Abstract
Identifying the molecular targets for the beneficial or detrimental effects of small-molecule drugs is an important and currently unmet challenge. We have developed a method, drug affinity responsive target stability (DARTS), which takes advantage of a reduction in the protease susceptibility of the target protein upon drug binding. DARTS is universally applicable because it requires no modification of the drug and is independent of the mechanism of drug action. We demonstrate use of DARTS to identify known small-molecule–protein interactions and to reveal the eukaryotic translation initiation machinery as a molecular target for the longevity-enhancing plant natural product resveratrol. We envisage that DARTS will also be useful in global mapping of protein–metabolite interaction networks and in label-free screening of unlimited varieties of compounds for development as molecular imaging agents.
Keywords: aging, label-free, proteomics, small molecules
Development of effective and safe therapies is the holy grail of medicine. For small-molecule drugs, which comprise most of today's medicines, a key challenge remains the identification of the molecular targets underlying drug therapeutic effects and/or adverse side effects. For small molecules discovered in phenotypic screens, which are increasingly popular in chemical genetics studies, identifying the biological (and potential therapeutic) targets, along with the off-targets, is a largely ad hoc affair; a systematic, widely applicable and robust approach is badly needed (1, 2). Current affinity-based target identification techniques are limited by the necessity to modify each drug individually (without losing bioactivity), whereas indirect, non-affinity-based approaches depend on the drug's ability to induce specific biochemical or cellular readouts (3, 4) (supporting information (SI) Text).
To overcome these limitations, we sought to develop a simple, universally applicable target identification approach that analyzes direct drug binding to targets. Given that a protein might become less susceptible to proteolysis when it is drug-bound than when it is drug-free (5–7), we hypothesized that this phenomenon could be exploited for target identification. This would allow the protein target of a drug to be revealed, without requiring modification or immobilization of the small molecule. Because our method, termed DARTS (drug affinity responsive target stability), is not limited by synthetic chemistry and is independent of any biological effects of the drug (save its binding to the target protein), it can potentially be used to identify the target for any small molecule.
Results
DARTS Strategy and Proof-of-Concept.
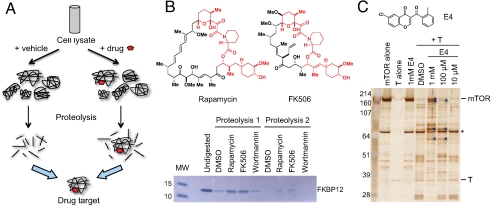
The basic strategy of DARTS is shown in Fig. 1A. Binding of drugs is proposed to stabilize target proteins, either globally or locally, e.g., in a specific conformation or by simply masking protease recognition sites, thereby reducing protease sensitivity of the target protein. This idea is analogous to several familiar concepts, from DNase resistance of DNA sites bound by transcription factors (8) to proteins protected from protease digestion through interacting with their natural ligands such as DNA (9) and carbohydrates (5). However, previous experiments used large (DNA) or high-affinity nanomolar hydrophilic (maltose) ligands, and both cases involve major conformational change in the host protein (10, 11). It remained unclear whether protease susceptibility of the target protein would be different in the absence of large conformational changes, e.g., upon binding of small hydrophobic drugs. Another question is whether the strategy would be amenable to lower-affinity ligands, e.g., clinically used drugs, which encompass a wide range of binding affinities, and hits identified from chemical genetic screens, which typically are in the micromolar range.
Fig. 1.
The DARTS method for drug target identification. (A) Scheme of DARTS. (B) Proof of principle. Recombinant human FKBP12 was incubated with indicated drugs and digested with subtilisin. (C) DARTS with a micromolar mTOR kinase inhibitor (E4). Purple arrow, recombinant human TOR fragments protected from thermolysin proteolysis; *, nonspecific band.
As a proof-of-principle, we examined the well-studied immunophilin FKBP12, which is the target for the nanomolar immunosuppressant drugs rapamycin and FK506 (12). Proteolysis of FKBP12 by the protease subtilisin was clearly decreased by the presence of rapamycin or FK506 (Fig. 1B). This protection is selective: Incubation with wortmannin, a drug that does not bind FKBP12, did not prevent proteolysis (Fig. 1B), and the drugs had no effect on subtilisin activity (Fig. S1A). Because X-ray cocrystal structures showed that binding of FK506 or rapamycin does not cause a conformational change in FKBP12 (12), our results above suggest that drug binding alone is likely sufficient to stabilize the bound protein in the protease-resistant state. This result could be due to a direct change in the protein folding–unfolding equilibrium upon ligand binding (5).
