Abstract
The 60-kDa heat shock protein (mHsp60) is a vital cellular complex that mediates the folding of many of the mitochondrial proteins. Its function is executed in cooperation with the co-chaperonin, mHsp10, and requires ATP. Recently, the discovery of a new mHsp60-associated neurodegenerative disorder, MitCHAP-60 disease, has been reported. The disease is caused by a point mutation at position 3 (D3G) of the mature mitochondrial Hsp60 protein, which renders it unable to complement the deletion of the homologous bacterial protein in Escherichia coli (Magen, D., Georgopoulos, C., Bross, P., Ang, D., Segev, Y., Goldsher, D., Nemirovski, A., Shahar, E., Ravid, S., Luder, A., Heno, B., Gershoni-Baruch, R., Skorecki, K., and Mandel, H. (2008) Am. J. Hum. Genet. 83, 30–42). The molecular basis of the MitCHAP-60 disease is still unknown. In this study, we present an in vitro structural and functional analysis of the purified wild-type human mHsp60 and the MitCHAP-60 mutant. We show that the D3G mutation leads to destabilization of the mHsp60 oligomer and causes its disassembly at low protein concentrations. We also show that the mutant protein has impaired protein folding and ATPase activities. An additional mutant that lacks the first three amino acids (N-del), including Asp-3, is similarly impaired in refolding activity. Surprisingly, however, this mutant exhibits profound stabilization of its oligomeric structure. These results suggest that the D3G mutation leads to entropic destabilization of the mHsp60 oligomer, which severely impairs its chaperone function, thereby causing the disease.
Type I chaperonins are essential molecular chaperones of the Hsp60 family found in eubacteria, mitochondria, and chloroplasts (1). They are key players in mediating the correct folding of newly translated, translocated, and stress-denatured proteins. The folding function of chaperonins is executed by the coordinated action of two oligomeric proteins, Hsp60 (also named cpn60 and in bacteria GroEL) and its co-chaperonin Hsp10 (also named cpn10 and in bacteria GroES). The GroEL molecule is composed of 14 subunits that form a barrel-like structure that consists of two back-to-back stacked heptameric rings with a large cavity at each end termed the “Anfinsen cage.” GroES is a heptameric ring formed by ∼10-kDa subunits. The co-chaperonin binds to the chaperonin in the presence of ATP and Mg2+ via a short, unstructured, but highly conserved region known as the mobile loop (2, 3). Due to the stability of the GroEL/ES oligomers and the ease with which they can be purified from bacteria, they have become the primary targets for study in the field of chaperonins. As a result, almost all of our knowledge concerning the structure and mechanism of chaperonins, both Hsp60 and Hsp10, is based largely on data from experiments on these E. coli proteins. The chaperonin reaction cycle starts when a non-folded protein substrate binds to the surface of the cavity of one of the GroEL rings. ATP-dependent GroES binding to that (cis) ring causes a dramatic conformational change that leads to a doubling of the cavity size and a switch in the cavity surface from being hydrophobic to hydrophilic. These conformational changes trigger the release of the encapsulated substrate protein into the cavity, where it can fold in a protected environment (4–9). Subsequent binding of ATP and GroES to the opposite (trans) ring promotes the release of GroES, ADP, and protein substrate from the cis ring into bulk solution.
The homologous mitochondrial chaperonin system was shown to be responsible for refolding proteins imported into the mitochondria (10, 11). The mitochondrial Hsp60 (mHsp60)3 is similar to GroEL in that it is made up of heptameric rings (12–14) that can refold denatured substrates in vitro with the assistance of a co-chaperonin and ATP (15). However, the mHsp60 oligomer is less stable than the bacterial homolog, and it exhibits unique nucleotide binding properties and specificity for co-chaperonin (13, 14, 16).
In addition to their essential function in mediating protein folding, the mammalian mitochondrial chaperonins were also suggested to be involved in extramitochondrial activities. A number of reports have suggested that mHsp60 can stimulate human leukocytes and vascular endothelial cells to produce proinflammatory cytokines (17). Furthermore, it has been reported that mHsp60 has proapoptotic and antiapoptotic roles, depending on its cellular localization (18, 19). Finally, mHsp60 and mHsp10 were found to change their expression pattern in tumor cells (20, 21).
