Abstract
The analysis of the great extent of data generated by using DNA microarrays technologies has shown that the transcriptional response to radiation can be considerably different depending on the quality, the dose range and dose rate of radiation, as well as the timing selected for the analysis. At present, it is very difficult to integrate data obtained under several experimental conditions in different biological systems to reach overall conclusions or build regulatory models which may be tested and validated. In fact, most available data is buried in different websites, public or private, in general or local repositories or in files included in published papers; it is often in various formats, which makes a wide comparison even more difficult. The Radiation Genes Database (http://www.caspur.it/RadiationGenes) collects microarrays data from various local and public repositories or from published papers and supplementary materials. The database classifies it in terms of significant variables, such as radiation quality, dose, dose rate and sampling timing, as to provide user-friendly tools to facilitate data integration and comparison.
Introduction
Mammalian cells respond to ionizing radiation through several regulatory pathways involved in the control of several cellular functions such as cell cycle arrest, DNA repair and apoptosis (1). An important component of this biological response is mediated by genes transcription modulation (2). In recent years the importance of functional genomic approaches to simultaneously analyze the transcriptional modulation of thousands of genes in response to radiation has been shown in mammalian cells (3–6). The analysis of the mass of data generated from DNA microarray technologies has shown that the transcriptional response to radiation can be considerably different depending on the quality (7), the dose range (8) and dose rate of radiation (9) as well as timing window selected for the analysis. It is very difficult to integrate data obtained under very different conditions and from several biological systems to reach much more general conclusions and build regulatory models which then could be tested and validated (10).
Hence, one of the main difficulties is the fact that most available data is to be found on different websites, public or local repositories or additional files. Moreover, this data appears in various formats (i.e. simple ratio versus log2 ratio between conditions and controls) thus making it very difficult to compare.
Most published studies on transcriptional response to ionizing radiation in mammalian cells involved extremely high, even supralethal, doses to ensure strong gene activation and only few reports focused on clinically relevant doses (<2 Gy). A recent review shows that extrapolation from high doses to low doses of ionizing radiation (IR) is inaccurate for most regulatory pathways activation (11).
There is a growing interest in the assessment of the biological effects of low doses of IR, and particularly when they are delivered at low dose rate, a situation which is relevant to most environmental exposures. It would be very important to collect all the available studies on transcriptional effects of low doses of irradiation in order to facilitate the comparison with a larger collection of data obtained at doses >2Gy. Doses >10 Gy are lethal for most cell types (12,13) and their effects on gene expression in surviving cells are prevalently indirect, being mediated by cell-cycle blockage.
The issue of radiation quality is also very relevant. We can distinguish between low- and high-linear energy transfer (LET) radiations. Low-LET radiation is comprised of X-, β-and γ-rays, while neutrons, high charge (Z) and energy (E) nuclei (HZE) and α-particles are high-LET radiation. There are qualitative differences between high- and low-LET radiation both in the induction and repair of DNA damage (14,16,17). Although high-LET radiation is widely used in medical applications and knowledge of its biological effects is crucial for human protection during space flights (14), at present, very few studies are available on its impact on gene transcription modulation. We consider it a priority to collect all the available microarray studies applied to high-LET radiation as we do believe that this will facilitate the search for high-LET versus low-LET-specific target genes. This will be possible only by an extensive comparison of the patterns of modulated genes obtained in the same cell types with different doses of a high- and low-LET radiation.
Finally, the time course of transcriptional response is also very relevant to understand the biological consequences of regulatory pathways activation.
We introduce a new database of radiation modulated genes that groups most of the available data, classifying it in terms of important variables, such as radiation quality, dose, dose rate and sampling timing. It provides user friendly tools to facilitate data integration and comparison. These user-friendly search tools enable the users to search for the database by gene name, dose quality, dose range and sampling time. Although input data was not statistically filtered (see below), an arbitrary cut-off can be chosen by users to select relevant data whose statistical reliability can be experimentally controlled. Up till date no similar database on transcriptome alterations induced by ionizing radiation exists freely accessible on the web. The introduction of this database allows researchers working on the transcriptional response of mammalian cells to radiation to raise important questions such as following:
— Which genes show a general transcriptional response to irradiation in different biological systems in vitro and in vivo?
