Purification of Rsc4(1–340) from Yeast
Rsc4(1–340) preceded by 10 × HIS and 2 × NLS sequences expressed in yeast was nickel affinity purified prior to examining acetylation. Whole-cell extracts were prepared as previously described (Saha et al., 2005) except in modified breaking buffer (12% glycerol, 50 mM Tris [pH 7.5], 0.1% Triton X-100, 500 mM NaCl, 1.5 mM β-mercaptoethanol, protease inhibitors). Extracts (3 mg) were incubated with 100 μl bed volume of Ni-NTA agarose (QIAGEN) for 3 hr at 4°C, poured into a small column, washed three times (1.2 ml) with wash buffer (50 mM NaPhosphate [pH 8.0], 150 mM NaCl, 10% glycerol) containing increasing amounts of imidazole (from 10 mM to 40 mM). Bound protein was eluted with wash buffer containing 300 mM imidazole and protease inhibitors prior to western blot analysis.
rsc4 K25A Fitness Determination
Fitness determinations compared isogenic strains, differing only at rsc4 K25A mutation, which were prepared by standard yeast gene transplacement methods. Comparisons used a method based on (Thatcher et al., 1998). Competition cocultures were established by inoculating 2 ml of YPD (rich media) or SD supplemented as needed for auxotrophies (minimal media) with 32 μl each from overnight cultures of YBC2898 and YBC2899 grown in YPD or SD. Equal numbers of cells (by OD reading) from each strain were inoculated, and cultures were incubated at 30°C or 37°C and were back diluted into fresh media daily. Each day, 1 ml of the competition coculture was collected and used to prepare genomic DNA using the YeaStar Genomic DNA kit (Zymo Research). Quantitative PCR (qPCR) was utilized to determine RSC4 genotype frequencies. Primers were designed so that the 3′ ends recognize either the K25 or the K25A codon allowing these primers to differentiate between the two RSC4 alleles. Genomic DNA harvested periodically from the competition cocultures was used in separate qPCR procedures essentially as previously described (Roberts et al., 2003) to determine the quantity of each allele individually. Primer for RSC4 K25 detection was the following: (BC3550) GCCTAAATACTTGCCGGGAAAA. Primer for rsc4 K25A detection was the following: (BC3551) CTAAATACTTGCCGGGAGCC. RSC4 reverse primer used for detection of either allele was the following: (BC3539) TGTATTTGTCGATAAGAACATCCAAAGTG. The average of three PCR replicates was taken for each genomic DNA analyzed. The change in allele ratio with time is given by the equation ln(Rt) = ln(R0) − st (where R0 is the initial genotype ratio, Rt is the ratio after t generations, and s is the selection coefficient) (Thatcher et al., 1998). The selection coefficient for the rsc4 K25A mutant relative to WT in each growth condition was calculated by fitting the above equation to the qPCR frequency data.
Supplementary Material
Supplemental Data include Supplemental Experimental Procedures, four figures, four tables, and Supplemental References and can be found with this article online at http://www.molecule.org/cgi/content/full/27/5/817/DC1/.
Acknowledgments
We thank Chad Nelson and the University of Utah Mass Spectrometry Core for mass spectrometry analysis; Bob Schackmann and the University of Utah Biotechnology Core Facility for DNA sequencing and oligo and peptide synthesis; Miles Pufall and Jack Skalicky for advice and assistance with NMR measurements; Tim Formosa, Jacqui Wittmeyer, Jeff Lenkart, and Alisha Schlichter for critical comments on the manuscript; Jon Seger and Fred Adler for advice on the fitness determination experiments; Derick Holt and Tim Parnell for assistance with transcriptional profiling; and Heather Szerlong for the Gcn5 expression construct. Operations at the National Synchrotron Light Source (NSLS) are supported by the Department of Energy, Office of Basic Energy Sciences, and by the National Institutes of Health (NIH). Data collection at the NSLS was funded by the National Center for Research Resources. This work was supported by NIH grants GM076242 (C.P.H.), GM60415 (B.R.C.), and CA20414 (for core facilities) and by American Cancer Society grant PF0304001GMC (A.P.V.). M.M.K. and B.R.C. are supported by the Howard Hughes Medical Institute.
Footnotes
Accession Numbers
Protein Data Bank entry codes are 2R0S for Rsc4(36-340), 2R0Y for Rsc4(36-340) peptide soak, 2R0V for acetylated Rsc4(1-340), and 2R1O for acetylated histone-Rsc4(1-321) chimera.
