Abstract
Apical membrane antigen-1 is a candidate for inclusion in a vaccine for the human malaria parasite Plasmodium vivax. We collected 231 complete sequences of the gene encoding this antigen (pvama-1) from three regions of Thailand, the most extensive collection to date of sequences at this locus. The domain II loop (previously mentioned as a potential vaccine component) was almost completely conserved, with a single amino acid variant (I313R) observed in a single sequence. The 3′ portion of the gene (domain II through the stop codon) showed significantly lower nucleotide diversity than the 5′ portion (start codon through domain I); and a given domain I sequence might be found in a haplotype with more than one domain II sequence. These results imply a hotspot of recombination between domains I and II. We found significant geographic subdivision among the three regions of Thailand (NW, East, and South) in which collections were made in 2007. Numbers of P. vivax infections have experienced overall declines since 1990 in all three regions; but the decline has been most recent in the NW, and there has been a rebound in numbers of infections in the South since 2000. Consistent with population history, amino acid sequence diversity was greatest in the NW. The South, which had by far the lowest sequence diversity of the three regions, showed signs of a population that has expanded from a small number of founders after a bottleneck.
Plasmodium vivax is the most widely distributed geographically of the malaria parasites infecting humans and is responsible for 100-300 million cases annually, especially in South America, Asia and Oceania; yet researchers have comparatively neglected this parasite (Baird 2007). As with other malaria parasites, several of the surface antigens of P. vivax have been considered as potential vaccine candidates (Webster and Hill 2003). One of these is apical membrane antigen 1 (AMA-1), which is expressed on the apical surface of the merozoite, the stage that invades host erythrocytes, and is believed to play a role in erythrocyte invasion (Hodder et al. 2001).
A number of studies have addressed polymorphism at the locus encoding AMA-1 of P. vivax (the pvama-1 locus) in various parts of the world (Figtree et al. 2000; Grynberg et al. 2008; Gunasakera et al. 2007; Rajesh et al. 2007). Understanding this polymorphism is a necessary prerequisite for development of an effective vaccine based on AMA-1 (Gunasakera et al. 2007). For example, a highly polymorphic antigen might still be used as the basis for a vaccine if polymorphism in local populations is limited, allowing the development of population-specific vaccines (Tanabe et al. 2004). Alternatively, if certain domains of a polymorphic protein show little polymorphism yet elicit an immune response, such domains might be potential vaccine targets (Gunasakera et al. 2007).
The AMA-1 proteins of both Plasmodium falciparum (PfAMA-1) and P. vivax (PvAMA-1) include three extracellular domains, designated domains I, II, and III in N- to C-terminal order (Pizarro et al. 2005). In the case of P. falciparum, there is strong evidence that amino acid sequence polymorphism in domains I, II, and III is selectively maintained, as indicated by a statistically significant excess of nonsynonymous nucleotide substitutions over synonymous substitutions in the regions of the gene encoding those domains (Verra and Hughes 2000).
In a sample of 23 pvama-1 sequences from Sri Lanka, nonsynonymous nucleotide diversity (πN) was significantly greater than synonymous (πS) nucleotide diversity in domain II, suggesting that polymorphism in the latter domain is selectively maintained (Gunasakera et al. 2007). On the other hand, these authors found no amino acid sequence polymorphism in an unstructured loop within domain II that is known to be a target for host antibodies (Pizarro et al. 2005), leading to the suggestion that this loop might provide a vaccine component (Gunasakera et al. 2007). Gunasakera et al. (2007) also suggested that there may be certain recombinational hotspots within the pvama-1 coding region. However, most pvama-1 sequences that have so far been sampled from natural populations have been partial; thus the action of natural selection and interallelic recombination on different domains of the protein remains poorly understood.
Comparison of a pvama-1 sequences from domain I of 320 isolates from around the world showed a striking contrast between Old World populations and those from Brazil (Grynberg et al. 2008). In the Brazilian Amazon, where P. vivax presumably is of post-Columbian origin, pvama-1 sequences showed little evidence of population subdivision, consistent with a rapid recent expansion of the parasite (Grynberg et al. 2008). By contrast, there was evidence of substantial subdivision in Asia, with high FST statistics between pvama-1 sequences collected from different Southeast Asian countries (Grynberg et al. 2008). However, previous studies have not provided evidence regarding the degree of population subdivision at a finer geographic scale. .