— Which genes show a general response to a wide range of doses and to a different quality of irradiation (i.e. low or high LET)?
— Which genes show an early and transient response and which show, instead, a long range modulation? These questions are very relevant for radiobiologists.
Methods
RadiationGenes (interactively accessible at http://www.caspur.it/RadiationGenes) is a relational database of transcription modulation data obtained from microarray experiments. The database is implemented on a Linux server (Suse Enterprise Server 9) running Apache, and the management system is MySQL 5.0. PHP scripts and GD libraries are used for graphical output display. The data is available through several web service (RGretrieval NCBIgene; on the site the WSDL pages are published) giving functionalities similar to the web interface for submission and data retrieval. The web service NCBIgene utilizes the eUtils (www.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html) to query NCBI databases.
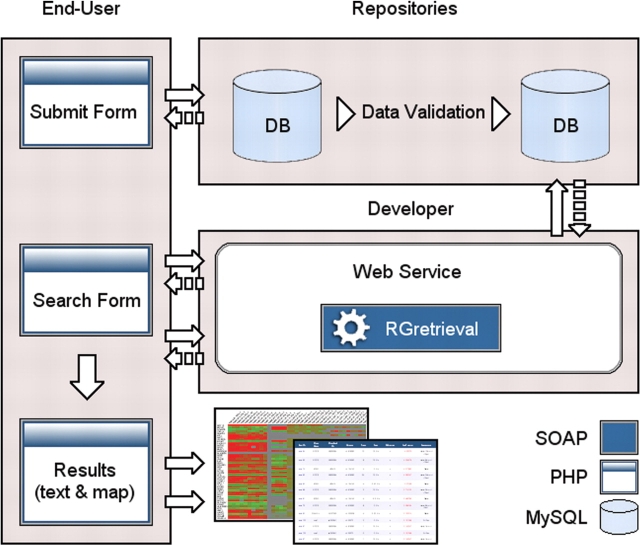
The software architecture of the system is shown in Figure 1 and in the web site at the URL: http://www.caspur.it/RadiationGenes/architecture.php
Figure 1.
The software architecture of the system.
Data set
The main purpose of the database is to allow an easy comparison between modulation patterns obtained with different doses of irradiation with low- or high-LET radiation. For this reason we designed a simplified and modified MIAME protocol for data import which requires a detailed description of irradiation conditions (Figure 2).
Figure 2.
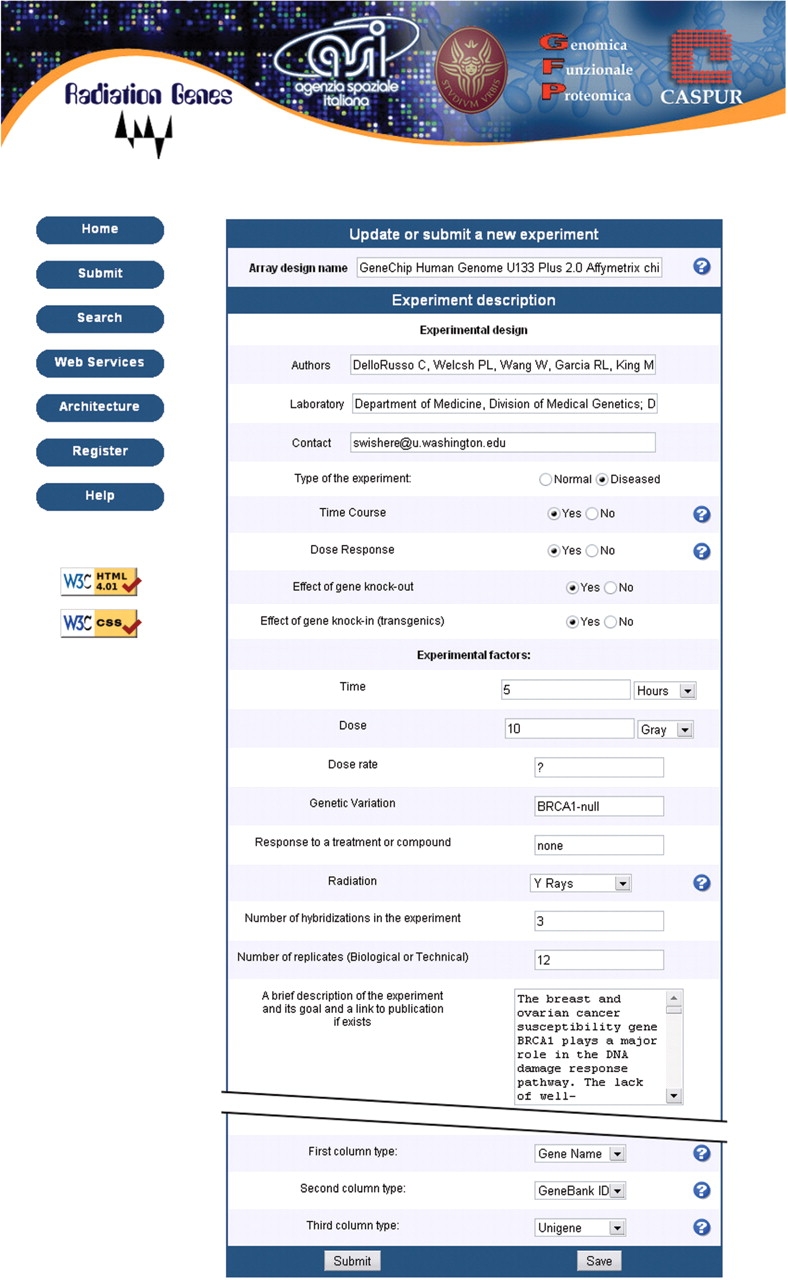
Screenshot of the submission form prepared according to a simplified and modified MIAME protocol. Notice that dose, dose rate and quality of radiation have been added to the experimental factors to be specified.
The database focuses on the early effects of sublethal doses. Therefore, we collected data obtained with irradiation doses up to 10 Gy and 72 h from irradiation. All imported data has already been normalized by the original Authors with the exception of experiments from 30 to 40. In these cases we applied Loess normalization (15). All the imported data was produced in at least two replicated experiments and the imported values represent the average of the replicated values. On the home page a dynamic table shows the number of expression data and genes stored in the database.
Currently there are 49 imported experiments, representing 72 distinct experimental conditions of cell type, time/dose or radiation quality. Data was retrieved from the GEO public repository or published papers supplementary materials (7,8,16).
Web interface
The web interface has two main functions: the submission and retrieval of data. Before submission, the user has to register on the site. The registration data allows the team in charge of the Radiation Genes to contact the submitter if they find inconsistencies in the data. Each submission is processed before loading it into the repository (e.g. if only hugo names are provided for genes, the NCBIgene web service supplies and stores the corresponding RefSeq, Genbank, Unigene, Entrez and alias for genes).
