Abstract
Stromal-derived hepatocyte growth factor (HGF) acting through its specific proto-oncogene receptor c-Met has been suggested to play a paracrine role in the regulation of tumor cell migration and invasion. The transition from pre-invasive ductal carcinoma in situ (DCIS) to invasive breast carcinoma is marked by infiltration of stromal fibroblasts and the loss of basement membrane. We hypothesized that HGF produced by the infiltrating fibroblasts may alter proteolytic pathways in DCIS cells and to study this hypothesis established 3D reconstituted basement membrane overlay co-cultures with two human DCIS cell lines: MCF10.DCIS and SUM102. Both cell lines formed large dysplastic structures in 3D cultures that resembled DCIS in vivo and occasionally developed invasive outgrowths. In co-culture with HGF-secreting mammary fibroblasts, the percentage of DCIS structures with invasive outgrowths was increased. Activation of c-Met with conditioned media from HGF-secreting fibroblasts or with recombinant HGF increased the percentage of DCIS structures with invasive outgrowths, their degradation of collagen IV and their secretion of urokinase plasminogen activator and its receptor. In agreement with the in vitro findings, co-injection with HGF-secreting fibroblasts increased invasiveness of MCF10.DCIS xenografts in SCID mice. Our study demonstrates that paracrine HGF/c-Met signaling between fibroblasts and pre-invasive DCIS cells enhances the transition to invasive carcinomas and suggests that 3D co-cultures are appropriate models for testing therapeutics that target tumor microenvironment-enhanced invasiveness.
Keywords: DCIS, HGF, uPA, invasion, fibroblast, 3D culture
Introduction
An altered stroma surrounding a pre-neoplastic lesion may play a critical role in neoplastic progression and may also affect treatment strategies as has been shown for breast cancer (1, 2). Fibroblasts accumulate during progression of human breast carcinomas, including in pre-invasive ductal carcinoma in situ (DCIS) lesions (3). In several tumor types, infiltrating fibroblasts have been shown to undergo alterations in secretion of growth factors, e.g., transforming growth factor-β1 and hepatocyte growth factor (HGF) (4). The hepatocyte growth factor (HGF)/c-Met pathway has effects on proliferation, motility, invasion and angiogenesis linking it to cancer progression (5). Together HGF and c-Met form a simple paracrine signaling loop linking HGF in stromal cells and c-Met in epithelial cells (6). In breast cancer, staining for HGF and c-Met increases with progression from normal breast/benign hyperplasia to DCIS to invasive carcinoma. Higher levels of HGF and c-Met staining in DCIS are associated with other aggressive tumor markers including comedo histology, high nuclear grade, p53 positivity, and bcl-2 negativity (7). Elevated expression of c-Met at the advancing margins (8, 9) suggests a strong association between c-Met signaling and transition to invasive carcinoma (10, 11). Thus, targeting the HGF/c-Met pathway with small molecule inhibitors, decoy receptors, receptor antagonists and humanized antibodies is an area of active investigation (12, 13).
HGF/c-Met signaling regulates downstream proteolytic pathways involved in tumor growth, progression and extracellular matrix degradation, including urokinase-type plasminogen activator (uPA) and its receptor (uPAR) (14, 15). The role of the plasminogen cascade in invasion and metastasis is well-established (for review, see (16); however, uPA and uPAR also support cell migration and invasion by plasmin-independent mechanisms, including interactions between uPA, uPAR, extracellular matrix proteins, integrins, endocytosis receptors, and growth factors such as HGF (17).
Elegant studies by the Bissell and Brugge laboratories have established that growing breast epithelial cells in 3D reconstituted basement membrane (rBM) overlay cultures distinguishes between normal and malignant epithelial cells and identifies pathways that mediate morphogenesis and oncogenesis of normal epithelial cells (for review, see (18, 19)). We have shown that incorporation of fibroblasts into 3D rBM cultures can recapitulate effects of the tumor microenvironment on cell growth and progression, including increases in proteolysis (20). Here we used 3D rBM overlay cultures to mimic DCIS growth and progression in vivo. Fibroblasts expressing HGF and recombinant HGF were able to induce the development of invasive outgrowths from 3D DCIS cultures and to increase their degradation of collagen IV and their secretion of uPA and uPAR. In vivo xenograft studies recapitulated the increase in invasiveness observed in vitro.
Materials and Methods
Cell lines
MCF-10A human mammary epithelial cells were maintained in DMEM/F12 (Sigma, St. Louis, MO) supplemented with 5% horse serum (Invitrogen, Carlsbad, CA), 5 ng/ml EGF and 100 μg/ml insulin (Sigma). MCF10.DCIS (21) and SUM102 (22) human mammary DCIS cell lines were maintained in DMEM/F12 (Sigma) supplemented with 5% horse serum and Hams F-12 media (Sigma) supplemented with 10% fetal bovine serum (FBS), respectively. Normal mammary fibroblasts (MF) (23) and normal mammary fibroblasts engineered to secrete hepatocyte growth factor (MF:HGF) (24) were maintained in DMEM (Sigma) supplemented with 10% FBS.
Homotypic and heterotypic 3D rBM overlay cultures of DCIS cells and fibroblasts
For homotypic cultures, 6 well plates were coated with 10 mg/ml Cultrex without phenol red (Trevigen, Gaithersburg, MD) and allowed to solidify for 20 min at 37 °C. MCF10.DCIS or SUM102 cells (3.0 × 105) were seeded as single cells onto the solidified rBM. For heterotypic co-cultures, 1.5 × 105 fibroblasts were mixed with the rBM prior to coating the wells. Cultures were grown in M171 Mammary Epithelial Media (Invitrogen) supplemented with Mammary Epithelial Growth Supplement (Invitrogen) and 2% rBM. Where indicated, concentrated, conditioned media from fibroblasts, 100 ng/ml recombinant HGF (rHGF; R&D, Minneapolis, MN), 2 μM SU11274 (Calbiochem, San Diego, CA) or rHGF plus SU11274 were added to culture media. Media were replenished every 3 days. The HGF concentration was based on preliminary studies establishing an increase in expression of uPA/uPAR, a response known to occur in parallel with HGF-induced tubulogenesis of MDCK cells (19).
