Abstract
Duplexes constituted by closed or open RNA circles paired to single-stranded oligonucleotides terminating with 3′-CCAOH form resected pseudoknots that are substrates of yeast histidyl-tRNA synthetase. Design of this RNA fold is linked to the mimicry of the pseudoknotted amino acid accepting branch of the tRNA-like domain from brome mosaic virus, known to be charged by tyrosyl-tRNA synthetases, with RNA minihelices recapitulating accepting branches of canonical tRNAs. Prediction of the histidylation function of the new family of minimalist tRNA-like structures relates to the geometry of resected pseudoknots that allows proper presentation to histidyl-tRNA synthetase of analogues of the histidine identity determinants N-1 and N73 present in tRNAs. This geometry is such that the analogue of the major N-1 histidine determinant in the RNA circles faces the analogue of the discriminator N73 nucleotide in the accepting oligonucleotides. The combination of identity elements found in tRNAHis species from archaea, eubacteria, and organelles (G-1/C73) is the most efficient for determining histidylation of the duplexes. The inverse combination (C-1/G73) leads to the worst histidine acceptors with charging efficiencies reduced by 2–3 orders of magnitude. Altogether, these findings open new perspectives for understanding evolution of tRNA identity and serendipitous RNA functions.
An unsolved problem in biology is the origin and evolution of tRNA aminoacylation systems that dictate correct expression of the genetic code at the translational level. It is at present known that identity of tRNAs is given by sets of a few nucleotides (1–3). Identity residues, properly located in several regions of the tRNAs, trigger specific recognition and charging by the cognate aminoacyl-tRNA synthetases. An intriguing finding concerns RNAs not involved in protein synthesis that are specifically recognized and even charged by synthetases (reviewed in ref. 4). Residues mimicking tRNA identity elements have been characterized in several viral tRNA-like domains (4–7), in the regulatory region of threonyl-tRNA synthetase mRNA in Escherichia coli (8), and in tmRNAs (9). From another viewpoint, the structural complexity of contemporary RNAs recognized by synthetases suggests that they have evolved from simplified RNA versions containing the primordial recognition elements. Minimalist structures of aminoacylatable RNAs have already been derived from contemporary tRNAs (reviewed in ref. 10) and represent ideal tools for searching relics of the primitive RNA-protein recognition code embodied in their structures (11).
In this paper we describe minimalist RNA structure derived from the tyrosine accepting tRNA-like domain of brome mosaic virus (BMV) RNA (12). We demonstrate that this structure is a substrate of yeast histidyl-tRNA synthetase (HisRS). Its design and prediction of its aminoacylation properties are linked to the mimicry between the major N-1 histidine identity nucleotide in tRNAHis (13, 14) and a residue from the pseudoknot forming the amino acid accepting branch of the tRNA-like domain from BMV (7, 15, 16) known to be a substrate of tyrosyl-tRNA synthetases (17). Structural investigations combined with functional studies on variants mutated at the positions expected to confer histidine identity allowed to propose an RNA fold, we call a resected pseudoknot. Implications for the evolution of RNA recognition by synthetases and peculiarities of the histidine systems are presented.
MATERIALS AND METHODS
Generation of RNA Molecules.
The 10-mers with the 3′-CCAOH accepting sequence (wild-type and three variants at the discriminator position, i.e., the fourth 3′-position) were prepared by chemical synthesis (Nucleic Acids Products Supply, Göttingen, Germany). The linear 24-mers (wild-type and 3 variants at position 117), from which the circular RNAs were made, were prepared according to established in vitro procedures by using T7 RNA polymerase (18). For transcriptional reasons and to facilitate subsequent ligation of the RNAs, the synthetic genes start from residue G122 (in red) and terminate at A123 (see Fig. 1b). Closed circles were obtained by ligation with T4 RNA ligase at 37°C (from Pharmacia) essentially as described (19) and closure was verified by PAGE separations under denaturing conditions (urea 8 M) of the ligation products (data not shown). Optimal ligation occurs at 37°C, which is the optimal temperature for the enzyme but not for self-hybridization of the RNA. Further evidence of the existence of circular RNAs was brought by phosphodiesterase treatment of the molecules (with Crotalus adamanteus venom phosphodiesterase from United States Biochemical, deprived of endonuclease activity and thus unable to cleave circular RNAs). Whatever the phosphodiesterase exonuclease was present or not, only bands corresponding to the circular conformer are observed after RNA ligase treatment at 37°C; in contrast, if the 24-mers are incubated without the ligase, phosphodiesterase treatment leads to degradations of the linear forms (not shown).