Given that rapamycin and FK506 are among the most potent and specific drugs available, we decided to test whether DARTS would work similarly with a much weaker inhibitor. E4 is a mid-micromolar kinase inhibitor of mTOR identified from a phenotype-based chemical genetic screen. Indeed, proteolysis of mTOR by thermolysin was decreased by E4 in a dose-dependent manner (Fig. 1C and Fig. S1B).
DARTS Using Complex Protein Mixtures.
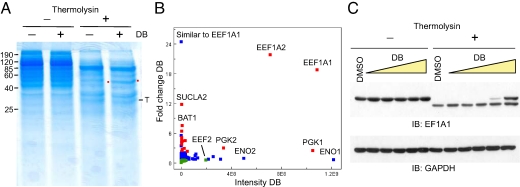
The experiments above established that DARTS can efficiently test, screen, or verify drug–protein interactions when the protein is available in relatively pure form. For DARTS to be generally useful as a discovery tool, however, applicability to complex protein mixtures (such as cell lysates) would be desirable. To demonstrate feasibility, we performed DARTS using human Jurkat cells treated with didemnin B (DB), an anticancer marine natural product whose binding to EF-1α had previously been well characterized (13). Given that EF-1α is a highly abundant protein, we first tested whether in the DARTS protocol DB would protect EF-1α from proteolysis and result in a detectable difference. Indeed, DARTS revealed a strong protected band at ≈50 kDa in the proteolysed extracts of DB-treated cells (Fig. 2A), whereas no detectable difference was observed in the same samples that underwent mock digestion.
Fig. 2.
DARTS using whole-cell lysate. (A) Intact Jurkat cells were treated with DB (1 μg/mL), and lysates were subjected to thermolysin digestion and Coomassie (SimplyBlue)-staining. (B) Enrichment of EF-1α isoforms in the protected band from A revealed by mass spectrometry analysis (SI Text and Fig. S6). Red, protein enriched >2-fold with P value <0.001; green, protein depleted >2-fold with P value <0.001; blue, unchanged protein. (C) DARTS detection via immunoblotting. GAPDH was resistant to thermolysin under the condition and served as a loading indicator.
Examination of the protected band and the matching gel region of the control lane by mass spectrometry confirmed that EF-1α was the primary protein present at higher abundance in the DB-treated sample (Fig. 2B). This analysis does not exclude the possibility of other protected targets of lower abundance that were not evident by eye on the gel. DB-concentration-dependent proteolytic protection of EF-1α was also observed by immunoblotting, both when intact cells were treated with DB (Fig. 2C) and when the lysates of untreated cells were incubated with DB in vitro (Fig. S2). The generality of this approach is further supported by experiments using diverse protein–drug pairs ranging from nano- to micromolar: mTOR-rapamycin, COX-2–celecoxib, and SCF E3 ubiquitin ligase-inhibitor (Fig. S3). Furthermore, DARTS is not enzyme specific, and much higher overall digestion efficiency can be achieved by using other proteases while retaining protection of the target protein (SI Text and Fig. S4).
Identification of a Molecular Target for Resveratrol Using DARTS.
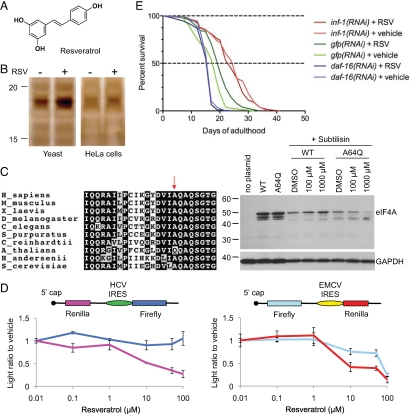
Next, we applied DARTS to identify a molecular target of resveratrol, a compound in red grapes and wine known for various health benefits including lifespan extension (14). Although resveratrol influences the activities of many proteins, no direct molecular target has been demonstrated. Low specific binding affinity as suspected from its modest size and structure (Fig. 3A), poor potency, and potential requirement of the polyphenol groups for its activity have discouraged generation of affinity reagents for target identification. Also, even at saturating concentrations, resveratrol inhibits yeast growth only very weakly if at all (SI Text), making it a poor candidate for target identification using fitness profiling strategies.