Immunoblots
3D structures were harvested from rBM by incubation with cold PBS supplemented with 5 mM EDTA, 1 mM NaVO4 and 1.5 mM NaF on ice for 45 minutes. Conditioned media were collected and concentrated 10× in Ultrafree-0.5 PBGC Centrifugal Filter Units with 10 kDa MWCO Biomax Membranes (Millipore, Billerica, MA). Cell lysates were prepared in lysis buffer [250 mM sucrose, 25 mM 2(N-morpholino)ethanesulfonic acid, 1 mM EDTA, 0.1% Triton X-100, pH 6.5], cleared by centrifugation at 14,000 × g for 10 minutes at 4 °C and the supernatant used for all subsequent procedures. Supernatants and conditioned media were separated by SDS-PAGE, transferred to nitrocellulose membranes and immunoblotted with polyclonal goat anti-HGF (R&D), rabbit anti-uPA (Abcam, Cambridge, MA) or rabbit anti-uPAR (Abcam) or monoclonal mouse anti-β-actin (AC-15; Sigma) or mouse anti-GAPDH (MAB374; Millipore).
Fibroblast-conditioned media
Equal numbers of MF or MF:HGF were cultured in M171 media for 4 days. Conditioned media were centrifuged at 100 × g to pellet any floating cells and centrifuged again at 800 × g to remove debris. Cleared media were concentrated to 1/10th of their original volume with a Centriprep Ultracell YM-10 (Millipore), stored at -20 °C and diluted 1:2 with fresh media before use.
Quantification of invasive outgrowths
Day 9 cultures were imaged in triplicate for development of invasive outgrowths by differential interference contrast imaging (DIC) on an Ultraview ERS (Perkin Elmer) utilizing a 10× objective. Invasive outgrowths were defined as consisting of two or more cells migrating away from their structure of origin. A minimum of 10 images was captured and analyzed for each experimental condition.
Live-cell proteolysis assay
Assays for quantification of degradation of DQ™-collagen IV substrate (Invitrogen) by live cells were performed as described previously (25). Where indicated, conditioned media, 100 ng/ml rHGF or 1 μM aprotinin (Calbiochem) were added to culture media. For the overlaid DQ-collagen IV degradation assay, MCF10.DCIS cells were grown in 3D rBM overlay culture on round glass coverslips for 4 days. Coverslips containing preformed structures were then incubated with CellTracker Orange (Invitrogen) and inverted onto rBM containing 25 μg/ml DQ-collagen IV, which had been diluted to 10 mg/ml with fibroblast conditioned medium. These cultures were imaged live on a Leica TCS SP5 confocal microscope with a 20× PL APO N.A. 0.7 objective.
Invasion assays
Transwell invasion assays were conducted over 5 days to allow formation of 3D structures. Briefly, 0.8 μm BioCoat control inserts (BD Biosciences, San Jose, CA) were coated with 20 μl of 5 mg/ml rBM and the rBM allowed to polymerize. MCF10.DCIS cells (1.0 × 104) in M171 media were seeded on rBM-coated inserts and cultured in a 24-well plate in the absence or presence of 1.0 × 104 MF or MF:HGF. Non-invasive cells were removed with a cotton swab, invasive cells fixed with 3.7% formaldehyde (Polysciences, Warrington, PA) and nuclei stained with DAPI (Invitrogen). Images, obtained with a Zeiss LSM510 META NLO confocal microscope, were scored for the number of cells (nuclei) that had invaded by two independent observers.
Phalloidin staining for 3D reconstructions
3D rBM overlay cultures on coverslips were fixed with 3.7% formaldehyde, permeabilized with PBS containing 0.1% Triton X-100, blocked with PBS containing 2% BSA and then incubated with PBS containing 0.1% Triton X-100 and 4 U/ml Alexa Fluor 546 phalloidin (Invitrogen). Cultures were imaged on a Zeiss LSM 510 META using an Acroplan 10×/0.3 water immersion objective and LSM AIM software. Z-stack data were imported into Volocity software (Improvision) to generate 3D reconstructions and movies.
Xenograft Studies
Xenografts were generated by injecting 1.0 × 106 MCF10.DCIS cells ± 5.0 × 105 MF or MF:HGF in 0.1 ml Cultrex subcutaneously at the base of the nipple of gland #5 of female ICRSC-M mice (Taconic Farms, Germantown, PA). Mice were maintained under aseptic conditions according to Institutional Animal Care and Use Committee guidelines. Four weeks post-injection mice were sacrificed and xenografts removed, fixed with Z-FIX (Anatech, Battle Creek, MI), embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E).
Results
Co-culture with HGF-secreting mammary fibroblasts increases invasiveness of MCF10.DCIS cells
To mimic increases in stromal fibroblasts (3) and stromal-derived HGF levels (7), we introduced MF or MF:HGF into 3D MCF10.DCIS rBM overlay cultures. We had previously established by immunoblot analysis of conditioned media from MCF10.DCIS, MF and MF:HGF cells that only the MF:HGF cells secreted HGF (105-210 ng/ml; n = 3; data not shown). Over a period of 9 days in either homotypic or heterotypic 3D rBM overlay cultures with MF cells, MCF-10A cells were observed to form acinar structures (Fig. 1A) and MCF10.DCIS cells to form irregular aggregates (Fig. 1A and Supplementary Movies 1 & 2). In heterotypic cultures with MF:HGF cells, MCF-10A acini were unchanged (Fig. 1A), but many of the MCF10.DCIS structures exhibited ‘invasive outgrowths’ consisting of two or more cells migrating into the surrounding extracellular matrix (Fig. 1A and Supplementary Movie 3). To assess the reproducibility of this effect, we scored cultures of MCF10.DCIS ± fibroblasts labeled with cytoplasmic dyes to facilitate discrimination of invasive outgrowths (Fig. 1A). In co-cultures of MF:HGF and MCF10.DCIS cells, a significantly higher percentage of 3D structures were found to display invasive outgrowths than did MCF10.DCIS cells alone or MCF10.DCIS cells in co-culture with MF cells (Fig. 1B). Thus, the pre-invasive DCIS cells, but not the immortalized MCF-10A cells, exhibited a paracrine response to HGF-secreting mammary fibroblasts.
Figure 1. HGF-secreting mammary fibroblasts increase invasive outgrowths from DCIS 3D structures and their invasion through rBM.