The importance of mHsp60 for human cell function has been demonstrated through the autosomal dominant hereditary spastic paraplegia SPG13, a neurodegenerative disorder associated with two independent mutations in the gene encoding mHsp60 (22, 23). Recently, a large kindred including 23 patients suffering from MitCHAP-60 disease, an autosomal recessive neurodegenerative disorder, has been identified (24). Magnetic resonance imaging of the brains of the patients showed diffuse hypomyelination and leukodystrophy, in which myelin is not formed properly. The disease-causing mutation was identified to be a homozygous missense mutation in the human HSPD1 gene encoding the mHsp60 protein (24), namely D3G in the mature protein. Initial studies showed that, in contrast to wild-type mHsp60, the mutant, together with mHsp10, was not able to fully complement a deletion of the bacterial homologues, GroEL and GroES, in E. coli (24). The mechanism by which the D3G mutation may compromise the function of mHsp60 has not been reported. In this study, we suggest that the D3G mutation impedes the function of mHsp60 by entropic destabilization of the oligomeric structure of the molecule.
EXPERIMENTAL PROCEDURES
Proteins
mHsp10 from mouse (25) carrying a hexahistidine tag was purified using a nickel agarose column in 25 mm potassium phosphate buffer (pH 8) with 100 mm NaCl and 5 mm imidazole and then further purified using a gel filtration column (Superdex 200, GE Healthcare) equilibrated with 50 mm Tris-HCl (pH 7.7) buffer containing 300 mm NaCl, 10 mm MgCl2, and 5% glycerol (supplemental Fig. 1S). GroEL was purified as described previously (26).
Purification of mHsp60
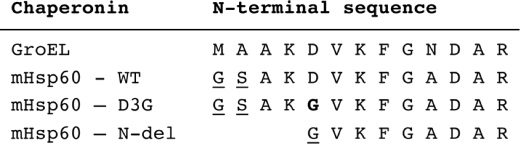
The nucleotide sequence encoding for the mature human mHsp60 was amplified using PCR and inserted between restriction sites for BamHI and NotI into a modified pET21d (27). The engineered plasmid overexpresses mHsp60 containing an octa-histidine tag at its amino terminus that can be removed after treatment with tobacco etch virus protease. The Rosetta™ E. coli strain (Novagen) was used as an overexpression host for all constructs in this study. The complete purification protocol will be described elsewhere. Briefly, mHsp60 monomers were first purified using a nickel-agarose column. Following removal of the histidine tag and further purification on an ion exchange column, oligomers were reconstituted as described previously (28) (supplemental Fig. 1S). Protein concentrations mentioned in this study refer to monomer concentrations and were determined using the bicinchoninic acid kit (Sigma catalog number BCA1). Due to cloning constraints, the various constructs used in this study contain minor differences at their amino termini. Fig. 1 details the various constructs examined in this study and the sequence of their amino termini.
FIGURE 1.
Amino-terminal sequences of chaperonins used in this study. The mutated amino acid that causes the MitCHAP-60 disease is marked in boldface type. The underlined letters signify amino acids that have been added due to cloning constraints. WT, wild type.
Cross-linking of mHsp60 Oligomers
Cross-linking with glutardialdehyde was carried out to verify the oligomeric state of mHsp60. For this purpose, a given concentration of mHsp60 was incubated with 0.1% glutardialdehyde for 10 min at 25 °C in buffer A (50 mm Na-HEPES (pH 7.5) 100 mm KCl, 30 mm MgCl2) or buffer B (100 mm Na-HEPES (pH 7.5), 10 mm KCl, 10 mm MgCl2). The cross-linking reaction was stopped by the addition of 3× SDS sample buffer containing 1 m urea and boiled for 5 min. The cross-linking products, containing 9 μg of protein, were separated on 2.4–12% acrylamide gradient SDS-PAGE and stained with Coomassie Blue.
Protein Folding Assay
The refolding assay of HCl-denatured MDH was carried out as described previously (29).
ATPase Assays
Initial (steady-state) rates of ATP hydrolysis were measured by monitoring time-resolved changes in fluorescence emission at 465 nm of coumarin-labeled phosphate-binding protein upon excitation at 430 nm using an ISS PCI spectrofluorometer (ISS, Inc.) (30, 31). The reactions were carried out in 50 mm Tris-HCl buffer (pH 7.5) containing 10 mm MgCl2, 10 mm KCl, and 1 mm dithiothreitol at 25 °C.
