Abstract
Background
We have previously identified an atherosclerosis quantitative trait locus (QTL) on mouse chromosome (Chr) 12 in an F2-intercross of atherosclerosis-resistant FVB and atherosclerosis-susceptible C57BL/6 (B6) mice on the LDL-receptor deficient background. The aim of the present study was to identify potentially causative genes at this locus.
Methods and Results
Expression QTL (eQTL) analysis of candidate genes in livers of F2-mice revealed that a disintegrin and metalloproteinase 17 (ADAM17) mRNA expression mapped to the physical position of ADAM17 on proximal Chr12 (21.6 Mb, LOD 3.3) and co-localized with the atherosclerosis QTL. The FVB allele was associated with significantly higher ADAM17 mRNA expression (39%) than the B6 allele. Likewise, ADAM17 mRNA levels in the parental strains were significantly elevated in FVB.LDLR−/− compared to B6.LDLR−/− mice in liver, macrophages, and aorta (68%, 58%, and 32% respectively). Reporter gene assays revealed a genetic variant that might explain these expression differences. Moreover, FVB.LDLR−/− macrophages showed 5-fold increased PMA-induced shedding of TNF-alpha and 32% increased release of TNF-receptor I compared to B6.LDLR−/−. The atherosclerosis locus and expression differences were confirmed in Chr12 interval-specific congenic mice.
Conclusion
Our data provide functional evidence for ADAM17 as a candidate gene of atherosclerosis susceptibility at the murine Chr12 QTL.
Keywords: Atherosclerosis, genetics, ADAM17, mouse models, expression QTL
INTRODUCTION
Atherosclerosis is a complex disease caused by the interplay of genetic and environmental factors.1 Heritability estimates indicate that ~50% of the risk of developing atherosclerosis is genetically determined.2 Mouse models have been extremely helpful in the identification of genetic factors that modify atherosclerosis. To date, experiments with transgenic and knockout mice have revealed several hundred genes affecting lesion development.1 In addition, differences in the susceptibility of inbred mouse strains in the development of atherosclerosis have been noted3 which have been used to identify atherosclerosis modifying genes4 (for a review see5).
In previous work, we have identified differences in atherosclerosis susceptibility between the mouse strains FVB and C57BL/6 (B6). When crossed onto the LDL-receptor deficient background, FVB.LDLR−/− mice were atherosclerosis resistant while B6.LDLR−/− mice were relatively atherosclerosis susceptible in spite of comparable hypercholesterolemia.6 In subsequent work, we have used these strains to identify genetic loci responsible for disease susceptibility by quantitative trait locus (QTL) mapping in a total of 459 F2s derived from the parental FVB.LDLR−/− and B6.LDLR−/−. To account for potential lineage effects, the F2-mice were generated in two sub-crosses: In cross “mB6xfFVB”, male B6.LDLR−/− mice were crossed to female FVB.LDLR−/− mice to generate 108 female and 114 male F2s. In cross ”mFVBxfB6”, male FVB.LDLR−/− mice were crossed to female B6.LDLR−/− mice to generate 119 female and 118 male F2s. Animals were phenotyped for atherosclerotic lesion area at the aortic root and a genome scan was carried out with 192 polymorphic microsatellite markers. QTL mapping revealed significant loci of atherosclerosis susceptibility on chromosomes (Chr) 3, 10 and 12.7 The Chr12 QTL showed lineage- and gender-dependency with a maximal LOD score of 3.9 (D12Mit82, 3cM) in female mice of cross mB6xfFVB and a maximal LOD score of 4.8 (D12Mit189, 24cM) in male mice of cross mFVBxfB6, respectively. It should be noted that the map position of the locus in female and male mice were not identical indicating that the loci were likely independent. Moreover, for females of cross mFVBxfB6 as well as for males of cross mB6xfFVB, no significant LOD score was identified on proximal Chr12, suggesting a complex interaction pattern of permissive cofactors influencing atherosclerosis susceptibility at this locus.
The aim of the present study was to identify potentially causal gene(s) underlying the atherosclerosis QTL at Chr12.
MATERIALS AND METHODS
An expanded methods section is provided in the online supplement (Methods).
Animals and Tissue Preparation
In previous work, 459 F2-mice had been generated in a reciprocal intercross of atherosclerosis-resistant FVB.LDLR−/− mice and atherosclerosis-susceptible B6.129S7-LdlrtmHer/J (henceforce called B6.LDLR−/−).7 Livers from F1- and F2-mice had been harvested at sacrifice and stored −80°C. In addition, in the present study a total of 19 parental FVB.LDLR−/− and 22 parental B6.LDLR−/− mice were used. Congenic animals carrying the Chr12 interval (0–28 cM) from B6 on the FVB.LDLR−/− background were generated by backcrossing B6.LDLR−/− mice to FVB.LDLR−/−. For atherosclerosis studies, these mice (designated FVB.LDLR−/−Chr12B6/FVB) were intercrossed to generate FVB.LDLR−/−Chr12FVB/FVB (n=37), FVB.LDLR−/−Chr12B6/FVB (n=38), and FVB.LDLR−/−Chr12B6/B6 (n=45) mice. All animals were treated and sacrificed like the F2-mice previously described.7
Bone marrow was isolated and cryo-preserved for further study. Animal care and experimental procedures involving animals were approved by the Rockefeller University’s Institutional Animal Care and Use Committee and the responsible authorities of the state of Saxony (Regierungspräsidium Sachsen, N 4/07).