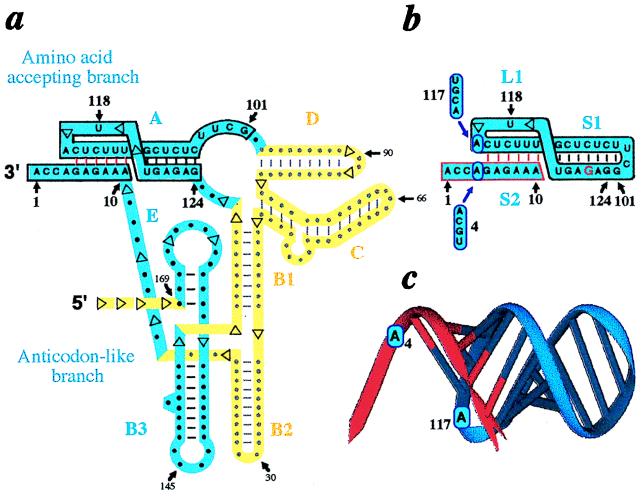
Figure 1.
Design of a resected RNA pseudoknot by analogy with the tRNA-like structure of BMV RNA. (a) Folding of the tRNA-like structure emphasizing mimicry with canonical tRNA (in blue) and domains maintaining the architecture of the tRNA-like core but not directly involved in tRNA mimicry (12) (in yellow). Numbering starts at the 3′-end. Domains A–E are denoted as in ref. 10. Explicit sequence is only displayed for domain A from which the RNA duplex is derived. (b) Sequence of a RNA duplex with histidine accepting capacity. Potential identity elements are circled in blue and mutations at N4 and N117 are indicated. Notice that the circular RNA (boxed in black) was designed by joining positions 101 and 124 of the complete tRNA-like molecule. The three domains S1, S2, and L1 required to built the resected pseudoknot are assigned according to refs. 25 and 26; the missing domain L3 corresponds to the N11-N100 stretch in a. (c) Model of the duplex derived from the BMV tRNA-like structure with accepting 10-mer (red) and circle mediating histidylation (blue). Putative histidine identity elements are indicated. The ribbon diagram was drawn from a full-atom model of the duplex with the algorithm DRAWNA (27).
Structural Analysis.
Absorbance data at 260 nm and melting temperatures were determined in a Kontron (Zurich) Uvicon 941 spectrophotometer equipped with a temperature regulation. Measurements were done in 10 mM cacodylate buffer at pH 8.1 containing 7.5 mM MgCl2 under ionic strength conditions mimicking those of the aminoacylation media. Probing with RNAse V1 (12, 20) was done at 15°C for 8 min on 7 μM of 5′ end-labeled 24-mers with 0.2 units of nuclease (Pharmacia) in media containing 50 mM of Tris⋅HCl (pH 7.8), 5 mM of MgCl2, and 100 mM of KCl. Before nuclease treatment, RNAs were heated 5 min at 65°C and slowly cooled to room temperature to ensure optimal hybridization of the RNA duplexes.
Aminoacylation Reactions.