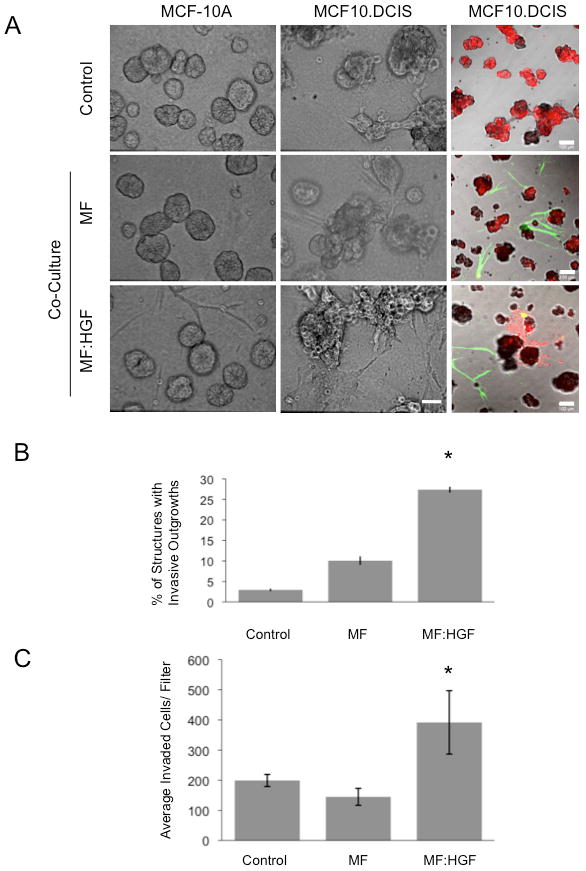
MCF-10A and MCF10.DCIS cells were grown in 3D rBM overlay culture in the absence (Control) or presence of mammary fibroblasts (MF) or HGF-secreting mammary fibroblasts (MF:HGF). A, Representative DIC images are illustrated in the 1st and 2nd column (bar, 50 μm) and representative merged images of green, red and DIC channels are shown in the 3rd column (bar, 100 μm). MCF10.DCIS cells were prelabeled with CellTracker Orange (red); fibroblasts were prelabeled with CellTracker Green (green) and embedded within the rBM. B, Images of MCF10.DCIS cells alone and in co-culture with fibroblasts were scored for the number of total structures and those with invasive outgrowths. Data in graphs are pooled from 14 fields of view in three independent experiments and are presented as the mean percentage of 3D structures with invasive outgrowths ± SD. *, p<0.02, (Student's t-test). C, Transwell invasion assay without fibroblasts (Control) or with fibroblasts grown in the well below the rBM-coated Transwell filter. Data in graphs are average number of cells that invaded through rBM coated Transwell filters, pooled from three independent experiments, and presented as mean ± SD. *, p<0.02, (Student's t-test).
To determine whether the increase in invasiveness of MCF10.DCIS cells required direct contact with the fibroblasts, we performed an invasion assay in which the two cell types were separated by a Transwell filter coated with a thick layer of rBM that allowed formation of 3D structures by the MCF10.DCIS cells. The presence of MF:HGF cells, but not MF cells, in the lower chamber resulted in a 2-fold increase in invasion of MCF10.DCIS cells (Fig. 1C), a finding consistent with soluble factors secreted by the MF:HGF cells inducing invasion of MCF10.DCIS cells.
Conditioned media from HGF-secreting fibroblasts increases invasion and development of invasive outgrowths by MCF10.DCIS 3D structures
The invasion assays suggested that a soluble factor, potentially pro-HGF secreted by the MF:HGF cells, stimulated MCF10.DCIS invasion. Fibroblast CM was therefore added to 3D rBM cultures of MCF10.DCIS cells and growth and development of invasive outgrowths monitored. At 24 hours after plating, we observed that fibroblast CM had stimulated migration of MCF10.DCIS cells; as a result, 3D structures in these cultures originated from clusters of cells rather than from single cells as in the control cultures. Consequently after 9 days, there were fewer 3D structures in cultures incubated with MF CM or MF:HGF CM than in control cultures (270 ± 40 and 231 ± 22, respectively, as compared to 325 ± 17; mean ± SEM). CM from the MF:HGF cells induced the formation of large invasive outgrowths from MCF10.DCIS 3D structures (Fig. 2A) and significantly increased (∼3-fold) the percentage of structures that developed invasive outgrowths (Fig. 2B). The observed comparable stimulation of invasive outgrowths by co-culture with MF:HGF cells and by addition of MF:HGF CM was consistent with soluble HGF secreted from the MF:HGF cells being the causative factor.
Figure 2. Conditioned media from HGF-secreting mammary fibroblasts stimulates invasive outgrowths of MCF10.DCIS cells, their invasion through rBM and degradation of DQ-collagen IV.
