Abstract
The nucleolus is a subnuclear compartment with multiple cellular functions, including ribosome biogenesis. USP36 is a deubiquitylating enzyme that localizes to nucleoli and plays an essential role in regulating the structure and function of the organelle. However, how the localization of USP36 is regulated remains unknown. Here, we identified a short stretch of basic amino acids (RGKEKKIKKFKREKRR) that resides in the C-terminal region of USP36 and serves as a nucleolar localization signal for the protein. We found that this motif interacts with a central acidic region of nucleophosmin/B23, a major nucleolar protein involved in various nucleolar functions. Knockdown of nucleophosmin/B23 resulted in a significant reduction in the amount of USP36 in nucleoli, without affecting the cellular USP36 level. This was associated with elevated ubiquitylation levels of fibrillarin, a USP36 substrate protein in nucleoli. We conclude that nucleophosmin/B23 recruits USP36 to nucleoli, thereby serving as a platform for the regulation of nucleolar protein functions through ubiquitylation/deubiquitylation.
The nucleolus is a subnuclear compartment involved in diverse cellular activities, including ribosome biogenesis, cell cycle control, and cellular stress response (1). The ubiquitin-proteasome system has been implicated in various aspects of nucleolar function. Proteins involved in rRNA processing during ribosome biogenesis, such as nucleophosmin (NPM)2/B23 and fibrillarin (FBL), undergo ubiquitylation-dependent degradation (2, 3). Inhibition of the proteasome activity affects the distribution of rRNA-processing proteins in nuclei, resulting in impaired rRNA processing (4). Ribosomal proteins that have failed to assemble into ribosomal subunits in nucleoli are also degraded by the proteasome (5). The nucleolar protein Arf, which regulates p53 function by sequestering the ubiquitin ligase Mdm2 to nucleoli, undergoes unconventional N-terminal ubiquitylation that directs Arf for proteasomal degradation (6).
Ubiquitylation is reverted by deubiquitylating enzymes that deconjugate ubiquitin from target proteins (7). USP36, a deubiquitylating enzyme of the ubiquitin-specific protease (USP) family, localizes to nucleoli in mammalian cells (8–10), where it deubiquitylates nucleolar proteins such as NPM and FBL and prevents their proteasomal degradation (10). RNA interference-mediated knockdown of USP36 causes loss of the nucleolar structure and retardation in the rates of rRNA transcription and processing, suggesting a crucial role for protein deubiquitylation in nucleoli (10). However, how the localization and activity of USP36 are regulated in nucleoli remains unknown.
NPM is a multifunctional protein that localizes mainly in nucleoli, although it shuttles between the nuclei and cytoplasm and exhibits diverse functions in and out of nucleoli (11). With its endoribonuclease activity, NPM processes the 32 S rRNA precursor to the 28 S mature rRNA in nucleoli (12). It also plays roles as a molecular chaperone in preventing the aggregation of assembling rRNAs and ribosomal proteins in nucleoli (13) and in transporting assembled ribosomal subunits to the cytoplasm (14). NPM also regulates the p53 checkpoint pathway through association with Arf in nucleoli (15, 16). Finally, the biological importance of NPM is underscored by observations that overexpression or alteration of the NPM gene is frequently associated with human cancer (11).
In this study, we show that NPM recruits USP36 and regulates the level of protein ubiquitylation in nucleoli. Our results demonstrate a novel role for multifunctional NPM and provide a molecular basis for the regulation of protein ubiquitylation in nucleoli.
MATERIALS AND METHODS
DNA Constructs
Expression vectors for FLAG-tagged human wild-type USP36, USP36-(1–420), USP36-(421–800), and USP36-(801–1121), as well as those for FLAG-FBL and glutathione S-transferase (GST)-USP36-(921–1121), were constructed as described (10). cDNAs for USP36ΔBM1, USP36ΔBM2, USP36ΔBM3, USP36ΔBM4, and USP36ΔBM5 were generated using the QuikChange site-directed mutagenesis system (Stratagene, La Jolla, CA) and cloned into the mammalian expression vector pME-FLAG. The enhanced green fluorescent protein (EGFP) fusion construct was generated by inserting chemically synthesized double-stranded oligonucleotides into pEGFP-N1 (Clontech). FLAG-tagged NPM constructs were provided by Dr. Charles J. Sherr (St. Jude Children's Research Hospital, Memphis, TN). The His-tagged NPM construct was generated by inserting the full-length NPM cDNA into pET-26b(+) (EMD Chemicals, Gibbstown, NJ). Small interfering RNA (siRNA) expression vectors for human NPM were constructed by inserting chemically synthesized double-stranded oligonucleotides into pSilencer1.0-U6 (Ambion, Austin, TX). The mRNA target sites were nucleotides 819–837 (5′-GAATTGCTTCCGGATGACT-3′) and 670–688 (5′-GGACAAGAATCCTTCAAGA-3′) from the translation initiation codon for NPM siRNAs 1 and 2, respectively.