Fig. 3.
DARTS identifies a molecular target of resveratrol. (A) Chemical structure of resveratrol. (B) Yeast cell lysates and human HeLa cell lysates were each treated with resveratrol in vitro, followed by thermolysin digestion and silver staining. Protected bands of similar size were detected. (C) Resveratrol protects the wild-type eIF4A, but not the A64Q-substituted eIF4A mutant protein, from proteolysis. (D) Resveratrol inhibits eIF4A-dependent translation in HEK 293 cells as assayed by bicistronic translation reporters. The EMCV IRES requires the eIF4A and eIF4G subunits of eIF4F, whereas the HCV IRES does not (55). (E) eIF4A is required for longevity in resveratrol-treated animals. Resveratrol (50 μM) lengthens the lifespan of wild-type N2 worms fed control (gfp) RNAi (green), but not worms fed eIF4A (inf-1) RNAi (red) or daf-16 RNAi (blue). gfp(RNAi), mVeh = 19 (n = 74), mRSV = 20 (n = 78), ***, P = 0.0006; inf-1(RNAi), mVeh = 26 (n = 76), mRSV = 24 (n = 79), P = 0.4687; daf-16(RNAi), mVeh = 17 (n = 78), mRSV = 17 (n = 76), P = 0.3305. m, mean lifespan (days of adulthood); n, number of animals tested.
DARTS with resveratrol-dosed yeast cell lysates revealed two silver-stained bands between the 15- and 20-kDa MW markers that were more intense in the resveratrol-treated lysate postproteolysis compared with vehicle control (Fig. 3B). Mass spectrometry analysis of both bands showed that eIF4A, along with several ribosomal proteins, were enriched in the resveratrol-treated sample (Table S1 and Dataset S1). This enrichment was confirmed by Western blot analysis using the TAP-tagged (15) eIF4A yeast strain (Fig. S5). This finding suggests that resveratrol might directly bind to one or more proteins comprising the protein translation machinery. Potential direct binding was further supported by a target mutation analysis, where a Tif1 A64Q point mutant confers resistance to resveratrol (Fig. 3C). Although the alanine is conserved throughout fungi and animals, plants have a glutamine at this position, and the bulkier side chain is hypothesized to protect plant eIF4A from resveratrol inhibition by minimizing self binding.
The molecular mechanisms underlying resveratrol's lifespan effect have been controversial (16, 17), and whether Sir2 serves as a direct target for resveratrol is an interesting problem that is being pursued. On the other hand, it is interesting to note that multiple genome-wide studies in S accharomyces cerevisiae and C aenorhabditis elegans have found knockouts or knockdowns of eIF4A and several ribosomal proteins to have significant increases in lifespan (18). Our finding of resveratrol-mediated protection of eIF4A and ribosomal proteins by DARTS suggested that the protein translation machinery may be a molecular target of resveratrol in lifespan extension. To test this notion, we first asked whether resveratrol has a specific effect on protein translation. Using bicistronic dual-luciferase reporters to monitor cap-dependent translation (which requires initiation factors) and translation mediated by IRESs (which exhibit differing requirements for initiation factors), we found that cap-dependent translation and EMCV IRES-mediated translation, both of which require eIF4A, were inhibited in a dose-dependent manner by resveratrol, whereas translation from the eIF4A-independent, HCV IRES was unaffected (Fig. 3D). These results indicate that resveratrol specifically inhibits eIF4A- or eIF4G-dependent translation initiation and does not impinge on other translation initiation factors or on translation elongation.
Finally, we asked whether eIF4A is required for resveratrol's longevity effect. Whereas resveratrol lengthens the lifespan of wild-type worms (Fig. 3E), as reported previously (14), this longevity effect is lost in eIF4A knockdown worms (Fig. 3E), consistent with eIF4A being a physiological target of resveratrol. Interestingly, the longevity effect of resveratrol appears to require daf-16 (Fig. 3E), the Forkhead transcription factor that mediates lifespan extension by the insulin/IGF-1 pathway (19), reminiscent of its requirement for longevity in eIF4G knockdown animals (20). Taken together, it is plausible that resveratrol increases lifespan by direct inhibition of translation initiation, through binding to eIF4A and/or one or more ribosomal proteins in the preinitiation complex. However, this interpretation should be taken with an important caveat because the eIF4A-knockdown worms show a significantly enhanced lifespan (beyond the extension produced by resveratrol in wild-type worms). Furthermore, it is possible that knocking down an initiation factor like eIF4A will affect the expression level of many other proteins that could be targets. Our findings also point to eIF4A (and possibly other translation factors) as a previously uncharacterized druggable target for antiaging therapy. Several potent eIF4A inhibitors have recently been identified (21), and it will be interesting to test them for potential longevity effects.
