Abstract
Gonadotropin-releasing hormone (GnRH) acts via 7 transmembrane region receptors on gonadotrophs to stimulate synthesis and secretion of the luteinizing hormone and follicle-stimulating hormone. It is secreted in pulses, and its effects depend on pulse frequency, but decoding mechanisms are unknown. Here we have used (nuclear factor of activated T-cells 2 (NFAT2)-emerald fluorescent protein) to monitor GnRH signaling. Increasing [Ca2+]i causes calmodulin/calcineurin-dependent nuclear NFAT translocation, a response involving proteins (calmodulins and NFATs) that decode frequency in other systems. Using live cell imaging, pulsatile GnRH caused dose- and frequency-dependent increases in nuclear NFAT2-emerald fluorescent protein, and at low frequency, translocation simply tracked GnRH exposure (albeit with slower kinetics). At high frequency (30-min intervals), failure to return to basal conditions before repeat stimulation caused integrative tracking, illustrating how the relative dynamics of up- and downstream signals can increase efficiency of GnRH action. Mathematical modeling predicted desensitization of GnRH effects on [Ca2+]i and that desensitization would increase with dose, frequency, and receptor number, but no such desensitization was seen in HeLa and/or LβT2 cells possibly because pulsatile GnRH did not reduce receptor expression (measured by immunofluorescence). GnRH also caused dose- and frequency-dependent activation of αGSU, luteinizing hormone β, and follicle-stimulating hormone β luciferase reporters, effects that were blocked by calcineurin inhibition. Pulsatile GnRH also activated an NFAT-responsive luciferase reporter, but this response was directly related to cumulative pulse duration. This together with the lack of desensitization of translocation responses suggests that NFAT may mediate GnRH action but is not a genuine decoder of GnRH pulse frequency.
Introduction
Gonadotropin-releasing hormone (GnRH)2 acts via seven-transmembrane region receptors to stimulate the synthesis and secretion of luteinizing hormone (LH) and follicle-stimulating hormone (FSH) and thereby mediates control of reproduction. It acts via type I GnRH receptors (GnRHR) to stimulate phospholipase C, activating protein kinases C, and mobilizing Ca2+. Consequent activation of mitogen-activated protein kinase pathways and Ca2+ effectors such as calmodulin mediates its effects on exocytotic gonadotropin secretion as well as its effects on expression of many genes including those for the gonadotropin subunits (1–3). GnRH is secreted in brief pulses, with pulse frequency varying under different physiological conditions. For example, frequency varies over the menstrual cycle with pulses on average every 6 h in mid- to late-luteal phases and every 90 min during follicular and early luteal phases (4). Frequency is higher in rats and mice with physiological pulse intervals of 8–240 min (5). GnRH effects depend upon pulse frequency as illustrated by early studies showing that constant GnRH suppresses LH and FSH secretion, whereas restoration of GnRH pulses restores gonadotropin secretion (6). Similarly, expression of genes encoding rodent LHβ, FSHβ, and the GnRHR are all increased more effectively at low or intermediate GnRH pulse frequency (pulses at 30–120 min) than at high frequency (pulses at 8–30 min) or with sustained stimulation (5, 7–13). Pulsatile stimulation with GnRH agonists is used to stimulate gonadotropin secretion, whereas sustained treatment ultimately reduces gonadotropin secretion, and this underlies agonist efficacy against steroid hormone-dependent cancers (14, 15). Given its physiological and pharmacological relevance, there is much interest in the mechanisms by which gonadotrophs decode GnRH pulse frequency, and this provides a particularly attractive model for exploring pulsatile hormone signaling because of the unique structural and functional characteristics of GnRHR. Type I mammalian GnRHRs lack C-terminal tails, structures that are phosphorylated in many seven-transmembrane receptors to facilitate arrestin binding, arrestin-dependent desensitization, and arrestin-mediated signaling (16–19). Consequently, decoding of pulsatile signals can be explored in this system without the complications of rapid homologous receptor desensitization or G-protein-independent signaling.
Pulsatile signals are used in many biological systems. In the simplest situation a train of brief stimuli elicits a series of corresponding responses in a process known as digital tracking (20). However, where downstream responses have slower inactivation kinetics, responses may not have returned to the basal level before repeat stimulation, and this can cause cumulative (or saw-tooth) responses (5, 20, 21). This process of integrative tracking can amplify signaling but cannot alone explain the bell-shaped frequency-response relationships seen in many systems. These require more complex systems involving positive or negative feedback or feed-forward circuits (21).
GnRHR-mediated activation of the Ca2+/calmodulin pathway can influence gonadotropin subunit gene expression (22–24), and mechanisms by which calmodulins interpret frequency-encoded Ca2+ signals are well established (25–28). More recently, the nuclear factor of activated T-cells (NFAT), a transcription factor activated by Ca2+/calmodulin-dependent activation of the protein phosphatase calcineurin (which dephosphorylates NFAT), has been implicated in transcriptional regulation by GnRH (29–31). This is of particular interest in light of the well established role of NFATs as frequency decoders in other systems (32–35) and the fact that NFATs often generate combinatorial signals with mitogen-activated protein kinases (36, 37), which also control gonadotropin transcription (1–3).
With regard to feedback mechanisms, it is known that type I mammalian GnRH do not desensitize, but agonists stimulate their internalization and thereby reduce cell surface GnRHR number (16–19, 38). GnRH also causes down-regulation of inositol 1,4,5-trisphosphate receptors (39, 40) and increases expression of regulator of G-protein signaling-2 (RGS2) that can inhibit Gq/11 signaling (41, 42). Similarly, GnRH increases expression of a number of dual-specificity phosphatases including DUSP1 and DUSP4 (mitogen-activated protein kinase phosphatases 1 and 2), both of which dephosphorylate (and inactivate) ERKs (43, 44). Any of these processes could generate negative feedback loops reducing Ca2+ signaling or ERK activity, thereby contributing to the frequency dependence of transcriptional regulation. Alternatively, it has been suggested that frequency decoding at the LHβ promoter involves interplay between Egr-1 and a co-regulator (Nab-2). In this model low GnRH pulse frequency causes transient Egr-1 expression, driving expression of Nab-2, which inhibits LHβ expression, but at high pulse frequency there is a more sustained increase in Egr-1, and this quenches Nab-2, increasing LHβ transcription (45). However, it is not clear whether this occurs in vivo (46). Similar interplay between c-Fos and the co-regulator TGIF has been proposed to underlie preferential activation of the FSHβ promoter at low GnRH pulse frequency (47). Here it is important to recognize that GnRH effects on gonadotropin secretion and gonadotropin subunit expression are both frequency-dependent (5, 7–13) and that frequency decoding mechanisms may well differ for these distinct GnRH effects. The upstream feedback mechanisms described above could underlie decoding for both exocytotic and transcriptional responses, whereas altered expression and/or activity of transcription factors could only directly influence transcriptional responses.
The models outlined above raise the fundamental question of whether feedback effects shaping cytoplasmic signals are actually pertinent to GnRH pulse frequency decoding. This in turn raises the concern that few studies have actually monitored GnRH signaling during pulsatile stimuli so that characteristics of signals passing from the cytoplasm to the transcriptome with pulsatile stimulation are largely unknown. Here we have used recombinant adenovirus (Ad) to express GnRHR and signaling reporters in HeLa and LβT2 cells. Using the nuclear translocation of NFAT2-EFP as a live cell readout for Ca2+/calmodulin/calcineurin activation, we have determined how its translocation is related to stimulus amplitude and frequency as well as GnRHR number. We have also used mathematical modeling to predict signaling and luciferase transcriptional reporters to monitor promoter activation in cells receiving pulsatile or sustained GnRH stimulation. We find that integrative tracking occurs with high frequency stimulation, but we find no evidence for desensitization of NFAT2-EFP responses during pulsatile stimulation. GnRH effects on an NFAT-responsive luciferase reporter were also dependent upon cumulative pulse duration, and together these data argue against a major role for negative feedback circuits in shaping frequency-response relationships through the Ca2+/calmodulin/calcineurin pathway.
EXPERIMENTAL PROCEDURES
Engineering of Plasmids and Viruses
Ad-expressing mouse (m) GnRHR were prepared, grown to high titer, and purified as described (16, 48). Ad-NFAT2-EFP (NFATc1-EFP) was from GE Healthcare. Plasmid luciferase reporters with promoters from human αGSU (−517/+44), rat FSHβ (−2000/+698), and rat LHβ (−791/+5) have been described (49, 50). The NFAT response element (NFAT-RE) luciferase reporter (containing three repeats of a 30-bp fragment of the human interleukin-2 promoter) was from Addgene (Cambridge, MA; plasmid 10959).