RESULTS AND DISCUSSION
The D3G Mutation Destabilizes the Oligomeric State of mHsp60
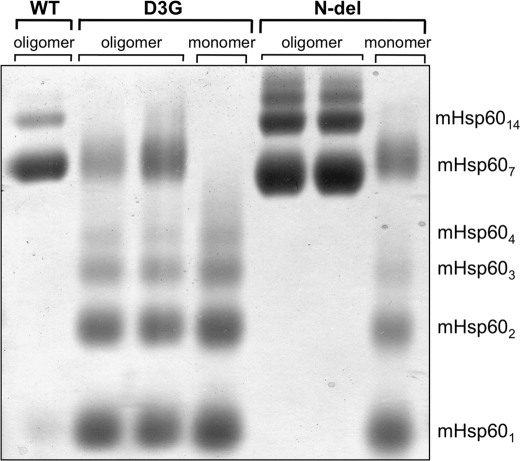
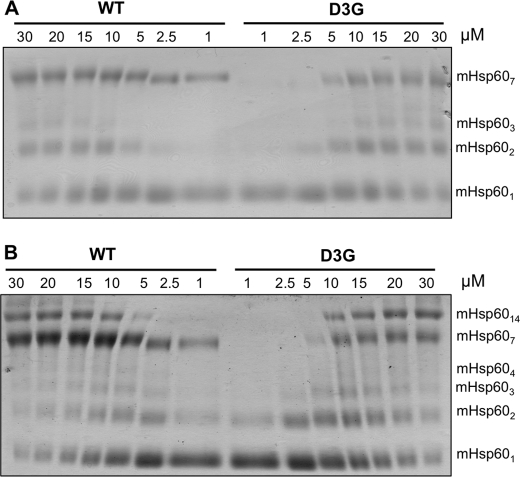
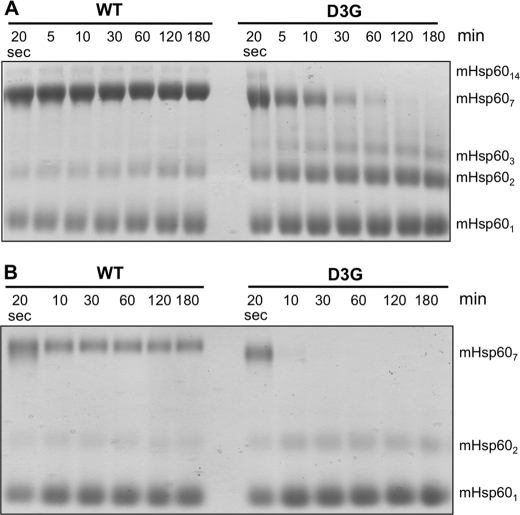
The mutation Asp-3 → Gly in the mature mHsp60 protein was recently suggested to be the cause for the MitCHAP-60 disease in humans (24). Previous works revealed the importance of the corresponding region in the bacterial GroEL for its stability (32–34). Consequently, we first sought to examine the effect of the D3G mutation on the stability of the mHsp60 oligomer. For this purpose, wild-type mHsp60 and the D3G mutant were expressed in bacteria and purified. In the last step of purification, we used gel filtration to separate active oligomeric forms of the protein from unassembled potentially inactive monomers (14, 28). All examined preparations eluted in two peaks corresponding to monomers and oligomers (supplemental Fig. 2S). The monomeric and oligomeric fractions were subjected to cross-linking following dilution to a protein concentration of 10 μm. It can be seen in Fig. 2 that wild-type mHsp60 is mainly a heptamer with a minor fraction of tetradecamers and monomers. In contrast, the D3G mutant is mainly in subheptameric forms with undetectable tetradecameric forms. Because both the wild-type mHsp60 and the D3G mutant eluted from the gel filtration column as oligomers, it appears that the mutant is less stable than the wild type and, therefore, dissociates more upon dilution. To further explore this possibility, we examined the oligomeric state of both proteins at different protein concentrations. The results presented in Fig. 3A demonstrate clearly that wild-type mHsp60 and the D3G mutant undergo dissociation when diluted to low concentrations, whereas at high protein concentrations, the oligomeric fraction is enriched. When ATP was included in the cross-linking reaction, the bands corresponding to the oligomeric forms were much stronger, and even the formation of tetradecamers could now be observed (Fig. 3B). These results are in accord with previously published results (14). Notably, the oligomers of the D3G mutant were much more sensitive to dilution than the wild-type oligomers. This is evident at the lowest concentrations examined (1 and 2.5 μm), in which the D3G mutant was completely dissociated into monomers, whereas a significant fraction of the wild-type mHsp60 was found as heptamers. As shown in Fig. 4, the dissociation of the D3G oligomer is also time-dependent. At 10 μm protein, complete dissociation of the D3G mutant was reached after 60 min, at which time only ∼30% of the wild-type protein was monomeric (Fig. 4A). At a protein concentration of 2.5 μm, dissociation of the oligomer to monomers was much more rapid. Ten minutes after dilution of proteins, all the D3G protein was found as monomers, whereas ∼30% of the wild-type protein was still oligomeric even after a 3-h incubation (Fig. 4B). Purified wild-type mHsp60 and the D3G mutant exhibited similar CD spectra (supplemental Fig. 4S), which suggests that the observed rapid dissociation of the D3G oligomers is not due to a drastic change in the secondary structure of the protein. We conclude that one main effect of the D3G mutation on mHsp60 is destabilization of the oligomeric structure of the protein.