Here we report the largest sample yet analyzed (231 sequences) of complete pvama-1 sequences from field isolates, isolated from three different geographic regions within a single Southeast Asian country, Thailand (Figure 1). Using comparative sequence analysis, we test for the role of natural selection in maintaining polymorphism at this locus, and the effects of inter-allelic recombination on the extent of polymorphism across the coding region. We analyze differences in allelic frequencies among the three regions in order to test for population subdivision at this locus. Finally, we present data on the cases of P. vivax malaria recorded over a thirty-year period in the three provinces from which our sequences were derived, enabling us to look for correlations between the observed pattern of pvama-1 sequence polymorphism and the population history of the parasite on a short time scale.
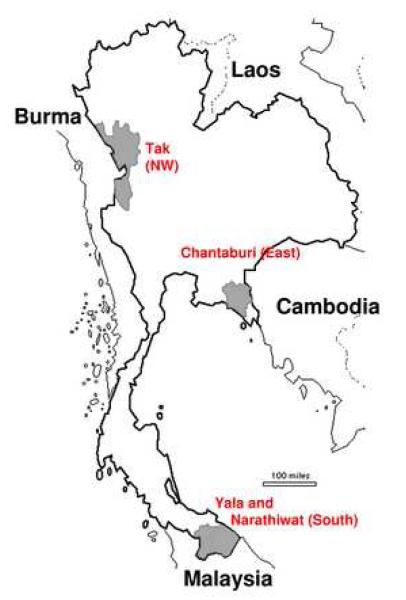
Figure 1.
Map of Thailand, showing the three collection areas (shaded).
Materials and Methods
P. vivax Samples
Pvama-1 sequences were collected from symptomatic malaria patients who acquired the infections from three endemic areas of Thailand, bordering Myanmar, Cambodia and Malaysia, respectively: (1) Tak Province (here designated Northwest or NW); (2) Chantaburi Province (designated East); and Yala and Narathiwat Provinces (designated South; Figure 1). A total of 173 isolates were collected in 2007: 44 from the NW, 56 from the East, and 73 from the South (25 from Yala and 48 from Narathiwat Provinces); and 58 isolates were collected from the NW in 1996. These isolates contained single infections based on genotyping at the highly variable domain of block 6 of the merozoite surface protein 1 locus (Putaporntip et al 2002). We compiled annual totals from 1979-2008 of the numbers of cases of P. vivax malaria in the three geographical regions where collections were made, using data from the Division of Vector-Borne Diseases, Ministry of Public Health, Thailand. The ethical aspects of this study have been approved by the Institutional Review Board of Faculty of Medicine, Chulalongkorn University.
Amplification of the PvAMA-1 gene
DNA of these malaria blood samples were extracted by using the QIAGEN DNA minikit (Hilden, Germany) following the manufacturer’s protocol. After the purification procedure, DNA samples were stored at −30°C until use. The complete coding region of PvAMA-1 was amplified by polymerase chain reaction (PCR) using forward and reverse primers, PvAMA1-F0: 5′-GCAGAGAGAGCAAACCAAATCG-3′ and PvAMA1-R0: 5′-GGCAAGCGAGTTGGCCAAGCAAA-3′, respectively. Thermal cycling profiles contained a preamplification denaturation at 94°C, 1 min; 35 cycles of denaturation at 94°C, 30 s; annealing at 60°C, 30 s; extension at 72°C, 2 min, and post amplification extension at 72°C, 5 min. DNA amplification was performed by using a GeneAmp 9700 PCR thermal cycler (Applied Biosystems, Foster City, CA). To minimize the error introduced in the sequences during PCR amplification, we used ExTaq DNA polymerase because it possesses efficient 5′→3′ exonuclease activity to increase fidelity and no strand displacement (Takara, Japan). The size of PCR product was examined by electrophoresis in a 1% agarose gel and visualized under a UV transilluminator (BioRad Molecular Imager ChemiDoc™ XRS, USA).