The submission form allows both the experimental irradiation conditions to be described and the previously stored experiment to be modified. In the latter case, the content of the experiment is automatically loaded on the text areas of the submission form. A screenshot of submission form is provided in Figure 2. So far, all submissions have been made online. The submission tool is primarily aimed at users with no substantial local bioinformatics support. No prior knowledge of the modified MIAME guidelines (RG-MIAME) is required, and help is available through links on the web interface.
Once the data is successfully loaded into the database, a summary page is displayed.
The database provides a flexible search menu. Searches can be made entering a gene name or accession number (Hugo, RefSeq, Genbank, Unigene, Entrez and alias) using the NCBIgene web service. More complex queries can be submitted by adding dose range or sampling time to the search form. The user can retrieve and visualize the gene expression values for multiple experiments. A list of genes that match the query ordered by log2 ratio is displayed. Links are provided to access the full annotation of raw data. Furthermore, the search can be limited to a particular kind of radiation X- or γ-rays, neutrons, heavy ions or alpha particles. A major goal of potential database users would probably be the identification of genes modulated in several irradiation conditions. There are different approaches to pursue such a goal. As an example ArrayExpress Atlas (http://www.ebi.ac.uk/microarray-as/atlas/) gives the opportunity to identify strong differential expression candidate genes in conditions of interest (i.e. irradiation conditions) but it adopts very stringent statistical filters. As a result it utilizes only a very limited number of the experiments contained in the ArrayExpress database and retrives a small number of candidates. We prefer to leave the user with the opportunity to fix an arbitrary fold change cut-off and retrieve a list of modulated genes whose reliability could be controlled on the original data, if necessary. An arbitrary cut-off of log2 ratio between intensities obtained for irradiated samples versus unirradiated controls can be fixed to extract a list of potentially modulated genes. The list can be filtered to those genes which pass the cut-off in a defined percent fraction of the conditions stored in the database. Since fold change values strongly depend upon the selected normalization method, we decided to introduce the option to filter the modulated genes on fold change percentile ranking. In this way it is possible to select the most modulated genes in each experiment without depending on a fold change absolute threshold.