Cell Culture and DNA Transfection
HeLa and COS-7 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Where described, cells were treated with 20 μm MG132 for 8 h prior to preparation of the lysate. For ectopic expression of proteins, expression vectors were transfected into cells for 48 h using FuGENE 6 transfection reagent (Roche Diagnostics). For RNA interference, siRNA expression vectors were transfected twice at 48-h intervals.
Immunoprecipitation and Immunoblotting
Cell lysates were prepared by solubilizing cells in lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 0.1% Tween 20, 1 mm EDTA, 10 mm N-ethylmaleimide, 50 mm NaF, 30 mm sodium pyrophosphate, 1 mm Na3VO4, and protease inhibitor mixture (1 mm phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A)) and collecting the supernatant after centrifugation. Immunoprecipitation and immunoblotting of the lysates were performed using standard procedures. For immunoprecipitation, anti-NPM (3 μg; Zymed Laboratories Inc., South San Francisco, CA), anti-FLAG (1 μg; clone M2, Sigma), and anti-GFP (1 μg; Invitrogen) antibodies were used. The primary antibodies used for immunoblotting were anti-USP36 (1:100) (10), anti-NPM (2 μg/ml), anti-FLAG (4 μg/ml), anti-GFP (4 μg/ml), anti-GST (1 μg/ml; Santa Cruz Biotechnology, Santa Cruz, CA), anti-α-tubulin (1:20,000; Sigma), and anti-ubiquitin (5 μg/ml; clone FK2, MBL, Nagoya, Japan). The secondary antibodies used were peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG (GE Healthcare). Immunoblots were detected using ECL reagent (GE Healthcare).
GST Pulldown
His-tagged NPM protein was expressed in Escherichia coli and purified using the nickel-nitrilotriacetic acid buffer kit (EMD Chemicals). GST fusion protein (∼5 μg) was also expressed in E. coli, coupled to glutathione-Sepharose beads (10 μl; GE Healthcare), and incubated with cell lysates or purified NPM (∼5 μg) for 16 h at 4 °C. After the beads were washed with lysis buffer, bound proteins were subjected to immunoblotting.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline for 10 min on ice, permeabilized with 0.2% Triton X-100 for 5 min, and stained consecutively with primary and secondary antibodies. The primary antibodies used were rabbit anti-FLAG (0.4 μg/ml), mouse anti-nucleolus (1:20; clone MAB1277, Millipore), mouse anti-NPM (10 μg/ml), and rabbit anti-USP36 (1:1000). The secondary antibodies used were Alexa 488- and Alexa 594-conjugated anti-mouse IgG and anti-rabbit IgG (1:1000; Invitrogen). To stain nuclei, cells were incubated with TO-PRO-3 iodide (642/661) (50 μm; Invitrogen) during incubation with secondary antibodies. Images were captured with a laser scanning confocal microscope (Axiovert 200M, Carl Zeiss, Oberkochen, Germany).
