Abstract
Telomerase is essential for telomere maintenance, and its activation is thought to be a critical step in cellular immortalization and tumorigenesis. Human telomerase reverse transcriptase (hTERT) is a major component of telomerase activity. We show here that hTERT is expressed soon after lymphocyte activation and that its expression is inhibited by rapamycin, wortmannin, and FK506, which was the most potent inhibitor. These results suggest a potential role for the transcription factor nuclear factor of activated T cells (NFAT) in the regulation of hTERT expression. Five putative NFAT-binding sites were identified in the hTERT promoter. In luciferase assays, the hTERT promoter was activated by overexpressed NFAT1. Moreover, serial deletions revealed that the promoter activation was mainly due to a −40 NFAT1-binding site flanked by two SP1-binding sites. Mutation of the −40 NFAT-binding site caused a 53% reduction in the transcriptional activity of hTERT promoter. Simultaneous mutations of the −40 NFAT-responsive element together with one or both SP1-binding sites led to a more dramatic decrease in luciferase activity than single mutations, suggesting a functional synergy between NFAT1 and SP1 in hTERT transcriptional regulation. NFAT1 overexpression in MCF7 and Jurkat cell lines induced an increase in endogenous hTERT mRNA expression. Inversely, its down-regulation was induced by NFAT1 silencing. Furthermore, chromatin immunoprecipitation assay demonstrated that NFAT1 directly binds to two sites (−40 and −775) in the endogenous hTERT promoter. Thus, we show for the first time the direct involvement of NFAT1 in the transcriptional regulation of hTERT.
Introduction
Telomeres are specialized structures located at the ends of linear mammalian chromosomes (1). The erosion of human telomeres at each cycle of cellular division is compensated for by de novo synthesis catalyzed by human telomerase reverse transcriptase (hTERT),3 the catalytic subunit of a ribonucleoprotein complex called telomerase (2). Telomerase maintains telomeres by protecting them from exonucleases and ligases and by preventing illegitimate recombination (3). However, hTERT is also implicated in cell immortalization and tumorigenesis (4) through its telomere-lengthening activity, as well as by a mechanism independent of telomere length (5).
Most normal human somatic tissues do not express hTERT, but germinal cells, several types of normally activated or proliferating cells, and tumor cells do (6–13). In particular, lymphocytes exhibit telomerase activity in response to stimulation (14). Regulation of telomerase expression in these cells is likely to occur in the G1 phase of the cell cycle as telomerase is inhibited by rapamycin, a compound that affects the mammalian target of rapamycin (mTOR) but is not inhibited by aphidicolin or hydroxyurea, substances that inhibit DNA synthesis (14–16). Phosphorylation of hTERT and the resulting effects on its nuclear translocation and telomerase activity have been well described (17, 18). These post-translational modifications may explain the discrepancies between hTERT expression (19) and the reduction of telomerase activity in response to several inhibitors of cell proliferation, such as rapamycin or wortmannin (17, 19, 20). In normal and malignant human cells, however, a positive correlation is consistently observed between telomerase activity and hTERT mRNA expression (6, 21–23), thereby highlighting the importance of the transcriptional regulation of hTERT.
hTERT promoter was cloned and characterized independently by two groups, Horikawa et al. (24) and Takakura et al. (25). The transcription initiation site (+1) was determined 55 bp upstream of the ATG (24). The hTERT promoter comprises three main regions. The first, consisting of a sequence of 258 bp (from −203 to +55), corresponds to the promoter core and is essential for transcriptional activation of the hTERT gene. The second, an activating region, is located between positions −1397 and −798. Finally the third, an inhibitory region, is located between positions −798 and −400 (24, 25). Several transcription factors have been identified as activators (c-Myc, Sp1, and activated estrogen receptor) (26, 27) and inhibitors (Mad1, WT1, p53, and MZF-2) (28–31) of hTERT mRNA expression. Some factors can act as activators or inhibitors depending on conditions such as which responsive element they bind to (Ets 1 and 2) (32) or whether the cells are normal or neoplastic (E2F protein family) (33, 34). Epigenetic regulation mechanisms such as methylation (35, 36) and acetylation (37, 38) also modulate hTERT expression.
Despite its importance for a basic understanding of cancer biology, the induction of hTERT mRNA expression after activation of peripheral blood lymphocytes remains poorly understood. A comparison of the metabolic pathways by which several immunosuppressors inhibit hTERT mRNA expression led us to investigate the role of the nuclear factor of activated T cells (NFAT). We demonstrate for the first time that NFAT stimulates the hTERT promoter mainly through a consensus binding site localized in the core promoter. In addition, we characterize the possible relationship of NFAT with SP1 and demonstrate the direct binding of NFAT to the promoter of hTERT in vivo.
EXPERIMENTAL PROCEDURES
Cell Culture
Normal human lymphocytes were isolated from heparinized peripheral blood from normal subjects on Ficoll-Hypaque gradients (d = 1.077, Sigma) by centrifugation (300 × g for 30 min). After separation of the monocytes, lymphocytes were grown in RPMI 1640 medium (1 × 106 ml−1) supplemented with 10% fetal calf serum (Invitrogen), 200 mm l-glutamine (Seromed), 100 IU/ml penicillin, and 50 μg/ml streptomycin (Invitrogen).
Jurkat cells were grown in RPMI 1640 medium, and GM847, HeLa, and MCF7 cells were grown in Dulbecco's modified Eagle's medium supplemented as indicated above.
Based on previous work by Horikawa et al. (24), hTERT promoter activity was studied in the telomerase-negative GM847 cell line, which expresses endogenous calcineurin and NFAT (data not shown). Regulation of the endogenous hTERT promoter was studied in the telomerase-positive MCF7 and Jurkat cell lines.
Peripheral Blood Lymphocyte Stimulation and Immunosuppressive Treatment
For the kinetic analysis of the induction of hTERT expression after lymphocyte activation, normal peripheral blood lymphocytes (PBL) were stimulated with PHA (1 μg/ml RPMI, Murex) and harvested before and after stimulation at the following times: 0, 2, 3, 6, 24, and 36 h. Flow cytometry analysis of immunological PBL phenotype was performed before and 36 h after PHA stimulation using anti-CD3-Cy7 (clone SK7, BD Biosciences), CD45 PercPCy5.5A (clone 2D1, BD Biosciences), CD19 PE-Cy7 (clone J4119, Beckman Coulter), CD56 PE (clone My31, BD Biosciences), CD14 FITC (clone RMO52, Beckman Coulter), CD4 Pacific Blue (clone RPA-T4, BD Biosciences), and CD8 AmCyan (clone SK1, BD Biosciences) antibodies, using a BD Biosciences FACSCanto II instrument and the FACSDiva analysis software. The PBL population was mainly composed and enriched of CD3+ cells after stimulation (supplemental Fig. S1).
To study the effects of several immunosuppressors, PBL were simultaneously stimulated with PHA and treated for 48 h with one of the following inhibitors: 10 or 100 nm FK506, which inhibits calcineurin (39) (Fujisawa Pharmaceuticals); 500 or 1000 nm rapamycin, which inhibits mTOR (Sigma); 5 or 10 nm wortmannin, which inhibits phosphatidylinositol 3′-kinase (40) (Sigma); or 4 or 8 μg/ml aphidicolin, which blocks DNA synthesis by inhibiting DNA polymerase α (Sigma). For FK506, stimulated PBL without any treatment were used as a control. For rapamycin, wortmannin, and aphidicolin, the same volume of dimethyl sulfoxide (Sigma) was added simultaneously with PHA to PBL as a control. Treatment efficiency was checked by cell cycle analysis (supplemental Fig. S2).
Cell-cycle Analysis
The distribution of cells in the cell cycle was determined by flow cytometry analysis of DNA content (Coulter EPIX XL2, Beckman) (41).