FIGURE 2.
Cross-linking of mHsp60 variants. Following their separation by gel filtration, fractions containing either oligomer or monomer of mHsp60 variants were subjected to cross-linking with glutardialdehyde in buffer A as described under “Experimental Procedures.” The protein concentration used in all the reactions was 10 μm (monomer). The oligomeric state of the cross-linking products is indicated at the right of the figure.
FIGURE 3.
The effect of protein concentration on the oligomeric state of wild-type (WT) mHsp60 and the D3G mutant. Oligomers of the wild type and the D3G mutant were incubated in the absence (A) or presence (B) of 2 mm ATP for 3 min at the indicated concentrations and then subjected to cross-linking with glutardialdehyde. The reactions were carried out in buffer B as described under “Experimental Procedures.” The indicated protein concentrations refer to monomer concentrations. The oligomeric state of the cross-linking products is indicated at the right of the figure.
FIGURE 4.
Time-dependent disassembly of wild-type (WT) mHsp60 and the D3G mutant oligomers. Oligomers of the wild type and the D3G mutant were subjected to cross-linking with glutardialdehyde after their dilution from a 330 μm stock solution to 10 (A) or 2.5 μm (B) at different times as indicated. The reactions were carried out in buffer B as described under “Experimental Procedures.” The indicated protein concentrations refer to monomer concentrations. The oligomeric state of the cross-linking products is indicated at the right of the figure.
The D3G Mutant Exhibits Reduced ATPase and Protein Refolding Activities
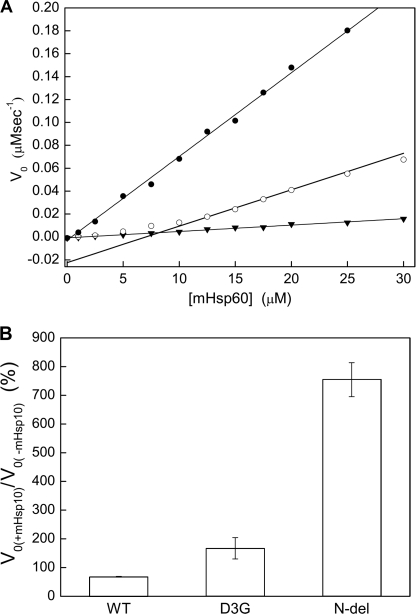
The function of chaperonin 60 requires hydrolysis of ATP that is dependent upon the oligomeric structure of the protein for optimal function. We, therefore, examined the ATPase activity of the D3G mutant when compared with that of the wild-type mHsp60. Taking into consideration the above mentioned observed effect of protein concentration on the oligomeric state of the protein, we examined the ATPase activity at different protein concentrations (Fig. 5A). Wild-type mHsp60 was found to have ATPase activity with a rate constant of 0.44 min−1. This rate was the same at all protein concentrations examined (from 1 to 30 μm) and independent of protein concentration. Notably, the ATPase activity of the D3G mutant was found to differ from that of the wild-type protein in (i) its significantly lower rate and (ii) the dependence of the rate on the protein concentration. Even at high protein concentrations, when the D3G mutant was expected to assemble into oligomers, its ATPase activity was only ∼40% of that observed for the wild-type protein. This result suggests that the D3G mutation severely impairs the ATPase activity of mHsp60 in addition to destabilizing the oligomeric structure of the protein.