Aminoacylation reactions of the RNA duplexes containing either closed or open circular 24-mer variants were performed at 15°C in 25 mM of Tris⋅HCl buffer (pH 8.1), 15 mM of MgCl2, 7.5 mM of ATP, 50 μM of l-[3H]histidine (58 Ci/mmol; 1 Ci = 37 GBq), the required concentrations of HisRS (≈30 to 100 enzymatic units/ml incubation mixture), and equimolar amounts of both 10-mer and 24-mer RNAs (in the μmol range). Before aminoacylation, RNAs were renatured as for structural probing (see above). A highly enriched yeast HisRS preparation was obtained by established procedures (21) with a specific activity of 660 units/mg (1 unit being the amount of protein charging 1 nmol histidine on tRNA in 1 min under optimal conditions at 37°C). Charging levels obtained with equivalent enzyme concentrations were calculated assuming that all 10-mer RNA strands are chargeable, i.e., are all hybridized with the circular RNA. Experimental errors on aminoacylation levels were ± 10%. Apparent Vmax/Km values for each variant were determined from Lineweaver-Burk plots and varied for replicate experiments at most 30%. Relative losses of catalytic efficiencies (L) are expressed as L = (Vmax/Km)duplex reference/(Vmax/Km)histidine acceptor. For double mutants, experimental L-values were compared with theoretical values calculated from experimental data obtained with single mutants. This permits to determine coupling factors defined as R = Lexperimental/RLcalculated (R ≈ 1 indicates additivity, R > 1 cooperativity and R < 1 anticooperativity) (22).
RESULTS AND DISCUSSION
Design of the RNAs.
The minimalist RNA structures derived from the tyrosine accepting tRNA-like domain of BMV RNA (Fig. 1a) are formed by short oligonucleotides hybridized to circular RNAs (Fig. 1 b and c). These duplexes contain an RNA fold we call “resected pseudoknot” because it is lacking the oligonucleotide stretch present in canonical pseudoknots that would connect the two RNAs forming the duplexes. These structures are expected to be charged by HisRS. The rationale underlying their design and prediction of their histidylation function are linked to a putative structural mimicry between the major histidine identity nucleotide located at position −1 in tRNAs (13, 14) and a residue (A117) from the pseudoknot forming the amino acid accepting branch of the BMV tRNA-like domain (7, 15, 16) (Fig. 1a). This residue (A117), likely is stacked over the amino acid accepting helix and faces A4 in the BMV RNA, the analogue of a tRNA discriminator N73 base, also involved in histidine identity. This view is based on chemical probing and modeling of the BMV tRNA-like structure (12) and finds support in the recently solved NMR structure of the pseudoknot found in the tRNA-like structure of turnip yellow mosaic virus RNA (23). The fact that minimalist RNA substrates containing these identity signals, either derived from tRNAHis (14) or from the tRNA-like domain of turnip yellow mosaic virus RNA (24), can be histidylable, was the argument that prompted us to study this RNA fold and its related aminoacylation properties. The unusual architecture of these small RNA constructs relies to the intricate fold of the tRNA-like core of BMV RNA (in blue) constituted by an amino acid accepting branch (domain A in boxed blue ribbon) assembled via a long-range interaction between the 3′-most terminal part of the RNA and an RNA stretch more than 100 nucleotides apart, linked to an anticodon-like branch (domains B3 and E) by the extension of the 3′-terminal sequence (Fig. 1a). The presence of the large extra-domain (in yellow) to form the amino acid accepting branch explains that minimalist pseudoknotted structures of unimolecular type cannot be derived from BMV RNA (unless replacing the extra-domain by a short L3 sequence). Consequently, an RNA circle was constructed by connecting G101 to G124, two residues in spatial proximity in the three-dimensional model of the tRNA-like domain (12). This generates a UUCG tetraloop of high stability (28). To favor a circular conformation of open circles, but also for transcriptional reasons, and if needed to facilitate closure of the circles by ligation, oligonucleotides starting at G122 (in red) were synthesized (Fig. 1b). The 10-mer oligonucleotides, mimicking the tRNA accepting 3′-end, are such to be able to hybridize to the closed or open RNA circles with their six first 5′-nucleotides complementary to residues 111–116 of the circles (Fig. 1b).
Solution Conformation of Resected RNA Pseudoknots.