Cell Culture Studies
RAW 264.7 macrophages were cultivated and used for transfection studies. Cryo-preserved bone marrow cells of B6.LDLR−/− and FVB.LDLR−/− were thawed and differentiated into macrophages in presence of L-cell-conditioned medium for 14 days. Macrophages were used for RNA isolation. For functional studies, macrophages were incubated for 1, 3, 6, and 24 hours with 100 nmol/L phorbol-12-myristate-13-acetate (PMA, Sigma-Aldrich, Munich, Germany). TNF-alpha and TNFR-I release into media was determined by enzyme-linked immunosorbent assay (BioRad, Munich, Germany) and concentrations were normalized to cell protein content determined by the method of Lowry as previously described.8
RNA Isolation and cDNA Synthesis
Total RNA was extracted from livers of 10 F1 and 400 F2-mice (59 from the total of 459 were not available), 9 FVB.LDLR−/− mice and 12 B6.LDLR−/− mice using TRIzol reagent. This method was also used for isolation of total RNA from cultivated bone marrow derived macrophages of FVB.LDLR−/− and B6.LDLR−/− mice. For RNA isolation from whole aortas, RNeasy Fibrous Tissue Mini (Qiagen, Hilden, Germany) was used. RNA was reverse transcribed into cDNA using SuperScript™ II Reverse Transcriptase (Invitrogen, Karlsruhe, Germany) and random hexamer primers.
Quantitative Fluorogenic RT-PCR (TaqMan)
Quantitative fluorogenic RT-PCR was performed in an ABI PRISM 7900 Sequence Detection System (Applied Biosystems, Darmstadt, Germany). Specific primers and probes for beta-actin, a disintegrin and metalloproteinase domain 17 (ADAM17), aryl-hydrocarbon receptor (AHR), TNF-alpha, TNFR-I, vascular cell adhesion molecule 1 (VCAM-1), and cytochrome P450, family 1, subfamily a, polypeptide 1 (CYP1A1) were selected to span two exons in order to avoid co-amplification of genomic DNA. mRNA expression levels were normalized to 103 copies of beta-actin as a housekeeping gene.
cDNA and Genomic DNA Sequencing
The promoter regions (~1.5 kb and ~2.7 kb for ADAM17) and coding regions of candidate genes were amplified using specific primers (online supplement Methods, Table II). Histone deacetylase 9 (HDAC9), AHR and NF-kappa-B inhibitor alpha (Iκ Bα) were sequenced from cDNA, ADAM17 was also sequenced from genomic DNA. Sequencing was performed by standard dye-terminator chemistry (Applied Biosystems, Darmstadt, Germany).
ADAM17 and Chr12 Genotyping
One single nucleotide polymorphism (SNP) in exon 19 of ADAM17 that we found to be polymorphic between FVB.LDLR−/− and B6.LDLR−/− mice was used as an additional marker to fine-map Chr12 in the F2-mice. Five additional SNPs evenly spaced across Chr12 were used for generation of congenic mice. Genotyping was performed using a homogenous fluorescent method as previously described.9
Allele-Specific Transcript Quantification (cis-trans test)
Allele-specific ADAM17 transcript quantification was performed in livers from 10 F1-mice using quantitative sequence analysis of cDNA of the A1098G sequence variant in exon 9 between FVB and B6. The allelic ratio in the F1 was determined by comparison of the peak heights of sequence analysis for the G-allele (FVB) and A-allele (B6) with peak heights for these alleles obtained in standard mixtures.
Transfection Study of ADAM17 Promoter Fragments
ADAM17 promoter fragments of FVB and B6 starting at −681 bp upstream of the start codon were amplified and cloned into the luciferase reporter vector pGl4.11[luc2CP] (Promega, Mannheim, Germany). Four sequence variations were identified in the 681 bp promoter fragment between FVB and B6. Mutagenesis was used to introduce each of the B6 variants (DEL-640CA, T-457G, C-216A, and T-25G) separately into the FVB promoter fragment. RAW 264.7 cells were co-transfected with 0.01 μg pGl4.74[hRluc/TK] vector (Invitrogen, Karlsruhe, Germany) for normalization and 1 μg of expression vectors using the diethylaminoethyl (DEAE)-dextran method.10 Firefly and renilla luciferase activities were measured sequentially using the Dual-Luciferase Reporter Assay System (Promega, Mannheim, Germany) in a Sirius Luminometer (Berthold Detection Systems, Pforzheim, Germany).
Statistical Analysis
All data are given as mean ± standard deviation unless otherwise indicated. Normality of distribution was assessed using the Kolmogorov–Smirnov test implemented in PRISM statistical software (GraphPad, San Diego, CA). Comparison of multiple groups was done using ANOVA and Tukey was performed as post-test. Comparison of two groups of normally distributed samples was done using the t-test. Linkage analysis for single QTLs was done using MAPMANAGER QTX B20, freely available at www.mapmanager.org.11 Levels of significance were determined empirically by permutation testing in 1 cM steps.
RESULTS
Selection and Sequencing of Genes at the Chr12 QTL
We screened the Ensembl database (www.ensembl.org) for potential atherosclerosis modifier genes underlying the atherosclerosis QTL on Chr12 identified in our previous work (Supplemental Table I).7 Four of these genes, ADAM17, HDAC9, AHR, and IkBα stood out because of their known functions and the possibility that variation between the strains might affect atherosclerosis susceptibility. ADAM17 (21.6 Mb) is a metalloproteinase responsible for the release of membrane bound substrates (for a review see12). HDAC9 (31.9 Mb) is a histone deacetylase that regulates transcription,13 the aryl-hydrocarbon receptor (AHR, 33.3 Mb) is a cytosolic receptor with a known association to cardiovascular risk,14 and IkBα (53.4 Mb) is a master-regulator of inflammatory signalling.15
In general, a QTL might be caused by genetic sequence variation leading either to differences in gene expression or gene function. We thus performed systematic DNA sequencing of the proximal promoter (~1.5 kB), the complete coding sequence, and the 3′-untranslated region (UTR) of the four candidate genes. The numbers of identified sequence variations between FVB and B6 mice are shown in Table 1. No sequence variations were present in HDAC9 and IkBα and those genes were thus excluded from further analysis.
Table 1.