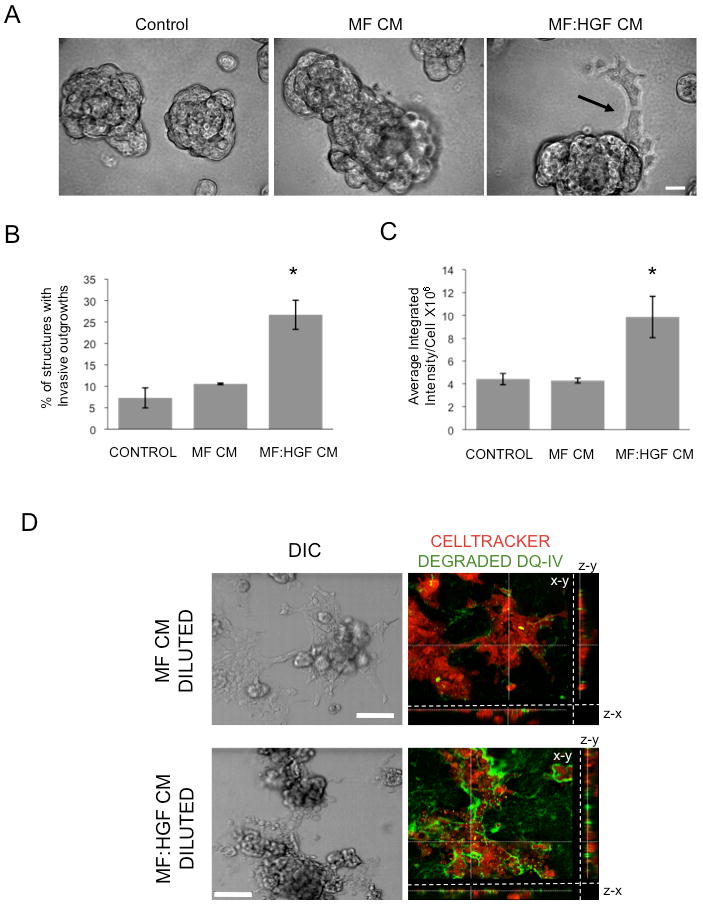
A, B and C: MCF10.DCIS cells were grown in 3D rBM culture in the absence of fibroblast-conditioned media (Control) or the presence of media conditioned by mammary fibroblasts (MF CM) or HGF-secreting mammary fibroblasts (MF:HGF CM). A, MF:HGF CM stimulates invasive outgrowths (arrow) of cells from 3D structures; bar, 20 μm. B, Images were scored for the percentage of structures with invasive outgrowths. Graphs represent data pooled from 14 fields of view from each of three separate experiments and are presented as mean percentage of 3D structures with invasive outgrowths ± SD. *, p<0.02, significant increase as compared to control and MF CM values (Student's t-test). C, MCF10.DCIS cells were grown in 3D rBM culture containing DQ-collagen IV without (Control) or with MF CM or MF:HGF CM for 4 days before imaging. Graphs represent average integrated fluorescence intensity per nuclei in relative fluorescence units (RFU) and are pooled from three separate experiments and presented as mean ± SEM (n=18). *, p<0.02, significant increase as compared to control and MF CM values (Student's t-test). D, MCF10.DCIS cells were grown in 3D rBM overlay culture on coverslips for 4 days, labeled with CellTracker Orange and the coverslips inverted onto glass bottom dishes coated with rBM, containing DQ-collagen IV, that had been diluted with either MF CM or MF:HGF CM. Cultures were imaged live following 18 hours. Representative DIC images and corresponding merged images of DIC, CellTracker Orange (red) and degraded DQ-collagen IV (green) channels are shown; bar, 100 μm. Each merged image represents a focal plane observed in x/y, y/z and x/z axes.
To examine whether the increased invasion was associated with increased degradation of the basement membrane, we used a live cell confocal microscopy-based assay to image and quantify degradation of a dye-quenched form of the basement membrane protein collagen type IV (25). Degradation of DQ-collagen IV by MCF10.DCIS cells was significantly increased by MF:HGF CM (∼2.5 fold; Fig. 2C), an increase similar to that in the percentage of structures with invasive outgrowths (Fig. 2B). In addition, we examined the effect of fibroblast-secreted HGF on DQ-collagen IV degradation by pre-formed MCF10.DCIS structures. Live imaging revealed a dramatic increase in degradation products surrounding the MCF10.DCIS structures that had been incubated with MF:HGF CM (Fig. 2D). Thus, HGF-enhanced collagen IV degradation was associated with invasion of pre-formed structures and not solely the result of proteolysis by growing and migrating DCIS cells.
Conditioned media from HGF-secreting fibroblasts activates c-Met and stimulates expression and secretion of uPA and uPAR
Binding of HGF, the only known ligand for the c-Met receptor, results in receptor activation and phosphorylation of tyrosine residues within the activation domain (26). Small molecule inhibitors that compete for ATP binding, such as SU11274, inhibit c-Met phosphorylation (27). To determine if HGF secreted by MF:HGF fibroblasts bound to and activated c-Met on DCIS cells, we collected cell lysates from 3D DCIS cultures treated with MF:HGF CM ± SU11274. To detect activation of c-Met, we immunoblotted cell lysates using antibodies specific for the double phosphorylation at phosphoepitopes Y1234/Y1235 of c-Met. Following MF:HGF CM treatment, we observed a sustained phosphorylation of c-Met in both MCF10.DCIS and SUM102 DCIS lines and SU11274 abrogated the phosphorylation (Fig. 3A). SU11274 also significantly decreased the development of invasive outgrowths induced when MCF10.DCIS cells were co-cultured with HGF-expressing fibroblasts (Suppl. Fig. 1), consistent with HGF activating c-Met in these co-cultures.
Figure 3. Phosphorylation of c-Met by MF:HGF CM is correlated with increased expression and secretion of uPA and uPAR by DCIS cells.
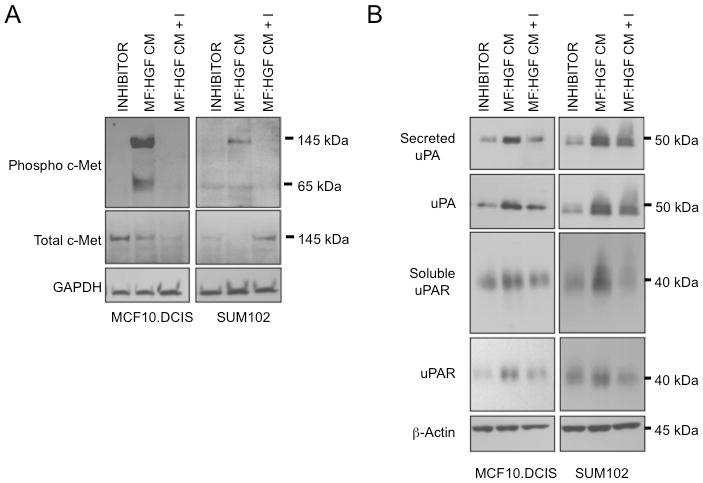
MCF10.DCIS and SUM102 cells were grown in 3D rBM cultures with either 2 μM SU11274 (INHIBITOR), MF:HGF conditioned media (MF:HGF CM) or MF:HGF conditioned media plus 2 μM SU11274 (MF:HGF CM + I). A, 3D rBM cultures were harvested and lysed in sample buffer, separated by 12% SDS-PAGE, transferred to nitrocellulose and analyzed with antibodies against phospho Y1234/Y1235 c-Met. Membranes were subsequently stripped and probed with antibodies against total c-Met and GAPDH as a loading control. B, Cell lysates and concentrated conditioned media from 3D rBM cultures were separated by 12% SDS-PAGE with loading based on protein concentration and relative volume, respectively. Proteins were transferred to nitrocellulose and analyzed with antibodies against uPA and uPAR.