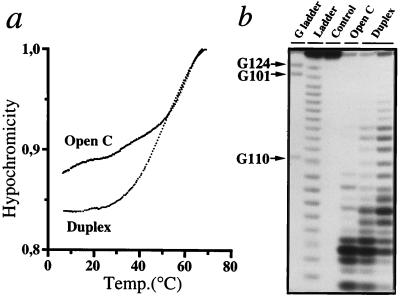
Existence of resected pseudoknots was demonstrated in solution by temperature melting and structural probing with RNase V1, an enzyme that recognizes helices or stacked nucleotides in RNA (20) (Fig. 2). Whereas circles have a broad and multimodal melting profile with hyperchromicity appearing at 10°C, duplexes exhibit hyperchromicity at temperatures above 20°C and show rather sharp cooperative transitions. For wild-type duplex with A4/A117, the melting temperature is 50°C (Fig. 2a); for duplex 4 with A4/G117 it is 47°C and for duplex 1 with C4/G117, 53°C (not shown). This melting behavior resembles to what was observed for a 9-mer RNA duplex chargeable with alanine (29). The high stability of the duplexes is probably due to the formation of a regular RNA helical stack after interaction of the 10-mers with the twisted circles, a conclusion supported by comparative probing of free circular RNA and duplex RNAs by RNase V1 (Fig. 2b). Hybridization of RNAs is evidenced by strong RNase V1 cuts between U111 and U115 (in “Duplex” lanes, Fig. 2b). The cleavage pattern indicates internal structure in the non base paired domain of the open circle (“Open C” lane), with possible formation of two additional base pairs (U111-U118 and U112-A117). In the duplex, the helical domain S1 is stabilized as shown by appearance of new RNase V1 cuts between C107 and G110.
Figure 2.
Resected RNA pseudoknot fold as evidenced by (a) temperature melting experiments and (b) structural probing with RNAse V1 specific of double-stranded or stacked nucleotides (20). Only results relevant to duplex 9 (see Table 1) with the wild-type A4/A117 combination and open circle with A117 are shown. In b, the autoradiogram compares RNase V1 cleavage patterns of free open 24-mer (“Open C” lane) and of open 24-mer/10-mer mixtures (“Duplex” lanes) having 1/1 (Left) and 1/4 (Right) stoichiometries.
The topology of resected pseudoknots (Fig. 1 b and c) derives from that of H-type (hairpin) pseudoknots (25, 26), but in contrast to such folds with two linkers L1 and L3 connecting double-strands S1 and S2 and crossing the major and minor grooves of the resulting co-axial RNA helix, resected pseudoknots are missing L3 and contain only one linker, analogous to L1 in canonical pseudoknots (L2 that could be present between G110 and U111 is missing). The short L1 sequence crosses the RNA major groove with residue A117, which may be stacked over the helical domain and therefore may mimic N-1 from tRNAHis. Stem S1 would be of canonical geometry and S2 is built by the bimolecular association between a hairpin loop and a free oligonucleotide (Fig. 1b). Note that circularity of RNA is not mandatory for establishment of the fold and that a similar type of topology was proposed for duplexes formed when targeting the TAR domain in HIV RNA by antisense RNAs (30).
Recognition of Resected Pseudoknots by Histidyl-tRNA Synthetase.
Functionality of the wild-type duplex with the histidine identity nucleotides A4 and A117 (duplex 9) and of 15 variants with closed 24-mers is given in Table 1. Kinetic parameters obtained with open circles (in brackets in Table 1) are essentially identical, indicating that single-stranded 24-mers fold per se in a circular conformation. Exceptions concern duplexes 5, 12, and 14, for which inversions of Km- and Vmax-effects occur. These inversions may reflect existence of mechanistic compensations leading to overall similar L-values, a phenomenon already observed in the yeast tRNA arginylation system when aminoacylation properties of tRNA variants were compared (34). The accepting 10-mers are histidylated to levels up to 4% for the wild-type construct and even ≈10% for duplexes 1 and 4. Charging is optimal at 15°C and is dependent upon the association of a 10-mer with its circular counterpart. Aminoacylation levels can be modulated by varying the concentrations of the RNA components of the duplex. If the [10-mer]/[circle] stoichiometry is increased, plateau’s are increased (up to 3-fold). When duplex formation is favored by increasing concentration of the circle, an inverse effect occur and may be due to binding of free circles on HisRS as revealed by competition experiments (slight inhibition of tRNAHis charging by free circles). As in the alanine system (29), charging can be amplified (2-fold) after a heating (50°C)/cooling (15°C) cycle that may facilitate exchange on the circular RNA co-factor of charged 10-mers by new acceptor RNAs. For long incubations and temperature above 15°C, charging levels decrease, probably as a result of deacylation (35). These peculiarities make the interpretation of aminoacylation data delicate, in particular because kinetic parameters can be affected by combined mechanistic effects in the enzymatic reaction due to the changes in identity signals and/or by subtle effects on the conformation of the duplex that may lead to transient dissociation/reassociation phenomena.