RESULTS
Nucleolar Localization Signal (NoLS) in USP36
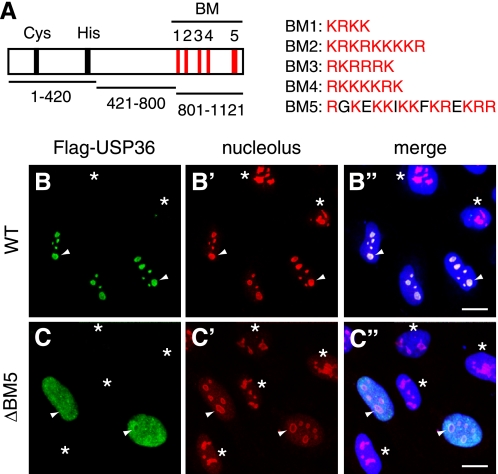
We have shown recently that the C-terminal third of USP36 (USP36-(801–1121)) (Fig. 1A) is required for the nucleolar localization of USP36 (10). Because this region contains five clusters of basic amino acids (BM1–5) (Fig. 1A), which are characteristic of the NoLS (17), we examined whether BM1–5 are required for the USP36 localization. We deleted the BM sequences individually from FLAG-tagged USP36 and expressed the mutants in HeLa cells. Each mutant protein was expressed at levels similar to those of wild-type USP36 as assessed by immunoblotting of the lysates of transfected cells (see Fig. 3C, middle panel) (data not shown). Immunofluorescence staining of transfected cells with anti-FLAG antibody showed that the mutants lacking each of BM1–4 (supplemental Fig. S1), as well as wild-type USP36 (Fig. 1, B–B"), distributed normally in nucleoli. By contrast, deletion of BM5 (1076RGKEKKIKKFKREKRR1091) abolished the nucleolar localization of USP36 and caused its leakage to the nucleoplasm (Fig. 1, C–C"), suggesting that BM5 is required for the nucleolar localization of USP36, whereas BM1–4 are dispensable.
FIGURE 1.
BM5 is required for the nucleolar localization of USP36. A, shown is the schematic structure of human USP36. Five basic motifs (BM1–5), the Cys and His boxes constituting the catalytic core, and the regions covered by the three truncated mutants used in this study (USP36-(1–420), USP36-(421–800), and USP36-(801–1121)) are indicated. Amino acid sequences of BMs are also shown. B–C″, HeLa cells were transfected with FLAG-tagged wild-type (WT) USP36 (B–B″) or USP36ΔBM5 (C–C″) and double-stained with anti-FLAG (B and C) and anti-nucleolus (B′ and C′) antibodies. B″ and C″ are merged images in which nuclei were also stained in blue. Arrowheads indicate typical nucleoli. Asterisks indicate untransfected cells. Scale bars = 20 μm.
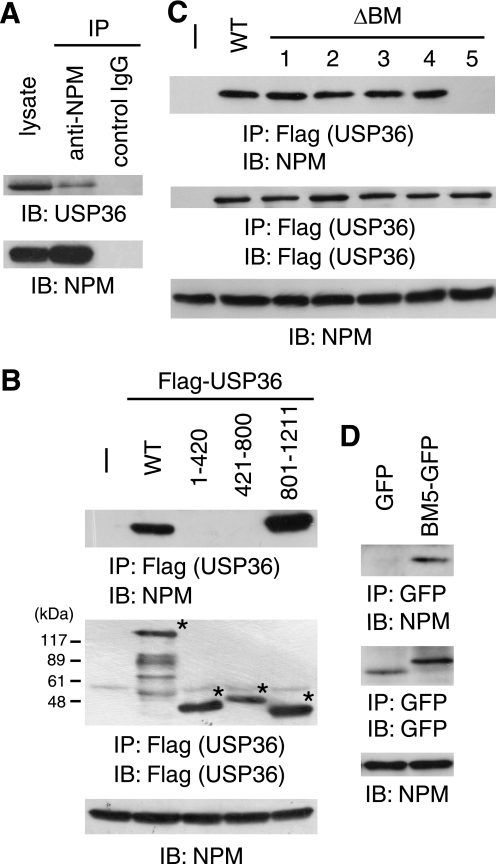
FIGURE 3.
USP36 binds to NPM via BM5. A, HeLa cell lysate was immunoprecipitated (IP) with anti-NPM or control IgG and immunoblotted (IB) with anti-USP36 and anti-NPM antibodies. B and C, lysates of COS-7 cells transfected with FLAG-tagged USP36 mutants were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-NPM (upper panel) and anti-FLAG (middle panel) antibodies. Total lysate was immunoblotted with anti-NPM antibody (lower panel). Asterisks in B indicate the positions of the USP36 mutants. WT, wild-type. D, lysates of COS-7 cells transfected with EGFP or BM5-EGFP were immunoprecipitated with anti-GFP antibody and immunoblotted with anti-NPM (upper panel) and anti-GFP (middle panel) antibodies. Total lysate was immunoblotted with anti-NPM antibody (lower panel).