HGF treatment in 2D monolayer cultures has been shown to increase expression and secretion of proteases capable of degrading extracellular matrices, including the uPA/uPAR system (15, 28, 29). We therefore determined whether treatment with MF:HGF CM (shown above to stimulate c-Met phosphorylation) increased the expression and secretion of uPA and uPAR in 3D rBM cultures of MCF10.DCIS and SUM102 cells. Cultures treated with MF:HGF CM exhibited increased levels of uPA and uPAR in cell lysates and conditioned media as compared to controls and SU11274 abrogated those increases in the MCF10.DCIS cells and partially in the SUM102 cells (Fig. 3B). As uPA and uPAR were not detected in concentrated MF:HGF CM itself (data not shown), the changes seen in the MF:HGF CM treated cultures reflected changes in expression and secretion by MCF10.DCIS and SUM102 cells.
Recombinant HGF increases development of invasive outgrowths from MCF10.DCIS and SUM102 3D structures
MF:HGF CM stimulated invasive outgrowths, invasion and degradation of DQ-collagen IV by MCF10.DCIS cells, consistent with an involvement of HGF. To confirm that HGF acting through c-Met activation was responsible for the increase in invasive outgrowths, we incubated the DCIS cell lines with rHGF and the c-Met inhibitor SU11274. Both MCF10.DCIS and SUM102 cell lines formed dysplastic 3D structures with few structures exhibiting invasive outgrowths in the presence of the DMSO vehicle control (Control) or SU11274 (I). Invasive outgrowths were observed in DCIS cell lines treated with rHGF, but not treated with both rHGF and the c-Met inhibitor (Fig. 4A). The percentage of structures with invasive outgrowths was significantly increased by rHGF (Fig. 4B). Stimulation of c-Met appeared to be responsible for the increase of invasive outgrowths since rHGF did not increase the percentage of structures with invasive outgrowths in the presence of SU11274 (Fig. 4B). Together these experiments suggest that HGF-secreting fibroblasts within the tumor microenvironment have the potential to increase invasiveness of DCIS lesions and thus their transition to invasive carcinomas.
Figure 4. Recombinant HGF acting through c-Met signaling stimulates invasive outgrowths and increased secretion of uPA and uPAR from DCIS cells.
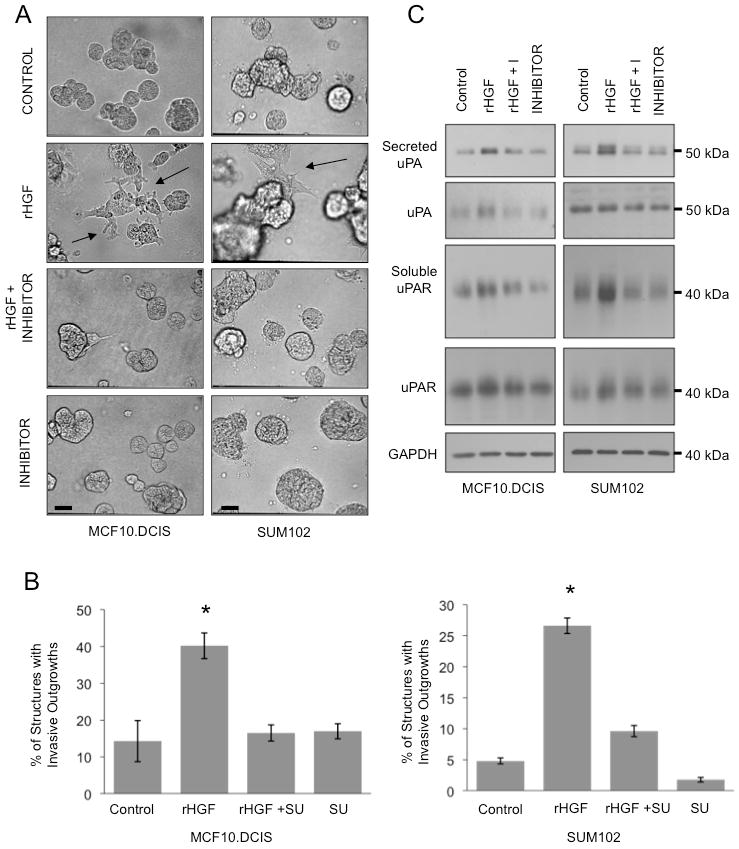
MCF10.DCIS and SUM102 cells were grown in 3D rBM culture with either 100 ng/ml HGF (rHGF), 100 ng/ml HGF + 2 μM SU11274 (rHGF + I), 2 μM SU11274 (I) or DMSO (Control). A, Representative DIC images depicting condition before harvesting cell lysates and conditioned media; bar, 200 μm. B, Images from MCF10.DCIS and SUM102 cell cultures were scored for the number of total structures and those with invasive outgrowths. C, Cell lysates and concentrated CM were separated by 12% SDS-PAGE under non-reducing conditions with lysates loaded based on protein concentration and CM loaded based on the protein concentration of the corresponding cell lysates. Proteins were transferred to nitrocellulose membranes and analyzed with antibodies against uPA or uPAR.
Recombinant HGF increases expression and secretion of uPA and uPAR
Since expression of uPA and uPAR was increased in 3D rBM cultures treated with MF:HGF CM (Fig. 3), we analyzed whether rHGF could elicit similar results. Total cell lysates and concentrated CM from cultures such as those shown in Fig. 4A were separated by SDS-PAGE and immunoblotted for uPA and uPAR. rHGF increased secretion of uPA and uPAR from DCIS cell lines (Fig. 4C). Immunoblotting of cell lysates with a c-Met phosphorylation specific antibody confirmed that c-Met was activated by rHGF and inhibited by SU11274 in this series of experiments (data not shown). Our results suggest that HGF-stimulated invasiveness was associated with increases in plasmin-generating potential through an imbalanced uPA/uPAR system (16).
Recombinant HGF increases degradation of DQ-collagen IV by MCF10.DCIS and SUM102 3D structures
HGF/c-Met-stimulated development of invasive outgrowths may be associated with increased proteolysis initiated by increased uPA secretion. Therefore, we used a live-cell proteolysis assay to image and quantify the degradation of DQ-collagen IV by MCF10.DCIS or SUM102 cells grown for 18 hours in the presence of 100 ng/ml rHGF ± 1 μM aprotinin, or untreated. We observed degradation products bordering the periphery of DCIS structures as well as intracellularly (Fig. 5A). rHGF increased the intensity of peripheral degradation products associated with MCF10.DCIS and SUM102 3D cultures (Fig. 5A and B; rHGF). We confirmed that the increase was significant by quantifying degradation products in the entire volume of the 3D cultures and normalizing to the number of cells contributing to degradation in that volume (Fig. 5C, data represented as average integrated intensity per cell). Aprotinin moderately decreased HGF-induced fluorescence surrounding DCIS structures (Fig. 5A and B; rHGF + A), but this reduction was not significant (Fig. 5C). Our results thus suggest an involvement of more than one family of proteases in the increases in degradation of type IV collagen associated with HGF/c-Met-induced invasive outgrowths in DCIS structures.