DNA sequencing
DNA sequences were determined directly and from both directions for each template using the Big Dye Terminator v3.1 Cycle Sequencing Kit on an ABI3100 Genetic Analyzer (Applied Biosystems, USA). Overlapping sequences were obtained by using sequencing primers. When singleton substitution was detected, sequence was re-determined using PCR products from two independent amplifications using the same DNA template. DNA sequences were submitted to the Genbank database under the accession numbers FJH784891-FJ785121.
Statistical Methods
Coding sequences were aligned using the CLUSTAL W program (Thompson et al. 1997). We used Nei and Gojobori’s (1986) method to estimate dN and the number of synonymous substitutions per synonymous site (dS). In preliminary analyses, the methods of Li (1993) and Yang and Nielsen (2000) yielded essentially identical results, as expected because the number of substitutions per site was low in this case (Nei and Kumar 2000). We computed the mean of all pairwise dS values, designated the synonymous nucleotide diversity (πS); and the mean of all pairwise dN values, designated the nonsynonymous nucleotide diversity (πN). Standard errors of πS and πN were estimated by the bootstrap method (Nei and Kumar 2000); 1000 bootstrap samples were used. We estimated πS and πN separately for the following regions (Gunasakera et al. 2007; Pizarro et al. 2005): (1) the 5′ region (including the signal peptide) from the start codon to the start of domain I (codons 1-42); (2) domain I (codons 43-248); domain II (codons 249-385), including the domain II loop (codons 293-334); domain III (codons 386-487); the 3′ region from the end of domain III to the stop codon, including the transmembrane and cytoplasmic regions (codons 488-563).
Pairwise FST among populations was estimated using the Arlequin 3.11 software (Excoffier et al. 2005). The gene diversity (heterozygosity) of amino acid sequence haplotypes in domains I and II was estimated by the formula:
where n is the number of alleles and xi is the population frequency of the ith allele (Nei 1987, p. 177) an expressed as a percentage. Tajima’s (1989) D test was used to test for balancing selection and/or other effects of population history within samples.
Because pairwise sequence comparisons are not independent, the significance of the correlation between the pairwise proportion of amino acid difference in domain I and that in domain II was tested by a randomization test. In the randomization test, 1000 pseudo data sets were created by sampling from the data with replacement; and the observed correlation coefficient was compared with the distribution of coefficients computed for the pseudo data sets.
Results
P. vivax Malaria 1979-2008
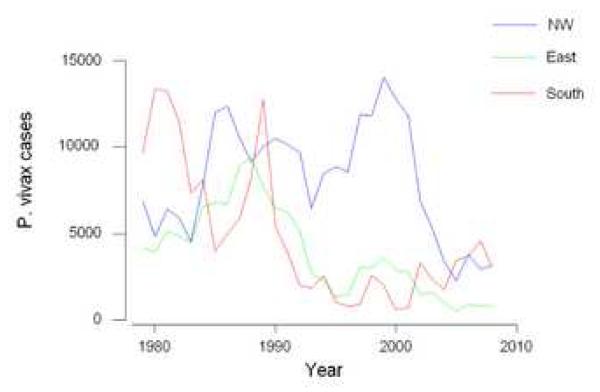
In all three of the regions where we obtained pvama-1 sequences (Figure 1), there was an overall decline in the number of reported cases of P. vivax malaria over the 30-year interval 1979-2008 (Figure 2). However, the patterns differed substantially among geographic regions. Around 1991, a precipitous decline in cases occurred in the East and South, but not in the NW (Figure 2). In the NW, 58.5% of the cases in the data set occurred after 1990, while in the East only 35.7% of cases occurred after 1990 and in the South only 28.3% of cases occurred after 1990 (Figure 2). This difference in proportions χ2 = 16471.6, 1 d.f. P < 0.001) and in the comparison between the NW and the South, (χ2 = 33388.5, 1 d.f. P < 0.001).
Figure 2.
Reported numbers of cases of P. vivax malaria in the three collection areas from 1979-2008 (source: Division of Vector-Borne Diseases, Ministry of Public Health, Thailand).
Beginning in about the year 2000, a decline in numbers of cases began in the NW (Figure 2). For the nine years 2000-2008, the number of cases in the NW was strongly negatively correlated with the year (r = −0.862; P = 0.003). Over the same time period, the number of cases in the East was also negatively correlated with the year (r = −0.856; P = 0.003). In the South, on the other hand, the number of cases, which had fallen since about 1991, began to increase (Figure 2). In fact, over the years 2000-2008, the number of cases in the South was significantly positively correlated with the year (r = 0.777; P = 0.014).