Ad-expressing luciferase reporters were made as follows. First, promoter regions were subcloned into pAd5-Luc2, an Ad shuttle vector containing a luciferase open reading frame downstream of a multiple cloning site (48), using HindIII for αGSU, BamHI/HindIII for LHβ, and KpnI/HindIII for NFAT-RE. Standard PCR-based techniques were used to amplify the promoter (−2000/+698) from FSHβ-Luc plasmid using the following primers: forward, ATG GTA CCG ATT GGT GGT TGA TCT CCC CAT GTT CC; reverse, CTT ATA AGC TTT GGC CAG GTA AGT CAA CAG C. PCR products were digested with KpnI/HindIII and subcloned into pAd5-Luc2. A blue fluorescent protein (BFP) nuclear marker was generated by the addition of three repeats of the nuclear localization sequence (NLS) from the simian virus 40 large T-antigen to the C terminus of BFP by PCR using the following primers with pRSET-BFP (Invitrogen): forward, CGA TGG GGA TCC GAA TTC GC; reverse, ACT CTA GAC CTC TAC CTT TCT CTT CTT TTT TGG ATC TAC CTT TCT CTT CTT TTT TGG ATC TAC CTT TCT CTT CTT TTT TGG ATC GGC TCG AGC CTT GTA CAG CTC GTC CAT GC. To generate Ad expressing NLS-BFP, PCR products were digested with EcoRI/XbaI and subcloned into pacAd5 CMV K-N pA (Gene Transfer Vector Core, University of Iowa, Iowa City, IA) shuttle vector. Shuttle vectors were verified by restriction digest analysis or DNA sequencing (Geneservice Ltd., Cambridge, UK). Ads were made from shuttle vectors as described (16, 48, 51) using NheI to cut the FSHβ shuttle vector and PacI for all others. Ad vectors were then grown to high titer and purified (16, 48). Recombinant Ad were named after their plasmid counterparts, i.e. Ad-αGSU-Luc, Ad-FSHβ-Luc, Ad-LHβ-Luc, Ad-NFAT-RE-Luc, and Ad-NLS-BFP.
Cell Culture, Transfection, and Transduction
HeLa cells were cultured in 10% fetal calf serum-supplemented DMEM. LβT2 cells were kindly provided by Prof. P Mellon (University of California, San Diego, CA) and cultured as described (52). For imaging, cells were cultured in Costar black-wall 96-well plates (Corning, Arlington, UK). They were transduced in DMEM containing 2% fetal calf serum. Ad-NFAT2-EFP was used at 2 pfu/nl, Ad-NLS-BFP was used at 75 pfu/nl, and Ad mGnRHR was used at 3 pfu/nl, except where indicated. For luciferase assays, Ad vectors were used at 1 pfu/nl. The Ad-containing medium was removed after 4–6 h and replaced with fresh DMEM with 0.1% fetal calf serum. The cells were then cultured for 16–24 h before GnRH stimulation. For plasmid transfections, cells were treated with Superfect (Qiagen, Crawley, UK) using 0.5 μg of DNA/well for 2 h before Ad transduction.
Semi-automated Image Acquisition and Analysis
Most imaging experiments were performed using an IN Cell Analyzer 1000 (GE Healthcare) high content imaging platform. Cells cultured in 96-well plates were transduced with Ad vectors as above. Images were acquired with a single field of view (0.6 mm2) and a 10× objective. Experiments were performed in duplicate or triplicate wells, and each field typically contained 300–500 cells.
Cell surface HA-mGnRHR expression was determined by immunostaining with monoclonal anti-HA, Alexa Fluor 488-conjugated goat anti-mouse antibody, and 4′,6-diamidino-2-phenylindole (400 nm) nuclear stain (38, 52). Images were acquired then analyzed with IN Cell Investigator software (Workstation 3.5, GE Healthcare) using a Dual Area Object Analysis algorithm and a filter to define the proportion of positively stained cells (cell surface HA-mGnRHR staining >10% above background). A cell surface expression index was then calculated by multiplying the proportion of stained cells (%+ve cells) by their mean fluorescence intensity (AFU).
For NFAT2-EFP assays, cells were stimulated as indicated, then fixed with 4% paraformaldehyde, permeabilized with methanol (−20 °C), and stained with 4′,6-diamidino-2-phenylindole. For live cell imaging, cells were plated as above using 12–14 wells in duplicate. Medium was replaced 25 min before imaging with phenol red-free DMEM-F-12 (with 100 μg/ml BSA and 10 μg/ml apotransferrin) and, if Ad-NLS-BFP was not included, contained 400 nm Hoechst nuclear stain (GE Healthcare). For imaging, cells were incubated in an environmental control chamber at 37 °C in a 5% CO2-humidified atmosphere. Images were acquired at the indicated time points, taking 80 s to capture 12 wells. Stimulation and washing (of individual wells) were staggered by 5–10 s to compensate for any delay in image acquisition. Cells were stimulated with GnRH either continuously or for 5 min (agonist removal by washing ×5 in DMEM- F-12). In some experiments, cells were subjected to repeat stimulation with GnRH (5 min stimulation, removal by washing as above) at the indicated frequency.
For measurement of [Ca2+]i using fluo-4, cells were incubated for 30 min at 37 °C in physiological salt solution (127 mm NaCl, 1.8 mm CaCl2, 5 mm KCl, 2 mm MgCl2, 0.5 nm NaH2PO4, 5 mm NaHCO3, 10 mm glucose, 0.1% bovine serum albumin, and 10 mm HEPES, pH 7.4) with 5 μm fluo-4/AM (Invitrogen). After dye loading, cells were washed (×2) and incubated in physiological salt solution with 400 nm Hoechst nuclear stain for 25 min. They were then imaged and stimulated with GnRH as described in the figure legends.
Image analysis and quantification of fluorescence intensity and localization were performed using IN Cell Analyzer Work station 3.5 software (IN Cell Investigator, GE Healthcare). Green channel (fluo-4, EFP, Alexa 488) and blue channel (BFP, Hoechst, 4′,6-diamidino-2-phenylindole) images were used to define whole-cell and nuclear regions, respectively. These fluorophores were used for calculation of population-averaged fluorescence intensities (with background subtracted) and ratios of nuclear to cytoplasmic fluorescence intensity (N:C).
Video Imaging of Cytosolic Ca2+ Using Fura-2
Measurement of [Ca2+]i by video imaging of fura-2-loaded cells was performed as described (53, 54). Cells were stimulated with GnRH as detailed in figure legends, and where indicated, GnRH was removed by washing (×5) with physiological salt solution.
Luciferase Assays
Cells were plated and transfected or transduced with luciferase reporters as above. After treatment as detailed in the figure legends, cells were washed in ice-cold phosphate-buffered saline and lysed, and luciferase activity was determined as described (43, 48, 55). Data are reported as relative light units normalized as -fold change over control, except where indicated.
Statistical Analysis, Data Presentation, and Mathematical Modeling
The figures show the mean ± S.E. of data pooled from at least three experiments. Data were normalized as described in the figure legends. Statistical analysis was by one or two-way ANOVA and post hoc tests (as detailed in figure legends) accepting p < 0.05 as statistically significant. Statistical analysis, curve fitting, and regression were performed using GraphPad Prism 4.0 (GraphPad Software Inc.). A published mathematical model was employed to predict cellular responses to GnRH by simultaneous solution of differential equations describing various aspects of the GnRH signaling to LH secretion (56). We solved these nonlinear equations numerically using XPP software to predict likely relationships between GnRH stimulation paradigm and responses as described in the supplemental data.