FIGURE 5.
Steady-state ATPase activities of mHsp60 variants. A, initial rate measurements of ATP hydrolysis by wild-type mHsp60 (●), D3G (○), and N-del (▾) mutants were performed at different concentrations of mHsp60 and a constant concentration of 2 mm ATP. The deviation from linearity observed for the D3G mutant reflects its dissociation into monomers at low protein concentrations. B, the effect of mHsp10 on the initial rates of ATP hydrolysis by wild-type (WT) mHsp60 and the D3G and N-del mutants was also determined and is presented as a percentage of change relative to the rate in the absence of mHsp10. These reactions were carried out in the presence of 30 μm mHsp60, 2 mm ATP, with or without 30 μm mHsp10.
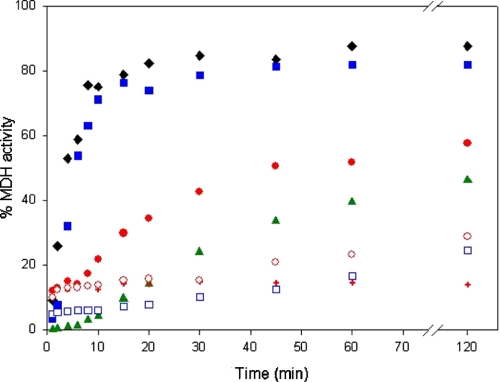
We next asked how the altered structural and ATP hydrolysis properties of the D3G mutant affect its ability to refold denatured proteins. For this purpose, we examined the chaperone properties of D3G and wild-type mHsp60 using MDH as a model substrate (Fig. 6). When HCl-denatured MDH was refolded with the help of either GroEL or wild-type mHsp60, in the presence of mHsp10, the yields of refolding after 2 h were 82 and 88%, respectively. The refolding yield in the presence of the D3G mutant and mHsp10 was found to be smaller (∼60%). Notably, although the refolding yield that is obtained with GroEL in the absence of co-chaperonin and ATP was close to background levels and did not increase over time (not shown), the yields obtained with wild-type mHsp60 and the D3G mutant, in the absence of ATP and mHsp10, were higher and increased with increasing incubation time. Thus, after a 2-h incubation, the yields in the case of the wild type and the D3G mutant were 25 and 30%, respectively. The latter result can be explained by a “minichaperone effect” (35, 36) due to the instability of the oligomeric structures of the D3G mutant and, although to a lesser extent, of the wild-type mHsp60. In other words, the chaperonins are able to bind unfolded proteins, but upon their dis-oligomerization, the unfolded substrate is released into solution in a form that is able, in vitro, to refold spontaneously. This explanation is supported by the observation that in the presence of ATP, which stabilizes the oligomeric state of mHsp60, the spontaneous refolding did not increase with increasing incubation time. Finally, another difference between the D3G mutant and wild-type mHsp60 is related to the rate of their protein folding activity. In the presence of both ATP and mHsp10, the maximum yield in the case of the wild-type protein was reached after 15 min, whereas in the case of the D3G mutant, it was reached after ∼45 min. In summary, the defective protein folding activity of the D3G mutant is reflected in both the yield and the rate when compared with the wild-type mHsp60.
FIGURE 6.
Time-dependent refolding activity of mHsp60 variants. Refolding of HCl-denatured MDH by GroEL (black diamonds), wild-type mHsp60 (blue squares), D3G (red circles), and N-del (green triangles) was carried out in the presence (filled symbols) or absence (empty symbols) of ATP and mHsp10. Red plus signs indicate refolding of MDH by the D3G mutant in the presence of ATP but without co-chaperonin.