Candidate Genes at Chr12 Atherosclerosis Susceptibility Locus
| Sequence variations |
||||||
|---|---|---|---|---|---|---|
| Gene | Map [Mb] | Description | Promoter | cDNA | coding | 3′UTR |
| ADAM17 | 21.6 | Metalloproteinase, protein shedding | 17 | 10 | 2 | 4 |
| HDAC9 | 31.9 | Histone deacetylase, gene transcription | 0 | 0 | 0 | 0 |
| AHR | 33.3 | Aryl-hydrocarbon- receptor, gene transcription | 8 | 10 | 4, Stop | 0 |
| IkBα | 53.4 | Inhibitor of NFkB, gene transcription | 0 | 0 | 0 | 0 |
In ADAM17, we identified 17 promoter variants, 10 coding variants and 4 additional sequence variants in the 3′-UTR. Of the 10 variants in the coding region, one changed an amino acid residue in the pro-domain (Asn113Asp) and the other in the EGF-like domain (Val593Ile). However, analysis with the Panther and SIFT software programs (freely available at www.pantherdb.org, http://blocks.fhcrc.org/sift/SIFT.html) suggested these amino acid exchanges were not likely to be functionally significant. For AHR, sequence comparison between FVB and B6 mice revealed 8 promoter variants and 10 SNPs within the coding sequence causing 4 amino-acid exchanges. In addition, a pre-terminal stop codon was identified in B6 leading to a 48 amino-acid truncation compared to FVB with potential functional significance.16
Fine Mapping of the Chr12 QTL
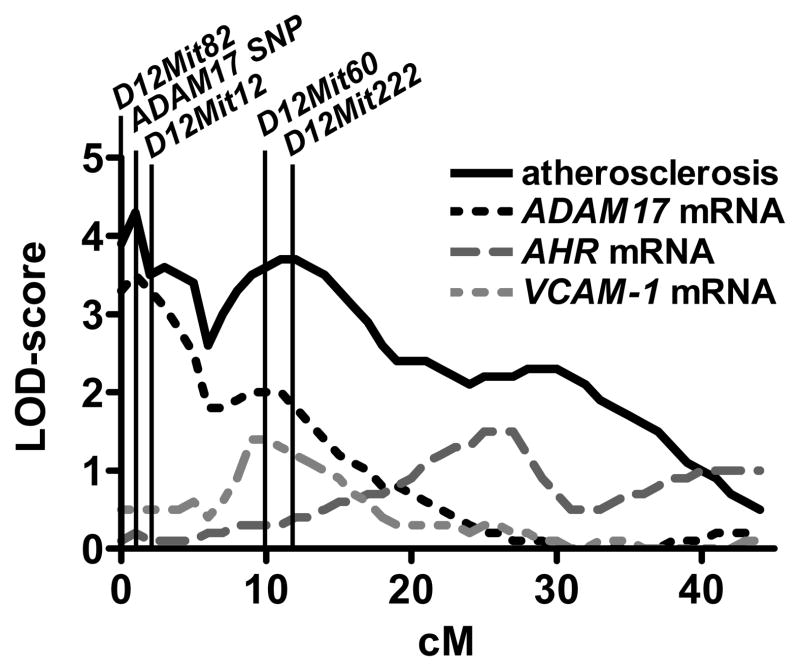
To improve the marker coverage at the chromosomal map-position of ADAM17, we genotyped all F2 animals with one of the SNPs identified in ADAM17 (exon 19). We have shown in our previous work that the atherosclerosis QTL at Chr12 was lineage specific.7 In female F2, the QTL was due to a subset of mice derived from male B6.LDLR−/− and female FVB.LDLR−/− (mB6xfFVB) parental mice while in male F2, it was due to a subset of mice derived from male FVB.LDLR−/− and female B6.LDLR−/− parental mice. Adding the ADAM17-SNP as an additional marker to the QTL map lead to a further increase of the maximal LOD score of atherosclerosis from 3.9 to 4.2 in this subset of mB6xfFVB female F2 (Figure 1). In male mice, however, the maximum LOD of atherosclerosis was more distal from the map-position of ADAM17 and adding this marker to the QTL map did not lead to significant changes of the LOD score plot for atherosclerosis. Since the chromosomal map-position of AHR was well covered with microsatellite markers, we did not perform additional genotyping using the newly identified SNPs in AHR. A full list of gene variants of ADAM17 and AHR is given in the online supplement, Figure I.
Figure 1.
Chr12 LOD score plot for atherosclerotic lesion area and for mRNA expression of ADAM17, AHR, and VCAM-1 for females cross mB6XfFVB. QTLs for atherosclerosis and ADAM17 mRNA expression co-localized at the physical position of ADAM17 indicating a regulation in cis.
Expression Quantitative Trait Locus (eQTL) Mapping
We next performed expression quantitative trait locus (eQTL) mapping to determine whether the identified promoter variants had an effect on mRNA expression levels of candidate genes. To this end, RNA from livers of the original F2 animals was isolated and mRNA expression levels of ADAM17 and AHR were determined by quantitative RT-PCR. mRNA expression levels were then used as phenotypes for QTL mapping: In female F2-mice derived from mB6xfFVB parental mice, the maximum LOD score of ADAM17 expression was 3.3 and co-localized with the physical map-position of ADAM17 as well as with the maximal LOD score for atherosclerosis (Figure 1). In contrast, in female F2 derived from mFVBxfB6 parentals, the LOD score at the map-position of ADAM17 was only 1.4 (no atherosclerosis QTL was present in this subcross). Likewise, in male F2 derived from mB6xfFVB parentals and mFVBxfB6 parentals, the LOD scores at the map-position of ADAM17 were lower, but still reached levels of 2.9 and 2.4, respectively. In summary, these data indicated a differential regulation of ADAM17 mRNA expression in cis with a high LOD of expression in the female mB6xfFVB subcross. This finding was confirmed by allele expression imbalance in the F1 livers (cis-trans test, data not shown). For AHR mRNA expression, no significant LOD score was identified in any of the subcrosses and no differences in mRNA expression levels of AHR between parental FVB.LDLR−/− and B6.LDLR−/− mice were observed.
Although there was no difference of AHR in expression between the strains, there were 4 amino acid exchanges and a truncation of the AHR protein present between FVB and B6. AHR is a cytosolic receptor playing a role in transcriptional regulation of target genes such as VCAM-1 and CYP1A1. If the coding differences in AHR between FVB and B6 indeed had functional consequences, one would expect co-localization of the maximum LOD scores of VCAM-1 and CYP1A1 expression with the physical map-position of AHR indicating a regulation in trans. However, no significant LOD score for VCAM-1 nor for CYP1A1 mapped to Chr12 (Figure 1). This and the lack of expression differences in AHR between the strains made AHR an unlikely candidate and this gene was not followed up in further analysis.