Figure 5. HGF increased degradation of DQ-collagen IV by MCF10.DCIS and SUM102 3D cultures.
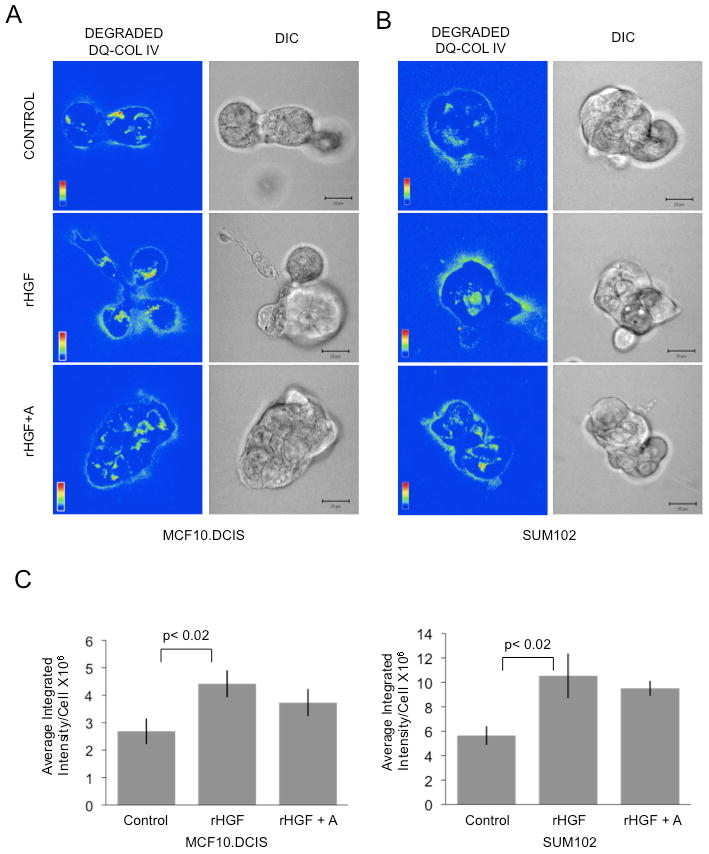
MCF10.DCIS or SUM102 cells grown in 3D rBM cultures containing DQ-collagen IV were treated with 100 ng/ml HGF (HGF), 100 ng/ml HGF plus aprotinin (HGF+A) or left untreated (Control). Integrated fluorescence due to proteolysis (green) was normalized to the number of cells (nuclei; blue). Representative fluorescence micrographs of one confocal plane of both the degraded DQ-collagen IV channel (degraded DQ-IV) and the corresponding DIC channel are illustrated for MCF10.DCIS (A) and SUM102 (B) 3D rBM cultures. Bar, 20 μm. C, Quantification of proteolysis in 3D structure volumes. Data were pooled from 3 representative experiments and are presented as mean ± SEM (n=18). Student's t-test, p<0.02.
Co-injection with HGF-secreting fibroblasts increases invasiveness of MCF10.DCIS xenografts
To determine whether HGF also affects invasiveness in vivo, we compared xenografts of MCF10.DCIS cells alone or coinjected with MF or MF:HGF cells. Tumor take was similar among the 3 groups of mice (n=8 per group), yet the median wet weight of the tumors was greatest in mice coinjected with DCIS cells and MF:HGF (680 mg; range: 70-869 mg), as compared to DCIS cells alone (99 mg; range: 66-124 mg) and those coinjected with DCIS cells and MF (171 mg; range: 90-502 mg). Strikingly, coinjection of DCIS cells and MF:HGF cells enhanced progression to invasive ductal carcinomas (Fig. 6). These results confirmed our in vitro findings that paracrine HGF/c-Met signaling between fibroblasts and pre-invasive DCIS enhances invasiveness.
Figure 6. Co-injection with HGF-secreting fibroblasts increases invasiveness of MCF10.DCIS xenografts.
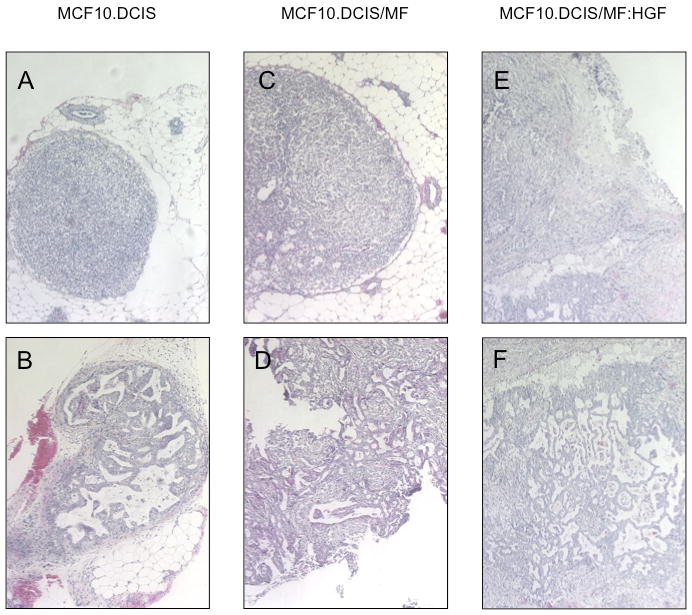
Representative 10× images of H&E stained MCF10.DCIS xenografts. MCF10.DCIS xenografts exhibited either atypical hyperplasias (A) or comedo DCIS lesions (B). MCF10.DCIS/MF xenografts exhibited primarily atypical hyperplasias (C) with a few progressing to an invasive phenotype (D). All of the MCF10.DCIS/MF:HGF xenografts had progressed to invasive ductal carcinoma (E and F).
Discussion
In this study, we established that DCIS cell lines grown in 3D rBM overlay cultures replicate characteristics of pre-neoplastic to neoplastic phenotypes that accompany the transition from pre-invasive to invasive. We observed that HGF/c-Met signaling increased the invasive phenotype of DCIS cells, including their ability to migrate, to degrade collagen type IV and their expression and secretion of uPA and uPAR. This appears to be generalizable as we obtained comparable results in two unrelated human DCIS cell lines [MCF10.DCIS (21) and SUM102 (22)]. Similar results were obtained in vivo in that HGF-secreting fibroblasts enhanced the invasive phenotype of MCF10.DCIS xenografts.