RESULTS
Image-based Assays for GnRH Signaling
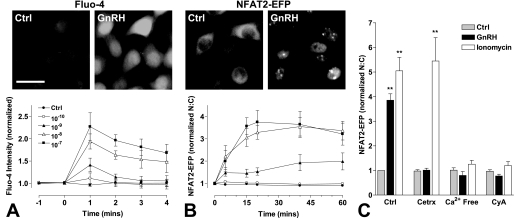
GnRH acts via Gq/11-coupled seven-transmembrane receptors, causing a dose-dependent increase in the cytoplasmic [Ca2+]i concentration in many models, including HeLa cells (54). Here, we assessed GnRH effects on [Ca2+]i using fluo-4 as a Ca2+ sensor (Fig. 1A) and found that GnRH caused a clear dose-dependent (pEC50 8.5 ± 0.4) and time-dependent (maximal at 1 min and then reduced over the next 3 min) increase in [Ca2+]i. We also assessed the ability of GnRH to alter the cellular distribution of NFAT, a Ca2+-sensitive transcription factor that translocates to the nucleus when [Ca2+]i is elevated in many systems. This reflects Ca2+/calmodulin-dependent activation of calcineurin, which in turn dephosphorylates, NFATs causing their translocation to the nucleus (36). When HeLa cells were transduced with NFAT2-EFP, the reporter was largely cytoplasmic in unstimulated cells, but GnRH caused a pronounced translocation to the nucleus (Fig. 1B and supplemental Fig. 1). We quantified this effect using an automated imaging system to define NFAT2-EFP fluorescence intensity in the nucleus and cytoplasm and the N:C NFAT2-EFP ratio for each individual cell (supplemental Fig. 1). As shown (Fig. 1B), GnRH caused an increase in NFAT2-EFP N:C ratio that was dose-dependent (pEC50 8.4 ± 0.1), slow in onset (maximal at 20–40 min for the highest doses), and sustained (for at least 1 h). This effect was blocked by pretreatment with a GnRH antagonist (cetrorelix, 10−7 m) or by the calcineurin inhibitor cyclosporin A (10−6 m), and GnRH failed to cause NFAT2-EFP translocation in Ca2+-free medium (Fig. 1C). The Ca2+ ionophore ionomycin (10−5 m) also caused NFAT2-EFP translocation to the nucleus, and this effect was also blocked by Ca2+-free medium or cyclosporin A but was not inhibited by cetrorelix. Accordingly, NFAT2-EFP translocation provides a readout for GnRHR-mediated Ca2+ mobilization and consequent calcineurin activation but has slower onset and offset than GnRH effects on [Ca2+]i. We calculated signal:noise ratios which revealed NFAT2-EFP translocation as a relatively robust readout, whereas this ratio was much lower with the fluo-4 assays (not shown). We also observed a time-dependent reduction in whole cell fluo-4 fluorescence (even in control cells) that was suggestive of dye washout and prevented longer experiments (not shown).
FIGURE 1.
Image-based readouts of GnRH-mediated calcium mobilization. HeLa cells were transduced with Ad-mGnRHR alone (panel A) or together with Ad-NFAT2-EFP (panels B and C). Panels A and B, cells were treated with fluo-4/AM (panel A only) and Hoechst stain before GnRH stimulation (from 0 min) at the indicated concentrations. Panel C, cells were pretreated with cetrorelix (Cetrx; 10−6 m, 5 min), cyclosporin A (CyA; 10−6 m, 15 min), or media replaced with Ca2+-free physiological salt solution (5 min) before stimulation with GnRH (10−7 m) or ionomycin (10−5 m) for 20 min. Cells were then washed with ice-cold phosphate-buffered saline, fixed with 4% paraformaldehyde, permeabilized, and stained with 4′,6-diamidino-2-phenylindole. Image acquisition with live (panels A and B) or fixed (C) cells and analysis were performed as described under “Experimental Procedures”. Representative images are shown before and after treatment with GnRH (10−7 m, peak response shown) acquired in the green (fluo-4, EFP) image channel. Scale bar, 30 μm. Data are normalized as the -fold change over control (at 0 or 20 min). Results shown are the mean ± S.E. of 3–4 independent experiments. Statistical analysis is by one-way ANOVA using the Bonferroni multiple comparison test, untreated control versus agonist treated; **, p < 0.01.
Effects of Brief or Continuous GnRH Treatment
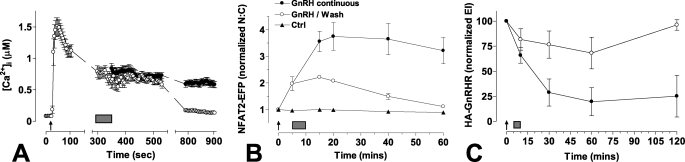
In the next experiments we compared the effects of brief (5 min) and sustained (15–120 min) stimulation with GnRH. Here, the key issue is whether the responses observed are simply triggered by the initial GnRHR activation or are dependent upon ongoing GnRHR activation. We loaded cells with the Ca2+-sensitive dye fura-2 and used a previously described (53, 54) ratiometric imaging system to monitor the effects on [Ca2+]i. As shown (Fig. 2A) GnRH caused a rapid increase in [Ca2+]i with a maximum response at ∼30 s and subsequent reduction to a plateau level that declined gradually from ∼5–15 min but remained elevated (∼10-fold above basal) at 15 min. In contrast, when cells were washed during the plateau phase (to remove GnRH), the [Ca2+]i declined to near basal values at 15 min. Accordingly, the wash was effective at removing GnRH, and as expected, the [Ca2+]i elevation was dependent on ongoing receptor activation. We also used a published mathematical model to estimate signaling parameters under these conditions. The model predicts a rapid increase in GnRHR occupancy, activation of an immediate effector (i.e. phospholipase C), inositol 1,4,5-trisphosphate concentration and [Ca2+]i, with maximal responses occurring within 2 min of stimulation (supplemental Fig. 3). With GnRH removal, after 5 min the model predicts rapid reversal of each of these parameters (supplemental Fig. 3). Consistent with published data (56), the model predicts that [Ca2+]i increases to a peak within 1–2 min of stimulation then reduces to a plateau that is sustained with continuous stimulation and rapidly reversed when GnRH is removed. Thus, the model accurately predicts characteristics of the [Ca2+]i response seen when mGnRHR are activated in HeLa cells (compare Fig. 2A and supplemental Fig. 3D).
FIGURE 2.
Effect of brief or sustained treatment on GnRH-mediated responses. Panel A, HeLa cells were transduced with Ad-mGnRHR and loaded with fura-2/AM before imaging. GnRH (10−7 m) was applied at the indicated time point (arrow) either continuously or for 5 min followed by repeated wash (the wash period indicated by a gray rectangle). Dynamic video imaging and measurement of [Ca2+]i was performed as described under “Experimental Procedures.” Breaks in the lines represent periods during which images were not collected. Panel B, cells were transduced with Ad-mGnRHR and Ad-NFAT2-EFP and stained with Hoechst nuclear stain before imaging. GnRH (10−7 m) was applied at the indicated time point either continuously or for 5 min followed by repeated wash (gray rectangle). Live cell image acquisition and analysis was performed as described under “Experimental Procedures.” Data shown are the N:C ratio of NFAT2-EFP fluorescence intensity (background-subtracted), normalized as the -fold change over control. Panel C, cells were transduced with HA-mGnRHR before stimulation with 10−7 m GnRH either continuously or for 5 min followed by repeated wash (gray rectangle). Cells were treated for the indicated time periods, washed with ice-cold phosphate-buffered saline, and then stained for cell surface HA-mGnRHR expression (initially as live intact cells) before image acquisition and analysis. The data shown are cell surface receptor expression in an expression index as defined under “Experimental Procedures.” All results shown are the mean ± S.E. of three independent experiments. Statistical analysis by two-way ANOVA revealed that treatment type (brief versus sustained) is a significant source of variation for all assays (p < 0.01). Post hoc analysis using the Bonferroni test revealed a statistically significant reduction in cell surface expression (p < 0.01) by 30 min of sustained GnRH treatment (panel C).
Brief (5 min) GnRH stimulation also caused NFAT2-EFP translocation to the nucleus, although the maximal response was reduced (by ∼60%) compared with sustained stimulation (Fig. 2B). Removal of GnRH after 5 min also reduced the NFAT2-EFP N:C ratio, although with markedly slower kinetics than the [Ca2+]i response. Thus, ongoing receptor activation is required for the sustained phase of the NFAT2-EFP response despite the slower reversal with this reporter compared with fura-2.
We also investigated the effects of brief and sustained GnRH treatment on cell surface receptor expression using an established method (38, 52) for quantification of GnRHR with an N-terminal HA tag (Fig. 2C). Continuous treatment reduced cell surface HA-mGnRHR expression to ∼25% of control. This effect was measurable at 30 min and maintained for 120 min. In contrast, when GnRH was removed after 5 min, it had no measurable effect on cell surface HA-mGnRHR expression (over the entire 120-min experiment).