DARTS Using Proteins Generated from cDNA.
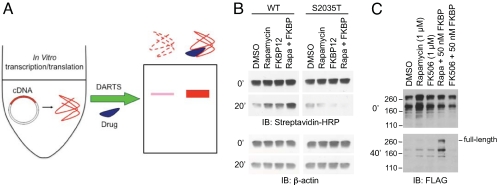
The utility of DARTS in complex mixtures suggests that potential drug targets can be identified by using a wide range of biological systems, and the method is unlimited by the availability and coverage of knockout (or knockdown) libraries and genome arrays for model organisms. The limiting aspect of DARTS analysis is likely to be sensitivity of detection by mass spectrometry or other potential methods (as in affinity chromatography). This limitation is being increasingly alleviated with the development of more sensitive analytical tools. Nonetheless, we tested whether DARTS can be applied to a complementary unbiased platform, namely using proteins generated from cDNAs by in vitro transcription/translation (IVT) (Fig. 4A).
Fig. 4.
DARTS using cDNAs. (A) Plasmid cDNA is used to program IVT for DARTS. (B) FKBP12-rapamycin protects translated mTOR fragment in DARTS. Streptavidin-HRP was used to detect biotin-Lys incorporated into the translation product. β-Actin was less susceptible to thermolysin under the condition and served as loading indicator. (C) DARTS with IVT FLAG-tagged mTOR.
Reticulocyte lysate IVT is a powerful technique routinely used in studies of protein function and is readily adaptable to express proteins in a high-throughput manner (22). As a test case, we used the human mTOR (mammalian target of rapamycin) protein, which is an important target (23, 24) and had been identified on the basis of its association with the FKBP12-rapamycin complex (see references in ref. 25). As shown in Fig. 4B, an IVT mTOR fragment containing the FKBP-rapamycin-binding domain (25) was protected from thermolysin digestion by the presence of FKBP12-rapamycin, whereas the S2035T-substituted mTOR that abolishes rapamycin binding (25) was not protected. IVT TOR proteins were labeled with biotin-Lys in this experiment, but other forms of amino acids could also be used. Alternatively, IVT proteins could be detected through an epitope tag without incorporation of any artificially labeled amino acids. For example, FLAG-tagged full-length mTOR protein was also a robust source of protein for the DARTS method (Fig. 4C). These results further demonstrate the versatility of DARTS and suggest nearly unlimited possibilities for screening cDNA libraries (26) and genome-wide collections of epitope-fused proteins (15, 27) using DARTS to systematically analyze small-molecule–protein interactions.
Discussion
Developing new methods for drug target identification is an area of intense interest, and both experimental and computational approaches have been developed (28, 29). Previous methods for drug target identification have had substantial success, but many limitations remain. Traditionally, affinity chromatography has played a major role in the identification of the binding targets for many biologically active small molecules and natural products (SI Text). In addition to affinity chromatography, many new methods for drug target identification have been developed, ranging from biochemistry to genetics, proteomics, and imaging (30–36) (SI Text). All current target identification methods are of two main categories: affinity-based methods, which detect the direct binding of the drug to its target(s); and phenotype-based methods, which infer drug targets/pathways from the physiological responses or biochemical signatures the drugs produce.
Affinity-Based Target Identification Methods.
Affinity-based methods include matrix-based affinity detection and matrix-free affinity labeling. Matrix-based affinity detection fuses the small molecule of interest to a solid support or capturable moiety such as biotin (SI Text). Such matrix-based methods must satisfy three basic conditions: (i) that the small molecule contains a derivatizable functionality, (ii) that bioactivity/binding specificity of the small molecule is unaffected by the derivatization, and (iii) that the matrix does not hinder the binding of target protein to drug. The latter two criteria cannot be predicted a priori. Matrix-free affinity labeling relies on the incorporation of radioisotope, photoreactive, or fluorescent labels into the small molecule of interest (SI Text) and must also satisfy criteria one and two above. In both affinity chromatography and matrix-free methods, proteins are incubated with the modified small molecule, and the binding proteins are revealed by mass spectrometry after gel electrophoresis. Genetic and other versions of matrix-based affinity chromatography, e.g., yeast three-hybrid (37) and phage display cloning (38), require tagged small molecules as well. Thus, all current affinity methods are limited to small molecules that contain derivatizable functionalities and whose bioactivity/binding is unaffected by the modification (SI Text). Because DARTS does not require labeled ligands and instead uses “native” (i.e., unmodified) small molecules for binding, it is not limited by chemistry and can potentially be used for any small molecule.