Data can be visualized using a display tool available with various search options. By clicking on the ‘Map’ button, the user can produce a heat-map of all modulation values resulting from a specific query. The list of selected genes can be exported into a table showing, for each gene, all the values stored in the database. We have implemented a tool which performs further hierarchical cluster analysis using the R language. This tool supports various types of distance functions (euclidean, maximum, manhattan, binary, minkowski) and various types of clustering functions (complete, average, single).
One of the applications is shown below.
In order to make the Radiation Genes remote programmatic interface accessible and integrated into workflows, we devised several methods for the submission and retrieval of data. The WSDL pages of the implemented web services, RGretrieval and NCBIgene are available on the web site to allow the interoperability of the data. Each web service is available both in Java and in Perl, to support users. A schematic diagram of the software architecture is shown on the website.
Application
Figure 3 shows an example of hierarchical clustering in which we have selected genes showing a log2 ratio between irradiated sample and control ≥1 in at least 20% of the experiments (this can be done by typing ‘1’ in the greater or equal than ‘cut off value’ text field and ‘20%’ as additional filter in the text field ‘show only genes which pass the cut off in’). We have obtained a list of 18 genes which passed the cut-off. Values were then clustered using the on line tool (by clicking on the ‘show clustering’ button). We performed a hierarchical clustering using Euclidean distance and complete linkage as parameters, producing a gene tree and a condition tree. An area of the heatmap can be zoomed by clicking with the cursor on the start and the end of the area of interest. To visualize the “condition tree” better it has been horizontally separated in three zoomed areas of Figure 3: the left portion (panel A); the central part (panel B) and the right portion (panel C). As it is evident, some of the genes show a similar modulation in different conditions and cell types. The most prominent feature is that doses >3 Gy tend to cluster together in the ‘condition tree’ (Figure 3A), and to show a higher level of induction for all the genes considered, indicating a clear dose dependency of their induction. Some of the genes like CDKN1A, GADD45A, GDF15, MR1, ACTA2, CXCR4, TNFSF9, PDE4B and DDR2 are also responsive to lower doses, although showing lower levels of induction (Figure 3B and C).
Figure 3.
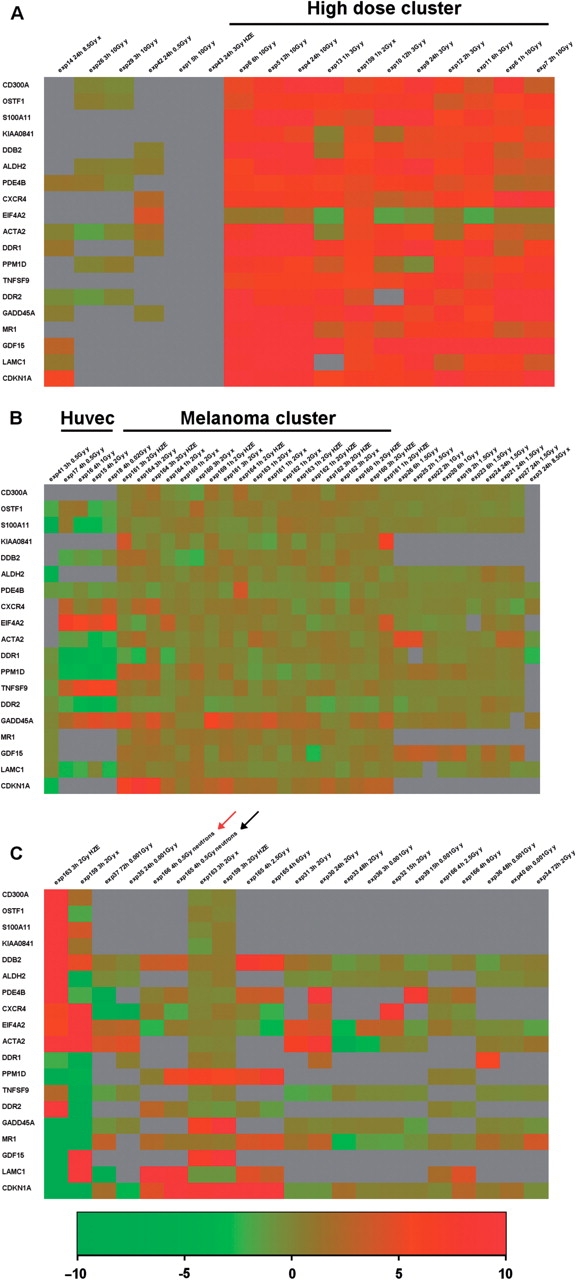
Hierarchical clustering of data for 18 genes which pass the log2 ratio cut-off: >1 in at least 20% of the experiments. Gene tree and conditions tree clustering were performed by selecting the following parameters: Euclidean distance and complete linkage. The condition tree has been zoomed by using the online tools and horizontally separated in three areas: the left portion (A) the central part (B) and the right portion (C).