Reverse Transcription and PCR Assay
Total RNA was extracted from PBL samples by a guanidinium-thiocyanate-phenol/chloroform procedure using RNA Plus (Bioprobe Systems). Semi-quantitative RT-PCR to examine hTERT and β-actin transcription levels was performed as previously described (41).
Real-time quantitative PCR was performed to quantify the relative abundance of hTERT using the QuantiTect SYBR Green PCR kit (Qiagen) as recommended by the manufacturers, on a MX3000P instrument from Stratagene (Agilent Technologies, Massy, France). Results were normalized to the expression of the reference β-actin gene or 18S RNA. Primers used for hTERT expression analysis were described previously (42). Primers used for β-actin, 18S RNA, and human NFAT1 expression analysis were QuantiTect Primer Assays QT01680476, QT00199367, and QT0053599, respectively, from Qiagen. The reactions proceeded for 1 cycle at 95 °C for 15 min, followed by 40 cycles at 95 °C for 30 s, 55 °C for 1 min, and 72 °C for 30 s. All samples for hTERT, β-actin, and 18S RNA analyses were processed in triplicate. To analyze the effect of NFAT1 overexpression on endogenous hTERT expression, forward 5′-TGCATCTAACCCCATCGAGTG-3′ and reverse 5′-TGAGGATCATTTGCTGGCG-3′ primers were used in the presence of 3% formamide for 1 cycle at 95 °C at 15 min, followed by 40 cycles at 95 °C for 30 s, 56 °C for 1 min, and 72 °C for 30 s.
TRAP Assay
Telomerase activity was assessed in PBL according to a telomeric repeat amplification protocol (TRAP) using a TRAPezeTM Telomerase Detection Kit (Oncor) following the manufacturer's recommendations with minor modifications as previously described (41).
Expression Vectors
The ΔCAM-AI gene expressing a constitutively active calcineurin (CN) cloned into an SRα expression vector, and the corresponding empty vector SRα, were kind gifts of Dr. C. V. Paya (43). The HA-tagged murine NFAT1(c2) (HA-NFAT1) expression vector encoding isoform C of NFAT1, pEFTAG-mNFAT1-c, was kindly provided by Dr. A. Rao (44). The corresponding empty vector was pEF-TAG.
Luciferase Reporter Gene Constructs
The constructs encoding luciferase downstream of an hTERT promoter (hereafter, hTERT-Luc reporter plasmids) are shown in Fig. 3. A 1787-bp fragment (−1652 to +135) of the 5′ region of the hTERT gene (accession number AF098956 in GenBankTM) was amplified by PCR. Briefly, genomic DNA from the Reh6 cell line was extracted using the Nucleon BACC2 kit (Amersham Biosciences) according to the supplier's protocol. The PCR reaction was performed with p-1652 and p+135(1) primers (Table 1) on 330 ng of extracted genomic DNA using the Expand Long Template PCR System (Roche Diagnostic, Mannheim, Germany) according to the manufacturer's instructions. The final product was inserted into the luciferase reporter vector pGL3-basic (Promega Corp., Madison, WI) using KpnI and NheI, such that the hTERT promoter fragment would drive transcription of the firefly luciferase. The resultant plasmid was named h-1652. The h-1399 construct was prepared as described previously using p-1399 and p+135(1) primers (Table 1). Due to the lack of restriction sites, the three other 5′-truncated fragments of the hTERT promoter region were generated by PCR reaction using sense primers containing an MluI restriction site and a common antisense primer (p+135) with a BglII restriction site (Table 1). The amplified fragments were then cloned into the pGL3-basic vector in the sense orientation relative to the luciferase coding sequence at the MluI and BglII sites.
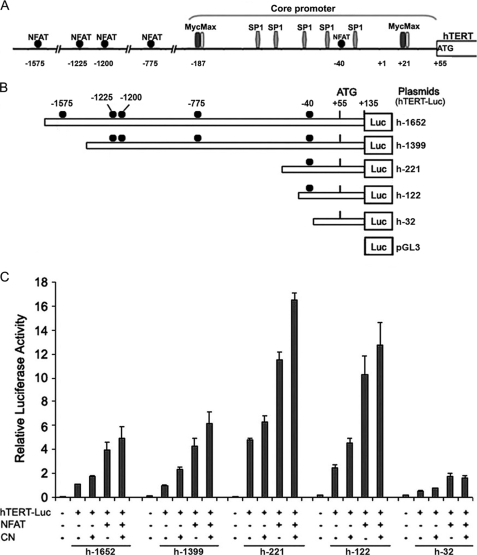
FIGURE 3.
Schematic representation of reporter plasmids and transcriptional activity of the hTERT promoter. A, schematic representation of the full-length hTERT promoter sequence used in this study. Potential NFAT-binding sites are indicated, as are E-boxes and SP1-binding sites in the promoter core. B, the hTERT-Luc reporter plasmids were generated by inserting the 1787-bp DNA and 5′-truncated fragments of the hTERT promoter upstream of the +1 transcription initiation site into the luciferase (Luc) reporter vector pGL3-basic in the sense orientation. The locations of NFAT-binding sites are indicated as the number of their first base upstream of the +1 transcription initiation site. The name of each reporter construct was assigned according to the nucleotide number at the 5′-end of the inserted promoter sequences. C, telomerase-negative GM847 cells were cotransfected with 1.5 μg of the different hTERT-Luc reporter plasmids in the absence (−) (3 μg of pEFTAG) or presence (+) of the HA-NFAT1 expression vectors (3 μg of pEFTAG-mNFAT1c), and in the absence (−) (0.6 μg of SRα) or presence (+) of the constitutively active calcineurin (CN) expression vectors (0.6 μg of ΔCAM-AI). For each transfection, luciferase activity was normalized to the Renilla luciferase activity due to cotransfected pRL-TK (0.25 μg). The relative activity of each construct is expressed as the ratio of its activity to that of h-1652 cotransfected with the empty versions of the NFAT and CN expression vectors (−). The negative control in these experiments involved cotransfection of the pGL3-basic vector with the empty expression vectors (−). The mean and standard deviation of at least three experiments are shown for each construct.
TABLE 1.
Primer sequences for luciferase reporter gene constructs
Forward primers were named according to the first nucleotide number at the 5′ end of the truncated hTERT promoter sequence. Reverse primers were named according to the last nucleotide of the hTERT promoter sequence. p+135(1) was used to amplify −1652 and −1399 hTERT promoter fragments; p+135 was used to amplify −221, −122, and −32 fragments. MluI restriction sites (forward primers) and BglII restriction sites (reverse primers) are indicated in bold. The addition of these restriction sites allows the cloning of the −221, −122, and −32 hTERT promoter fragments in sense orientation into pGL3-basic vector.
| Forward primers | |
| p-1652 | 5′-CAAAGACACACTAACTGCACC-3′ |
| p-1399 | 5′-CAGAGACAATTCACAAACACAGCC-3′ |
| p-221 | 5′-CCACGCGTATTCGCGGGCACAGACGCC-3′ |
| p-122 | 5′-CCCACGCGTCCGCGCGGACCCCGCCCCGT-3′ |
| p-32 | 5′-CCCACGCGTGCCCCGCCCTCTCCTC-3′ |
| Reverse primers | |
| p+135(1) | 5′-AACGTGGCCAGCGGCAGCACCT-3′ |
| p+135 | 5′-CCCAGATCTAACGTGGCCAGCGGCAGCACCT-3′ |
Primers containing substituted nucleotides were designed (Fig. 4A) to synthesize proximal promoter constructs with mutated NFAT1- and/or SP1-binding sites using the QuikChange site-directed mutagenesis method (Stratagene, Amsterdam, The Netherlands). All the constructs were confirmed by sequencing (Genome Express, Meylan, France).