Affinity-Free Target Identification Methods.
Indirect, non-affinity-based approaches, which infer drug targets/pathways from the physiological responses or biochemical signatures the drugs produce, have also been developed. For example, classical genetics relies on the isolation of drug-resistant mutations (39) or gene dosage effects (40), and several genome-wide methods also rely on fitness changes (3, 41–44). An inherent limitation of these methods is that they are applicable only to drugs that affect cell growth/viability. Another powerful approach, genome-wide expression profiling (4, 45), on the other hand, is applicable only to drugs that induce major transcriptome changes. These genetic and large-scale “omic” profiling approaches are also primarily limited to yeast or other simple, well-characterized model organisms. Moreover, the “readout” is often far downstream from the drug target.
Advantages of DARTS.
Like affinity chromatography, DARTS relies on the affinity between a drug molecule and its protein target and thereby is able to pinpoint direct binding partner(s) of the drug. The key advantage of DARTS, however, is that because it does not require labeled ligands and instead uses “native” (i.e., unmodified) small molecules for binding, it is not limited by chemistry and can potentially be used to identify binding targets for any small molecule. Additionally, unlike cell-based methods, DARTS is completely independent of any effects of the drug on the system, and is therefore compatible with any mechanism of action, making it useful for any small molecule of interest. Moreover, DARTS can be performed by using any cell or tissue type from any organism and is thus not limited by the availability and coverage of knockout (or knockdown) libraries and genome arrays for model organisms.
Once identified, potential drug targets can be confirmed by functional studies, and kinetics and affinities of the interactions can be measured by using a variety of analytical methods. Although biophysical methods (i.e., surface plasmon resonance, isothermal titration calorimetry, etc.) are traditionally used to analyze direct binding, DARTS proves to be a fast and robust method to determine direct binding of a small molecule (or metabolite) without requiring large amounts of pure protein and is even amenable to using whole-cell lysates.
Potential Limitations of the DARTS Method.
First, the binding affinity of the drug to its target may be a limiting factor. To date, our experiments suggest that DARTS is effective for molecules with inhibitory concentrations across many orders of magnitude, up to high-micromolar. Second, a potential fundamental limitation for DARTS is that a protein's susceptibility to proteolysis is determined by its conformational energy landscape, and it has been demonstrated that a small number of evolutionarily selected proteins (e.g., stress proteins) are quite refractory to protease digestion (46). Third, drug binding may change the protease susceptibility of nontarget proteins, such as those that interact with or are part of complexes containing the target. But this result could be an advantage of the DARTS approach as well, insofar as it would provide information about protein complexes that are dissociated (or formed) upon drug binding. Drug binding in vivo might also increase proteolytic susceptibility of the target protein (47) (see SI Text). This would—in the DARTS protocol—also identify target proteins of the small molecule being analyzed. A small-molecule effector that destabilizes a protein could, of course, also be identified by DARTS.
An extrinsic limiting aspect of DARTS analysis is likely to be sensitivity of detection by mass spectrometry (as in affinity chromatography). Although DB-mediated protection of EF-1α was visualized by eye on a stained gel in Fig. 2A, this will not necessarily be the case with many target proteins of lower abundance. Quantitative imaging or densitometry could prove useful with DARTS to assist in finding more subtle differences in protein abundance. Furthermore, proteomic techniques including 2D gels (48), DIGE (49), and gel-free approaches like MudPIT (50) would likely provide even greater sensitivity in conjunction with DARTS. Finally, the use of cDNA libraries to express proteins in cell culture or by IVT, as demonstrated in Fig. 4, also provides viable alternatives.
Effect of in Vivo Protein Stability on DARTS.