ADAM17 Expression Levels in F2 and Parental Mice
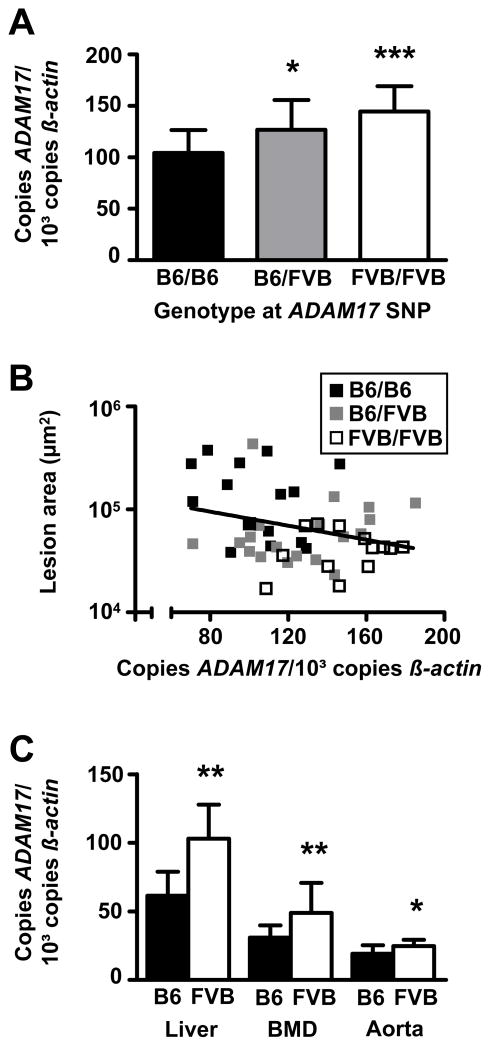
We next determined the allelic effect of the ADAM17-SNP on ADAM17 mRNA expression levels in the F2-mice. As shown in Figure 2A, data from female F2-mice derived from mB6xfFVB parentals indicate that ADAM17 expression in the livers of F2-mice homozygous for FVB at the ADAM17-SNP was 39% higher compared to mice homozygous for B6 (P < 0.001). Mice heterozygous at the ADAM17-SNP still had 22% higher ADAM17 expression levels than B6 mice (P < 0.05). The average expression variance associated with the FVB genotype was 19% per allele. As shown in Figure 2B, increased ADAM17 mRNA expression was significantly correlated with decreased atherosclerosis in female F2-mice derived from mB6xfFVB parentals (P = 0.04, r2 = 0.09). Differential expression of ADAM17 could also be replicated in the parentals: In livers of FVB.LDLR−/− mice, expression of ADAM17 was 68% increased compared to B6 mice (P < 0.01) (Figure 2C). To test whether expression was also different in other tissues, we determined ADAM17 mRNA in bone marrow derived macrophages (MΦ) and found 58% higher ADAM17 expression in FVB.LDLR−/− MΦ compared to B6.LDLR−/− MΦ (P < 0.01). Moreover, ADAM17 mRNA expression was also significantly increased (32%) in aorta from FVB.LDLR−/− compared to B6.LDLR−/− mice (P < 0.05, Figure 2C).
Figure 2.
ADAM17 mRNA expression in F2 and F0 animals. (A) Allelic effects of an ADAM17-SNP in exon 19 on ADAM17 mRNA expression in livers of mB6XfFVB F2-females. (B) Correlation of ADAM17 mRNA expression in livers of mB6XfFVB F2-females with atherosclerotic lesion size (P < 0.05, r2 = 0.09). (C) ADAM17 mRNA expression in liver (B6, n=11; FVB, n=7), bone marrow derived macrophages (B6, n=12; FVB, n=9), and aortas (B6, n=10; FVB, n=10) of parental FVB.LDLR−/− and B6.LDLR−/− mice. mRNA expression was determined in triplicates by TaqMan RT-PCR. * P < 0.05, ** P < 0.01, *** P < 0.001 compared to B6.LDLR−/−, respectively.
Reporter Gene Assay of ADAM17 Promoter Variants
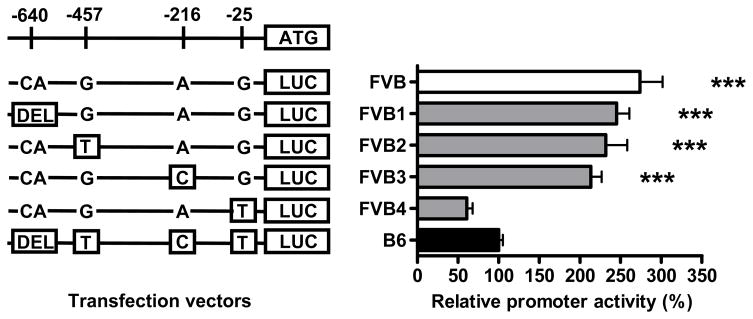
We next performed reporter gene assays to identify the causal variant(s) leading to differences of ADAM17 expression levels in FVB and B6 mice. Comparative sequence analysis of the ADAM17 promoter (http://rvista.dcode.org) (data not shown) revealed greatest sequence conservation between humans, rats and mice for a promoter region approximately spanning from −600 bp to the start codon suggesting that important regulatory elements might reside in this region. Therefore, we investigated a 681 bp fragment of the proximal promoter of ADAM17 containing 4 sequence variations between FVB and B6 at positions –640 (DEL/CA), -457 (T/G), -216 (C/A), and -25 (T/G). The FVB and B6 versions of this fragment were cloned into a luciferase reporter vector. Reporter activity was 2.7-fold increased in RAW macrophages transfected with the FVB promoter compared to cells transfected with the B6 promoter (p< 0.001) (Figure 3). In order to identify the genetic variant responsible for expression differences between strains, we generated four additional reporter constructs introducing each of the four B6 variants into the FVB promoter fragment by site-directed mutagenesis. As shown in Figure 3, the relative reporter activity was basically unchanged when the B6 variants were introduced into the FVB reporter construct at positions -640, -457 and -216. However, introduction of the B6 variant into the FVB reporter at position -25 lead to a dramatic reduction of reporter activity comparable to the B6 variant. The relevance of this finding was confirmed by mutating the FVB variant into the B6 promoter leading to a significant increase of promoter activity (data not shown). Analysis of transcription factor binding sites with AliBaba2.1 (www.gene-regulation.com/pub/programs/alibaba2/index.html) revealed that the -25 T/G variant led to an insertion of a C/EBP-alpha binding site in FVB which was absent in B6.