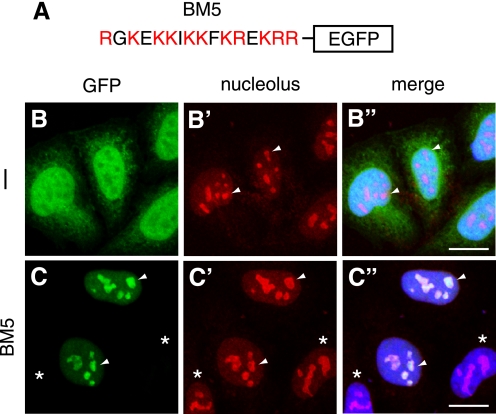
We next fused the BM5 sequence to the N terminus of EGFP (Fig. 2A) and expressed the fusion protein in HeLa cells (see Fig. 3D, middle panel). Fluorescence imaging showed that whereas EGFP is scattered in the nucleoplasm and cytoplasm (Fig. 2, B–B″), BM5-EGFP is highly concentrated in nucleoli (C–C″). These results suggest that BM5 is sufficient for the normal distribution of USP36. The results in Figs. 1 and 2 collectively indicate that BM5 serves as an NoLS for USP36.
FIGURE 2.
BM5 is sufficient for the nucleolar localization of USP36. A, shown is the schematic structure of BM5-EGFP. B–C″, HeLa cells were transfected with EGFP (B–B″) or BM5-EGFP (C–C″) and stained with anti-nucleolus antibody (B′ and C′). B and C show the fluorescent signal by EGFP. B″ and C″ are merged images in which nuclei were also stained in blue. Arrowheads indicate typical nucleoli. Asterisks indicate untransfected cells. Scale bars = 20 μm.
USP36 Binds to NPM via the NoLS
We reported previously the interaction between ectopically expressed USP36 and NPM (10). To elucidate its physiological relevance, the interaction of endogenous proteins was examined. Anti-NPM (but not control) antibody co-immunoprecipitated endogenous USP36 from untransfected HeLa cells, suggesting that these proteins associate with each other under normal conditions (Fig. 3A).
To examine the possibility that USP36 localizes to nucleoli via interaction with NPM, we mapped the NPM-binding site in USP36. We first examined the NPM-binding ability of three truncated USP36 mutants: USP36-(1–420), USP36-(421–800), and USP36-(801–1121) (Fig. 1A). Among the mutants, only FLAG-USP36-(801–1121) co-immunoprecipitated endogenous NPM when expressed in COS-7 cells, suggesting that the C-terminal region of USP36 containing BM1–5 harbors the NPM-binding site (Fig. 3B).
Because NPM contains two clusters of acidic Asp and Glu residues that potentially interact with basic amino acid sequences (Fig. 4A) (11), we next examined the effect of deleting BM1–5 from USP36 on NPM binding. FLAG-tagged mutants lacking each of BM1–5 were expressed in COS-7 cells and immunoprecipitated with anti-FLAG antibody. Immunoblotting of the precipitates with anti-NPM antibody showed that USP36ΔBM5 completely lacked NPM-binding ability, whereas the others bound to NPM as efficiently as wild-type USP36 (Fig. 3C), suggesting that BM5 is required for NPM binding.
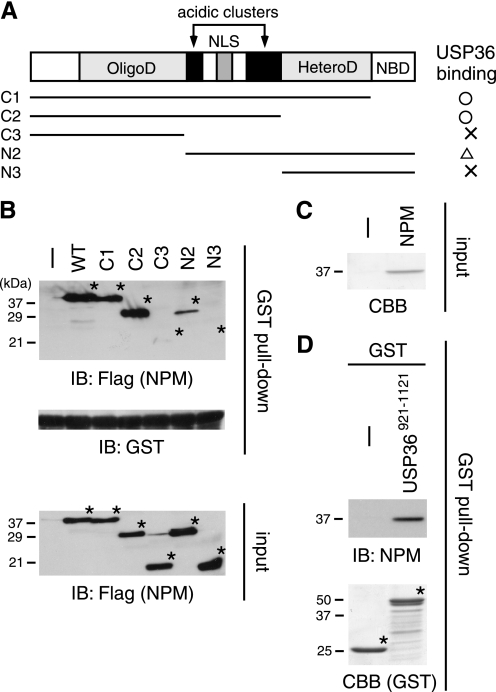
FIGURE 4.