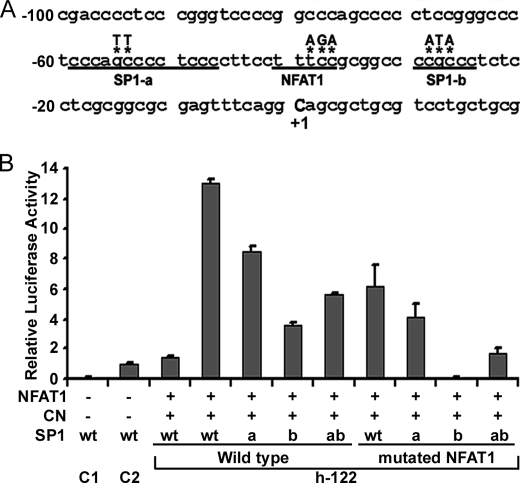
FIGURE 4.
NFAT1 and SP1 cooperation in the transcriptional activation of hTERT. A, part of the sequence of the hTERT promoter in the h-122 construct. The putative −40 NFAT1-binding site and two SP1 sites (SP1-a and SP1-b) are underlined. The mutated nucleotides are noted by asterisks. B, histogram of the relative luciferase activity of hTERT promoter reporter constructs. The h-122 wild-type construct (1.5 μg) or h-122 mutated in the NFAT-binding site (mutated NFAT) or distal (a) or proximal (b) SP1 sites was cotransfected with 3 μg of HA-NFAT1 expression vector (NFAT) and 0.6 μg of expression vector encoding a constitutively active form of CN. For each transfection, the luciferase activity was normalized to the Renilla luciferase activity of cotransfected pRL-TK (0.25 μg). The relative activity of each construct is expressed as a ratio of its activity to the activity of h-1652 cotransfected with the empty versions of the HA-NFAT1 and CN expression vectors. The mean and the standard deviation of at least three experiments are shown for each construct.
Luciferase Assays
Transient transfections of luciferase reporter plasmids were performed using the Max family (Eurogentec) according to the manufacturer's protocol. GM847 cells were plated at 35 × 105 cells/well in 6-well plates, cultured overnight before transfection, and harvested 72 h after transfection. The different hTERT-Luc reporter plasmids were transfected alone or cotransfected with HA-NFAT1 and/or CN expression vectors. HA-NFAT1 and CN expression vectors were transfected using a ratio of 5:1 based on the previous studies of Sheridan et al. (45) and on the results of preliminary transfections (supplemental Figs. S3 and S4). Negative controls were obtained by transfecting cells with empty versions of all three expression vectors (1.5 μg of pGL3 basic, 3 μg of pEFTAG, and 0.6 μg of SRα). pRL-TK encoding Renilla luciferase was cotransfected (0.25 μg) in all experiments. Luciferase assays were performed with the Dual-Luciferase Reporter Assay System (Promega), in which the Renilla luciferase activity was used as a control to standardize the transcription efficiency. Luminosity measurements were taken with a Turner Designs TD 20/20 luminometer (Turner Designs).
For the construct h-1652, the effects of NFAT1 and/or CN were expressed as a ratio relative to the basal activity. All assays were performed eight times. For assays with a deleted or mutated hTERT promoter, relative luciferase activity was calculated as a ratio relative to the basal luciferase activity of h-1652, which was used as an internal control in the experiments. All these assays were performed at least three times.
Expression Vector Transfection
MCF7 cells were transfected using Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer, with 3 μg of HA-NFAT1 expression vector (or the corresponding empty vector for the negative control), and with or without 0.6 μg of ΔCAM-AI expression vector. For ChIP assays, cells were harvested after 72 h of culture. For analysis of endogenous hTERT expression, the medium was replaced 24 h after transfection by medium without phenol red and supplemented with 5% fetal calf serum. Cells were then harvested after 30 h of culture.
Jurkat cells were transfected with HA-NFAT1 expression vector or the corresponding empty vector for the negative control using the MicroPorator MP-100 (Labtech, Palaiseau, France) following the manufacturer's recommendations. Cells were harvested after 48 h.
siRNA-induced NFAT1 Inhibition
Jurkat cells were transfected twice with a 24-h interval using an anti-NFAT1 siRNA (anti-NFATc2 ON-TARGETplus SMARTpool siRNA J-003606-07, Dharmacon) or a negative control (ON-TARGETplus Non-targeting siRNA#1 D-001810-01-05, Dharmacon), as described above. 24 h after the second transfection, cells were stimulated with phorbol 12-myristate 13-acetate (50 μg/ml) plus ionomycin (1 μm), for 6 h. The efficiency of phorbol 12-myristate 13-acetate/ionomycin treatment had been controlled on untransfected cells by flow cytometric DNA content analysis and semi-quantitative RT-PCR for IL-2 and c-myc mRNA expression analysis (supplemental Fig. S5).
ChIP
Transfected cells (50 × 106) were washed and scraped off in phosphate-buffered saline. Cell fragmentation was carried out as previously described (46). Nuclear proteins were then cross-linked to DNA by incubation with 1% formaldehyde for 10 min at room temperature. Nuclear DNA was sonicated on a Vibra Cell 72434 (Bioblock Scientific) at 50 watts and 50% amplitude; samples were sonicated six times for 1 min, with a 1-min rest on ice between sonications. A 30-μl fraction of the supernatant was set aside as an “Input” sample, and the remainder was diluted 1:3 in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl (pH 8.1), 167 mm NaCl). The chromatin solution was precleared twice for 30 min by incubation with 40 μl of salmon sperm DNA-protein G-agarose beads (Upstate Biotechnology). Afterward, 5 μg of monoclonal anti-HA (12CA5) antibody (Roche Applied Science) was added, and the mixture was incubated overnight at 4 °C, after which 60 μl of salmon sperm DNA-protein G-agarose beads was added for 1 h. The supernatant (“Unbound” fraction) was collected. Immune complexes were eluted from the beads with 1% SDS and 0.1 m NaHCO3 (“Bound” fraction). Cross-links were reversed by heating samples at 65 °C in 200 mm NaCl. DNA was recovered by proteinase K digestion, phenol extraction, and ethanol precipitation. On the resulting DNA, three sequences were analyzed by PCR to evaluate the binding of NFAT to different responsive elements (REs). The first sequence containing two putative NFAT-binding sites at −1225 and −1200 (named RE 1 + 2) was amplified using p-1271 and p-1153 primers generating a 118-bp fragment (Table 2). The second sequence encompassing a −775 putative responsive element (named RE 3) was amplified using p-829 and p-714 primers generating a 115-bp fragment (Table 2). The third sequence (257 bp) generated by primers p-122 and p+135 (Table 2) contained the −40-binding site (named RE 4) located in the core promoter. To check the specificity of the PCR, a duplex PCR was carried out simultaneously using primers p-122 and p+135 amplifying the third 257-bp sequence and primers specific to a promoter region without any putative NFAT-binding site (p-692 and p-523, Table 2) yielding a 169-bp fragment. The amount of template (70 ng) was optimized by determining the amplification efficiency of the unbound DNA at 25, 70, and 100 ng (supplemental Fig. S6). PCR was conducted for 35 cycles for Input and Unbound samples, and for 45 cycles for Bound samples.
TABLE 2.
Primer sequences for analysis of NFAT-binding sites after ChIP
| Forward primers | |
| p-1271 | 5′-GAGGGTGCGAGGCCTGTTCA-3′ |
| p-829 | 5′-GTTGTGGCTGGTGTGAGC-3′ |
| p-692 | 5′-GTCCTGCCCCTTCACCTT-3′ |
| p-122 | 5′-CCCACGCGTCCGCGCGGACCCCGCCCCGT-3′ |
| Reverse primers | |
| p-1153 | 5′-GCAAACACTGAAATGCTAAC-3′ |
| p-714 | 5′-GCAAACCACCCCAAATCTGT-3′ |
| p-523 | 5′-CAGCGCTGCCTGAAACTC-3′ |
| p+135 | 5′-CCCAGATCTAACGTGGCCAGCGGCAGCACCT-3′ |
Control of Transfection Efficiency
Transfection efficiency of NFAT constructs was evaluated by immunofluorescence, Western blotting as previously described (41), and/or real-time quantitative RT-PCR (supplemental Fig. S7).