Figure 3.
Luciferase pGl4.11 reporter gene assay of the proximal ADAM17 promoter (−681 bp). RAW 264.7 cells were transfected with vectors containing the FVB and B6 sequences and FVB promoter-based constructs containing the respective B6 variants DEL-640CA (FVB1), T-457G (FVB2), C-216A (FVB3) and T-25G (FVB4). Measurements were performed in quadruplicates. *** P < 0.001 compared to B6-transfected cells.
ADAM17 shedding activity in macrophages
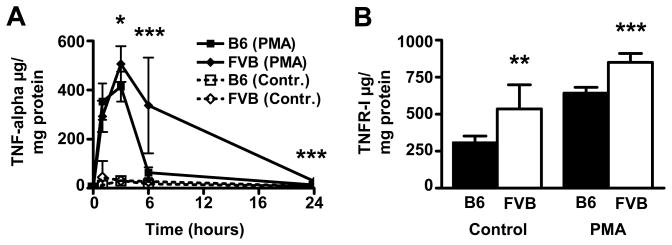
ADAM17 is responsible for shedding of membrane–bound substrates such as TNF-alpha and TNFR-I. To test, whether expression differences of ADAM17 between FVB.LDLR−/− and B6.LDLR−/− were reflected on the functional level, we determined release of TNF-alpha and TNFR-I from cultivated MΦ. As shown in Figure 4A, basal release of TNF-alpha was low and showed no significant differences between strains. Shedding activity of ADAM17 is known to increase upon incubation of cells with PMA without inducing transcription.17 Interestingly, we observed significantly higher peak concentrations and sustained release of TNF-alpha in FVB.LDLR−/− MΦ compared to B6.LDLR−/− MΦ upon activation with PMA (Figure 4A). We also investigated the release of TNFR-I, another substrate of ADAM17. TNFR-I release in native MΦ was elevated 73% in FVB.LDLR−/− compared to B6.LDLR−/− (P < 0.001) (Figure 4B). TNFR-I levels were increased upon stimulation with PMA and the significant difference between MΦ from FVB.LDLR−/− and B6.LDLR−/− was retained (32%, P < 0.01) (Figure 4B). Different substrate release was not due to an increased expression of TNF-alpha and TNFR-I in FVB.LDLR−/− cells as determined by quantitative RT-PCR. On the contrary, significantly lower levels of TNF-alpha and TNFR-I mRNA were found in FVB.LDLR−/− macrophages compared to B6.LDLR−/− (52%, P < 0.05 and 64%, P < 0.01, respectively) (data not shown).
Figure 4.
Release of ADAM17 substrates TNF-alpha and TNFR-I. (A) Time-course of TNF-alpha concentrations in cell culture supernatants from bone marrow-derived macrophages of FVB and B6 mice incubated with PMA (100nmol/L) and without PMA (Contr.). (B) TNFR-I concentrations in cell culture supernatants from bone marrow-derived macrophages of FVB and B6 with and without incubation with PMA for 6 h. Experiments were performed with macrophages from 7 FVB and 12 B6 mice on 48-well plates in duplicate dishes. *P < 0.05, ** P < 0.01, *** P < 0.001.
Confirmation of Chr12 QTL and ADAM17 Expression Differences in Congenic Mice
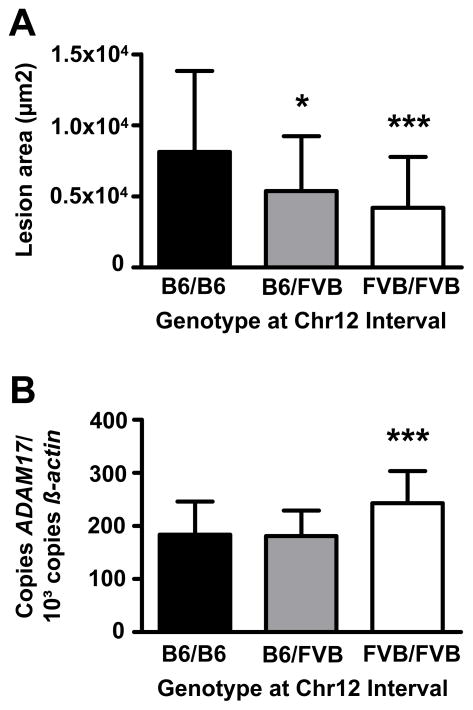
To further strengthen the association of the FVB allele with decreased lesion formation and increased ADAM17 expression, we generated congenic mice carrying the Chr12 interval (0–28 cM) from B6 on the FVB.LDLR−/− background. Atherosclerosis was significantly reduced in FVB.LDLR−/−Chr12FVB/FVB and FVB.LDLR−/−Chr12B6/FVB compared to FVB.LDLR−/−Chr12B6/B6 mice (Figure 5A). We could also replicate our finding of increased ADAM17 mRNA expression in livers of congenic animals homozygous for the FVB allele at the Chr12 interval compared to animals homozygous for B6 (Figure 5B).
Figure 5.
Atherosclerotic lesion size (A) at the aortic root and (B) ADAM17 expression in liver of Chr12 congenic animals carrying the B6/B6, B6/FVB and FVB/FVB genotype at the Chr12 QTL (0–28cM) on the FVB.LDL−/− background. * P < 0.05, *** P < 0.001 compared to FVB.LDLR−/−Chr12B6/B6 respectively.