Table 1.
Histidylation properties of resected RNA pseudoknots and comparison with those of the BMV tRNA-like domain and yeast tRNAHis
| RNAs, duplex no. | Charging level,* apparent % | Km, μM | Vmax, arbitrary units | L† |
|---|---|---|---|---|
| RNA/RNA duplexes | ||||
| Most efficient duplex | ||||
| 1 G117 C4 | 9.3 (9.3) | 0.7 (0.9) | 42.0 (45.6) | 1 (1) |
| Single mutants | ||||
| 2 U117 C4 | 6.7 (7.4) | 1.0 (0.6) | 29.6 (21.5) | 2 (2) |
| 3 A117 C4 | 7.1 (7.2) | 1.4 (0.8) | 26.2 (14.9) | 3 (3) |
| 4 G117 A4 | 10.7 (10.7) | 1.7 (1.7) | 29.6 (29.6) | 3 (3) |
| 5 G117 U4 | 6.2 (6.2) | 25.0 (1.0) | 252.0 (22.3) | 6 (3) |
| 6 C117 C4 | 2.0 (2.1) | 3.6 (1.2) | 6.1 (4.8) | 35 (15) |
| 7 G117 G4 | 4.0 (4.0) | 2.4 (2.4) | 3.6 (3.6) | 40 (40) |
| Double mutants | ||||
| 8 U117 U4 | 3.0 (3.3) | 0.8 (1.9) | 6.7 (7.6) | 7 (15) |
| 9 A117 A4 (wt) | 4.0 (3.3) | 1.8 (1.1) | 6.8 (5.6) | 16 (12) |
| 10 A117 U4 | 2.0 (2.0) | 4.2 (2.0) | 9.7 (5.7) | 26 (21) |
| 11 C117 U4 | 1.1 (1.1) | 1.9 (1.5) | 2.2 (2.2) | 52 (41) |
| 12 U117 G4 | 0.9 (1.3) | 2.3 (123.0) | 1.9 (24.2) | 73 (305) |
| 13 U117 A4 | 1.3 (1.0) | 1.8 (3.1) | 1.3 (1.8) | 83 (103) |
| 14 A117 G4 | 0.7 (1.0) | 3.0 (37.4) | 1.8 (6.4) | 100 (350) |
| 15 C117 A4 | 0.9 (1.0) | 2.2 (9.5) | 1.1 (4.5) | 120 (127) |
| 16 C117 G4 | 0.2 (0.5) | 0.9 (11.8) | 0.2 (0.5) | 270 (1416) |
| BMV tRNA-kike domain | ||||
| 17 A117 A4 (wt) | 40 | 1.3 | 170 | 0.46 |
| yeast tRNAHis transcript | ||||
| 18 G-1 A73 | 100 | 0.3 | 35 103 | 5.1 10−4 |
| 10-mers alone | ||||
| 19–22 A4, C4, G4, or U4 | 0.0 | ND | ND | — |
Experimental data are given for resected pseudoknots with closed and open (between brackets) circles.
Charging levels were obtained for 40 min incubation in the presence of equivalent amounts of enzyme (2.5-fold excess as compared to initial rate conditions for resected pseudoknot charging) and were calculated assuming that all 10-mers are chargeable (all hybridized). For initial rate determinations enzyme was 30 units/ml incubation mixture (except for tRNAHis where it was 150-fold decreased) and incubation times up to 5 min (variants 1–3, 17), 10 min (variants 4, 8–11), 20 min (variants 6, 7, 12–15), and 50 min (variant 16). All aminoacylations were performed at 15°C as described.