Deletion of the First Three Amino Acids of mHsp60 Stabilizes the Oligomeric State of mHsp60 but Impairs Its Refolding Activity
The most obvious explanation for the effect of the D3G mutation on the structural stability of mHsp60 is that Asp-3 participates in interactions that are important for the stabilization of the oligomeric structure of the protein (e.g. salt bridges). However, the known crystal structures of GroEL do not provide any support for such an explanation. An alternative explanation is that the amino terminus of mHsp60 becomes more unstructured and can adopt more conformations in the monomeric state when aspartate is replaced with glycine. The D3G mutation, therefore, increases the loss of conformational entropy upon oligomerization that leads to destabilization of the oligomeric form. We reasoned that a test for this explanation would be to examine the effect of deletion of the first three amino acids (including Asp-3) of the mature mHsp60. If the latter explanation is correct, then the deletion should not lead to destabilization of the mHsp60 oligomer, and its chaperone activity will be maintained. If, however, the former explanation is correct, then the deletion should result in a destabilizing effect, as in the case of the D3G mutation. The results presented in Fig. 2 show clearly that deletion of the first three amino acids of mHsp60 (N-del) creates a very stable oligomer, even when compared with the wild-type protein. Cross-linking products corresponding to subheptameric forms of the protein are barely observed. The N-del oligomer was also found to be more resistant to heat-induced aggregation in comparison with wild-type mHsp60 and the D3G mutant (supplemental Fig. 3S) and to have a CD spectrum exhibiting slightly more secondary structure than the wild type and the D3G mutant (supplemental Fig. 4S). Interestingly, the yield of refolded MDH that is obtained with the N-del protein was close to that obtained with the D3G mutant (Fig. 6). In summary, we found that deletion of the first three amino acids in mHsp60 leads to unexpected stabilization of the protein but reduced chaperone activity (∼50% of the wild-type activity), ruling out the possible participation of Asp-3 in stabilizing interactions.
The Deletion Mutant of mHsp60 Shows Unusual ATPase Activity
To gain more insight into the effect of the deletion mutant (N-del), we examined the ability of this mutant to hydrolyze ATP. When present alone, N-del was found to have a very low rate constant of ATP hydrolysis (Fig. 5A) of 0.036 min−1, which is ∼8 and 21% of those of the wild-type protein and the D3G mutant, respectively. Examining the ATPase activity of the wild type and the two mutants in the presence of mHsp10 yielded a very surprising result (Fig. 5B). In the case of the wild-type protein, the addition of mHsp10 resulted in 30% inhibition of its hydrolysis rate, whereas in the case of the D3G mutant, the ATPase activity was increased by 70% (although the co-chaperonin is expected to inhibit the ATPase activity by 50% (37)). Previous studies showed that mHsp10 stabilizes the oligomeric structure of the unstable mitochondrial and chloroplast chaperonins and, therefore, can cause an indirect activation of the ATPase activity as observed in the case of the D3G mutant (14, 29). Interestingly, the rate of ATP hydrolysis by the N-del mutant was enhanced by 650% when mHsp10 was included. As this mutant has a very stable oligomeric structure, this increase in ATPase activity cannot be explained by the stabilization effect of mHsp10 on the N-del mutant. How can deletion of the first three amino acids cause such a profound effect on the ATP hydrolysis properties of mHsp60? The allosteric regulation of ATP hydrolysis by mHsp60 requires a fine tuning of the strength of contacts between the subunits of the mHsp60 oligomer. Thus, it could be that the deletion leads to a very strong association between the subunits of the oligomer in a less active T state (38) that switches to the more active R state only upon the addition of ATP and mHsp10. Consequently, deletion of the three amino-terminal amino acids results in impaired ATP hydrolysis only in the absence of mHsp10.
Conclusions
The aim of this study was to determine the molecular basis of the MitCHAP-60 disease caused by the D3G mutation. For this purpose, we studied the structural and functional properties of the purified D3G mutant and compared them with those of wild-type mHsp60 and a deletion mutant, in which the first three amino acids were removed. We demonstrated that at high protein concentrations, the D3G mutant can assemble into oligomers (heptamers and tetradecamers). However, the D3G oligomers are unstable when compared with wild-type mHsp60 and dissociate into monomers rapidly upon dilution. Interestingly, the deletion mutant was found to be much more stable than both the wild type and the D3G mutant. Taken together, our results suggest that the D3G mutation leads to entropic destabilization of the oligomer that may cause the MitCHAP-60 disease. Given, however, that the very stable deletion mutant has altered ATPase and protein folding activities, it is likely that the D3G mutation also has allosteric effects that remain to be identified. In summary, we suggest that the D3G mutation impairs the protein folding activity of mHsp60 by its profound destabilization effect on the mHsp60 oligomer and by disrupting its ATPase function by a yet to be determined mechanism.