Synonymous and Nonsynonymous Nucleotide Diversity
For 231 pvama-1 sequences from Thailand, we estimated synonymous (πS) and nonsynonymous (πN) nucleotide diversity separately for the three extracellular domains and for the coding regions located 5′ and 3′ to these domains (Table 1). In the case of domain II, we analyzed the loop region and the remainder of the domain separately (Table 1). None of the regions showed πS significantly different from πN (Table 1). In the present data, πN was slightly greater than πS in domain II as a whole and in the portion of the domain excluding the loop, but the difference was not significant in either case (Table 1). For the entire coding region, πS was slightly but not significantly greater than πN (Table 2). Similar patterns were seen when the data for the four collections (NW 1996, NW 2007, East 2007, and South 2007) were analyzed separately (not shown).
Table 1.
Synonymous (πS) and nonsynonymous (πN) nucleotide diversity (± S.E.) in pvama-1 sequences (N = 231) from Thailand.
| Region | No. codons | πS | πN |
|---|---|---|---|
| 5′ remainder | 42 | 0.0046 ± 0.0048 | 0.0050 ± 0.0035 |
| Domain I | 206 | 0.0193 ± 0.0071 | 0.0127 ± 0.0034 |
| Domain II | |||
| Loop | 42 | 0.0000 ± 0.0000a | 0.0001 ± 0.0001b |
| Remainder | 95 | 0.0100 ± 0.0061 | 0.0105 ± 0.0042 |
| All | 137 | 0.0071 ± 0.0044 | 0.0072 ± 0.0028 |
| Domain III | 102 | 0.0000 ± 0.0000a | 0.0016 ± 0.0010b |
| 3′ remainder | 75 | 0.0000 ± 0.0000a | 0.0000 ± 0.0000b |
| All | 562 | 0.0091 ± 0.0027 | 0.0071 ± 0.0015 |
Z-tests of the hypothesis that πS or πN equals the corresponding value in Domain I:
P < 0.01
p < 0.001.
Table 2.
Synonymous (πS) and nonsynonymous (πN) nucleotide diversity (± S.E.) in 5′ and 3′ portions of pvama-1 from different collections.
| Collection | 5′ portion | 3′ portion | ||
|---|---|---|---|---|
| πS | πN | πS | πN | |
| NW 1996 | 0.0148 ± 0.0057 | 0.0121 ± 0.0033 | 0.0025 ± 0.0018a | 0.0014 ± 0.0014a |
| NW 2007 | 0.0156 ± 0.0057 | 0.0124 ± 0.0033 | 0.0036 ± 0.0021a | 0.0042 ± 0.0016a |
| East 2007 | 0.0193 ± 0.0066 | 0.0120 ± 0.0032 | 0.0058 ± 0.0031 | 0.0035 ± 0.0013a |
| South 2007 | 0.0080 ± 0.0041 | 0.0057 ± 0.0021 | 0.0000 ± 0.0000 | 0.0018 ± 0.0009 |
| Mean | 0.0148 ± 0.0057 | 0.0105 ± 0.0016 | 0.0030 ± 0.0012b | 0.0034 ± 0.0006b |
Z-tests of the hypothesis that πS or πN in the 3′ portion equals the corresponding value in the 5′ region: P < 0.05.
Paired t-tests of the hypothesis than mean πS or πN in the 3′ portion equals the corresponding value in the 5′ region: p < 0.001.
As reported by Gunasakera et al. (2007), the domain II loop was highly conserved at the amino acid sequence level, with πN about 1 in 10,000 (Table 1). However, unlike the data of Gunasakera et al. (2007), we did find one polymorphic amino acid site in the domain II loop. At amino acid position 313, one sequence collected in the NW in 1996 showed Ile instead of Arg. The near-absence of nonsynonymous polymorphism supported the hypothesis that this loop is subject to strong purifying selection. This hypothesis was further supported by the fact that πN in the loop (0.0001 ± 0.0001; Table 1) was significantly less than that in the remainder of the gene (0.0076 ± 0.0016; Z-test; P < 0.001). Likewise, although there was no synonymous polymorphism in the loop to serve as a basis for comparison, πN in the loop was significantly less than πS in the whole gene (0.0091 ± 0.0027; Table 1; Z-test; P < 0.001).