Effects of Pulsatile GnRH
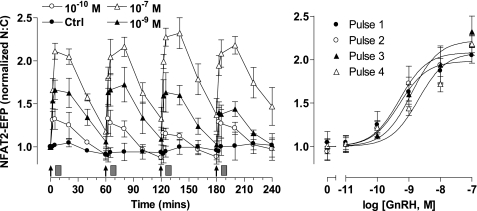
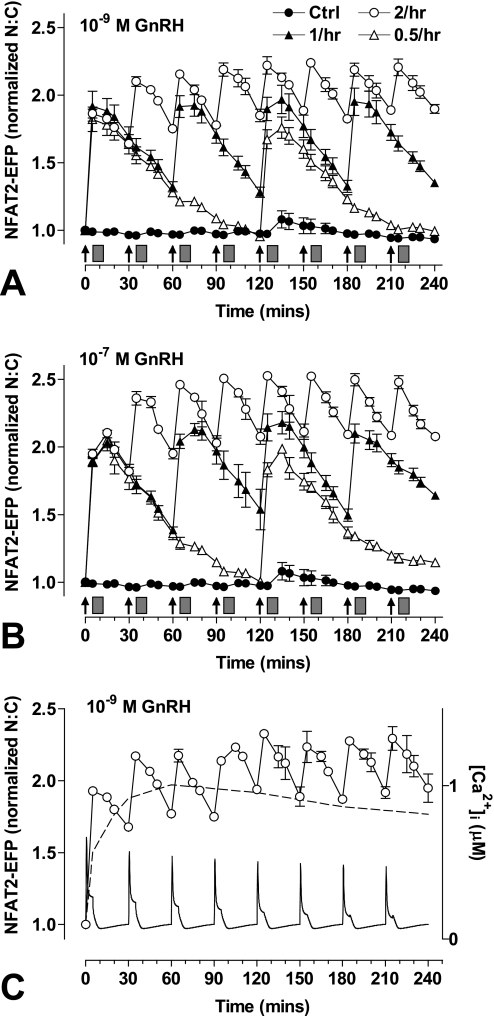
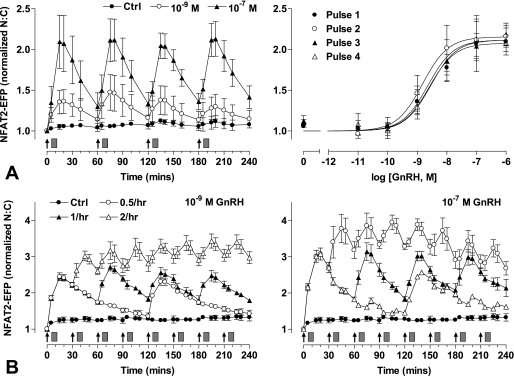
Modeling effector activation with 5 min of stimuli at varied dose and frequency predicted reproducible transient responses at low GnRH concentration and/or frequency (supplemental Fig. 4), but responses were predicted to desensitize as concentration and/or frequency were increased, approaching the marked desensitization seen with constant stimulation at 10−9 or 10−7 m GnRH. We next tested for dose- and frequency-dependent desensitization by monitoring NFAT2-EFP translocation in live cells receiving a train of 4 GnRH stimuli of 5 min duration and at 1-h intervals. As expected, GnRH caused a dose-dependent translocation of NFAT2-EFP to the nucleus (pEC50 8.7 ± 0.2 to 9.3 ± 0.2). These responses had comparable kinetics at all doses, increasing to maxima at 5–15 min and reducing toward basal values over the next 45 min. Repeat stimulation with GnRH resulted in recurrent NFAT2-EFP translocation, with each pulse of GnRH causing a corresponding translocation response (Fig. 3). No desensitization was seen, as ANOVA revealed the GnRH dose (p < 0.001) but not pulse number or pulse-dose interaction as a significant source of variation. We also explored frequency dependence by applying 5-min pulses of 10−7 or 10−9 m GnRH at 0.5-, 1-, or 2-h intervals (Fig. 4). As before, repeat stimulation caused recurrent NFAT2-EFP translocation with no evidence of desensitization. However, with the 30-min pulse frequency NFAT2-EFP failed to return to baseline between pulses so that a cumulative effect was seen in which maximal N:C seen with the first GnRH pulse were lower than those with subsequent pulses. We also performed similar experiments, activating the endogenous mGnRHR of NFAT2-EFP-expressing LβT2 cells, and similar results were obtained. Using 5 min of stimuli at 1-h intervals, GnRH caused a clear dose-dependent (pEC50 8.5 ± 0.2 to 8.8 ± 0.2) translocation of NFAT2-EFP with responses maximal at 5–15 min and reducing toward basal thereafter (Fig. 5). The responses were reproducible with repeat stimulation, and there was no evidence of desensitization during a train of four GnRH pulses. As with HeLa cells, repeat stimulation at 1- or 2-h intervals caused recurrent NFAT2-EFP translocation responses, but with the 30-min pulse frequency the NFAT2-EFP N:C did not return to baseline between pulses, and cumulative effects were seen (maximal N:C seen with the first GnRH pulses were lower than most subsequent pulses).
FIGURE 3.
Live cell imaging of NFAT2-EFP translocation during pulsatile GnRH treatment. Cells were transduced with Ad-mGnRHR and Ad-NFAT2-EFP and stained with Hoechst nuclear stain before imaging. GnRH was applied at the indicated concentrations at 0, 60, 120, and 180 min for 5 min followed by repeated wash steps as indicated (gray rectangles). Live cell image acquisition and analysis was performed as described under “Experimental Procedures.” The data shown are the N:C ratio of NFAT2-EFP fluorescence intensity (background-subtracted), normalized as the -fold change over control (at 0 min). The right panels show data 20 min after each GnRH addition, and curve-fitting revealed pEC50 values in the range of 8.7 ± 0.2 to 9.3 ± 0.2. The results shown are the mean ± S.E. of three independent experiments, performed in duplicate wells. Two-way ANOVA revealed that GnRH concentration is a significant source of variation (p < 0.001, F5,41 = 85.7), whereas pulse number was not (p = 0.926, F3,41 = 0.15).
FIGURE 4.
Live cell imaging with varied GnRH pulse frequency. Cells were transduced with Ad-mGnRHR, Ad-NLS-BFP, and Ad-NFAT2-EFP before imaging. Cells were treated with 10−7 or 10−9 m GnRH either continuously (panel C only) or for 5 min at 30-min intervals, 1-h intervals, or every 2 h as indicated. All wells were subject to 0.5-h washes (gray rectangles) 5 min after GnRH or control addition. Live cell image acquisition and analysis was performed as described under “Experimental Procedures.” An established mathematical model for GnRH signaling (56) was used to estimate [Ca2+]i in cells stimulated with 10−9 m GnRH for 5 min at 0.5-h intervals (panel C). For this analysis the initial receptor concentration was set at 10−11 m, and computation was performed as described under “Experimental Procedures.” The data shown are the N:C ratio of NFAT2-EFP fluorescence intensity (background subtracted), normalized as the -fold change over control (at 0 min). Results shown are the mean ± S.E. of three independent experiments or one representative experiment (panel C). Statistical analysis using repeated measures ANOVA and the Bonferroni multiple comparison test revealed significant differences between the first peak (at 5 or 15 min) and subsequent peak responses for 0.5-h GnRH treatment (p < 0.01 for 10−9 m GnRH, p < 0.001 for 10−7 m GnRH; panels A and B, respectively).
FIGURE 5.
Live cell imaging of GnRH-mediated NFAT2-EFP translocation in the gonadotroph-derived LβT2 cell line. LβT2 cells were transduced with Ad-NLS-BFP and Ad-NFAT2-EFP before imaging. Panel A, cells were treated with the indicated concentrations of GnRH at 0, 60, 120, and 180 min for 5 min followed by repeated wash steps, as indicated (gray rectangles). The right panel (A) shows data 20 min after each GnRH addition. Panel B, cells were treated (for 5 min) with 10−9 or 10−7 m GnRH at 30-min intervals, 1-h intervals, or every 2 h as indicated. All wells were subject to 0.5-h washes (gray rectangles) 5 min after GnRH or control addition. The data shown are the N:C ratio of NFAT2-EFP fluorescence intensity (background-subtracted), normalized as the -fold change over control (at 0 min). Results shown are the mean ± S.E. of three independent experiments, performed in duplicate wells. For panel A, curve fitting revealed pEC50 values in the range of 8.5 ± 0.2 to 8.8 ± 0.2. Two-way ANOVA revealed GnRH concentration as a significant source of variation (p < 0.001, F6,56 = 39.5), whereas pulse number was not (p = 0.936, F3,56 = 0.14). For panel B, statistical analysis using repeated measures of ANOVA and Bonferroni multiple comparison test revealed significant differences between the first peak (at 15 min) and subsequent peak responses for 0.5-h GnRH treatment (p < 0.001 for 10−9 m GnRH, p < 0.01 for peaks up to 135 min with 10−7 m GnRH).