We rely on in vitro proteolysis using exogenous proteases in our DARTS method. Protein stability in vivo on the other hand is a much more complicated problem. Because degradation of proteins inside the cell is predominantly carried out by supramolecular machines, known as the proteasomes and aggresomes, and is elaborately controlled by posttranslational modifications such as phosphorylation and ubiquitinylation, protein stability in vivo is largely unpredictable. Indeed, in vivo stability of proteins upon drug/ligand binding is highly idiosyncratic in the literature; drug binding has been shown to both increase and decrease proteolytic susceptibility of the target protein (6, 47, 51, 52). For instance, whereas unstable FRB domain and FKBP12 mutants are stabilized by the presence of ligands (6, 51) and topoisomerase-1 is destabilized by camptothecin (47), binding of estrogen receptor ligands each affects receptor stability differently (53). In any event, this information would be useful in conjunction with DARTS for elucidating the molecular mechanisms of action of drugs.
Additional Applications of DARTS.
Beyond drugs, we envisage that DARTS will also be useful for global mapping of protein–metabolite interaction networks and in elucidating potential protein targets for small molecules found in food or dietary supplements. DARTS may also be useful in identifying a wide range of small molecules that can be developed into a new genre of molecular imaging agents. Pharmaceutical agents almost always interact with the active sites of enzymes or the ligand-binding sites of receptors. The design of most small-molecule molecular imaging probes usually begins with modification of the structure of known drugs. However, enzyme active sites and ligand-binding sites represent only a very small percentage of the tertiary structures of target proteins. Small molecules that bind tightly and specifically to sites other than the active site or the ligand-binding site on a protein and are detectable by DARTS would provide initial “hits” from which probes that can stoichiometrically measure protein concentrations by fluorescence, positron emission tomography, single-photon emission tomography, and other molecular imaging technologies could be developed through conventional medicinal chemistry or secondary chemical library procedures.
Materials and Methods
Reagents and Plasmid Constructs.
See SI Text for additional information.
DARTS with Pure Proteins.
See SI Text for additional information.
DARTS with Complex Protein Mixtures.
For Fig. 2 A and C, intact Jurkat cells were treated with DB from 100 pg/mL to 1 μg/mL or DMSO control for 30 min. Cells were lysed (without washing, in these experiments) with M-PER (Pierce) supplemented with protease and phosphatase inhibitors. After centrifugation (14,000 rpm using Beckman Coulter Microfuge 22R with F241.5P rotor, 15 min), lysates were diluted to the same final volume and protein concentration with M-PER and proteolysed in reaction buffer [50 mM Tris·HCl (pH 8.0), 50 mM NaCl, 10 mM CaCl2]. All steps were performed on ice or at 4 °C to help prevent premature protein degradation. Each sample was then quickly warmed to room temperature and proteolysed with 1 μg of thermolysin for every 15 μg of lysate for 10 min. To stop proteolysis, 0.5 M EDTA (pH 8.0) was added to each sample at a 1:10 ratio, mixed well, and placed on ice.
For DARTS using yeast cell lysates incubated in vitro with resveratrol (Fig. 3B), S. cerevisiae BY4742 cells were used (see SI Text and Fig. S7).
Mass Spectrometry Analysis.
Gel bands were cut out and prepared for mass spectrometry analysis with trypsin digestion as described in SI Text. Peptides were analyzed by LC/MS/MS on a Thermo LTQ-Orbitrap mass spectrometer with an Eksigent LC pump. For quantitative comparison of protein and peptide abundances, MS spectra were analyzed by using the differential workflow of Rosetta Elucidator (Rosetta Inpharmatics) (54). Annotation was performed using PeptideTeller and ProteinTeller (see SI Text).
In Vivo Translation Assays.
See SI Text for additional information.
Lifespan Analysis.
C. elegans lifespan analysis was conducted with N2 worms at 20 °C (see SI Text).
DARTS Using Proteins Generated by Rabbit Reticulocyte Lysate In Vitro Transcription/Translation (IVT) System.
For Fig. 4B, IVT was performed by using Promega TnT T7 Quick Coupled Transcription/Translation System, with 0.5 μg of pcDNA3.1-hTOR1968C (encoding human mTOR amino acid 1968 to C-ter) or pcDNA3.1-hTOR1968C S2035T [encoding corresponding rapamycin-resistant mutant (25)] vectors (see SI Text), and 2 μL of ε-biotin-Lys-tRNA (Transcend tRNA; Promega) in 50-μL reaction at 30 °C for 3 h. DARTS was performed by using 5 μL of translated lysate in a 10-μL total reaction volume in 50 mM Tris·HCl (pH 8.0), 50 mM NaCl, 10 mM CaCl2, with 1 μM rapamycin, 50 nM FKBP12, or 50 nM FKBP12 + 1 μM rapamycin (preincubated on ice for 30 min to allow complex formation) and incubated on ice for 30 min. Proteolysis was performed with 2 ng of thermolysin at room temperature for 20 min, and stopped with 1 μL of 0.5 M EDTA (pH 8.0).