RESULTS
In Vitro Modulation of hTERT Expression in Normal, Stimulated Lymphocytes
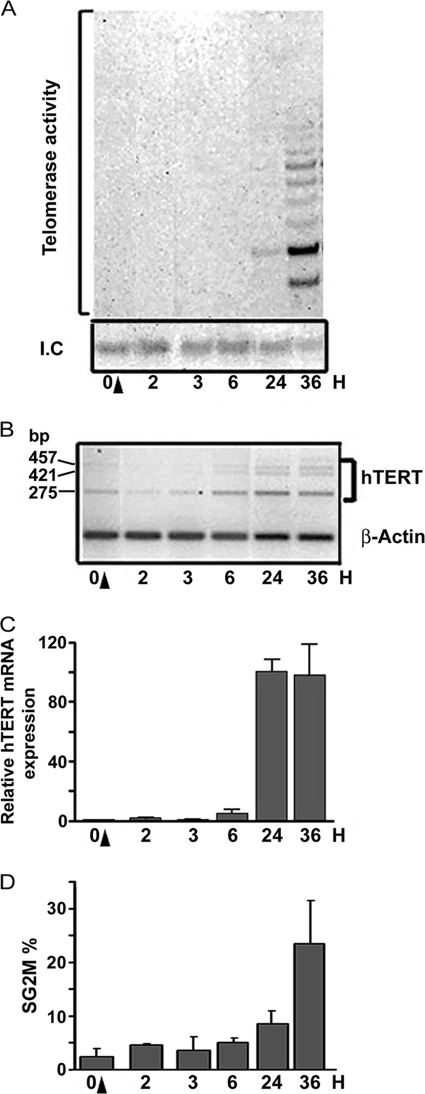
Low levels of telomerase activity induced by stimulation of PBL with PHA were observed after 24 h (Fig. 1A). Induced hTERT mRNA expression could clearly be observed starting 6 h after stimulation (Fig. 1, B and C). This increase in hTERT mRNA occurred with all three alternative transcripts (Fig. 1B). The early activation of the hTERT promoter after lymphocyte activation shown here is in agreement with the results of Liu et al. (10), who described an increase in hTERT expression as early as 12 h after stimulation.
FIGURE 1.
Variation in telomerase activity and hTERT mRNA expression during PBL stimulation. Telomerase activity and hTERT mRNA expression were measured at the indicated times in PBL during the first 36 h of PHA stimulation. A, telomerase activity was assessed by TRAP assay. Representative SYBRTM Green I-stained gel obtained after TRAP assay on lymphocyte protein samples at the indicated times. B, representative semi-quantitative RT-PCR showing the three alternative transcripts of hTERT (457, 421, and 275 bp, respectively) and β-actin. C, hTERT mRNA expression was assessed by real-time quantitative PCR. Histogram summarizing the results from at least three independent experiments (ratio of hTERT expression in stimulated versus unstimulated PBL (0 H), normalized to actin expression). D, lymphocyte proliferation levels after stimulation as evaluated by flow cytometry (I.C., internal control; H, hours; black arrows indicate the time of PHA stimulation).
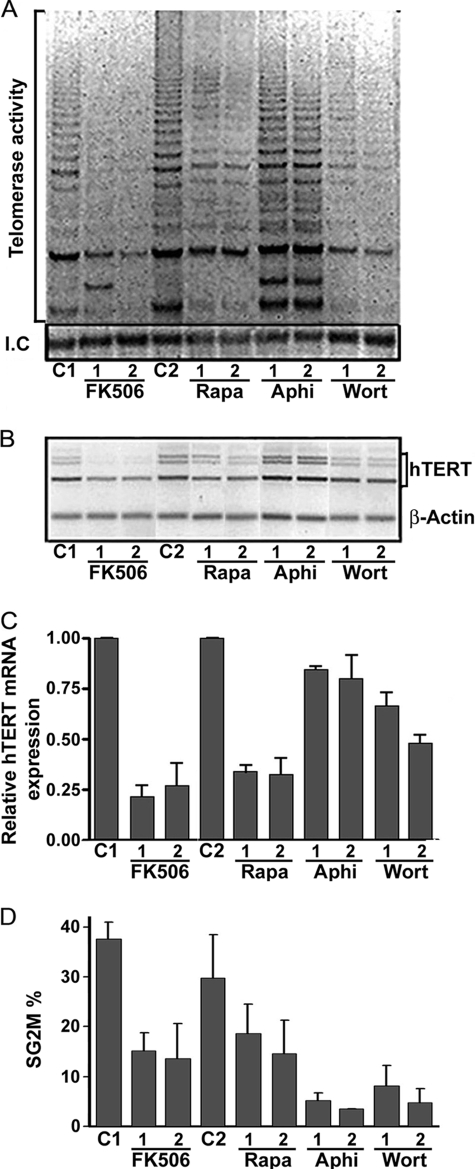
Simultaneous PHA stimulation and treatment with several inhibitors of lymphocyte activation or proliferation modulated the level of telomerase activity and of hTERT mRNA expression to different degrees, providing important new information about hTERT regulation. Aphidicolin, which blocks DNA synthesis, did not cause significant changes in telomerase activity (Fig. 2A) or hTERT mRNA expression (Fig. 2, B and C). The three inhibitors rapamycin (500 and 1000 nm), FK506 (10 and 100 nm), and wortmannin (5 and 10 nm) caused cell cycle arrest in G0/G1 phase (supplemental Fig. S2). Regardless of the dose, wortmannin produced the weakest inhibition of hTERT mRNA expression, although telomerase activity was sharply decreased. This decrease in telomerase activity may be due to the inhibition of Akt, which inhibits hTERT phosphorylation and thus its activity (19). The calcineurin inhibitor FK506 inhibited hTERT expression the most (∼50%) (Fig. 2B), and produced a profound drop in telomerase activity (Fig. 2A). Globally, the decrease in hTERT mRNA expression occurred with all three alternative transcripts of hTERT, as demonstrated in Fig. 2B.
FIGURE 2.
Telomerase activity and hTERT mRNA expression in PBL simultaneously stimulated and treated with different immunosuppressors. Lymphocytes were stimulated with 1 μg/μl PHA and simultaneously treated with one of several inhibitors of cell proliferation for 48 h as indicated. A, telomerase activity is shown in a representative illustration of a SYBRTM Green I-stained gel obtained after TRAP assay on lymphocyte protein samples. B, hTERT mRNA expression is shown in a representative illustration after semi-quantitative RT-PCR. C, hTERT mRNA expression was assessed by real-time quantitative PCR. Histogram summarizes the results from three independent experiments (ratio of hTERT expression in simultaneously treated and stimulated PBL to expression in untreated and stimulated PBL, normalized to actin expression). The strongest inhibitory effect was observed with FK506, while aphidicolin caused minimal inhibition. D, lymphocyte proliferation levels after stimulation or simultaneous stimulation and treatment as evaluated by flow cytometry. (C1, untreated and stimulated PBL used as a control for FK506; C2, PBL with simultaneous addition of PHA and DMSO used as control for rapamycin, aphidicolin, and wortmannin; FK506 1, FK506 at 10 nm; FK506 2, FK506 at 100 nm; Rapa 1, rapamycin at 500 nm; Rapa 2, rapamycin at 1000 nm; Aphi 1, aphidicolin at 4 μg/ml; Aphi 2, aphidicolin at 8 μg/ml; Wort 1, wortmannin at 5 nm; Wort 2, wortmannin at 10 nm.)
NFAT is a transcription factor that plays an important role in lymphocyte activation, a substrate of calcineurin, and a direct or indirect target of all three immunosuppressors used. The rapid induction of hTERT expression after lymphocyte activation and the varying transcriptional effects of the three immunosuppressors led us to wonder whether NFAT might regulate hTERT transcription.