We have previously demonstrated that, when grown in 3D rBM overlay culture, isogenic MCF-10A epithelial variants form an in vitro progression series for analysis of pre-neoplastic and neoplastic phenotypes (30). In 3D rBM overlay cultures, MCF10.DCIS cells grow into large dysplastic structures (30), a finding recapitulated here for another DCIS cell line: SUM102. Interestingly, the dysplastic structures formed by both DCIS cell lines resemble in morphology and size those depicted for MCF-10A cells that have been transfected with ErbB2 (31). In MCF10.DCIS cells, MAPK/ERK activity is elevated (30), whereas in SUM102 cells, EGFR is elevated and EGFR ligands are secreted (22). ErbB2, MAPK/ERK and EGFR all have been linked to kidney tubulogenesis (32). Our results are thus consistent with the pre-neoplastic invasive outgrowths from dysplastic DCIS structures having the same molecular underpinnings as tubular outgrowths formed during normal developmental processes.
Fibroblasts induce formation of capillary-like 3D networks by endothelial cells (33) and branching morphogenesis of mammary epithelial cells (34). Growth and angiogenesis of subcutaneous MDA-MB-231 human breast carcinoma tumors is increased by co-implantation of fibroblasts, an effect reduced by downregulating HGF in the fibroblasts or c-Met in the carcinoma cells (35). These latter in vivo results are consistent with those reported here for DCIS-fibroblast 3D rBM co-cultures and xenografts. We suggest that there may be interaction between c-Met signaling and aberrant ErbB2 signaling in the DCIS cell lines. Such interactions have been shown for MCF-10A cells that express an inducible ErbB2 receptor and are infected with a retrovirus expressing HGF cDNA. These cells develop into large dysplastic structures with enhanced protrusive behavior (36), resembling the ones formed by the two DCIS lines in response to HGF. Our results would thus be consistent with ErbB2 related alterations in MCF10.DCIS and SUM102 cells interacting with HGF/c-Met signaling to promote development of large dysplastic structures and invasive outgrowths. Interestingly, HGF/c-Met signaling also may contribute to the intrinsic resistance to EGFR tyrosine kinase inhibitors seen in breast cancers. In cooperation with c-Src, HGF/c-Met signaling has been shown to trans-activate EGFR even in the presence of EGFR tyrosine kinase inhibitors (37).
The accumulation of fibroblasts and increased HGF found in DCIS lesions in vivo in parallel with losses in basement membrane suggest a relationship for fibroblasts and HGF in progression of DCIS from pre-invasive to invasive. The 3D rBM overlay culture and co-culture models for DCIS replicated the morphology and progression of human DCIS in vivo and allowed us to demonstrate that paracrine HGF/c-Met signaling stimulated invasiveness, upregulated expression and secretion of uPA/uPAR and increased degradation of collagen IV. We suggest that the 3D DCIS rBM overlay cultures/co-cultures will be useful for analyzing the contributions of stromal-derived factors to DCIS progression and for screening therapeutics that target this progression, e.g., agents that target HGF/c-Met signaling. We are presently unable to distinguish DCIS lesions that will rapidly progress to invasive carcinomas from those that will not, thus subjecting many women to unnecessary biopsies and others who should be treated to extended periods of monitoring during which their disease may progress. This is of clinical relevance as nearly 30% of newly diagnosed breast cancers are found at the DCIS stage (21).
Supplementary Material
Acknowledgments
We thank Drs. Fred Miller and Stephen Ethier (Karmanos Cancer Institute, Detroit, MI), Kornelia Polyak (Dana Farber Cancer Institute, Boston, MA), and Charlotte Kuperwasser (Tufts University, Boston, MA) for their kind gifts of cell lines. We gratefully acknowledge Bruce Linebaugh and Mackenzie Herroon for their assistance with xenograft studies.
Financial Support: R01 CA56586 (BFS), DOD BC013005 (BFS) and DOD BC051230 (CJ). Imaging was performed in the Microscopy and Imaging Resources Laboratory supported, in part, by NIH Center Grants P30 CA22453 and P30 ES 06639 and by U54 RR02084330.
Footnotes
Disclosure of Potential Conflicts of Interest None of the authors report potential conflicts of interest.
References
- 1.Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597–601. doi: 10.4161/cc.5.15.3112. [DOI] [PubMed] [Google Scholar]
- 3.Pavlakis K, Messini I, Vrekoussis T, et al. The assessment of angiogenesis and fibroblastic stromagenesis in hyperplastic and pre-invasive breast lesions. BMC cancer. 2008;8:88. doi: 10.1186/1471-2407-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–9. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 5.Gao CF, Vande Woude GF. HGF/SF-Met signaling in tumor progression. Cell research. 2005;15:49–51. doi: 10.1038/sj.cr.7290264. [DOI] [PubMed] [Google Scholar]
- 6.Gentile A, Trusolino L, Comoglio PM. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008;27:85–94. doi: 10.1007/s10555-007-9107-6. [DOI] [PubMed] [Google Scholar]
- 7.Jin L, Fuchs A, Schnitt SJ, et al. Expression of scatter factor and c-met receptor in benign and malignant breast tissue. Cancer. 1997;79:749–60. doi: 10.1002/(sici)1097-0142(19970215)79:4<749::aid-cncr12>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 8.Edakuni G, Sasatomi E, Satoh T, Tokunaga O, Miyazaki K. Expression of the hepatocyte growth factor/c-Met pathway is increased at the cancer front in breast carcinoma. Pathology international. 2001;51:172–8. doi: 10.1046/j.1440-1827.2001.01182.x. [DOI] [PubMed] [Google Scholar]
- 9.Tuck AB, Park M, Sterns EE, Boag A, Elliott BE. Coexpression of hepatocyte growth factor and receptor (Met) in human breast carcinoma. Am J Pathol. 1996;148:225–32. [PMC free article] [PubMed] [Google Scholar]