The second notable feature is that there is no clear effect of the radiation quality. In fact, most of the genes show induction after irradiation with X- and γ-rays, and HZE particles and the only neutron irradiation condition (red arrow in Figure 3C) included in the 72 contained in the database shows induction of four out of the seven genes for which we have data (CDKN1A, PPM1D, DDR2 and DDB2). Also the sampling time does not appear to be crucial, at least in the range of our data which goes from 1 to 72 h from irradiation, since we see no tendency for early or late times to cluster together. This can be due to the small sample size and/or to the relative low number (15/72) of late (≥24 h) time conditions.
A clear cell type effect is instead evident: data from HUVEC cells (experiments 15–19) cluster together and show repression rather than induction of a group of genes (DDR1, PMD1D, ACTA2, DDR2, DDB2 and S100A11) upon irradiation with various doses of X-rays (Figure 3B). This probably reflects the peculiarity of the response of endothelial cells to irradiation which has been previously described in detail (17). Furthermore most of the experiments performed on melanoma cell lines (experiments 159–164) tend to cluster together independently from irradiation with X-rays or HZE particles.
Some of the 18 induced genes were already previously shown to have an important role in the response to irradiation. This is the case for two out of seven of the genes responding to a wide range of doses (CDKN1A and GADD45A) while much less is known about the possible rational for the induction of the other five genes of this group (GDF15, MR1, ACTA2, DDR2 and CXCR4). On the same line, the induction of PPM1D and DDB2 at high doses of radiation is somehow expected and previously described, while more puzzling is the involvement of the other nine genes. In conclusion, a master role for GADD45A, and CDKN1A (P21) in the general response to irradiation is confirmed but other interesting candidates have been identified whose role could be clarified in various cellular systems in the future. In particular GDF15, which is induced at least 1.5-fold in 15 out of the 23 conditions for which we have data, shows a general responsivity. A table which summarizes the biological role of the 18 genes and references which link them to the response to irradiation is included at the URL http://www.caspur.it/RadiationGenes/supplementary/supplementary_table1-1.xls.
Future enhancement
The mass of data contained in the database is still limited but the number of experiments in transcriptional modulation by ionizing radiation, especially low-dose irradiation and the number of published articles is rapidly increasing. We will continue to import published data and offer the opportunity to external groups to import their unpublished data, provided that they follow our modified MIAME protocol. A message board will allow us to receive users’ feedback to improve the usefulness of the database. The database is currently sponsored by the Italian Space Agency ‘MoMa’ project which plans to perform several extensive microarray studies on transcriptional effects of high LET radiation. The experimental results will be stored in the Radiation Genes database. The Italian Federation of Radiation Research (FIRR) has a link to Radiation Genes in its website (http://biotec.casaccia.enea.it/FIRR/) and we hope that the Italian community of radiobiologists will be encouraged to interact extensively with the database in the near future.
Supplementary data
Supplementary data are available at Database Online.
Funding
ASI MoMa program ‘From Molecules to Man: Space Research Applied to the improvement of the Quality of Life of the Ageing Population on Earth’ grant (2005–2009).