Effects of NFAT1 on hTERT Transcription
NFAT drives gene transcription by targeting GGAAA-binding sites or truncated sites (47). DNA sequence analysis of the hTERT promoter revealed five potential GGAAA-binding sites at positions −1575, −1225, −1200, −775, and −40 relative to the hTERT transcription initiation site (+1 position) (Fig. 3A). To identify whether NFAT binds to one or more of these sites to induce hTERT gene activation, we prepared a series of constructs carrying deletions in the hTERT promoter and containing the firefly luciferase gene as a reporter (Fig. 3B). Telomerase-negative GM847 cells were cotransfected with each of these different constructs and with HA-NFAT1 and/or CN. Analysis of the transcriptional activation of the construct h-1652 was performed in eight independent experiments and showed a significant activation by NFAT1 (3.95 ± 0.63-fold), CN (1.74 ± 0.1-fold), or both (4.28 ± 0.65-fold) (Mann-Whitney test, p = 0.0002, Fig. 3C). Construct h-1399 showed a basal level of transcriptional activation and a pattern of promoter activation similar to h-1652, suggesting a minimal role for the deleted NFAT-binding site at position −1575. The deletion construct h-221 showed the highest basal level of promoter activity (4.8 ± 0.09). Cotransfection of h-221 with HA-NFAT1 alone or together with CN produced an increase in the promoter activity relative to the h-221 basal activity (2.75 ± 0.32 and 3.68 ± 0.15, respectively); this increase was comparable to that observed for h-1652 (Fig. 3C). This high basal level is consistent with previous studies that described this sequence as the core of the hTERT promoter that uncovers two E-boxes required for c-Myc binding and five SP1-binding sites (Fig. 3A) (24, 25). To minimize the role of endogenous c-Myc, the distal E-box was deleted, resulting in construct h-122 (Fig. 3B). Although still high, the basal activity of this promoter construct was lower than that of h-221 (2.45 ± 0.28-fold). NFAT1 overexpression alone or with CN induced the greatest increase (4.18 ± 0.62- and 5.2 ± 0.78-fold, respectively) as compared with the basal activity of h-122 (Fig. 3C), suggesting that the putative NFAT-binding site in this region is important for hTERT promoter regulation.
In contrast, the h-32 construct containing the proximal E-box in position +21 (48) showed a very weak level of basal activity (0.49 ± 0.04-fold). NFAT1 still slightly activates this part of the promoter (1.58 ± 0.24-fold), indicating a potential role for the truncated GGAA-binding site in this sequence (Fig. 3C) and/or an activation by NFAT of c-myc acting through the +21 E-box (49).
Together, these results suggest that the overexpression of NFAT1 triggers hTERT transcription and that CN enhances this effect. Moreover, the putative −40 NFAT1-binding site seems to be particularly important in this process.
Cooperation between NFAT1 and SP1 in Transcriptional Activation of hTERT
To confirm that the effects of NFAT1 are at least partly due to binding at the −40 site, we compared the transcriptional activity of mutated h-122 constructs to the activity of the native h-122 construct in response to overexpression of NFAT1 and CN (Fig. 4A). Mutation of the −40 NFAT-binding site caused a 53% reduction in hTERT promoter transcriptional activity (Fig. 4B). Because SP1-binding sites flank the putative −40 NFAT-binding site, we explored a potential interaction between NFAT1 and SP1 by making site-directed mutations in the h-122 construct (Fig. 4A). Mutation of the upstream (SP1-a), the downstream (SP1-b), or both SP1-binding sites reduced hTERT promoter activity by 35%, 73, and 57%, respectively (Fig. 4B). Simultaneous mutations of the −40 NFAT responsive element together with SP1-a, SP1-b, or both led to a more dramatic decrease in luciferase activity (69%, 100, and 87%, respectively) than single mutations. These results suggest a functional synergy between NFAT1 and SP1.
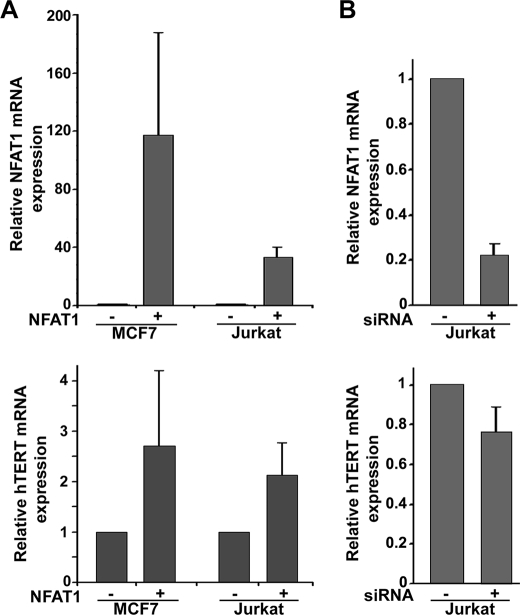
Variations of Endogenous hTERT mRNA Levels According to NFAT1 Expression
We analyzed the effect of HA-NFAT1 overexpression on endogenous hTERT transcription in the MCF7 cell line. To minimize the role of other activators of hTERT transcription, cells were cultured in medium without phenol red, which is known to present an E2-like activity (50) and supplemented with 5% fetal calf serum. We observed an increase in hTERT expression with large variations (Fig. 5A). A similar increase in hTERT expression was observed in Jurkat cells, although NFAT1 transfection efficiency in these cells was lower than in MCF7 cells. This increase of hTERT mRNA expression upon NFAT overexpression is consistent with the variations in hTERT expression induced by the functional modulations of NFAT in PBL either stimulated by PHA or treated with immunosuppressors.
FIGURE 5.
Effects of NFAT1 expression on endogenous hTERT mRNA levels. hTERT mRNA and NFAT1 mRNA expressions were analyzed by real-time quantitative PCR and expressed as a ratio to 18S RNA expression. A, variations of endogenous hTERT mRNA expression when NFAT1 is overexpressed. MCF7 and Jurkat cells were transfected with the HA-NFAT1 expression vector. NFAT1 mRNA expression was assessed to control the efficiency of the transfection. Results are presented as the ratio of hTERT or NFAT1 mRNA expression in HA-NFAT1 transfected cells to their expression in cells transfected with the corresponding empty vector, normalized to RNA 18S expression. Four independent experiments in MCF7 and two in Jurkat cells were performed. B, variations of endogenous hTERT mRNA expression when NFAT1 expression is inhibited. Jurkat cells were transfected with a siRNA anti-NFAT1. Results of three independent experiments are presented as the ratio of hTERT or NFAT1 mRNA expression in siRNA anti-NFAT1 transfected cells to their expression in control cells transfected with a non-targeting siRNA normalized to RNA 18S expression.
We next verified that the silencing of NFAT1 by siRNA induced a decrease in hTERT mRNA expression in the Jurkat cell line (Fig. 5B). The inhibition of hTERT expression occurred when NFAT1 silencing exceeded 70%. An inhibition of NFAT1 expression over 80% decreased hTERT mRNA level by 40%.