For Fig. 4C, N-terminal FLAG-tagged full-length human TOR protein was synthesized by IVT using 0.5 μg of pcDNA3.1-FLAG-hTOR vector in a 50-μL reaction at 30 °C for 3 h (Promega TnT T7 Quick Coupled Transcription/Translation System). DARTS was performed by using 6 μL of translated lysate in a 10-μL total reaction volume in reaction buffer [50 mM Tris·HCl (pH 8.0), 50 mM NaCl, 10 mM CaCl2] with 1 μM rapamycin or 1 μM FK506 or cotreatment with 50 nM FKBP12 (preincubated on ice for 30 min to allow complex formation) and incubated on ice for 45 min. Proteolysis was performed with 1 ng of thermolysin at room temperature for 40 min and stopped with 1 μL of 0.5 M EDTA (pH 8.0).
Supplementary Material
Acknowledgments.
We thank Ken Houk and Ray Deshaies for comments and discussions and David Sinclair and Matt Kaeberlein for advice on lifespan assays. This work was partially supported by National Institutes of Health National Cancer Institute Grant R01 CA124974 and by American Cancer Society Grant RSG-07-035-01-CCG. B.L. and M.A. were trainees of the National Institutes of Health UCLA Chemistry–Biology Interface Predoctoral Training Program T32 GM008496.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910040106/DCSupplemental.
References
- 1.Inglese J, et al. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 2.Burdine L, Kodadek T. Target identification in chemical genetics: The (often) missing link. Chem Biol. 2004;11:593–597. doi: 10.1016/j.chembiol.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Giaever G, et al. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet. 1999;21:278–283. doi: 10.1038/6791. [DOI] [PubMed] [Google Scholar]
- 4.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 5.Park C, Marqusee S. Pulse proteolysis: A simple method for quantitative determination of protein stability and ligand binding. Nat Methods. 2005;2:207–212. doi: 10.1038/nmeth740. [DOI] [PubMed] [Google Scholar]
- 6.Stankunas K, et al. Conditional protein alleles using knockin mice and a chemical inducer of dimerization. Mol Cell. 2003;12:1615–1624. doi: 10.1016/s1097-2765(03)00491-x. [DOI] [PubMed] [Google Scholar]
- 7.Tucker CL, Fields S. A yeast sensor of ligand binding. Nat Biotechnol. 2001;19:1042–1046. doi: 10.1038/nbt1101-1042. [DOI] [PubMed] [Google Scholar]
- 8.Maniatis T, Ptashne M. Multiple repressor binding at the operators in bacteriophage lambda. Proc Natl Acad Sci USA. 1973;70:1531–1535. doi: 10.1073/pnas.70.5.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J, Weintraub H, Kedes L. Intramolecular regulation of MyoD activation domain conformation and function. Mol Cell Biol. 1998;18:5478–5484. doi: 10.1128/mcb.18.9.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma PC, Rould MA, Weintraub H, Pabo CO. Crystal structure of MyoD bHLH domain-DNA complex: Perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 11.Sharff AJ, Rodseth LE, Spurlino JC, Quiocho FA. Crystallographic evidence of a large ligand-induced hinge-twist motion between the two domains of the maltodextrin binding protein involved in active transport and chemotaxis. Biochemistry. 1992;31:10657–10663. doi: 10.1021/bi00159a003. [DOI] [PubMed] [Google Scholar]
- 12.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 13.Crews CM, Collins JL, Lane WS, Snapper ML, Schreiber SL. GTP-dependent binding of the antiproliferative agent didemnin to elongation factor 1 alpha. J Biol Chem. 1994;269:15411–15414. [PubMed] [Google Scholar]
- 14.Wood JG, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 15.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 16.Howitz KT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 17.Kaeberlein M, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 18.Smith ED, et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008;18:564–570. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol. 2008;20:126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen M, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 21.Clardy J. Stopping trouble before it starts. ACS Chem Biol. 2006;1:17–19. doi: 10.1021/cb0600029. [DOI] [PubMed] [Google Scholar]
- 22.King RW, Lustig KD, Stukenberg PT, McGarry TJ, Kirschner MW. Expression cloning in the test tube. Science. 1997;277:973–974. doi: 10.1126/science.277.5328.973. [DOI] [PubMed] [Google Scholar]
- 23.Bjornsti MA, Houghton PJ. The TOR pathway: A target for cancer therapy. Nat Rev Cancer. 2004;4:335–348. doi: 10.1038/nrc1362. [DOI] [PubMed] [Google Scholar]