USP36 binds to the central region of NPM. A, shown are the schematic structures of NPM and its mutants used in this study. OligoD, oligomerization domain; NLS, nuclear localization signal; HeteroD, heterodimerization domain; NBD, nucleic acid-binding domain. The USP36-binding ability of each NPM mutant (results in B) is summarized on the right. B, lysates of COS-7 cells transfected with FLAG-tagged NPM mutants were incubated with GST-USP36-(921–1121) coupled to glutathione beads, and bound NPM proteins were immunoblotted (IB) with anti-FLAG antibody (upper panel). Amounts of GST-USP36-(921–1121) used were assessed by anti-GST immunoblotting of the pulldown samples (middle panel). Inputs of the NPM mutants were assessed by anti-FLAG immunoblotting of the total lysate used for the pulldown assay (lower panel). Asterisks indicate the positions of the NPM mutants. WT, wild-type. C, His-tagged NPM was purified from transformed E. coli and stained with Coomassie Brilliant Blue (CBB). D, purified NPM was incubated with GST or GST-USP36-(921–1121) coupled to glutathione beads, and bound NPM was immunoblotted with anti-NPM antibody (upper panel). Amounts of GST proteins (asterisks) were assessed by Coomassie Brilliant Blue staining (lower panel).
To test whether BM5 alone is sufficient for the interaction, the binding of EGFP-fused BM5 to NPM was examined in co-immunoprecipitation experiments. We found that BM5-EGFP, but not EGFP, is capable of binding to endogenous NPM (Fig. 3D). The results in Fig. 3 indicate that BM5 also serves as an NPM-binding site.
USP36 Binds to the Central Region of NPM
We next mapped the USP36-binding site in NPM by examining the interaction of truncated NPM mutants (Fig. 4A) (15) with USP36 in GST pulldown experiments. The C-terminal portion of USP36 containing BM5 (residues 921–1121) was bacterially expressed and purified as a GST fusion protein. GST-USP36-(921–1121) coupled to glutathione beads was incubated with lysates of COS-7 cells transfected with FLAG-tagged NPM mutants, and bound NPM proteins were detected by immunoblotting with anti-FLAG antibody. The binding ability of the C1 and C2 mutants to GST-USP36-(921–1121) was not significantly reduced compared with that of wild-type NPM (Fig. 4B). By contrast, C3 and N3, both of which lack the central region containing the acidic clusters, almost completely lost the binding ability (Fig. 4B). USP36 binding of the N2 mutant was also affected considerably (Fig. 4B). GST did not pull down wild-type NPM by itself (data not shown). These results suggest that NPM binds to USP36 via the central region and that the N-terminal region is also involved in the interaction in some way.
To confirm that USP36 interacts with NPM directly, His-tagged wild-type NPM was bacterially expressed, purified using Ni2+ affinity beads (Fig. 4C), and incubated with GST-USP36-(921–1121) in the GST pulldown assay. Immunoblotting of the pulldown precipitates with anti-NPM antibody showed that purified NPM bound to USP36 but not to control GST (Fig. 4D), demonstrating direct interaction between USP36 and NPM.
NPM Is Required for the Nucleolar Localization of USP36
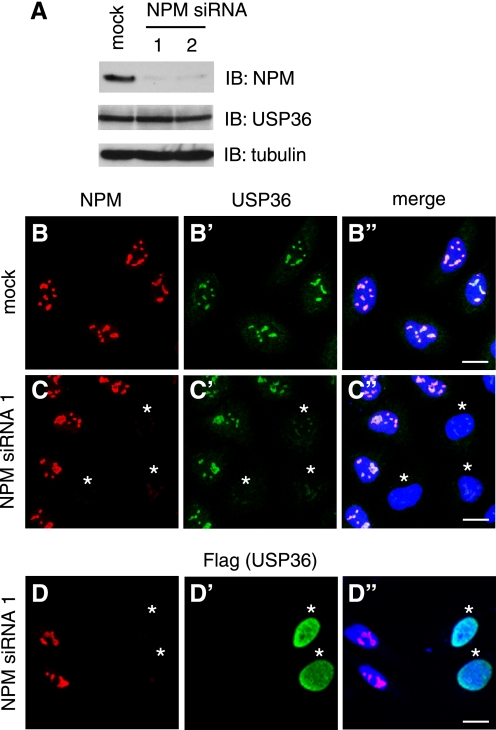
The above results suggested that USP36 localizes to nucleoli via interaction with NPM. To test this possibility, we examined the effect of RNA interference-mediated knockdown of NPM on USP36 localization in HeLa cells. Immunoblotting showed that, upon the transfection of two independent siRNAs for NPM (siRNAs 1 and 2), the level of endogenous NPM protein was significantly reduced (Fig. 5A). The level of USP36 was not affected by NPM depletion (Fig. 5A), suggesting that the interaction does not stabilize USP36 protein. However, immunofluorescence staining showed that, in cells transfected with NPM siRNA 1 or 2 and exhibiting mostly undetectable levels of nucleolar NPM, USP36 staining was drastically reduced in nucleoli (Fig. 5, C–C″, asterisks) (data not shown). This effect was observed in >90% of ∼300 NPM-depleted cells examined and was not due merely to uneven antibody staining because nuclear staining with TO-PRO-3 was normal in these cells (Fig. 5, C–C″, blue).