Another neurodegenerative disorder associated with a point mutation (V72I) in the gene encoding mHsp60 is hereditary spastic paraplegia SPG13 (22). However, in contrast to the recessive inheritance of the disease studied here, the SPG13 disease was also observed in a heterozygote background. How can this difference in the inheritance pattern be explained? The dominant negative effect of the V72I mutation suggests that subunits with the V72I mutation form stable oligomers and produce inactive chaperonin 60 complexes, even when incorporated into mixed oligomers with wild-type subunits (39). In contrast, in the case of the MitCHAP-60 disease, we hypothesize that the mutated subunits (carrying the D3G mutation) do not form stable homo- or hetero-oligomers; consequently, they are rapidly degraded in the cell. As a result, a sufficient amount of wild-type oligomers is formed in the mitochondria. An alternative explanation is that mixed oligomers do retain activity. Hence, no dominant negative phenotype is observed in a heterozygote background.
Supplementary Material
Acknowledgment
We thank Dr. Celeste Weiss-Katz for useful discussions.
This work was supported by a grant from the Morasha program of the Israel Science Foundation (Grant 1902/08) (to A. A. and H. M.) and by an Eshkol Fellowship from the Ministry of Science (to A. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1S–4S and supplemental text.
- Hsp60
- 60-kDa heat shock protein
- mHsp60
- mitochondrial Hsp60
- MDH
- malate dehydrogenase
- MitCHAP-60
- mHsp60-associated neurodegenerative disorder.
REFERENCES
- 1.Levy-Rimler G., Bell R. E., Ben-Tal N., Azem A. (2002) FEBS Lett. 529, 1–5 [DOI] [PubMed] [Google Scholar]
- 2.Landry S. J., Taher A., Georgopoulos C., van der Vies S. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11622–11627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landry S. J., Zeilstra-Ryalls J., Fayet O., Georgopoulos C., Gierasch L. M. (1993) Nature 364, 255–258 [DOI] [PubMed] [Google Scholar]
- 4.Hartl F. U. (1996) Nature 381, 571–579 [DOI] [PubMed] [Google Scholar]
- 5.Horwich A. L., Farr G. W., Fenton W. A. (2006) Chem. Rev. 106, 1917–1930 [DOI] [PubMed] [Google Scholar]
- 6.Horovitz A. (1998) Curr. Opin. Struct. Biol. 8, 93–100 [DOI] [PubMed] [Google Scholar]
- 7.Lin Z., Rye H. S. (2006) Crit. Rev. Biochem. Mol. Biol. 41, 211–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horwich A. L., Fenton W. A., Chapman E., Farr G. W. (2007) Annu. Rev. Cell Dev. Biol. 23, 115–145 [DOI] [PubMed] [Google Scholar]
- 9.Horovitz A., Willison K. R. (2005) Curr. Opin. Struct. Biol. 15, 646–651 [DOI] [PubMed] [Google Scholar]
- 10.Cheng M. Y., Hartl F. U., Martin J., Pollock R. A., Kalousek F., Neupert W., Hallberg E. M., Hallberg R. L., Horwich A. L. (1989) Nature 337, 620–625 [DOI] [PubMed] [Google Scholar]
- 11.Ostermann J., Horwich A. L., Neupert W., Hartl F. U. (1989) Nature 341, 125–130 [DOI] [PubMed] [Google Scholar]
- 12.Picketts D. J., Mayanil C. S., Gupta R. S. (1989) J. Biol. Chem. 264, 12001–12008 [PubMed] [Google Scholar]
- 13.Nielsen K. L., Cowan N. J. (1998) Mol. Cell 2, 93–99 [DOI] [PubMed] [Google Scholar]
- 14.Levy-Rimler G., Viitanen P., Weiss C., Sharkia R., Greenberg A., Niv A., Lustig A., Delarea Y., Azem A. (2001) Eur. J. Biochem. 268, 3465–3472 [DOI] [PubMed] [Google Scholar]
- 15.Rospert S., Glick B. S., Jenö P., Schatz G., Todd M. J., Lorimer G. H., Viitanen P. V. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10967–10971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson A., Schwager F., Landry S. J., Georgopoulos C. (2001) J. Biol. Chem. 276, 4981–4987 [DOI] [PubMed] [Google Scholar]