References
- Angus-Hill ML, Schlichter A, Roberts D, Erdjument-Bromage H, Tempst P, Cairns BR. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol Cell. 2001;7:741–751. doi: 10.1016/s1097-2765(01)00219-2. [DOI] [PubMed] [Google Scholar]
- Baetz KK, Krogan NJ, Emili A, Greenblatt J, Hieter P. The ctf13–30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol Cell Biol. 2004;24:1232–1244. doi: 10.1128/MCB.24.3.1232-1244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, Peterson CL. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell. 2002;10:935–942. doi: 10.1016/s1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Cairns BR. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr Opin Genet Dev. 2005;15:185–190. doi: 10.1016/j.gde.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CR, Wu CS, Hom Y, Gartenberg MR. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 2005;19:3031–3042. doi: 10.1101/gad.1356305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da G, Lenkart J, Zhao K, Shiekhattar R, Cairns BR, Marmorstein R. Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc Natl Acad Sci USA. 2006;103:2057–2062. doi: 10.1073/pnas.0510949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Awad S, Prochasson P. The Swi2/Snf2 bromodomain is required for the displacement of SAGA and the octamer transfer of SAGA-acetylated nucleosomes. J Biol Chem. 2006;281:18126–18134. doi: 10.1074/jbc.M602851200. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, Kowanetz K, Breitling R, Mann M, Stenmark H, Dikic I. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol. 2006;8:163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- Howe L, Auston D, Grant P, John S, Cook RG, Workman JL, Pillus L. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 2001;15:3144–3154. doi: 10.1101/gad.931401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J Mol Biol. 2000;304:355–370. doi: 10.1006/jmbi.2000.4207. [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyan J, Cowburn D. Modular peptide recognition domains in eukaryotic signaling. Annu Rev Biophys Biomol Struct. 1997;26:259–288. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- Mujtaba S, He Y, Zeng L, Farooq A, Carlson JE, Ott M, Verdin E, Zhou MM. Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol Cell. 2002;9:575–586. doi: 10.1016/s1097-2765(02)00483-5. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DN, Stewart AJ, Huff JT, Cairns BR. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc Natl Acad Sci USA. 2003;100:14695–14700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas JR, Trievel RC, Zhou J, Mo Y, Li X, Berger SL, Allis CD, Marmorstein R. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature. 1999;401:93–98. doi: 10.1038/43487. [DOI] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- Shen W, Xu C, Huang W, Zhang J, Carlson JE, Tu X, Wu J, Shi Y. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry. 2007;46:2100–2110. doi: 10.1021/bi0611208. [DOI] [PubMed] [Google Scholar]
- Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, Soularue P, Navarro F, Cairns BR, Lefebvre O, Werner M. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol. 2006;26:4920–4933. doi: 10.1128/MCB.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sun H, Liu J, Zhang J, Shen W, Huang H, Xu C, Dai H, Wu J, Shi Y. Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4. Biochem Biophys Res Commun. 2007;358:435–441. doi: 10.1016/j.bbrc.2007.04.139. Published online May 2, 2007. [DOI] [PubMed] [Google Scholar]
- Syntichaki P, Topalidou I, Thireos G. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature. 2000;404:414–417. doi: 10.1038/35006136. [DOI] [PubMed] [Google Scholar]
- Thatcher JW, Shaw JM, Dickinson WJ. Marginal fitness contributions of nonessential genes in yeast. Proc Natl Acad Sci USA. 1998;95:253–257. doi: 10.1073/pnas.95.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trievel RC, Rojas JR, Sterner DE, Venkataramani RN, Wang L, Zhou J, Allis CD, Berger SL, Marmorstein R. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc Natl Acad Sci USA. 1999;96:8931–8936. doi: 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Li Y, Ishizuka T, Guenther MG, Lazar MA. A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J. 2003;22:3403–3410. doi: 10.1093/emboj/cdg326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukawa M, Katoh S, Miyakawa T, Tsuchiya E. Nps1/Sth1p, a component of an essential chromatin-remodeling complex of Saccharomyces cerevisiae, is required for the maximal expression of early meiotic genes. Genes Cells. 1999;4:99–110. doi: 10.1046/j.1365-2443.1999.00242.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data include Supplemental Experimental Procedures, four figures, four tables, and Supplemental References and can be found with this article online at http://www.molecule.org/cgi/content/full/27/5/817/DC1/.