FIGURE 5.
NPM is required for the nucleolar localization of USP36. A, lysates of HeLa cells transfected with the mock, NPM siRNA 1, or NPM siRNA 2 vector were immunoblotted (IB) with anti-NPM, anti-USP36, and anti-α-tubulin antibodies. B–C″, HeLa cells transfected with the mock (B–B″) or NPM siRNA 1 (C–C″) vector were double-stained with anti-NPM (B and C) and anti-USP36 (B′ and C′) antibodies. D–D″, HeLa cells cotransfected with FLAG-USP36 and NPM siRNA 1 were double-stained with anti-NPM (D) and anti-FLAG (D′) antibodies. B"–D″ are merged images in which nuclei were also stained in blue. Asterisks in C–D″ indicate cells in which NPM was efficiently depleted. Scale bars = 20 μm.
We also examined the localization of ectopically expressed USP36 in NPM knockdown cells. HeLa cells were transfected with FLAG-USP36 together with NPM siRNA 1 or 2 and double-stained with anti-FLAG and anti-NPM antibodies. In NPM-depleted cells, FLAG-USP36 was leaked to the nucleoplasm from nucleoli (Fig. 5, D–D″, asterisks) (data not shown) similarly to FLAG-USP36ΔBM5 in normal cells (Fig. 1, C–C″). In mock-transfected cells, FLAG-USP36 correctly localized to nucleoli (data not shown). Collectively, these results suggest that NPM is required for the nucleolar retention of USP36. Failure to detect the nucleoplasmic leakage of endogenous USP36 in NPM knockdown cells (Fig. 5, C–C″) was probably due to a low level of endogenous USP36, which did not allow its immunofluorescent detection when dispersed in the nucleoplasm.
NPM Regulates the Level of FBL Ubiquitylation
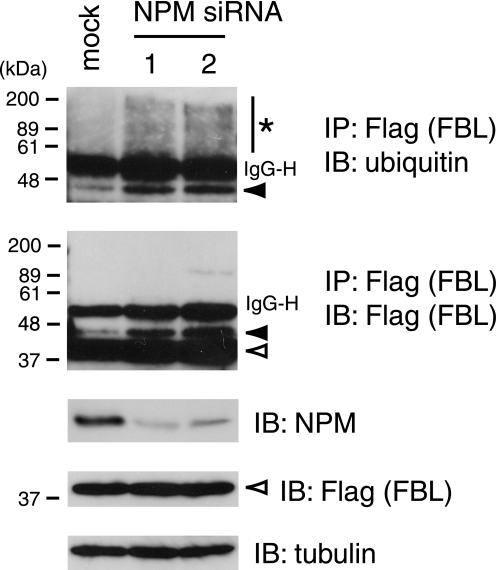
Knockdown of USP36 results in elevated levels of poly- and monoubiquitylated FBL, a nucleolar protein involved in rRNA processing, suggesting that FBL is one of the substrate proteins for USP36 (10). To test whether NPM is required for USP36 to exert the catalytic function, we examined the effect of NPM depletion on the level of FBL ubiquitylation. FLAG-tagged FBL was expressed in HeLa cells transfected with NPM siRNAs. After treatment of the cells with MG132, FBL was immunoprecipitated from their lysates with anti-FLAG antibody and immunoblotted with anti-ubiquitin antibody. In NPM-depleted cells, the levels of polyubiquitylated (Fig. 6, top panel, asterisk) and monoubiquitylated (Fig. 6, first and second panels, closed arrowheads) FBL were significantly elevated, suggesting that the nucleolar recruitment of USP36 regulates the level of FBL ubiquitylation.