- 10.Beviglia L, Matsumoto K, Lin CS, Ziober BL, Kramer RH. Expression of the c-Met/HGF receptor in human breast carcinoma: correlation with tumor progression. Int J Cancer. 1997;74:301–9. doi: 10.1002/(sici)1097-0215(19970620)74:3<301::aid-ijc12>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 11.Elliott BE, Hung WL, Boag AH, Tuck AB. The role of hepatocyte growth factor (scatter factor) in epithelial-mesenchymal transition and breast cancer. Canadian journal of physiology and pharmacology. 2002;80:91–102. doi: 10.1139/y02-010. [DOI] [PubMed] [Google Scholar]
- 12.Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov. 2008;7:504–16. doi: 10.1038/nrd2530. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Yao W, Newton RC, Scherle PA. Targeting the c-MET signaling pathway for cancer therapy. Expert opinion on investigational drugs. 2008;17:997–1011. doi: 10.1517/13543784.17.7.997. [DOI] [PubMed] [Google Scholar]
- 14.Pepper MS, Matsumoto K, Nakamura T, Orci L, Montesano R. Hepatocyte growth factor increases urokinase-type plasminogen activator (u-PA) and u-PA receptor expression in Madin-Darby canine kidney epithelial cells. J Biol Chem. 1992;267:20493–6. [PubMed] [Google Scholar]
- 15.Jeffers M, Rong S, Vande Woude GF. Enhanced tumorigenicity and invasion-metastasis by hepatocyte growth factor/scatter factor-met signalling in human cells concomitant with induction of the urokinase proteolysis network. Mol Cell Biol. 1996;16:1115–25. doi: 10.1128/mcb.16.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dano K, Behrendt N, Hoyer-Hansen G, et al. Plasminogen activation and cancer. Thromb Haemost. 2005;93:676–81. doi: 10.1160/TH05-01-0054. [DOI] [PubMed] [Google Scholar]
- 17.Mazar AP. Urokinase plasminogen activator receptor choreographs multiple ligand interactions: implications for tumor progression and therapy. Clin Cancer Res. 2008;14:5649–55. doi: 10.1158/1078-0432.CCR-07-4863. [DOI] [PubMed] [Google Scholar]
- 18.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nature methods. 2007;4:359–65. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–88. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 20.Sameni M, Cavallo-Medved D, Dosescu J, et al. Imaging and quantifying the dynamics of tumor-associated proteolysis. Clin Exp Metastasis. 2008 doi: 10.1007/s10585-008-9218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller FR, Santner SJ, Tait L, Dawson PJ. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J Natl Cancer Inst. 2000;92:1185–6. doi: 10.1093/jnci/92.14.1185a. [DOI] [PubMed] [Google Scholar]
- 22.Sartor CI, Dziubinski ML, Yu CL, Jove R, Ethier SP. Role of epidermal growth factor receptor and STAT-3 activation in autonomous proliferation of SUM-102PT human breast cancer cells. Cancer Res. 1997;57:978–87. [PubMed] [Google Scholar]
- 23.Hu M, Yao J, Carroll DK, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13:394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuperwasser C, Chavarria T, Wu M, et al. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci U S A. 2004;101:4966–71. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jedeszko C, Sameni M, Olive MB, Moin K, Sloane BF. Visualizing protease activity in living cells: from two dimensions to four dimensions. Current protocols in cell biology. 2008;Chapter 4(Unit 420) doi: 10.1002/0471143030.cb0420s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furge KA, Zhang YW, Vande Woude GF. Met receptor tyrosine kinase: enhanced signaling through adapter proteins. Oncogene. 2000;19:5582–9. doi: 10.1038/sj.onc.1203859. [DOI] [PubMed] [Google Scholar]
- 27.Koon EC, Ma PC, Salgia R, et al. Effect of a c-Met-specific, ATP-competitive small-molecule inhibitor SU11274 on human ovarian carcinoma cell growth, motility, and invasion. Int J Gynecol Cancer. 2007 doi: 10.1111/j.1525-1438.2007.01135.x. [DOI] [PubMed] [Google Scholar]
- 28.Qiu D, Owen K, Gray K, Bass R, Ellis V. Roles and regulation of membrane-associated serine proteases. Biochemical Society transactions. 2007;35:583–7. doi: 10.1042/BST0350583. [DOI] [PubMed] [Google Scholar]
- 29.Tacchini L, Matteucci E, De Ponti C, Desiderio MA. Hepatocyte growth factor signaling regulates transactivation of genes belonging to the plasminogen activation system via hypoxia inducible factor-1. Exp Cell Res. 2003;290:391–401. doi: 10.1016/s0014-4827(03)00348-3. [DOI] [PubMed] [Google Scholar]
- 30.Li Q, Mullins SR, Sloane BF, Mattingly RR. p21-Activated kinase 1 coordinates aberrant cell survival and pericellular proteolysis in a three-dimensional culture model for premalignant progression of human breast cancer. Neoplasia. 2008;10:314–29. doi: 10.1593/neo.07970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–92. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakurai H, Barros EJ, Tsukamoto T, Barasch J, Nigam SK. An in vitro tubulogenesis system using cell lines derived from the embryonic kidney shows dependence on multiple soluble growth factors. Proc Natl Acad Sci U S A. 1997;94:6279–84. doi: 10.1073/pnas.94.12.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velazquez OC, Snyder R, Liu ZJ, Fairman RM, Herlyn M. Fibroblast-dependent differentiation of human microvascular endothelial cells into capillary-like 3-dimensional networks. FASEB J. 2002;16:1316–8. doi: 10.1096/fj.01-1011fje. [DOI] [PubMed] [Google Scholar]
- 34.Simian M, Hirai Y, Navre M, Werb Z, Lochter A, Bissell MJ. The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development. 2001;128:3117–31. doi: 10.1242/dev.128.16.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang WG, Grimshaw D, Martin TA, et al. Reduction of stromal fibroblast-induced mammary tumor growth, by retroviral ribozyme transgenes to hepatocyte growth factor/scatter factor and its receptor, c-MET. Clin Cancer Res. 2003;9:4274–81. [PubMed] [Google Scholar]
- 36.Witt AE, Hines LM, Collins NL, et al. Functional proteomics approach to investigate the biological activities of cDNAs implicated in breast cancer. Journal of proteome research. 2006;5:599–610. doi: 10.1021/pr050395r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mueller KL, Hunter LA, Ethier SP, Boerner JL. Met and c-Src cooperate to compensate for loss of epidermal growth factor receptor kinase activity in breast cancer cells. Cancer Res. 2008;68:3314–22. doi: 10.1158/0008-5472.CAN-08-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