Single Cell Analysis
The data above were derived from average responses observed in large populations of cells, but the assays used also provide measures on each individual cell facilitating single cell analysis. Having established that GnRH causes a dose-dependent increase in N:C NFAT2-EFP ratio (population analysis, Fig. 1), we sought to determine whether this response is switch-like (all or nothing) or graded in individual cells. Frequency distribution plots (supplemental Fig. 2A) revealed that the N:C NFAT2-EFP ratio was <1.1 for >98.4% of control cells and >1.1 for >95.6% of GnRH (10−7 m)-stimulated cells. Using this as a cutoff to define activated cells, we found that GnRH caused a dose-dependent increase in the proportion of cells in which NFAT2-EFP was activated and also caused a dose-dependent increase in the N:C ratio within these activated cells (supplemental Fig. 2B). Thus, GnRH increases the N:C NFAT2-EFP ratio in cell populations (Fig. 1) by increasing the proportion of cells in which NFAT2-EFP is activated and the extent of its activation in these cells (supplemental Figs. 2, A and B).
The frequency distribution analysis also revealed a wide range of responses to GnRH (i.e. N:C varying from 0.9 to >2 in GnRH-stimulated cells), raising the question of whether response magnitude is characteristic for a single cell. To test this we compared the effects of GnRH on N:C NFAT2-EFP ratio for individual cells receiving repeated pulses of GnRH. This revealed a strong correlation between amplitude of NFAT2-EFP translocation responses in the first and second stimulus with GnRH (supplemental Fig. 2C). Thus, although the response is extremely variable from cell to cell, there is considerable reproducibility over time within individual cells. Similar variability from cell to cell but reproducibility from one stimulus to the next within an individual cell is seen when fura-2 is used to monitor [Ca2+]i in these cells and in gonadotroph cell lines (not shown).
Relevance of GnRH Receptor Number
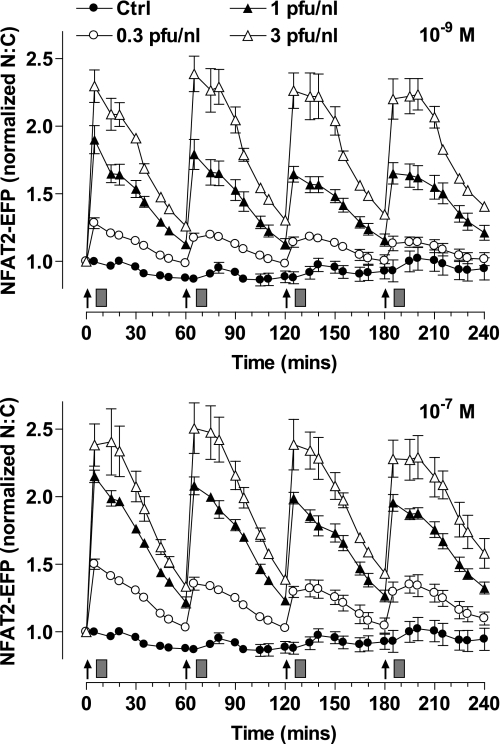
GnRHR number varies under physiological conditions (i.e. through estrus and puberty), and GnRH effects are dependent upon receptor number in many models (50, 57). To simulate this mathematically, we set GnRHR concentration at 10−11 m (the value used above) or at 0.5 or 2 × 10−11 m and solved the published model (56) to predict effector activation and [Ca2+]i responses using 5-min pulses with 10−7 m GnRH at hourly intervals. This predicted little desensitization of the [Ca2+]i response at low receptor concentration but a pronounced desensitization at the highest receptor concentration (supplemental Fig. 5). Accordingly, we sought to determine the effects of GnRHR number on the NFAT2-EFP translocation response. To do so HeLa cells were transduced with Ad mGnRHR at 0.3, 1, or 3 pfu/nl. In radioligand binding experiments (not shown) these titers gave cell surface expression of ∼40,000, 80,000, and 160,000 sites/cell and were selected to give low physiological, high physiological, and super-physiological levels of expression as compared with the physiological range (∼20,000–85,000 sites per cell) in rat gonadotroph and gonadotroph-derived cell lines (57, 58). As shown (Fig. 6), the magnitude of the NFAT2-EFP translocation response was dependent upon GnRHR number with the greatest response at the highest Ad titer. However, the kinetics of the responses were comparable at all titers (increasing to maxima at 5–15 min of stimulation and reducing toward basal over the following 45 min), and there was no evidence for signal integration or desensitization with either dose of GnRH (10−9 and 10−7 m GnRH). Accordingly, in this model receptor number influences the NFAT2-EFP translocation response to hourly GnRH pulses quantitatively but not qualitatively.
FIGURE 6.
Influence of GnRH receptor number on NFAT2-EFP translocation responses to pulsatile stimulation. Cells were transduced with Ad-NFAT2-EFP and Ad-NLS-BFP and 0.3, 1, or 3 pfu/nl of Ad-mGnRHR. Cells were treated with the indicated concentrations of GnRH at 0, 60, 120, and 180 min for 5 min followed by repeated washing as indicated (gray rectangles). Live cell image acquisition and analysis was performed as described under “Experimental Procedures.” The data shown are the N:C ratio of NFAT2-EFP fluorescence intensity (background-subtracted), normalized as the -fold change over control (at 0 min). Results shown are the mean ± S.E. of three independent experiments performed in duplicate wells.
Effects of Sustained and Pulsatile GnRH on Transcription Reporters
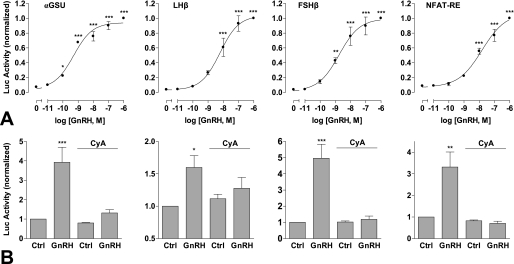
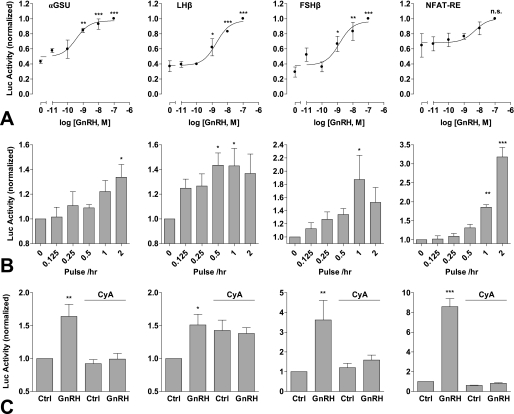
Gonadotropin gene expression is differentially regulated by GnRH pulse frequency and amplitude (5). Therefore, in a further series of experiments we sought to determine transcriptional effects of pulsatile GnRH using αGSU, FSHβ, and LHβ luciferase reporters. Continuous treatment with GnRH for 8 h resulted in dose-dependent increases in αGSU, FSHβ, and LHβ luciferase activity, with pEC50 values of 9.3 ± 0.2, 8.8 ± 0.2, and 8.2 ± 0.2, respectively (Fig. 7A). Pulsatile treatment (5 min hourly) also elicited dose-dependent increases in luciferase activity, with comparable potencies (pEC50 values of 9.5 ± 0.3, 8.9 ± 0.3, 8.7 ± 0.2, respectively) (Fig. 8A). Basal luciferase activity was noticeably greater in cells undergoing pulsatile treatment than continuous (∼40% of the maximum response compared with <10%), most likely as a result of mechanical stimulation during the repeated wash steps. To investigate frequency response relationships, cells were stimulated with 10−9 m GnRH for 5 min at 0.5-, 1-, and 2-h intervals (Fig. 8B). Maximal αGSU reporter activity was observed at the highest frequency (0.5-h stimulation). In contrast, maximal LHβ and FSHβ responses were seen at slower pulse frequencies (2 and 1 h, respectively).
FIGURE 7.
Transcriptional responses to sustained GnRH treatment. Cells were transfected with αGSU-Luc, LHβ-Luc, FSHβ-Luc, or NFAT-RE-Luc plasmids and transduced with Ad-mGnRHR (panel A) or transduced with Ad-αGSU-Luc, Ad-LHβ-Luc, Ad-FSHβ-Luc, or Ad-NFAT-RE-Luc and Ad-mGnRHR (panel B). Cells were treated with the indicated concentrations of GnRH (panel A) or with 10−7 m GnRH (panel B) for 8 h continuously. Where indicated (panel B), cells were pretreated with 10−5 m cyclosporin A (CyA) for 15 min before stimulation. The data shown are luciferase activity in relative luminescence units normalized to the maximum response (A) or to control (B). Results shown are the mean ± S.E. of at three or four independent experiments, performed in triplicate wells. Significant differences are indicated comparing untreated control versus agonist treated using one-way ANOVA and the Bonferroni multiple comparison test; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
FIGURE 8.