Direct Interaction of NFAT1 and hTERT Promoter in Vivo
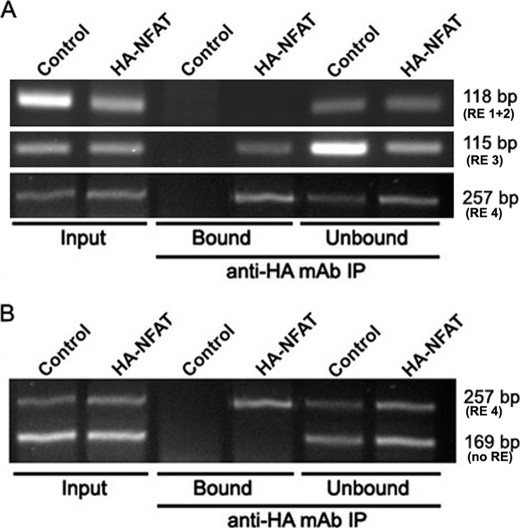
We then tested whether NFAT1 can bind to the hTERT promoter in vivo by overexpressing HA-NFAT1 in MCF7 cells. A ChIP assay was performed using an anti-HA antibody. Promoter sequences that co-precipitated with the HA-tagged NFAT1 were detected by PCR (Fig. 6). For the negative control trial, the experiment was performed with MCF7 cells transfected with the empty expression vectors. The primers encompassing the putative −1225 and −1200 (RE 1 + 2), −775 (RE 3), and −40 (RE 4) NFAT-binding sites allowed bands of the correct size to be amplified from the Input and Unbound fractions of all samples (118, 115, and 257 bp, respectively). We could not observe any amplified band corresponding to the sequence containing the putative −1225 and −1200 responsive elements in the Bound samples. Inversely, bound samples from cells overexpressing HA-NFAT1 gave amplified DNA products of the expected size for the −775 and −40 NFAT-binding sites, whereas Bound samples from the negative control did not (Fig. 6A). Conditions were established to perform duplex PCR that allowed semi-quantitative analysis of two amplicons simultaneously, one containing the −40 NFAT-binding site and the other without any NFAT-binding site (supplemental Fig. S6). As expected, the primers amplifying the hTERT promoter fragment without any NFAT putative consensus site failed to amplify any band in the Bound fractions, but they did yield a band of the expected size in all Input and Unbound fractions (Fig. 6B). These data demonstrated the coprecipitation of HA-NFAT1 and the hTERT promoter sequences containing the putative NFAT-binding sites, thereby confirming that NFAT1 binds to at least two consensus sites.
FIGURE 6.
ChIP of endogenous hTERT promoter. MCF7 cells were cotransfected with expression vectors encoding HA-NFAT1 and a constitutively active form of calcineurin. A ChIP assay was performed with 5 μg of anti-HA monoclonal antibody (12CA5). As a negative control, the same experiment was performed with MCF7 cells cotransfected with the empty version of the HA-NFAT1 expression vector. Immunoprecipitated DNA was analyzed by PCR using hTERT promoter-specific primers that amplify three different sequences containing putative NFAT-binding sites. PCR products were visualized on a 3% agarose gel. A, illustration of DNA PCR amplification sequences obtained in the different ChIP fractions and corresponding to the 118-, 115-, and 257-bp bands, which contain the RE 1 + 2 (−1225 and −1220), RE 3 (−775), or RE 4 (−40) responsive elements, respectively. B, illustration of the duplex PCR amplification in the different ChIP fractions. Immunoprecipitated DNA was analyzed by PCR using hTERT promoter-specific primers that amplify a 257-bp region containing the putative −40 (RE 4) and no others. To verify the specificity of the results, a control was performed using hTERT promoter-specific primers that amplify a 169-bp region lacking any putative NFAT-binding site (no RE). “Input” bands were obtained from DNA purified from chromatin not yet immunoprecipitated, “Bound” corresponds to DNA co-immunoprecipitated with HA-NFAT1 proteins, and “Unbound” to DNA in the supernatant prior to elution.
DISCUSSION
When stimulated, lymphocytes can induce telomerase activity (15). Telomerase activation is linked to the induction of transcription of the hTERT and hTR genes (15) and takes place in the G1 phase of the cell cycle (14, 15). We have shown that hTERT expression is induced very early after PBL stimulation, within the first 6 h. Furthermore, we have observed in PBL that rapamycin, wortmannin, and FK506 inhibited hTERT mRNA expression to varying degrees, with FK506 causing the strongest inhibition. FK506 binds to the membrane receptor FKBP. FK506-FKBP forms a complex with calmodulin and calcineurin subunits A and B, leading to inhibition of calcineurin phosphatase activity (51). This phosphatase activity is responsible for dephosphorylating transcription factors such as NFAT (52).
NFAT proteins comprise a family of transcription factors with at least five members, NFAT1 through NFAT5 (47, 53, 54). The NFAT proteins possess Rel homology regions responsible for binding to DNA at GGAAA consensus sites (47, 55). In quiescent cells, NFAT proteins are phosphorylated in part by the glycogen synthase kinase-3β (GSK-3β) and remain in the cytoplasm and inactive as transcription factors. Stimulation inhibits GSK-3β kinase activity and leads to the mobilization of calcium, which activates calcineurin. This enzyme rapidly dephosphorylates NFAT, which then translocates into the nucleus and acts as a transcription factor (56). The different NFAT family members perform many complex functions (57). NFAT regulates the expression of genes implicated in early activation of human T cells (58, 59), is involved in the activation of the IL-2 promoter (47, 58), and is important for FasL induction, which plays a major role in cell death by apoptosis (55).
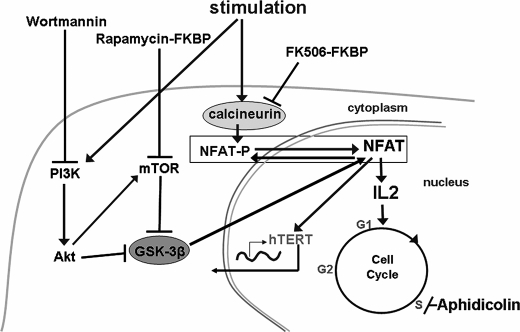
Rapamycin blocks lymphocyte stimulation by inhibiting mTOR (39). It has also been reported to inhibit hTERT expression and phosphorylation in endothelial cancer cells (20). Similarly, wortmannin blocks lymphocyte stimulation and has been shown to inhibit telomerase activity in part by inhibiting Akt via phosphatidylinositol 3-kinase, thereby preventing the phosphorylation of hTERT (17). Both of these inhibitors activate GSK-3β through mTOR and Akt kinase, respectively (60). Thus, all three immunosuppressors, wortmannin, rapamycin, and FK506, may inhibit NFAT's function as a transcription factor, the first two by favoring NFAT phosphorylation by GSK-3β and the last by directly inhibiting NFAT dephosphorylation by calcineurin. This led us to hypothesize that NFAT participates in the transcriptional regulation of hTERT (Fig. 7).
FIGURE 7.
Diagram representing lymphocyte stimulation and inhibition pathways. Schematically, lymphocyte stimulation leads to two different activation pathways. The first pathway consists in the activation of the calcineurin phosphatase. This enzyme rapidly dephosphorylates NFAT, which then translocates into the nucleus and acts as a transcription factor. NFAT regulates the expression of genes implicated in early activation of human T cells, and is involved in the activation of the IL-2 promoter. The second pathway consists in activation of the phosphatidylinositol 3′-kinase/AKT/mTOR pathway. AKT and mTOR activation inhibits glycogen synthase kinase-3β (GSK-3β) activity. GSK-3β ensures the phosphorylation of NFAT, which then remains inactive as transcription factor in cytoplasm of quiescent cells. GSK-3β inhibition by AKT and mTOR allows NFAT dephosphorylation and its nuclear translocation. FK506 binds to the membrane receptor FKBP and FK506-FKBP forms a complex with calmodulin and calcineurin subunits A and B, leading to the inhibition of calcineurin phosphatase activity. NFAT remains phosphorylated and sequestrated in cytoplasm. Rapamycin blocks lymphocyte stimulation by inhibiting mTOR, and wortmannin blocks lymphocyte stimulation by inhibiting AKT. These two immunosuppressors thereby lead to the activation of GSK-3β and induce the inactivation of NFAT by its phosphorylation and nuclear export. Aphidicolin, which blocks DNA synthesis, did not cause changes in telomerase activity. Thus, all three immunosuppressors, FK506, rapamycin, and wortmannin, may inhibit NFAT as a transcription factor, and we showed that all of these compounds provoke the inhibition of hTERT mRNA expression. We could then postulate that NFAT may induce hTERT transcription.