FIGURE 6.
NPM regulates the level of FBL ubiquitylation. HeLa cells were transfected with FLAG-FBL together with the mock, NPM siRNA 1, or NPM siRNA 2 vector. Their lysates were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted (IB) with anti-ubiquitin (upper panel) and anti-FLAG (second panel) antibodies. Total lysate was immunoblotted with anti-NPM (third panel), anti-FLAG (fourth panel), and anti-α-tubulin (lower panel) antibodies. The asterisk, closed arrowheads, and open arrowheads indicate poly-, mono-, and non-ubiquitylated FBL, respectively. Polyubiquitylated NPM was below the detectable level in the anti-FLAG immunoprecipitation-immunoblotting experiment (second panel). IgG-H indicates the IgG heavy chain used for immunoprecipitation.
DISCUSSION
Basic amino acid stretches of ∼30 residues have been identified as NoLSs for a variety of nucleolar proteins (17). No consensus sequence has been identified in these NoLSs, and they are believed to serve as binding sites for specific residential nucleolar elements (i.e. ribosomal DNAs, rRNAs, or proteins) but not for a common nucleolus-targeting machinery. Weber et al. (18) and Horke et al. (19) pointed out, however, that NoLSs of ∼10 proteins harbor a single or multiple copies of the (K/R)(K/R)X(K/R) sequence, where X represents any amino acid. The NoLS of USP36, identified in this study and referred to as BM5 (1076RGKEKKIKKFKREKRR1091) (Fig. 1), contains three partially overlapping (K/R)(K/R)X(K/R) sequences: KKIK, KKFK, and KREK. Therefore, each of these sequences may individually serve as an NoLS. Consistently, parts of the USP36 NoLS, 1076RGKEKKIKKFKR1087 and 1083KKFKREKRR1091, were still functional as an NoLS when fused to EGFP, although somewhat less efficiently (data not shown). Triplication of the (K/R)(K/R)X(K/R) sequence in the NoLS might enhance the affinity of USP36 for nucleoli. Although BM1–4 also consist of 4–9 K/R residues (Fig. 1), they were all dispensable for the nucleolar localization of USP36 (supplemental Fig. S1). Thus, BM1–4 may not be exposed to the surface of the USP36 molecule and not be able to interact with nucleolar elements.
The NoLS of USP36 interacted with NPM (Fig. 3). Previous studies showed that the tumor suppressor Arf (16) and the Rex protein of human T-cell leukemia virus-1 (20) harbor NoLSs containing the (K/R)(K/R)X(K/R) motif and bind NPM via the NoLSs. This tetrapeptide motif therefore might constitute the NPM-binding core for at least several proteins. The central region of NPM containing two acidic clusters was essential for USP36 binding of NPM (Fig. 4), suggesting that an electrostatic interaction between the NoLS of USP36 and the acidic regions in NPM mediates the interaction of these proteins. Together, the requirement of NPM for the nucleolar localization of USP36 (Fig. 5) indicates that NPM recruits USP36 to nucleoli.
NPM forms a homo-oligomer in vitro (21) and composes a large complex of 2–5 MDa in the cell (15). In addition, NPM interacts with various proteins (11). Based on these observations, it was proposed recently that NPM serves as a hub that mediates the retention of multiple proteins in nucleoli (17). The nucleolar localization of such NPM-binding proteins regulates their activities positively or negatively. When nucleolar localization of USP36 was perturbed by NPM depletion, ubiquitylation levels of its substrate protein FBL were elevated (Fig. 6). These results suggest that nucleolar retention facilitates, but does not sequester, the catalytic activity of USP36. Our previous study suggests that NPM itself is also stabilized by USP36-mediated deubiquitylation (10). Therefore, once recruited to nucleoli by NPM, USP36 may in turn regulate the function of NPM as well.
Recent reports showed that NPM also binds to desumoylating enzymes SENP3 and SENP5, which deconjugate SUMO (small ubiquitin-like modifier) proteins from target proteins (22, 23). NPM is required for the nucleolar localization of SENP3/5 and desumoylation of nucleolar proteins (23). Because a proteome analysis suggests that USP36 deubiquitylates multiple proteins in nucleoli (10), the present study, in combination with others (22, 23), demonstrates that NPM is a key regulator of nucleolar functions that serves as a platform for protein deubiquitylation and desumoylation and controls the levels of post-translational modification of various nucleolar proteins. Finally, as NPM is deeply implicated in human cancer (11), it is of interest in future studies to elucidate how this NPM function relates to cellular transformation.