Transcriptional responses to pulsatile GnRH treatment. Cells were transfected with αGSU-Luc, LHβ-Luc, FSHβ-Luc, or NFAT-RE-Luc plasmids and transduced with Ad-mGnRHR (panel A) or transduced with Ad-αGSU-Luc, Ad-LHβ-Luc, Ad-FSHβ-Luc, or Ad-NFAT-RE-Luc and Ad-mGnRHR (panels B and C). Cells were also transduced with Ad-NFAT2-EFP to increase signal strength (far right, panels B and C only). Panel A, cells were briefly treated (5 min followed by repeated wash steps) with the indicated concentrations of GnRH at 1-h intervals. Panel B, cells were briefly treated with 10−9 m GnRH at the indicated frequency (with 0.5-h washes as a control) for 8 h. Panel C, cells were briefly treated with 10−7 m GnRH at 1-h intervals. Where indicated, cells were pretreated for 15 min with 10−5 m cyclosporin A (CyA), which remained during all wash steps. The data shown are luciferase activity in relative luminescence units normalized to the maximum response (A) or to control (panels B and C). Results shown are the mean ± S.E. of three-seven independent experiments, performed in triplicate wells. Significant differences are indicated comparing untreated control versus agonist-treated using one-way ANOVA and the Bonferroni multiple comparison test; *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Similar experiments were undertaken with an NFAT-RE luciferase reporter as readout for Ca2+/calmodulin/calcineurin activation. Sustained treatment with GnRH caused dose-dependent NFAT-RE-Luc activity with a pEC50 value of 7.8 ± 0.2 (Fig. 7A). Intermittent treatment also dose-dependently increased NFAT-RE-Luc activity, but this was not statistically significant (most probably because of the high basal activity seen with pulsatile stimulation) so frequency dependence was assessed in cells transduced with NFAT2-EFP (as well as the mGnRHR and NFAT-RE-Luc). This revealed that pulsatile GnRH can, indeed, cause a significant stimulation of NFAT-RE-Luc activity, with the greatest response at the highest frequency (Fig. 8B).
The data outlined above reveal that pulsatile GnRH stimulation increases transcription of all four reporters tested, although the highest frequency (2 pulses/h) was optimal for the αGSU and NFAT-RE reporters, whereas lower pulse frequencies (0.5 or 1 pulse/h) were optimal for the LHβ and FSHβ reporters. In a final series of experiments we used the calcineurin inhibitor cyclosporin A to test for a possible involvement of NFAT in mediating the observed transcriptional responses. Treatment with cyclosporin A (10−5 m) markedly inhibited αGSU, LHβ, FSHβ, and NFAT-RE transcriptional responses to sustained GnRH stimulation (Fig. 7B) and also inhibited these responses to pulsatile treatment (Fig. 8C), although basal LHβ transcription was elevated by cyclosporin A addition under these conditions.
DISCUSSION
Gonadotropin-releasing hormone is secreted from hypothalamic neurones to control synthesis and secretion of the pituitary gonadotropins LH and FSH. It is secreted in brief pulses, with pulse frequency varying under different physiological conditions. GnRH effects on gonadotropin synthesis and secretion are dependent upon pulse frequency, and although this has clear physiological and pharmacological relevance, the molecular and cellular mechanism of frequency decoding by gonadotrophs is essentially unknown. To address this issue we considered it essential to monitor key aspects of GnRHR signaling during pulsatile and sustained stimulation. Accordingly, we have used an ERK2-green fluorescent protein translocation assay as a live cell readout for ERK activation3 and the NFAT2-EFP translocation assay as a downstream readout for cytoplasmic Ca2+ (data herein). With this assay GnRH caused a robust translocation response, increasing both the proportion of cells with high levels of NFAT2-EFP in the nucleus and the N:C NFAT2-EFP ratio in those cells (Fig. 1 and supplemental Figs. 1 and 2). Its effect was dose-dependent with a pEC50 of 8.4, which is comparable to the pEC50 of 8.5 for [Ca2+]i elevation as measured with fluo-4 (Fig. 1). Its effect was blocked by a GnRH antagonist or by cyclosporin A and was mimicked by stimulation with a Ca2+ ionophore (Fig. 1), consistent with GnRH acting via its receptors to increase [Ca2+]i, causing a Ca2+/calmodulin-dependent activation of calcineurin that dephosphorylates cytoplasmic NFAT and thereby facilitates its translocation to the nucleus. We noted that the NFAT2-EFP translocation response was reversible (i.e. the response was more sustained with a constant GnRH stimulus than with a 5-min stimulus), although as expected for a downstream effector, the NFAT2-EFP translocation response was slower in onset and reversal than the effects of GnRH on [Ca2+]i (Fig. 2). These features are important as reversibility is expected for a frequency-decoding system, and the slow downstream response provides the possibility for integrative tracking.
A mathematical model has been developed (56) for GnRH effects on [Ca2+]i and LH secretion, and this predicts that pulsatile GnRH stimulation will elicit reproducible [Ca2+]i responses at low frequency and dose but that desensitization will occur as dose and frequency are increased, approaching the pronounced desensitization seen with sustained stimulation (supplemental Fig. 4). However, when we used live cell imaging to monitor the effects of GnRH pulses (5-min pulses at 60-min intervals) on NFAT2-EFP in mGnRHR-expressing HeLa cells we saw clear dose-dependent translocation without desensitization (i.e. response amplitude and pEC50 were comparable during the first and fourth pulse with GnRH) (Fig. 3). We also saw no evidence for desensitization when pulse frequency was varied (Fig. 4), although pronounced integrative tracking was seen in cells stimulated every 30 min. The dose- and time-dependent desensitization of GnRH signaling anticipated from the mathematical modeling (supplemental Fig. 4) is largely due to an assumed reduction in cell surface GnRHR number, so we tested for this by expressing mGnRHR with N-terminal (exofacial) HA tags in HeLa cells and using an imaging assay that quantifies cell surface HA-GnRHR (38, 52). This revealed that although sustained GnRH (10−7 m) stimulation caused a pronounced down-regulation of cell surface GnRHR, no such reduction was seen with a 5-min stimulation. Accordingly, the lack of dose- or frequency-dependent desensitization of the NFAT2-EFP translocation response could simply reflect the absence of GnRHR down-regulation. An alternative concern is that the absence of desensitization could be due to the use of a heterologous expression system so we performed similar experiments activating GnRHR in LβT2 cells transduced with Ad NFAT2-EFP. The results obtained were remarkably similar (in terms of dose dependence and kinetics) to those in HeLa cells (compare Figs. 3, 4, and 5) with no evidence for dose- or frequency-dependent desensitization but clear integrative tracking at the highest pulse frequency in both models. LβT2 are gonadotroph-lineage cells that express endogenous mGnRHR as well as all three gonadotropin subunits (59, 60). Importantly, GnRHR-mediated effects on transcription of numerous endogenous genes has been found to be dependent upon pulse frequency (45), and bell-shaped frequency-response curves are seen for GnRH effects on LHβ-, FSHβ-, and GnRH-Luc reporters in these cells (7). Thus, we see no evidence for negative feedback regulation of the NFAT-EFP translocation response in a relatively mature gonadotroph model where GnRH frequency decoding is well established.
We also explored the possible relationship between receptor number and NFAT2-EFP translocation response because mathematical modeling predicts greater desensitization for responses to pulsatile GnRH at higher receptor numbers (supplemental Fig. 5) and because the effects of GnRH on gonadotropin subunit promoter activity have been shown to be related to receptor number (50). When Ad titer was varied to set the GnRHR number to low, high, or super-physiological levels (57, 58), GnRH-stimulated NFAT2-EFP translocation increased as the receptor number increased, but the responses to hourly GnRH pulses were qualitatively similar at all receptor densities. Importantly, no desensitization was observed (comparing the first and last pulse) with either dose (10−9 or 10−7 m) or at any receptor expression level (Fig. 6). Again, it is possible that the discrepancy between the experimental data (Fig. 6) and the mathematical model predictions (supplemental Fig. 5) reflects the absence of GnRHR down-regulation with 5 min of stimulation (Fig. 2).
We also used a number of luciferase reporters to monitor GnRH effects on transcription. As expected, we found that sustained stimulation (8 h) with GnRH caused robust dose-dependent increases in activity of αGSU-Luc, LHβ-Luc, and FSHβ-Luc and also increased NFAT-RE-Luc activity (Fig. 7). Similar dose-dependent stimulation was seen with pulsatile GnRH (5-min pulses at hourly intervals for 8 h) but for each reporter basal activity was greatly increased (compare Figs. 7A and 8A). This increase in basal luciferase activity may well be due to the mechanical stimulation (the repeated stimulation and washing) with the pulsatile protocols because more robust responses were observed when superfusion systems were used to activate similar gonadotropin subunit reporters with GnRH pulses (7). Nevertheless, we were able to use our static culture system to test for pulse frequency dependence of GnRH effects, and this revealed maximal activation of LHβ-Luc and FSHβ-Luc at 60- or 120-min intervals, as compared with maximum activation of αGSU-Luc and NFAT-RE-Luc at the highest pulse frequency (30-min intervals). Thus, GnRH exerts frequency-dependent and reporter-specific effects on transcription in this model, and frequency decoding of GnRH effects on transcription are clearly not restricted to gonadotrophs or gonadotroph-lineage cell lines. We also found that cyclosporin A inhibited the effects of GnRH on the αGSU-Luc, FSHβ-Luc, and LHβ-Luc reporters irrespective of whether sustained or pulsatile stimulation was used. This is consistent with a possible role for NFAT in mediating the GnRH effects on these promoters, although it is important to recognize that calcineurins target proteins other than NFAT so the roles of NFATs in GnRH signaling remain to be explored in detail.