DISCUSSION
In the present study, we provide evidence for the metalloproteinase ADAM17 as a novel genetic factor of atherosclerosis susceptibility. Increased activity of ADAM17 in atherosclerosis-resistant FVB.LDLR−/− mice was associated with decreased lesion formation and elevated release of TNF-alpha and TNFR-I compared to atherosclerosis-susceptible B6.LDLR−/− mice. Differential expression of ADAM17 between FVB.LDLR−/− and B6.LDLR−/− mice might be due to a genetic variant at position -25 (T/G) leading to the insertion of a putative C/EBP-alpha binding site in the FVB promoter which was absent in the B6 promoter. The atherosclerosis locus and expression differences were confirmed in Chr12 interval-specific congenic mice.
ADAM17 was first characterized by two independent groups as the metalloproteinase responsible for releasing soluble TNF-alpha from the cell membrane and thus designated TACE (tumor necrosis factor-alpha converting enzyme).18, 19 It was later recognized that ADAM17 activity is also responsible for proteolytic cleavage of numerous other transmembrane proteins such as growth factors, growth factor receptors, cytokines, cytokine receptors, and adhesion molecules. This process is also known as “ectodomain shedding” (for review see12). ADAM17 is expressed ubiquitously18 and has also been demonstrated in atherosclerotic lesions of ApoE−/− mice.20
Our data now provide first evidence for an atheroprotective role of ADAM17 because increased expression and activity was found in atherosclerosis-resistant FVB.LDLR−/− mice (Figure 2). This was surprising since the prototypical substrate of ADAM17, TNF-alpha, is a proinflammatory cytokine that might have the potential to induce atherosclerosis.21 However, data in mice deficient for TNF-alpha or its receptors on atherosclerosis susceptibility are inconsistent.22–24 In addition to its effect on modulating the TNF-alpha pathway, ADAM17 is also responsible for shedding a number of cell-adhesion molecules such as ICAM-1 25, VCAM-126, and CX3CL1 (fractalkine).27 Data about their role in atherosclerosis are less ambiguous since mice deficient for these proteins have decreased atherosclerosis.28–30 Indeed, this would be quite consistent with an atheroprotective role of ADAM17. One could speculate that increased ADAM17 activity might result in decreased adhesion of inflammatory cells to the vessel wall, one of the initial steps in atherogenesis. A reduction of atherosclerosis has also been shown in mice deficient for interleukin-1 receptor, another substrate of ADAM17.31 Taken together, ADAM17 is responsible for modulating the abundance of a number of proteins on the cell surface with pro- and anti-atherogenic properties and a net anti-atherogenic effect might be the result of the sum of their actions.
It should be mentioned that Mu et al have previously also mapped an atherosclerosis locus, designated Ath-6, to proximal Chr12. This locus was identified in a cross between C57BL/6J and C57BLKS/J mice (in this cross, the more atherosclerosis sensitive strain is the latter) with a suggestive LOD score of 2.5 at D12mit49.32 In subsequent work, Ath6 was narrowed to a 0.26cM interval and ADAM17 was excluded as a candidate gene.33 However, this does not necessarily rule out ADAM17 as a the responsible gene for the Chr12 QTL in our cross since we have shown that complex lineage effects have to be considered at this locus.7 In addition, we have been using FVB mice on the LDLR−/− background7 in contrast to wild-type B6BLKS and SPRET/Ei mice in a model of diet-induced atherosclerosis33 and the genetic basis of atherosclerosis in these models might be quite different.
In our initial paper, we observed a strong linage dependency of atherosclerosis at the Chr12 QTL. Significant linkage was only present in female mice derived from mB6xfFVB parental mice and male mice derived from mFVBxfB6 parentals.7 In the present study, co-localization of the peak LOD score of ADAM17 mRNA expression to the peak LOD score of atherosclerosis at proximal Chr12 suggested that both phenotypes might be related. This was also reflected at the level of ADAM17 mRNA expression in the different subcrosses. The QTL of ADAM17 mRNA expression was significantly higher (LOD 3.3) in female F2 derived from mB6xfFVB parental mice and compared to females F2 derived from mFVBxfB6 (LOD 1.4), thus corresponding to the lineage effects of atherosclerosis in female mice. In male mice, linkage for ADAM17 expression was suggestive for ADAM17 expression. However, the peak atherosclerosis LOD score mapped more distally in male mice. It thus appeared that a gene different from ADAM17 might be more relevant for differences in atherosclerosis susceptibility in male animals, even though there was evidence for differential regulation of ADAM17 mRNA (LOD 2.9 and 2.4, in male sub-crosses). We would like to argue that this does not necessarily rule out ADAM17 as a candidate gene since its effect might depend on the interaction with additional permissive co-factors present only in specific female lineages. Unfortunately, we are just at the beginning of understanding these lineage effects which might not only depend on the distribution of the sex-chromosomes, but also on the maternal mode of inheritance of mitochondria and epigenetic factors.34
Differential expression of ADAM17 between FVB and B6 mice was of particular interest because the gene is expressed constitutively18 and little is known about its regulation.35 In addition, it was important to identify the variant responsible for differential expression as a prerequisite for generating induced mutant mice to confirm its effect on atherosclerosis susceptibility. Overlap of the maximum LOD score of ADAM17 expression with its physical position at Chr12 indicated differential regulation in cis (Figure 1). This was confirmed by allele expression imbalance in the F1 livers (cis/trans test) suggesting that a genetic variant in the ADAM17 promoter might be responsible for differences in ADAM17 expression between FVB and B6. To identify the causal variant, we performed reporter gene assays of the 681 bp proximal promoter region containing potentially important regulatory sites as determined by analysis of sequence conservation between species (rVista). Systematic mutation of the 4 promoter variants between FVB and B6 indicated that a polymorphism at position -25 (T/G) was responsible for expression differences (Figure 3). This nucleotide exchange resulted in the insertion of a putative C/EBP-alpha binding site in FVB, absent in B6. This finding was particularly important because the identified variant might be causative for differences in atherosclerosis susceptibility between the two strains and thus be a target for production of knock-in mice for confirmation of differences of lesion susceptibility.