- 17.Osterloh A., Meier-Stiegen F., Veit A., Fleischer B., von Bonin A., Breloer M. (2004) J. Biol. Chem. 279, 47906–47911 [DOI] [PubMed] [Google Scholar]
- 18.Knowlton A. A., Gupta S. (2003) Cardiovasc. Toxicol. 3, 263–268 [DOI] [PubMed] [Google Scholar]
- 19.Xanthoudakis S., Roy S., Rasper D., Hennessey T., Aubin Y., Cassady R., Tawa P., Ruel R., Rosen A., Nicholson D. W. (1999) EMBO J. 18, 2049–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deocaris C. C., Kaul S. C., Wadhwa R. (2006) Cell Stress Chaperones 11, 116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cappello F., Czarnecka A. M., La Rocca G., Di Stefano A., Zummo G., Macario A. J. (2007) Cancer Biol. Ther. 6, 487–489 [DOI] [PubMed] [Google Scholar]
- 22.Hansen J. J., Dürr A., Cournu-Rebeix I., Georgopoulos C., Ang D., Nielsen M. N., Davoine C. S., Brice A., Fontaine B., Gregersen N., Bross P. (2002) Am. J. Hum. Genet. 70, 1328–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen J., Svenstrup K., Ang D., Nielsen M. N., Christensen J. H., Gregersen N., Nielsen J. E., Georgopoulos C., Bross P. (2007) J. Neurol. 254, 897–900 [DOI] [PubMed] [Google Scholar]
- 24.Magen D., Georgopoulos C., Bross P., Ang D., Segev Y., Goldsher D., Nemirovski A., Shahar E., Ravid S., Luder A., Heno B., Gershoni-Baruch R., Skorecki K., Mandel H. (2008) Am. J. Hum. Genet. 83, 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dickson R., Larsen B., Viitanen P. V., Tormey M. B., Geske J., Strange R., Bemis L. T. (1994) J. Biol. Chem. 269, 26858–26864 [PubMed] [Google Scholar]
- 26.Bonshtien A. L., Weiss C., Vitlin A., Niv A., Lorimer G. H., Azem A. (2007) J. Biol. Chem. 282, 4463–4469 [DOI] [PubMed] [Google Scholar]
- 27.Iosefson O., Levy R., Marom M., Slutsky-Leiderman O., Azem A. (2007) Protein Sci. 16, 316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viitanen P. V., Lorimer G., Bergmeier W., Weiss C., Kessel M., Goloubinoff P. (1998) Methods Enzymol. 290, 203–217 [DOI] [PubMed] [Google Scholar]
- 29.Bonshtien A. L., Parnas A., Sharkia R., Niv A., Mizrahi I., Azem A., Weiss C. (2009) Cell Stress Chaperones 14, 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brune M., Hunter J. L., Howell S. A., Martin S. R., Hazlett T. L., Corrie J. E., Webb M. R. (1998) Biochemistry 37, 10370–10380 [DOI] [PubMed] [Google Scholar]
- 31.Brune M., Hunter J. L., Corrie J. E., Webb M. R. (1994) Biochemistry 33, 8262–8271 [DOI] [PubMed] [Google Scholar]
- 32.Horovitz A., Bochkareva E. S., Girshovich A. S. (1993) J. Biol. Chem. 268, 9957–9959 [PubMed] [Google Scholar]
- 33.Horovitz A., Bochkareva E. S., Kovalenko O., Girshovich A. S. (1993) J. Mol. Biol. 231, 58–64 [DOI] [PubMed] [Google Scholar]
- 34.White Z. W., Fisher K. E., Eisenstein E. (1995) J. Biol. Chem. 270, 20404–20409 [DOI] [PubMed] [Google Scholar]
- 35.Altamirano M. M., Golbik R., Zahn R., Buckle A. M., Fersht A. R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3576–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buckle A. M., Zahn R., Fersht A. R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3571–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandrasekhar G. N., Tilly K., Woolford C., Hendrix R., Georgopoulos C. (1986) J. Biol. Chem. 261, 12414–12419 [PubMed] [Google Scholar]
- 38.Yifrach O., Horovitz A. (1995) Biochemistry 34, 5303–5308 [DOI] [PubMed] [Google Scholar]
- 39.Bross P., Naundrup S., Hansen J., Nielsen M. N., Christensen J. H., Kruhøffer M., Palmfeldt J., Corydon T. J., Gregersen N., Ang D., Georgopoulos C., Nielsen K. L. (2008) J. Biol. Chem. 283, 15694–15700 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.