Both πS and πN values were significantly greater in domain I than in the domain II loop, in domain III, or in the 3′ remainder (Table 1). In fact, no synonymous differences were observed in any of the latter regions, and the level of nonsynonymous diversity was likewise low (Table 1). This pattern suggested that interallelic recombination may have homogenized the 3′ portion of the gene, starting with domain II, leading to reduced diversity in comparison to the 5′ portion of the gene. We tested this hypothesis by examining synonymous and nonsynonymous nucleotide diversity in each of these portions of the pvama-1 coding region separately for each of the four collections (Table 2). In every collection except South 2007, πN in the 3′ portion was significantly lower than that in the 5′ portion (Table 2). Furthermore, in both NW collections, πS in the 3′ portion was significantly lower than that in the 5′ portion; and the overall mean values of πS and πN in the 3′ portion for all four collections were both significantly less than the corresponding values for the 5′ portion (Table 2). These results suggest that within a given pvama-1 haplotype, the 5′ and 3′ portions (and thus domain I and domain II) may have different evolutionary histories a result of recombination.
Domain I and II Haplotypes
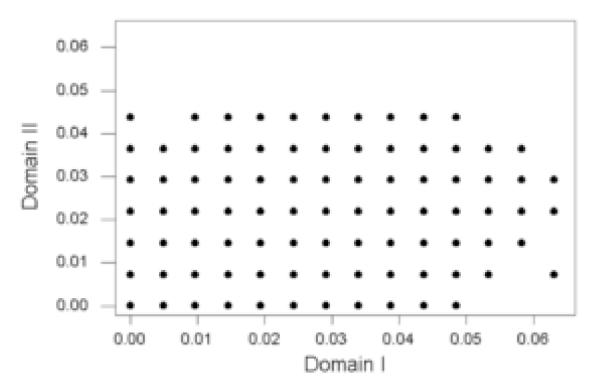
In order to assess the potential impact of recombination on the antigenic properties of domains I and II, we examined the patterns of amino acid sequence difference in domains I and II. When we plotted the proportion of amino acid difference (p) in domain II versus that in domain I for all pairwise comparisons among the 231 sequences, there was a weak but significant positive correlation between p in domain II and that in domain I (r = 0.298; P < 0.001; randomization test; Figure 3). However, there were certain comparisons with very high p in domain I but with few or no differences in domain II (Figure 3). Conversely, there were certain comparisons with very high p in domain II but with few or no differences in domain I (Figure 3). There were 823 comparisons with no amino acid differences in domain II yet at least one amino acid difference in domain I. Of these, 69 (8.4%) comparisons showed p in domain I greater than 0.04; and 514 (62.5%) showed p in domain I greater than 0.02. Likewise, there were 468 comparisons with no amino acid differences in domain I yet at least one amino acid difference in domain II. Of these, 4 (0.9%) comparisons showed p in domain II greater than 0.04; and 123 (26.3%) showed p in domain II greater than 0.02.
Figure 3.
Plot of the proportion of amino acid difference (p) in domain II vs. that in domain I for all pairwise comparisons among 231 pvama-1 sequences (r = 0.298; P < 0.001; randomization test).
Table 3 summarizes the numbers of distinct amino acid haplotypes observed in domain I and in domain II in each of the collections. The gene diversity (“heterozygosity”) of haplotypes in both domains was very high (over 85%) in all collections except South 2007 (Table 3). Although more sequences (73) were obtained in the South 2007 collection than in any of the other collections, in that collection only 4 haplotypes were observed in domain I and only 4 haplotypes in domain II (Table 3). Moreover, in the South 2007 collection, each domain I haplotype was observed in combination with just one domain II haplotype; as a result there were just 4 haplotypes for domains I and II combined (Table 3). By contrast, in all of the other collections, there were more domain I haplotypes than domain II haplotypes, resulting in a slightly higher gene diversity of domain I haplotypes than of domain II haplotypes (Table 3). Moreover, in these collections, certain domain I haplotypes were seen in combination with more than one domain II haplotype; as a result, the total number of haplotypes in both regions combined substantially exceeded that of domain I haplotypes alone (Table 3). Thus, recombination events between domain I and domain II played a role in increasing overall diversity across the two domains.