In summary, our data provide evidence for a role of ADAM17 in modulating atherosclerosis susceptibility. Increased activity of ADAM17 was associated with decreased lesion formation and elevated release of ADAM17 substrates. We conclude that ADAM17 might represent an interesting novel target for the prevention of atherosclerosis.
Supplementary Material
Acknowledgments
We are indebted to Franziska Jeromin, Claudia Weise, Wolfgang Wilfert and Marietta Tan for their excellent technical assistance. This study was supported by a grant of the Deutsche Forschungsgemeinschaft to Daniel Teupser (Te 342/2-1) and a doctoral student fellowship of the Medical Faculty of the University Leipzig to Lesca M. Holdt. Part of the manuscript has been published in abstract form (AHA Scientific Sessions, Orlando, 2007).
Footnotes
Author Disclosures
Lesca M Holdt:
Research Grant: Research Grant from Medical Faculty, University Leipzig, Amount: < $10,000
Joachim Thiery: No disclosures
Jan L Breslow: No disclosures
Daniel Teupser:
Research Grant: Resarch grant from German Research Foundation (DFG), Amount: >= $10,000
Disclaimer: The manuscript and its contents are confidential, intended for journal review purposes only, and not to be further disclosed.
References
- 1.Lusis AJ, Fogelman AM, Fonarow GC. Genetic basis of atherosclerosis: part I: new genes and pathways. Circulation. 2004;110:1868–1873. doi: 10.1161/01.CIR.0000143041.58692.CC. [DOI] [PubMed] [Google Scholar]
- 2.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 3.Paigen B, Morrow A, Brandon C, Mitchell D, Holmes P. Variation in susceptibility to atherosclerosis among inbred strains of mice. Atherosclerosis. 1985;57:65–73. doi: 10.1016/0021-9150(85)90138-8. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Ria M, Kelmenson PM, Eriksson P, Higgins DC, Samnegard A, Petros C, Rollins J, Bennet AM, Wiman B, de Faire U, Wennberg C, Olsson PG, Ishii N, Sugamura K, Hamsten A, Forsman-Semb K, Lagercrantz J, Paigen B. Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat Genet. 2005;37:365–372. doi: 10.1038/ng1524. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Ishimori N, Korstanje R, Rollins J, Paigen B. Identifying novel genes for atherosclerosis through mouse-human comparative genetics. Am J Hum Genet. 2005;77:1–15. doi: 10.1086/431656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teupser D, Persky AD, Breslow JL. Induction of atherosclerosis by low-fat, semisynthetic diets in LDL receptor-deficient C57BL/6J and FVB/NJ mice: comparison of lesions of the aortic root, brachiocephalic artery, and whole aorta (en face measurement) Arterioscler Thromb Vasc Biol. 2003;23:1907–1913. doi: 10.1161/01.ATV.0000090126.34881.B1. [DOI] [PubMed] [Google Scholar]
- 7.Teupser D, Tan M, Persky AD, Breslow JL. Atherosclerosis quantitative trait loci are sex- and lineage-dependent in an intercross of C57BL/6 and FVB/N low-density lipoprotein receptor−/− mice. Proc Natl Acad Sci U S A. 2006;103:123–128. doi: 10.1073/pnas.0509570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teupser D, Thiery J, Walli AK, Seidel D. Determination of LDL- and scavenger-receptor activity in adherent and non-adherent cultured cells with a new single-step fluorometric assay. Biochim Biophys Acta. 1996;1303:193–198. doi: 10.1016/0005-2760(96)00094-x. [DOI] [PubMed] [Google Scholar]
- 9.El Housni H, Heimann P, Parma J, Vassart G. Single-nucleotide polymorphism genotyping by melting analysis of dual-labeled probes: examples using factor V Leiden and prothrombin 20210A mutations. Clin Chem. 2003;49:1669–1672. doi: 10.1373/49.10.1669. [DOI] [PubMed] [Google Scholar]
- 10.Escher G, Hoang A, Georges S, Tchoua U, El-Osta A, Krozowski Z, Sviridov D. Demethylation using the epigenetic modifier, 5-azacytidine, increases the efficiency of transient transfection of macrophages. J Lipid Res. 2005;46:356–365. doi: 10.1194/jlr.D400014-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Manly KF, Cudmore RH, Jr, Meer JM. Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome. 2001;12:930–932. doi: 10.1007/s00335-001-1016-3. [DOI] [PubMed] [Google Scholar]
- 12.Mezyk R, Bzowska M, Bereta J. Structure and functions of tumor necrosis factor-alpha converting enzyme. Acta Biochim Pol. 2003;50:625–645. [PubMed] [Google Scholar]
- 13.Petrie K, Guidez F, Howell L, Healy L, Waxman S, Greaves M, Zelent A. The histone deacetylase 9 gene encodes multiple protein isoforms. J Biol Chem. 2003;278:16059–16072. doi: 10.1074/jbc.M212935200. [DOI] [PubMed] [Google Scholar]
- 14.Korashy HM, El-Kadi AO. The role of aryl hydrocarbon receptor in the pathogenesis of cardiovascular diseases. Drug Metab Rev. 2006;38:411–450. doi: 10.1080/03602530600632063. [DOI] [PubMed] [Google Scholar]
- 15.de Winther MP, Kanters E, Kraal G, Hofker MH. Nuclear factor kappaB signaling in atherogenesis. Arterioscler Thromb Vasc Biol. 2005;25:904–914. doi: 10.1161/01.ATV.0000160340.72641.87. [DOI] [PubMed] [Google Scholar]
- 16.Thomas RS, Penn SG, Holden K, Bradfield CA, Rank DR. Sequence variation and phylogenetic history of the mouse Ahr gene. Pharmacogenetics. 2002;12:151–163. doi: 10.1097/00008571-200203000-00009. [DOI] [PubMed] [Google Scholar]