The saw-tooth NFAT2-EFP translocation response seen with high frequency GnRH stimulation (Figs. 4 and 5) illustrates how integrative tracking can amplify signaling. Assuming that the transcriptional effect of NFAT2 is related to the integral of nuclear NFAT2 (i.e. to the areas under the curves in Figs. 4 and 5), transcription would be activated as effectively with 5-min pulses at 30-min intervals as it is with continuous stimulation (Fig. 4C). In vivo a small number of neurons (hundreds) release GnRH into the hypothalamo-pituitary portal circulation from which it is rapidly cleared, so continuous GnRHR stimulation would require continuous secretion. Extrapolation of our data to the in vivo system suggests that this effector could be maximally activated, with pulsatile secretion increasing biological efficiency of the system (as compared with continuous secretion for continuous activation).
Although integrative tracking can increase the efficiency of signaling (as above), frequency decoding requires involvement of feedback or feed-forward pathways. A number of potential feedback mechanisms have been identified (e.g. down-regulation of cell surface GnRHR or intracellular inositol 1,4,5-trisphosphate receptors and induction of RGS-2) that could underlie GnRH frequency decoding (5, 21), but these would all be expected to influence the GnRH effects on [Ca2+]i and thereby influence its effects on NFAT2-EFP translocation. The fact that we have not seen any evidence for desensitization of the NFAT2-EFP translocation response under any of the experimental conditions used in HeLa or LβT2 cells clearly argues against a role for NFATs in GnRH frequency decoding despite the fact that NFATs clearly do act as a frequency decoder in other systems.
In exploring frequency decoding it is important to distinguish between dependence upon pulse duration and frequency. Where increasing frequency increases responses, this could reflect either the increased cumulative pulse duration or the reduced interpulse interval. True frequency decoders sense interpulse intervals independently of cumulative pulse duration (21) as illustrated by the GnRH effects on rodent LHβ and FSHβ expression, both of which are increased more effectively at low or intermediate (30–120 min) pulse frequency than at high (8–30 min) frequency or with sustained stimulation (5, 7–13). In this regard it is important that NFAT-RE-Luc activity increased as GnRH pulse frequency increased and that the responses were directly proportional to cumulative GnRH pulse duration (Fig. 8B). These data, therefore, reinforce the conclusion that NFATs (and their upstream activators) do not act as genuine GnRH pulse frequency decoders in this model. The alternative possibilities are that frequency decoding occurs within other signaling pathways (such as the ERK cascade) and/or that it occurs downstream of these pathways. The latter possibility is implicit in models where differential regulation of FSHβ and LHβ expression is attributed to the interplay of transcription factors and coactivators (45, 47). Moreover, a recent study has showed how DUSP effects on ERK activation could underlie frequency decoding with pulsatile GnRH (61). This is consistent with our own data as this mechanism could generate frequency decoding without negative upstream adaptive responses (i.e. GnRHR down-regulation or RGS activation) that would also affect Ca2+/calmodulin/calineurin/NFAT signaling.
In summary, we show that NFAT2-EFP translocation provides a robust readout for GnRHR-mediated and Ca2+-dependent activation of calmodulin/calcineurin signaling and that the GnRH effect on NFAT2-EFP location is reversible but is slower in onset and offset than the underlying change in [Ca2+]i. Pulsatile GnRH causes dose- and frequency-dependent NFAT2-EFP translocation, and at low pulse frequency NFAT2-EFP translocation effectively tracks GnRHR occupancy. In contrast, integrative tracking is seen at high GnRH frequency (pulses every 30 min), illustrating how the relative dynamics of upstream and downstream signals can increase efficiency of GnRH action. Mathematical modeling predicted desensitization of GnRHR-mediated effects on [Ca2+]i and that such desensitization would increase with dose, pulse frequency, and receptor number, but no such desensitization was seen in HeLa or LβT2 cells, possibly because pulsatile GnRH did not reduce cell surface GnRHR expression. GnRHR activation also caused dose- and pulse frequency-dependent activation of αGSU, LHβ, and FSHβ luciferase reporters, and each of these responses was prevented by cyclosporin A, indicating dependence upon the Ca2+/calmodulin/calcineurin pathway. Pulsatile GnRH also activated an NFAT-responsive luciferase reporter, but in this case the response was directly related to cumulative pulse duration. This together with the fact that we saw no desensitization of the NFAT2-EFP translocation responses argues that although NFATs may mediate GnRH action, they are not genuine decoders of GnRH pulse frequency.
This work was supported by Wellcome Trust Grants 084588 and 078407 (to C. A. M.) and a Biotechnology and Biological Sciences Research Council, Swindon, United Kingdom doctoral training grant award (to the University of Bristol Laboratories for Integrative Neuroscience and Endocrinology).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
S. P. Armstrong, C. J. Caunt, and C. A. McArdle, manuscript in preparation.
- GnRH
- gonadotropin-releasing hormone (also known as GnRH I = pGlu-His-Trp-Ser-Tyr-Gly-Leu-Arg-Pro-Gly-NH2)
- GnRHR
- GnRH receptor
- mGnRHR
- mouse GnRHR
- LH
- luteinizing hormone
- FSH
- follicle-stimulating hormone
- NFAT
- nuclear factor of activated T cells
- ERK
- extracellular signal-regulated kinase
- Ad
- recombinant adenovirus
- pfu
- plaque-forming units
- EFP
- emerald fluorescent protein
- DMEM
- Dulbecco's modified Eagle's medium
- RE
- response element
- Ctrl
- control
- GSU
- gonadotropin subunit
- BFP
- blue fluorescent protein
- NLS
- nuclear localization sequence
- ANOVA
- analysis of variance
- HA
- hemagglutinin
- N:C
- nuclear:cytoplasmic
- pEC50
- negative log10 EC50.