Table 3.
Diversity of amino acid sequence haplotypes in Domain I and Domain II.
| Domain I | Domain II | Domains I & II | |||||
|---|---|---|---|---|---|---|---|
| Collection | No. sequences |
No. haplotypes |
Gene diversity |
No. haplotypes |
Gene diversity |
No. haplotypes |
Gene diversity |
| NW 1996 | 58 | 40 | 93.7% | 29 | 90.7% | 52 | 96.1% |
| NW 2007 | 44 | 25 | 96.6% | 19 | 94.6% | 31 | 97.7% |
| East 2007 | 56 | 18 | 86.4% | 17 | 85.1% | 24 | 87.8% |
| South 2007 | 73 | 4 | 43.6% | 4 | 43.6% | 4 | 43.6% |
| All | 231 | 56 | 90.6% | 39 | 89.3% | 90 | 92.4% |
Population Differences
Tajima’s D computed for the 1996 NW sample was 0.106, whereas that for the 1996 NW sample was 1.069. D for the 2007 East sample was 0.481; and that for the 2007 South sample was 0.462. None of these three values was significantly different from zero. Likewise, no significant D values were obtained when this statistic was calculated separately for the different domains (not shown). We estimated pairwise FST between each of the three regions in the 2007 collections (Table 4). The results showed substantial subdivision, as indicated by highly significant FST values (Table 4). The values of FST were similar whether based on the 5′ portion of the gene (the start of the coding region through domain I) alone, or the 3′ portion of the gene (domain II through the end of the coding region; Table 4).
Table 4.
Pairwise FST estimates between populations in 2007 collections.
| NW vs. East | NW vs. South | East vs. South | |
|---|---|---|---|
| 5′ portion | 0.059*** | 0.291*** | 0.344*** |
| 3′ portion | 0.076*** | 0.327*** | 0.315*** |
P < 0.001.
Discussion
In the most extensive collection to date of complete sequences of the pvama-1 gene of Plasmodium vivax, overall nucleotide diversity at both synonymous (πS) and nonsynonymous (πN) sites was less than 1% (Table 1). The overall nonsynonymous nucleotide diversity (0.0071) was over half that observed on the ama-1 gene of Plasmodium falciparum (Verra and Hughes 2000; Escalante et al. 2001). In our data for P. vivax, the highest πN values were seen in domain I (0.0127) and in the non-loop portion of domain II (0.010%); but these values were much lower than the over 3 % value of πN observed in P. falciparum (Verra and Hughes 20000; Escalante et al. 2001).
Given the available data, it does not seem possible to decide definitively whether or not polymorphism at the pvama-1 locus is selectively maintained. Indeed, obtaining an unambiguous signal regarding natural selection is difficult when the level of nucleotide diversity is as low as in the case of pvama-1. Gunasakera et al. (2007) found that πN was significantly greater than πS in domain II of pvama-1 in a sample of 23 sequences from Sri Lanka. In our tenfold larger sample, πN was slightly greater than πS in domain II; but the difference was not significant (Table 1). Likewise, Tajima’s D did not show a significantly positive value, which would be suggestive of balancing selection, in any of our collections. Thus, we found no clear indication that the polymorphism at the pvama-1 locus is maintained by balancing selection, at least in Thailand. It is possible of course that the selective environment is different in Sri Lanka. In any event, our results agree with those of Gunasakera et al. (2006) in revealing a substantially lower level of nonsynonymous polymorphism at this locus in P. vivax than in P. falciparum, implying that natural selection acts differently at this locus in the two species.
Confirming the important result of Gunasakera et al. (2007), we found a high degree of amino acid sequence conservation in the domain II loop. Unlike Gunasakera et al. (2007), we did find one polymorphic amino acid position in the domain II loop. However, the polymorphic variant (I313R) at this site was evidently quite rare, being found in just one of 231 sequences. The fact that the domain II loop shows a much lower level of nonsynonymous nucleotide diversity than either the remainder of domain II or the remainder of the gene supports the hypothesis that the amino acid sequence of this loop is subject to strong purifying selection. The existence of one or a few very rare variants would not necessarily argue against Gunasakera et al.’s (2007) suggestion that the domain II loop of AMA-1 might be used as a component of a vaccine for P. vivax.