We identified five GGAAA putative NFAT-binding sites on the hTERT promoter at positions −1575, −1225, −1200, −775, and −40 relative to the +1 transcription initiation site. The studies presented here indicate that overexpression of NFAT1 activates the 1787-bp hTERT promoter region transfected in telomerase-negative GM847 cells. This activation is increased by the coexpression of a constitutively active calcineurin. The relatively low basal activity of our constructs h-1652 and h-1399, may be explained by the presence of the inhibitory region located between positions −798 and −400 and containing four already demonstrated MZF-2 inhibitor responsive elements (30). This does not implicate an inhibitory putative effect of NFAT1 through the −775 site. In contrast, the h-221 construct showed a relatively high basal activity. This fragment lacked the inhibitory sequences and included a 59-bp region of the core promoter (−208 to −150 upstream +1 site) that contains an E-box at −187 and five SP1 consensus sites necessary for the promoter's maximal activity (24).
The relative degree of promoter activation by NFAT1 alone or in the presence of calcineurin with the h-221 construct, which contains a −40 NFAT1 consensus site, was identical to that observed with the h-1652 or h-1399 constructs. These results suggest that the putative NFAT1 sites at positions −1575, −1225, −1200, and −775 play only a minimal role in the activation of the hTERT promoter by NFAT1.
The importance of the −40 NFAT-binding site was confirmed by the strong transcriptional activation of the construct h-122 by NFAT. This NFAT consensus site is flanked by two SP1 sites. Cooperation between SP1 and NFAT as transcription factors has already been described in the context of FasL and p21waf1 transcriptional regulation (61, 62). The present study also supports a possible cooperation between SP1 and NFAT in the transcriptional regulation of hTERT expression. Experiments carried out using constructs with mutations in the NFAT and/or one of two SP1 sites indicate that the proximal SP1 site appears to be the most important for this cooperation. The function of the distal site remains to be more closely examined. These results demonstrate the ability of NFAT to activate the hTERT promoter in vitro.
The role of NFAT1 on hTERT transcription in vivo was suggested by the variations of hTERT mRNA expression shown in PBL after stimulation or immunosuppressive treatments. This hypothesis was validated by the increase in endogenous hTERT expression induced by NFAT1 overexpression in MCF7 and Jurkat cell lines, as well as by endogenous hTERT down-regulation after NFAT1 silencing in Jurkat cells.
We show, using ChIP assay, that NFAT1 binds the endogenous hTERT promoter at the −40 and −775 NFAT-binding sites but not at the −1225 and −1200 ones. Several partners were described for NFAT transcriptional activity such as AP-1 (63) and SP1 (61, 62). Furthermore, we have shown a possible cooperation between NFAT and SP1 for the regulation of hTERT expression through the −40 NFAT-binding site. The presence of SP1 responsive elements about 100 bp near the −775 and −40 NFAT1-binding sites (26, 27) might favor its recruitment and link to the promoter sequence.
Altogether our results demonstrate for the first time the direct activation of the hTERT promoter by NFAT1. Furthermore, it has been previously reported that ectopic activation of NFAT in pancreatic cancer cells activates c-Myc, a major activator of hTERT transcription (49). NFAT can thus play both direct and indirect roles in the activation of hTERT transcription. This discovery may allow us to understand more fully the link between lymphocyte activation and hTERT expression and to better comprehend the anti-telomerase effects of immunosuppressors.
Acknowledgment
In particular, we thank Prof. E. Gilson for fruitful suggestions and discussions.
This work was supported by the Ligues Contre le Cancer du Rhône, et de Saône et Loire, by the Region Rhônes Alpes (Contract 00 81 60 45), and by the Cancéropôle Lyon Auvergne Rhône-Alpes.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- hTERT
- human telomerase reverse transcriptase
- mTOR
- mammalian target of rapamycin
- NFAT
- nuclear factor of activated T cells
- PBL
- peripheral blood lymphocyte
- RT
- reverse transcription
- TRAP
- telomeric repeat amplification protocol
- CN
- calcineurin
- HA
- hemagglutinin
- PHA
- phytohemagglutinin
- FKBP
- FK506-binding protein
- ChIP
- chromatin immunoprecipitation
- siRNA
- small interference RNA
- RE
- responsive element
- GSK-3β
- glycogen synthase kinase-3β.
REFERENCES
- 1.Harley C. B., Futcher A. B., Greider C. W. (1990) Nature 345, 458–460 [DOI] [PubMed] [Google Scholar]
- 2.Morin G. B. (1989) Cell 59, 521–529 [DOI] [PubMed] [Google Scholar]
- 3.Blackburn E. H. (1991) Nature 350, 569–573 [DOI] [PubMed] [Google Scholar]
- 4.Hahn W. C., Counter C. M., Lundberg A. S., Beijersbergen R. L., Brooks M. W., Weinberg R. A. (1999) Nature 400, 464–468 [DOI] [PubMed] [Google Scholar]
- 5.Stewart S. A., Hahn W. C., O'Connor B. F., Banner E. N., Lundberg A. S., Modha P., Mizuno H., Brooks M. W., Fleming M., Zimonjic D. B., Popescu N. C., Weinberg R. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12606–12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D., Ho P. L., Coviello G. M., Wright W. E., Weinrich S. L., Shay J. W. (1994) Science 266, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 7.Broccoli D., Young J. W., de Lange T. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9082–9086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Counter C. M., Gupta J., Harley C. B., Leber B., Bacchetti S. (1995) Blood 85, 2315–2320 [PubMed] [Google Scholar]
- 9.Hiyama K., Hirai Y., Kyoizumi S., Akiyama M., Hiyama E., Piatyszek M. A., Shay J. W., Ishioka S., Yamakido M. (1995) J. Immunol. 155, 3711–3715 [PubMed] [Google Scholar]
- 10.Liu K., Schoonmaker M. M., Levine B. L., June C. H., Hodes R. J., Weng N. P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 5147–5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsiao R., Sharma H. W., Ramakrishnan S., Keith E., Narayanan R. (1997) Anticancer Res. 17, 827–832 [PubMed] [Google Scholar]
- 12.Yasumoto S., Kunimura C., Kikuchi K., Tahara H., Ohji H., Yamamoto H., Ide T., Utakoji T. (1996) Oncogene 13, 433–439 [PubMed] [Google Scholar]
- 13.Fujimoto R., Kamata N., Taki M., Yokoyama K., Tomonari M., Nagayama M., Yasumoto S. (2003) Oral Oncol. 39, 445–452 [DOI] [PubMed] [Google Scholar]
- 14.Buchkovich K. J., Greider C. W. (1996) Mol. Biol. Cell 7, 1443–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodnar A. G., Kim N. W., Effros R. B., Chiu C. P. (1996) Exp. Cell Res. 228, 58–64 [DOI] [PubMed] [Google Scholar]