REFERENCES
- 1.Millar R. P., Lu Z. L., Pawson A. J., Flanagan C. A., Morgan K., Maudsley S. R. (2004) Endocr. Rev. 25, 235–275 [DOI] [PubMed] [Google Scholar]
- 2.Burger L. L., Haisenleder D. J., Dalkin A. C., Marshall J. C. (2004) J. Mol. Endocrinol. 33, 559–584 [DOI] [PubMed] [Google Scholar]
- 3.Naor Z. (2009) Front. Neuroendocrinol. 30, 10–29 [DOI] [PubMed] [Google Scholar]
- 4.Crowley W. F., Jr., Filicori M., Spratt D. I., Santoro N. F. (1985) Recent Prog. Horm. Res. 41, 473–531 [DOI] [PubMed] [Google Scholar]
- 5.Ferris H. A., Shupnik M. A. (2006) Biol. Reprod. 74, 993–998 [DOI] [PubMed] [Google Scholar]
- 6.Belchetz P. E., Plant T. M., Nakai Y., Keogh E. J., Knobil E. (1978) Science 202, 631–633 [DOI] [PubMed] [Google Scholar]
- 7.Bédécarrats G. Y., Kaiser U. B. (2003) Endocrinology 144, 1802–1811 [DOI] [PubMed] [Google Scholar]
- 8.Dalkin A. C., Haisenleder D. J., Ortolano G. A., Ellis T. R., Marshall J. C. (1989) Endocrinology 125, 917–924 [DOI] [PubMed] [Google Scholar]
- 9.Haisenleder D. J., Dalkin A. C., Ortolano G. A., Marshall J. C., Shupnik M. A. (1991) Endocrinology 128, 509–517 [DOI] [PubMed] [Google Scholar]
- 10.Shupnik M. A. (1990) Mol. Endocrinol. 4, 1444–1450 [DOI] [PubMed] [Google Scholar]
- 11.Weiss J., Jameson J. L., Burrin J. M., Crowley W. F., Jr. (1990) Mol. Endocrinol. 4, 557–564 [DOI] [PubMed] [Google Scholar]
- 12.Yasin M., Dalkin A. C., Haisenleder D. J., Kerrigan J. R., Marshall J. C. (1995) Endocrinology 136, 1559–1564 [DOI] [PubMed] [Google Scholar]
- 13.Kaiser U. B., Jakubowiak A., Steinberger A., Chin W. W. (1993) Endocrinology 133, 931–934 [DOI] [PubMed] [Google Scholar]
- 14.Conn P. M., Crowley W. F., Jr. (1994) Ann. Rev. Med. 45, 391–405 [DOI] [PubMed] [Google Scholar]
- 15.Schally A. V. (1999) Peptides 20, 1247–1262 [DOI] [PubMed] [Google Scholar]
- 16.Caunt C. J., Finch A. R., Sedgley K. R., Oakley L., Luttrell L. M., McArdle C. A. (2006) J. Biol. Chem. 281, 2701–2710 [DOI] [PubMed] [Google Scholar]
- 17.Heding A., Vrecl M., Hanyaloglu A. C., Sellar R., Taylor P. L., Eidne K. A. (2000) Endocrinology 141, 299–306 [DOI] [PubMed] [Google Scholar]
- 18.Hislop J. N., Caunt C. J., Sedgley K. R., Kelly E., Mundell S., Green L. D., McArdle C. A. (2005) J. Mol. Endocrinol. 35, 177–189 [DOI] [PubMed] [Google Scholar]
- 19.Willars G. B., Heding A., Vrecl M., Sellar R., Blomenröhr M., Nahorski S. R., Eidne K. A. (1999) J. Biol. Chem. 274, 30146–30153 [DOI] [PubMed] [Google Scholar]
- 20.Berridge M. J. (2008) Cell Signalling Biology, www.cellsignallingbiology.org, Portland Press Ltd., London, UK [Google Scholar]
- 21.Krakauer D. C., Page K. M., Sealfon S. (2002) J. Theor. Biol. 218, 457–470 [PubMed] [Google Scholar]
- 22.Burger L. L., Haisenleder D. J., Aylor K. W., Marshall J. C. (2008) Biol. Reprod. 79, 947–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haisenleder D. J., Ferris H. A., Shupnik M. A. (2003) Endocrinology 144, 2409–2416 [DOI] [PubMed] [Google Scholar]
- 24.Haisenleder D. J., Burger L. L., Aylor K. W., Dalkin A. C., Marshall J. C. (2003) Endocrinology 144, 2768–2774 [DOI] [PubMed] [Google Scholar]
- 25.Hanson P. I., Meyer T., Stryer L., Schulman H. (1994) Neuron 12, 943–956 [DOI] [PubMed] [Google Scholar]
- 26.De Koninck P., Schulman H. (1998) Science 279, 227–230 [DOI] [PubMed] [Google Scholar]
- 27.Craske M., Takeo T., Gerasimenko O., Vaillant C., Török K., Petersen O. H., Tepikin A. V. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4426–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mermelstein P. G., Deisseroth K., Dasgupta N., Isaksen A. L., Tsien R. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 15342–15347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardner S., Pawson A. J. (2009) Neuroendocrinology 89, 241–251 [DOI] [PubMed] [Google Scholar]
- 30.Lim S., Luo M., Koh M., Yang M., bin Abdul Kadir M. N., Tan J. H., Ye Z., Wang W., Melamed P. (2007) Mol. Cell. Biol. 27, 4105–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oosterom J., van Doornmalen E. J., Lobregt S., Blomenröhr M., Zaman G. J. R. (2005) Assay Drug Dev. Technol. 3, 143–154 [DOI] [PubMed] [Google Scholar]
- 32.Berridge M. J. (2006) Biochem. Soc. Trans. 34, 228–231 [DOI] [PubMed] [Google Scholar]
- 33.Tomida T., Hirose K., Takizawa A., Shibasaki F., Iino M. (2003) EMBO J. 22, 3825–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dolmetsch R. E., Xu K., Lewis R. S. (1998) Nature 392, 933–936 [DOI] [PubMed] [Google Scholar]
- 35.Li W., Llopis J., Whitney M., Zlokarnik G., Tsien R. Y. (1998) Nature 392, 936–941 [DOI] [PubMed] [Google Scholar]
- 36.Macian F. (2005) Nat. Rev. Immunol. 5, 472–484 [DOI] [PubMed] [Google Scholar]
- 37.Molkentin J. D. (2004) Cardiovasc. Res. 63, 467–475 [DOI] [PubMed] [Google Scholar]
- 38.Finch A. R., Caunt C. J., Armstrong S. P., McArdle C. A. (2009) Am. J. Physiol. Cell Physiol. 297, C591–C600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willars G. B., Royall J. E., Nahorski S. R., El-Gehani F., Everest H., McArdle C. A. (2001) J. Biol. Chem. 276, 3123–3129 [DOI] [PubMed] [Google Scholar]
- 40.Wojcikiewicz R. J., Xu Q., Webster J. M., Alzayady K., Gao C. (2003) J. Biol. Chem. 278, 940–947 [DOI] [PubMed] [Google Scholar]
- 41.Wurmbach E., Yuen T., Ebersole B. J., Sealfon S. C. (2001) J. Biol. Chem. 276, 47195–47201 [DOI] [PubMed] [Google Scholar]
- 42.Karakoula A., Tovey S. C., Brighton P. J., Willars G. B. (2008) Eur. J. Pharmacol. 587, 16–24 [DOI] [PubMed] [Google Scholar]
- 43.Armstrong S. P., Caunt C. J., McArdle C. A. (2009) Mol. Endocrinol. 23, 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang T., Roberson M. S. (2006) J. Mol. Endocrinol. 36, 41–50 [DOI] [PubMed] [Google Scholar]
- 45.Lawson M. A., Tsutsumi R., Zhang H., Talukdar I., Butler B. K., Santos S. J., Mellon P. L., Webster N. J. G. (2007) Mol. Endocrinol. 21, 1175–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burger L. L., Haisenleder D. J., Aylor K. W., Marshall J. C. (2009) Biol. Reprod. biolreprod.109.079426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsutsumi R., Webster N. J. (2009) Endocr. J. 56, 729–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caunt C. J., Armstrong S. P., Rivers C. A., Norman M. R., McArdle C. A. (2008) J. Biol. Chem. 283, 26612–26623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holdstock J. G., Aylwin S. J., Burrin J. M. (1996) Mol. Endocrinol. 10, 1308–1317 [DOI] [PubMed] [Google Scholar]
- 50.Kaiser U. B., Sabbagh E., Katzenellenbogen R. A., Conn P. M., Chin W. W. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 12280–12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson R. D., Haskell R. E., Xia H., Roessler B. J., Davidson B. L. (2000) Gene Ther. 7, 1034–1038 [DOI] [PubMed] [Google Scholar]
- 52.Finch A. R., Sedgley K. R., Caunt C. J., McArdle C. A. (2008) J. Endocrinol. 196, 353–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McArdle C. A., Bunting R., Mason W. T. (1992) Mol. Cell. Neurosci. 3, 124–132 [DOI] [PubMed] [Google Scholar]
- 54.Hislop J. N., Everest H. M., Flynn A., Harding T., Uney J. B., Troskie B. E., Millar R. P., McArdle C. A. (2001) J. Biol. Chem. 276, 39685–39694 [DOI] [PubMed] [Google Scholar]
- 55.Maru B. S., Tobias J. H., Rivers C., Caunt C. J., Norman M. R., McArdle C. A. (2009) Bone 44, 102–112 [DOI] [PubMed] [Google Scholar]
- 56.Washington T. M., Blum J. J., Reed M. C., Conn P. M. (2004) Theor. Biol. Med. Model. 1, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Finch A. R., Green L., Hislop J. N., Kelly E., McArdle C. A. (2004) J. Clin. Endocrinol. Metab. 89, 1823–1832 [DOI] [PubMed] [Google Scholar]
- 58.Hislop J. N., Madziva M. T., Everest H. M., Harding T., Uney J. B., Willars G. B., Millar R. P., Troskie B. E., Davidson J. S., McArdle C. A. (2000) Endocrinology 141, 4564–4575 [DOI] [PubMed] [Google Scholar]
- 59.Turgeon J. L., Kimura Y., Waring D. W., Mellon P. L. (1996) Mol. Endocrinol. 10, 439–450 [DOI] [PubMed] [Google Scholar]
- 60.Graham K. E., Nusser K. D., Low M. J. (1999) J. Endocrinol. 162, R1–R5 [DOI] [PubMed] [Google Scholar]
- 61.Lim S., Pnueli L., Tan J. H., Naor Z., Rajagopal G., Melamed P. (2009) PLoS ONE 4, e7244. [DOI] [PMC free article] [PubMed] [Google Scholar]