Our results suggested that recombination, occurring between domains I and II, has been a recurrent feature of the evolution of pvama-1. As a result of this recombination, and subsequent events of either genetic drift or directional selection, the 3′ portion of the gene (from domain II through the end of the coding region) has been homogenized relative to the 5′ portion of the gene (from the start of the coding region through domain I). Thus, both synonymous and nonsynonymous nucleotide diversity in the 3′ portion of the gene were found to be significantly lower in the 3′ portion of the gene than in the 5′ portion of the gene. An additional effect of this recombination has to create new combinations of domain I and domain II amino acid sequence motifs. However, the number of such combinations was still limited in comparison to the theoretical maximum. Overall, we observed 90 different amino acid sequence haplotypes in domains I and II combined; this amounts to only 4.1% of the possible combinations of the 56 domain I haplotypes and the 39 domain II haplotypes (Table 3).
Pairwise FST estimates among geographic regions (NW, East, and South) provided significant evidence of population subdivision. This degree of population subdivision is in marked contrast to the Brazilian Amazon, where there was no population subdivision at the pvama-1 locus between populations as geographically distant as out Thai populations or even more so (Grynberg et al. 2008). These results thus support the hypothesis that P. vivax has infected human populations of Southeast Asia for a long time, in contrast to its post-Columbian introduction and recent spread in Brazil (Grynberg et al. 2008). The long-term history of this parasite in human populations in Southeast Asia has evidently allowed for genetic differentiation of local populations, contrary to what is seen in South America.
Using amino acid haplotypes in domains I and II as markers, the most diverse of the populations sampled in the present study was the NW (Table 3). The East was somewhat less diverse than the NW, and the South substantially less diverse than either NW or East (Table 3). The higher domain I and II haplotype diversity in the NW than in the East is consistent with the more recent decline in P. vivax infection in the NW than in the East, resulting in higher mean levels of human infection in the NW than in the East in both 1996 and 2007 (Figure 1). The fact that haplotype diversities in the NW did not differ substantially between 1996 and 2007 may seem surprising given the sharp decline in infections in the NW over that period (Figure 1). However, the absence of an observable effect of the population decline in the NW may be explained by the fact that the decline has been very rapid, and a prolonged bottleneck is required to produce a substantial reduction in gene diversity (Nei et al. 1975). P. falciparum infections also declined in the NW over the same time span, with little change in diversity at the pfmsp-2 and pfcsp antigen loci, although the former showed substantial allele frequency differences (Putaporntip et al. 2008, 2009).
The greatly reduced domain I and II haplotype diversity seen in the South may at least in part reflect a population history including a sharp decline in the early 1990’s followed by a rebound after 2000 (Figure 2). The limited number of pvama-1 allelic variants found in the South, in spite of the fact that more sequences (73) were obtained there in 2007 than in either of the other regions (Table 3), suggests a population expanding rapidly from a small number of founders. It would be of interest to test whether other polymorphic markers besides pvama-1 likewise show indications of rapid expansion from a small number of founders, in order to obtain further evidence regarding the factors responsible for the unusual pattern seen at pvama-1 in this region.