- 16.Greider C. W., Blackburn E. H. (1985) Cell 43, 405–413 [DOI] [PubMed] [Google Scholar]
- 17.Kang S. S., Kwon T., Kwon D. Y., Do S. I. (1999) J. Biol. Chem. 274, 13085–13090 [DOI] [PubMed] [Google Scholar]
- 18.Liu K., Hodes R. J., Weng N. p. (2001) J. Immunol. 166, 4826–4830 [DOI] [PubMed] [Google Scholar]
- 19.Kawauchi K., Ihjima K., Yamada O. (2005) J. Immunol. 174, 5261–5269 [DOI] [PubMed] [Google Scholar]
- 20.Zhou C., Gehrig P. A., Whang Y. E., Boggess J. F. (2003) Mol. Cancer Ther. 2, 789–795 [PubMed] [Google Scholar]
- 21.Shay J. W., Bacchetti S. (1997) Eur. J. Cancer 33, 787–791 [DOI] [PubMed] [Google Scholar]
- 22.Ulaner G. A., Hu J. F., Vu T. H., Giudice L. C., Hoffman A. R. (1998) Cancer Res. 58, 4168–4172 [PubMed] [Google Scholar]
- 23.Meyerson M., Counter C. M., Eaton E. N., Ellisen L. W., Steiner P., Caddle S. D., Ziaugra L., Beijersbergen R. L., Davidoff M. J., Liu Q., Bacchetti S., Haber D. A., Weinberg R. A. (1997) Cell 90, 785–795 [DOI] [PubMed] [Google Scholar]
- 24.Horikawa I., Cable P. L., Afshari C., Barrett J. C. (1999) Cancer Res. 59, 826–830 [PubMed] [Google Scholar]
- 25.Takakura M., Kyo S., Kanaya T., Hirano H., Takeda J., Yutsudo M., Inoue M. (1999) Cancer Res. 59, 551–557 [PubMed] [Google Scholar]
- 26.Kyo S., Takakura M., Taira T., Kanaya T., Itoh H., Yutsudo M., Ariga H., Inoue M. (2000) Nucleic Acids Res. 28, 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kyo S., Takakura M., Kanaya T., Zhuo W., Fujimoto K., Nishio Y., Orimo A., Inoue M. (1999) Cancer Res. 59, 5917–5921 [PubMed] [Google Scholar]
- 28.Oh S., Song Y., Yim J., Kim T. K. (1999) J. Biol. Chem. 274, 37473–37478 [DOI] [PubMed] [Google Scholar]
- 29.Kanaya T., Kyo S., Hamada K., Takakura M., Kitagawa Y., Harada H., Inoue M. (2000) Clin. Cancer Res. 6, 1239–1247 [PubMed] [Google Scholar]
- 30.Fujimoto K., Kyo S., Takakura M., Kanaya T., Kitagawa Y., Itoh H., Takahashi M., Inoue M. (2000) Nucleic Acids Res. 28, 2557–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Günes C., Lichtsteiner S., Vasserot A. P., Englert C. (2000) Cancer Res. 60, 2116–2121 [PubMed] [Google Scholar]
- 32.Xiao X., Athanasiou M., Sidorov I. A., Horikawa I., Cremona G., Blair D., Barret J. C., Dimitrov D. S. (2003) Exp. Mol. Pathol. 75, 238–247 [DOI] [PubMed] [Google Scholar]
- 33.Crowe D. L., Nguyen D. C., Tsang K. J., Kyo S. (2001) Nucleic Acids Res. 29, 2789–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Won J., Yim J., Kim T. K. (2002) FASEB J. 16, 1943–1945 [DOI] [PubMed] [Google Scholar]
- 35.Guilleret I., Benhattar J. (2003) Exp. Cell Res. 289, 326–334 [DOI] [PubMed] [Google Scholar]
- 36.Guilleret I., Benhattar J. (2004) Biochem. Biophys. Res. Commun. 325, 1037–1043 [DOI] [PubMed] [Google Scholar]
- 37.Cong Y. S., Bacchetti S. (2000) J. Biol. Chem. 275, 35665–35668 [DOI] [PubMed] [Google Scholar]
- 38.Takakura M., Kyo S., Sowa Y., Wang Z., Yatabe N., Maida Y., Tanaka M., Inoue M. (2001) Nucleic Acids Res. 29, 3006–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fruman D. A., Klee C. B., Bierer B. E., Burakoff S. J. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 3686–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arcaro A., Wymann M. P. (1993) Biochem. J. 296, 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chebel A., Chien W. W., Gerland L. M., Mekki Y., Bertrand Y., Ffrench P., Galmarini C. M., Ffrench M. (2007) Leuk. Res. 31, 1649–1658 [DOI] [PubMed] [Google Scholar]
- 42.Poncet D., Belleville A., t'kint de Roodenbeke C., Roborel de Climens A., Ben Simon E., Merle-Beral H., Callet-Bauchu E., Salles G., Sabatier L., Delic J., Gilson E. (2008) Blood 111, 2388–2391 [DOI] [PubMed] [Google Scholar]
- 43.Frantz B., Nordby E. C., Bren G., Steffan N., Paya C. V., Kincaid R. L., Tocci M. J., O'Keefe S. J., O'Neill E. A. (1994) EMBO J. 13, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo C., Shaw K. T., Raghavan A., Aramburu J., Garcia-Cozar F., Perrino B. A., Hogan P. G., Rao A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8907–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheridan C. M., Heist E. K., Beals C. R., Crabtree G. R., Gardner P. (2002) J. Biol. Chem. 277, 48664–48676 [DOI] [PubMed] [Google Scholar]
- 46.Urbanowicz-Kachnowicz I., Baghdassarian N., Nakache C., Gracia D., Mekki Y., Bryon P. A., Ffrench M. (1999) Int. J. Cancer 82, 98–104 [DOI] [PubMed] [Google Scholar]
- 47.Rooney J. W., Sun Y. L., Glimcher L. H., Hoey T. (1995) Mol. Cell Biol. 15, 6299–6310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenberg R. A., O'Hagan R. C., Deng H., Xiao Q., Hann S. R., Adams R. R., Lichtsteiner S., Chin L., Morin G. B., DePinho R. A. (1999) Oncogene 18, 1219–1226 [DOI] [PubMed] [Google Scholar]
- 49.Buchholz M., Schatz A., Wagner M., Michl P., Linhart T., Adler G., Gress T. M., Ellenrieder V. (2006) EMBO J. 25, 3714–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welshons W. V., Wolf M. F., Murphy C. S., Jordan V. C. (1988) Mol. Cell Endocrinol. 57, 169–178 [DOI] [PubMed] [Google Scholar]
- 51.McKeon F. (1991) Cell 66, 823–826 [DOI] [PubMed] [Google Scholar]
- 52.Shaw K. T., Ho A. M., Raghavan A., Kim J., Jain J., Park J., Sharma S., Rao A., Hogan P. G. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 11205–11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Northrop J. P., Ho S. N., Chen L., Thomas D. J., Timmerman L. A., Nolan G. P., Admon A., Crabtree G. R. (1994) Nature 369, 497–502 [DOI] [PubMed] [Google Scholar]
- 54.Trama J., Go W. Y., Ho S. N. (2002) J. Immunol. 169, 5477–5488 [DOI] [PubMed] [Google Scholar]
- 55.Holtz-Heppelmann C. J., Algeciras A., Badley A. D., Paya C. V. (1998) J. Biol. Chem. 273, 4416–4423 [DOI] [PubMed] [Google Scholar]
- 56.Loh C., Shaw K. T., Carew J., Viola J. P., Luo C., Perrino B. A., Rao A. (1996) J. Biol. Chem. 271, 10884–10891 [DOI] [PubMed] [Google Scholar]
- 57.Monticelli S., Rao A. (2002) Eur. J. Immunol. 32, 2971–2978 [DOI] [PubMed] [Google Scholar]
- 58.Shaw J. P., Utz P. J., Durand D. B., Toole J. J., Emmel E. A., Crabtree G. R. (1988) Science 241, 202–205 [DOI] [PubMed] [Google Scholar]
- 59.Rao A., Luo C., Hogan P. G. (1997) Annu. Rev. Immunol. 15, 707–747 [DOI] [PubMed] [Google Scholar]
- 60.Dong J., Peng J., Zhang H., Mondesire W. H., Jian W., Mills G. B., Hung M. C., Meric-Bernstam F. (2005) Cancer Res. 65, 1961–1972 [DOI] [PubMed] [Google Scholar]
- 61.Xiao S., Matsui K., Fine A., Zhu B., Marshak-Rothstein A., Widom R. L., Ju S. T. (1999) Eur. J. Immunol. 29, 3456–3465 [DOI] [PubMed] [Google Scholar]
- 62.Santini M. P., Talora C., Seki T., Bolgan L., Dotto G. P. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9575–9580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Macián F., López-Rodríguez C., Rao A. (2001) Oncogene 20, 2476–2489 [DOI] [PubMed] [Google Scholar]