- 17.Doedens JR, Mahimkar RM, Black RA. TACE/ADAM-17 enzymatic activity is increased in response to cellular stimulation. Biochem Biophys Res Commun. 2003;308:331–338. doi: 10.1016/s0006-291x(03)01381-0. [DOI] [PubMed] [Google Scholar]
- 18.Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 19.Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, Carter HL, Chen WJ, Clay WC, Didsbury JR, Hassler D, Hoffman CR, Kost TA, Lambert MH, Leesnitzer MA, McCauley P, McGeehan G, Mitchell J, Moyer M, Pahel G, Rocque W, Overton LK, Schoenen F, Seaton T, Su JL, Becherer JD, et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature. 1997;385:733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- 20.Canault M, Peiretti F, Kopp F, Bonardo B, Bonzi MF, Coudeyre JC, Alessi MC, Juhan-Vague I, Nalbone G. The TNF alpha converting enzyme (TACE/ADAM17) is expressed in the atherosclerotic lesions of apolipoprotein E-deficient mice: possible contribution to elevated plasma levels of soluble TNF alpha receptors. Atherosclerosis. 2006;187:82–91. doi: 10.1016/j.atherosclerosis.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 21.Branen L, Hovgaard L, Nitulescu M, Bengtsson E, Nilsson J, Jovinge S. Inhibition of tumor necrosis factor-alpha reduces atherosclerosis in apolipoprotein E knockout mice. Arterioscler Thromb Vasc Biol. 2004;24:2137–2142. doi: 10.1161/01.ATV.0000143933.20616.1b. [DOI] [PubMed] [Google Scholar]
- 22.Chandrasekharan UM, Mavrakis L, Bonfield TL, Smith JD, DiCorleto PE. Decreased atherosclerosis in mice deficient in tumor necrosis factor-alpha receptor-II (p75) Arterioscler Thromb Vasc Biol. 2007;27:e16–17. doi: 10.1161/01.ATV.0000255551.33365.22. [DOI] [PubMed] [Google Scholar]
- 23.Blessing E, Bea F, Kuo CC, Campbell LA, Chesebro B, Rosenfeld ME. Lesion progression and plaque composition are not altered in older apoE−/− mice lacking tumor necrosis factor-alpha receptor p55. Atherosclerosis. 2004;176:227–232. doi: 10.1016/j.atherosclerosis.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 24.Schreyer SA, Vick CM, LeBoeuf RC. Loss of lymphotoxin-alpha but not tumor necrosis factor-alpha reduces atherosclerosis in mice. J Biol Chem. 2002;277:12364–12368. doi: 10.1074/jbc.M111727200. [DOI] [PubMed] [Google Scholar]
- 25.Tsakadze NL, Sithu SD, Sen U, English WR, Murphy G, D’Souza SE. Tumor necrosis factor-alpha-converting enzyme (TACE/ADAM-17) mediates the ectodomain cleavage of intercellular adhesion molecule-1 (ICAM-1) J Biol Chem. 2006;281:3157–3164. doi: 10.1074/jbc.M510797200. [DOI] [PubMed] [Google Scholar]
- 26.Singh RJ, Mason JC, Lidington EA, Edwards DR, Nuttall RK, Khokha R, Knauper V, Murphy G, Gavrilovic J. Cytokine stimulated vascular cell adhesion molecule-1 (VCAM-1) ectodomain release is regulated by TIMP-3. Cardiovasc Res. 2005;67:39–49. doi: 10.1016/j.cardiores.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 27.Garton KJ, Gough PJ, Blobel CP, Murphy G, Greaves DR, Dempsey PJ, Raines EW. Tumor necrosis factor-alpha-converting enzyme (ADAM17) mediates the cleavage and shedding of fractalkine (CX3CL1) J Biol Chem. 2001;276:37993–38001. doi: 10.1074/jbc.M106434200. [DOI] [PubMed] [Google Scholar]
- 28.Kitagawa K, Matsumoto M, Sasaki T, Hashimoto H, Kuwabara K, Ohtsuki T, Hori M. Involvement of ICAM-1 in the progression of atherosclerosis in APOE-knockout mice. Atherosclerosis. 2002;160:305–310. doi: 10.1016/s0021-9150(01)00587-1. [DOI] [PubMed] [Google Scholar]
- 29.Cybulsky MI, Iiyama K, Li H, Zhu S, Chen M, Iiyama M, Davis V, Gutierrez-Ramos JC, Connelly PW, Milstone DS. A major role for VCAM-1, but not ICAM-1, in early atherosclerosis. J Clin Invest. 2001;107:1255–1262. doi: 10.1172/JCI11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teupser D, Pavlides S, Tan M, Gutierrez-Ramos JC, Kolbeck R, Breslow JL. Major reduction of atherosclerosis in fractalkine (CX3CL1)-deficient mice is at the brachiocephalic artery, not the aortic root. Proc Natl Acad Sci U S A. 2004;101:17795–17800. doi: 10.1073/pnas.0408096101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chi H, Messas E, Levine RA, Graves DT, Amar S. Interleukin-1 receptor signaling mediates atherosclerosis associated with bacterial exposure and/or a high-fat diet in a murine apolipoprotein E heterozygote model: pharmacotherapeutic implications. Circulation. 2004;110:1678–1685. doi: 10.1161/01.CIR.0000142085.39015.31. [DOI] [PubMed] [Google Scholar]
- 32.Mu JL, Naggert JK, Svenson KL, Collin GB, Kim JH, McFarland C, Nishina PM, Levine DM, Williams KJ, Paigen B. Quantitative trait loci analysis for the differences in susceptibility to atherosclerosis and diabetes between inbred mouse strains C57BL/6J and C57BLKS/J. J Lipid Res. 1999;40:1328–1335. [PubMed] [Google Scholar]
- 33.Purcell MK, Mu JL, Higgins DC, Elango R, Whitmore H, Harris S, Paigen B. Fine mapping of Ath6, a quantitative trait locus for atherosclerosis in mice. Mamm Genome. 2001;12:495–500. doi: 10.1007/s00335001-0006-9. [DOI] [PubMed] [Google Scholar]
- 34.Wood AJ, Oakey RJ. Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet. 2006;2:e147. doi: 10.1371/journal.pgen.0020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizui Y, Yamazaki K, Sagane K, Tanaka I. cDNA cloning of mouse tumor necrosis factor-alpha converting enzyme (TACE) and partial analysis of its promoter. Gene. 1999;233:67–74. doi: 10.1016/s0378-1119(99)00155-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.