Conflict of interest. None declared.
Acknowledgements
The authors are grateful to Bhupesh Singh and Lisa Gabrieli for their help in manuscript editing.
References
- 1.Weichselbaum RR, Hallahan DE, Sukhatme V, et al. Biological consequences of gene regulation after ionizing radiation exposure. J. Natl Cancer Inst. 1991;83:480–484. doi: 10.1093/jnci/83.7.480. [DOI] [PubMed] [Google Scholar]
- 2.Holbrook NJ, Liu Y, Fornace A.JJr. Signalling events controlling the molecular response to genotoxic stress. In: Feige U, Morimoto RI, Yahara I, et al., editors. Stress-Inducible Cellular Responses. Basel: Birkhauser Verlag; 1996. [DOI] [PubMed] [Google Scholar]
- 3.Amundson AA, Bittner M, Chen Y, et al. Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene. 1999;18:3666–3672. doi: 10.1038/sj.onc.1202676. [DOI] [PubMed] [Google Scholar]
- 4.Amundson SA, Bittner M, Meltzer P, et al. Induction of gene expression as a monitor of exposure to ionizing radiation. Radiat. Res. 2001;156:657–661. doi: 10.1667/0033-7587(2001)156[0657:iogeaa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 5.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dent P, Yacoub A, Contessa J, et al. Stress and radiation-induced activation of multiple intracellular signalling pathways. Radiat. Res. 2003;159:283–300. doi: 10.1667/0033-7587(2003)159[0283:sariao]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 7.Amundson SA, Do KT, Vinikoor L, et al. Stress-specific signatures: expression profiling of p53 wild-type and –null human cells. Oncogene. 2005;24:4572–4579. doi: 10.1038/sj.onc.1208653. [DOI] [PubMed] [Google Scholar]
- 8.Lanza V, Pretazzoli V, Olivieri G, et al. Transcriptional response of human umbilical vein endothelial cells to low doses of ionizing radiation. J. Radiat. Res. 2005;46:265–276. doi: 10.1269/jrr.46.265. [DOI] [PubMed] [Google Scholar]
- 9.Wang B, Ohyama H, Haginoya K, et al. Adaptive response in embryogenesis III. Relationship to radiation-induced apoptosis and TP53 status. Radiat. Res. 2000;154:277–282. doi: 10.1667/0033-7587(2000)154[0277:arieir]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 10.Amundson SA. Functional genomics in radiation biology: a gateway to cellular systems-level studies. Radiat. Environ. Biophys. 2007;47:25–31. doi: 10.1007/s00411-007-0140-1. [DOI] [PubMed] [Google Scholar]
- 11.Criswell T, Leskov K, Miyamoto S, et al. Transcription factors activated in mammalian cells after clinically relevant doses of ionizing radiation. Oncogene. 2003;22:5813–5827. doi: 10.1038/sj.onc.1206680. [DOI] [PubMed] [Google Scholar]
- 12.Chapman JD. Single-hit mechanism of tumour cell killing by radiation. Int. J. Radiat. Biol. 2003;79:71–81. [PubMed] [Google Scholar]
- 13.Vorobiev AI. Acute radiation disease and biological dosimetry in 1993. Stem Cells. 1997;15(Suppl. 2):269–274. doi: 10.1002/stem.5530150736. [DOI] [PubMed] [Google Scholar]
- 14.Durante M, Cucinotta FA. Heavy ion carcinogenesis and human space exploration. Nat. Rev. Cancer. 2008;8:465–472. doi: 10.1038/nrc2391. [DOI] [PubMed] [Google Scholar]
- 15.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 16.DelloRusso C, Welcsh PL, Wang W, et al. Functional characterization of a novel BRCA1-null ovarian cancer cell line in response to ionizing radiation. Mol. Cancer Res. 2007;5:33–45. doi: 10.1158/1541-7786.MCR-06-0234. [DOI] [PubMed] [Google Scholar]
- 17.Lanza V, Fadda P, Iannone C, et al. Low dose of ionizing radiation stimulates endothelin transcription and production by human vein endothelial cells. Radiation Res. 2007;168:193–198. doi: 10.1667/RR0780.1. [DOI] [PubMed] [Google Scholar]