- 24.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci USA. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rolfs A, et al. A biomedically enriched collection of 7000 human ORF clones. PLoS ONE. 2008;3:e1528. doi: 10.1371/journal.pone.0001528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu H, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 28.Terstappen GC, Schlupen C, Raggiaschi R, Gaviraghi G. Target deconvolution strategies in drug discovery. Nat Rev Drug Discov. 2007;6:891–903. doi: 10.1038/nrd2410. [DOI] [PubMed] [Google Scholar]
- 29.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 30.Bharucha N, Kumar A. Yeast genomics and drug target identification. Comb Chem High Throughput Screen. 2007;10:618–634. doi: 10.2174/138620707782507340. [DOI] [PubMed] [Google Scholar]
- 31.Sleno L, Emili A. Proteomic methods for drug target discovery. Curr Opin Chem Biol. 2008;12:46–54. doi: 10.1016/j.cbpa.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 32.Huang J, et al. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc Natl Acad Sci USA. 2004;101:16594–16599. doi: 10.1073/pnas.0407117101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perlman ZE, et al. Multidimensional drug profiling by automated microscopy. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 34.Eggert US, et al. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2004;2:e379. doi: 10.1371/journal.pbio.0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: From enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 36.Ong SE, et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci USA. 2009;106:4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Licitra EJ, Liu JO. A three-hybrid system for detecting small ligand–protein receptor interactions. Proc Natl Acad Sci USA. 1996;93:12817–12821. doi: 10.1073/pnas.93.23.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sche PP, McKenzie KM, White JD, Austin DJ. Display cloning: Functional identification of natural product receptors using cDNA-phage display. Chem Biol. 1999;6:707–716. doi: 10.1016/s1074-5521(00)80018-6. [DOI] [PubMed] [Google Scholar]
- 39.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 40.Rine J, Hansen W, Hardeman E, Davis RW. Targeted selection of recombinant clones through gene dosage effects. Proc Natl Acad Sci USA. 1983;80:6750–6754. doi: 10.1073/pnas.80.22.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lum PY, et al. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell. 2004;116:121–137. doi: 10.1016/s0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- 42.Parsons AB, et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 43.Luesch H, et al. A genome-wide overexpression screen in yeast for small-molecule target identification. Chem Biol. 2005;12:55–63. doi: 10.1016/j.chembiol.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 44.Butcher RA, et al. Microarray-based method for monitoring yeast overexpression strains reveals small-molecule targets in TOR pathway. Nat Chem Biol. 2006;2:103–109. doi: 10.1038/nchembio762. [DOI] [PubMed] [Google Scholar]
- 45.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 46.Park C, Zhou S, Gilmore J, Marqusee S. Energetics-based protein profiling on a proteomic scale: Identification of proteins resistant to proteolysis. J Mol Biol. 2007;368:1426–1437. doi: 10.1016/j.jmb.2007.02.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen AA, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008 doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 48.Shevchenko A, et al. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: A single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 50.Wolters DA, Washburn MP, Yates JR., III An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 51.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishiya Y, et al. Drug-target identification from total cellular lysate by drug-induced conformational changes. Anal Biochem. 2009;385:314–320. doi: 10.1016/j.ab.2008.11.034. [DOI] [PubMed] [Google Scholar]
- 53.Wijayaratne AL, McDonnell DP. The human estrogen receptor-alpha is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J Biol Chem. 2001;276:35684–35692. doi: 10.1074/jbc.M101097200. [DOI] [PubMed] [Google Scholar]
- 54.Neubert H, et al. Label-free detection of differential protein expression by LC/MALDI mass spectrometry. J Proteome Res. 2008;7:2270–2279. doi: 10.1021/pr700705u. [DOI] [PubMed] [Google Scholar]
- 55.Bordeleau M,E, et al. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc Natl Acad Sci USA. 2005;102:10460–10465. doi: 10.1073/pnas.0504249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.