Supplementary Material
Acknowledgment
We thank C. Sherr for generously providing the NPM expression constructs.
This work was supported by Grant-in-aid 19570178 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- NPM
- nucleophosmin
- FBL
- fibrillarin
- USP
- ubiquitin-specific protease
- GST
- glutathione S-transferase
- BM
- basic motif
- EGFP
- enhanced green fluorescent protein
- siRNA
- small interfering RNA
- NoLS
- nucleolar localization signal.
REFERENCES
- 1.Boisvert F. M., van Koningsbruggen S., Navascués J., Lamond A. I. (2007) Nat. Rev. Mol. Cell Biol. 8, 574–585 [DOI] [PubMed] [Google Scholar]
- 2.Chen M., Rockel T., Steinweger G., Hemmerich P., Risch J., von Mikecz A. (2002) Mol. Biol. Cell 13, 3576–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Itahana K., Bhat K. P., Jin A., Itahana Y., Hawke D., Kobayashi R., Zhang Y. (2003) Mol. Cell 12, 1151–1164 [DOI] [PubMed] [Google Scholar]
- 4.Stavreva D. A., Kawasaki M., Dundr M., Koberna K., Müller W. G., Tsujimura-Takahashi T., Komatsu W., Hayano T., Isobe T., Raska I., Misteli T., Takahashi N., McNally J. G. (2006) Mol. Cell. Biol. 26, 5131–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lam Y. W., Lamond A. I., Mann M., Andersen J. S. (2007) Curr. Biol. 17, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo M. L., den Besten W., Bertwistle D., Roussel M. F., Sherr C. J. (2004) Genes Dev. 18, 1862–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. (2005) Cell 123, 773–786 [DOI] [PubMed] [Google Scholar]
- 8.Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. (2005) Nature 433, 77–83 [DOI] [PubMed] [Google Scholar]
- 9.Catic A., Fiebiger E., Korbel G. A., Blom D., Galardy P. J., Ploegh H. L. (2007) PLoS ONE 2, e679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Endo A., Matsumoto M., Inada T., Yamamoto A., Nakayama K. I., Kitamura N., Komada M. (2009) J. Cell Sci. 122, 678–686 [DOI] [PubMed] [Google Scholar]
- 11.Grisendi S., Mecucci C., Falini B., Pandolfi P. P. (2006) Nat. Rev. Cancer 6, 493–505 [DOI] [PubMed] [Google Scholar]
- 12.Savkur R. S., Olson M. O. (1998) Nucleic Acids Res. 26, 4508–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szebeni A., Olson M. O. (1999) Protein Sci. 8, 905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maggi L. B., Jr., Kuchenruether M., Dadey D. Y., Schwope R. M., Grisendi S., Townsend R. R., Pandolfi P. P., Weber J. D. (2008) Mol. Cell. Biol. 28, 7050–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertwistle D., Sugimoto M., Sherr C. J. (2004) Mol. Cell. Biol. 24, 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korgaonkar C., Hagen J., Tompkins V., Frazier A. A., Allamargot C., Quelle F. W., Quelle D. E. (2005) Mol. Cell. Biol. 25, 1258–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emmott E., Hiscox J. A. (2009) EMBO Rep. 10, 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber J. D., Kuo M. L., Bothner B., DiGiammarino E. L., Kriwacki R. W., Roussel M. F., Sherr C. J. (2000) Mol. Cell. Biol. 20, 2517–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horke S., Reumann K., Schweizer M., Will H., Heise T. (2004) J. Biol. Chem. 279, 26563–26570 [DOI] [PubMed] [Google Scholar]
- 20.Adachi Y., Copeland T. D., Hatanaka M., Oroszlan S. (1993) J. Biol. Chem. 268, 13930–13934 [PubMed] [Google Scholar]
- 21.Herrera J. E., Correia J. J., Jones A. E., Olson M. O. (1996) Biochemistry 35, 2668–2673 [DOI] [PubMed] [Google Scholar]
- 22.Haindl M., Harasim T., Eick D., Muller S. (2008) EMBO Rep. 9, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yun C., Wang Y., Mukhopadhyay D., Backlund P., Kolli N., Yergey A., Wilkinson K. D., Dasso M. (2008) J. Cell Biol. 183, 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.