Acknowledgments
We are grateful to all patients who donated their blood samples for this study and to Thongchai Hongsrimuang, Sunee Seethamchai, Pannadhat Areekul and the staff of the Bureau of Vector Borne Disease, Department of Disease Control, Ministry of Public Health, Thailand, for assistance in field work. This research was supported by grants from the National Research Council of Thailand and the Thai Government Research Budget to S.J and C.P.; The Thailand Research Fund (RMU5080002) to C.P.; grants from the National Institutes of Health GM43940 to A.L.H and D43TW006571 to C.P. and L.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baird JK. Neglect of Plasmodium vivax malaria. Trends Paraditol. 2007;23:533–539. doi: 10.1016/j.pt.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Escalante A,A, Grebert HM, Chaiyaroj SC, Magris M, Biswas S, Nahlen BL, Lal AA. Polymorphism in the gene encoding the apical membrane antigen-1 (AMA-1) of Plasmodium falciparum. X. Asembo Bay Cohort Project. Mol. Biochem. Parasitol. 2001;113:279–287. doi: 10.1016/s0166-6851(01)00229-8. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin version 3.0: an integrated software package for population genetics data analysis. Evol. Bioinfor. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Figtree M, Pasay CJ, Slade R, Cheng Q, Cloonan N, Walker J, Saul A. Plasmodium vivax synonymous substitution frequencies, evolution and population structure deduced from diversity in AMA-1 and MSP-1 genes. Mol. Biochem. Parasitol. 2000;108:53–66. doi: 10.1016/s0166-6851(00)00204-8. [DOI] [PubMed] [Google Scholar]
- Gunasekera AM, Wickramarachchi T, Neafsey DE, Ganguli I, Perera L, Premaratne PH, Hartl D, Handunnetti SM, Udagama-Randeniya PV, Wirth DF. Genetic diversity and selection at the Plasmodium vivax apical membrane antigen-1 (PvAMA-1) locus in a Sri Lankan population. Mol. Biol. Evol. 2007;24:939–947. doi: 10.1093/molbev/msm013. [DOI] [PubMed] [Google Scholar]
- Grynberg P, Fontes CJ, Hughes AL, Braga ÉM. Polymorphism at the apical membrane antigen I locus reflects world population history of Plasmodium vivax. BMC Evol. Biol. 2008;8:123. doi: 10.1186/1471-2148-8-123. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodder AN, Crewther PE, Anders RF. Specificity of the protective antibody response to apical membrane antigen 1. Infect. Immun. 2001;69:3286–3294. doi: 10.1128/IAI.69.5.3286-3294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H. Unbiased estimates of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York: 1987. [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford University Press; New York: 2000. [Google Scholar]
- Nei M, Maruyama T, Charkraborty R. The bottleneck effect and genetic variability in populations. Genet. Res. 1975;35:65–83. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- Pizarro JC, Vulliez-Le Normand B, Chesne-Seck ML, Collins CR, Withers-Martinez C, Hackett F, Blackman MJ, Faber BW, Remarque EJ, Kocken CH, Thomas AW, Bentley GA. Crystal structure of the malaria vaccine candidate apical membrane antigen 1. Science. 2005;308:408–411. doi: 10.1126/science.1107449. [DOI] [PubMed] [Google Scholar]
- Putaporntip C, Jongwutiwes S, Sakihama N, Ferreira MU, Kho WG, Kaneko A, Kanbara H, Hattori T, Tanabe K. Mosaic organization and heterogeneity in frequency of allelic recombination of the Plasmodium vivax merozoite surface protein-1 locus. Proc. Natl. Acad. Sci. USA. 2002;99:16348–16353. doi: 10.1073/pnas.252348999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putaporntip C, Jongwutiwes S, Hughes AL. Differential selective pressures on the merozoite surface protein 2 locus of Plasmodium falciparum in a low endemic area. Gene. 2008;427:51–57. doi: 10.1016/j.gene.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putaporntip C, Jongwutiwes S, Hughes AL. Natural selection maintains a stable polymorphism at the circumsporozoite protein locus of Plasmodium falciparum in a low endemic area. Infec. Genetic. Evol. 2009 doi: 10.1016/j.meegid.2009.02.006. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajesh V, Elamaran M, Vidya S, Gowrishankar M, Kochar D, Das A. Plasmodium vivax: Genetic diversity of the apical membrane antigen-1 (AMA-1) in isolates from India. Exp. Parasitol. 2007;116:252–256. doi: 10.1016/j.exppara.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Sakihama N, Kaneko A. Stable SNPs in malaria antigen genes in isolated populations. Science. 2004;303:493. doi: 10.1126/science.1092077. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Diggins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verra F, Hughes AL. Evidence for ancient balanced polymorphism at the apical membrane antifen-1 (AMA-1) locus of Plasmodium falciparum. Mol. Biochem. Parasitol. 2000;105:149–153. doi: 10.1016/s0166-6851(99)00162-0. [DOI] [PubMed] [Google Scholar]
- Webster D, Hill AV. Progress with new malaria vaccines. Bull. World Health Org. 2003;81:902–909. [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]