Relative losses in catalytic efficiencies of histidine acceptors compared to efficiency of duplex 1 are expressed as L = (Vmax/Km)duplex 1/(Vmax/Km)histidine acceptor. ND, not detectable. The A117/A4 combination found in the BMV tRNA-like structure and other bromovirus RNAs (31) is in bold (duplex 9). Combinations present in RNAs from phylogenetically related cucumo- and bromoviruses (31) are underlined. Duplexes 3, 4 and 13 contain identity elements present in natural substrates of eukaryotic HisRSs (A117/C4 in the tRNA-like domains of the RNAs from tobacco mosaic virus and its satellite virus, G117/A4 in yeast and other eukaryotic tRNAHis species, and U117/A4 in the tRNA-like domain of the RNA from TYMV; for sequence data see refs. 4, 32 and 33).
The wild-type resected pseudoknotted RNA substrate (duplex 9) is charged 35-fold less efficiently than a control BMV RNA and 30,000-fold less than a tRNAHis transcript. This compares well with charging efficiencies of canonical tRNA minihelices (10, 14, 36). Among the variants tested, six behave better than wild-type duplex (Table 1). The best substrate (duplex 1) with the identity combination G117/C4 mimicking G-1/C73 present in tRNAHis species from archaea, eubacteria, and organelles (31), is only 2,000-fold less efficiently charged than tRNAHis. The worst one (duplex 16) has a N117/N4 combination never found in histidine accepting RNAs. Noticeable in this context are the different melting temperatures found for three typical duplexes: highest (53°C) for duplex 1, the G117/C4 variant, medium (50°C) for duplex 9, the wild-type A117/A4 construct and lowest (47°C) for duplex 4, the G117/A4 variant, whereas the aminoacylation plateaus level at 9.3%, 4.0%, and 10.7%, respectively. The high plateau with duplex 4 is likely explained by the combined effect of an easier recycling of the accepting 10-mer because of the lower melting temperature of this duplex and the occurrence of the identity nucleotide combination as in eukaryotes. The four single 10-mers are totally inactive.
When the most efficient RNA minisubstrate for histidylation is taken as the reference (duplex 1), it appears that six duplexes are single mutants and nine are double mutants (Table 1). For single mutants, significant losses of catalytic efficiency occur only for duplexes 6 and 7. Effects are more pronounced for most double mutants. A phenomenological analysis of the kinetic data indicates that the double mutants behave essentially additively (Table 2). This is reminiscent to what observed in the aspartate system, where double identity mutations located in the same structural domain of the tRNA behave additively (22). A significant exception is double-mutant 13, which shows strong cooperativity. This cooperativity may originate from the presence of a N-1 histidine identity mimic of weak strength (U117 instead of a purine at N-1 in tRNAHis species) and especially from a constrained structure of the histidine accepting end with a U117-A4 pair that likely hinders optimal adaptation of the RNA to HisRS, instead of more loose pairs in other efficient histidine acceptors that permit easier adaptations.
Table 2.
Mechanistic relationship between N4 and N117 histidine identity determinants in histidylation of resected RNA pseudoknots
| RNAs, duplex no. | Ldm, experimental | Lsm1 | Lsm2 | Ldm, calculated | R |
|---|---|---|---|---|---|
| 1 G117 C4 | 1 | — | — | — | — |
| 10 A117 U4 | 26 | 3 | 6 | 18 | 1.4 |
| 9 A117 A4 (wt) | 16 | 3 | 3 | 9 | 1.8 |
| 14 A117 G4 | 100 | 3 | 40 | 120 | 0.8 |
| 8 U117 U4 | 7 | 2 | 6 | 12 | 0.6 |
| 13 U117 A4 | 83 | 2 | 3 | 6 | 13.8 |
| 12 U117 G4 | 73 | 2 | 40 | 80 | 0.9 |
| 11 C117 U4 | 52 | 35 | 6 | 20 | 0.25 |
| 15 C117 A4 | 120 | 35 | 3 | 105 | 1.1 |
| 16 C117 G4 | 270 | 35 | 40 | 1400 | 0.2 |
Activities of double-mutants are compared to that of the most active HisRS substrate, namely duplex 1. Only data obtained with closed circles are displayed. Ldm are losses of catalytic efficiency for double mutants, Lsm for single-mutants, and Ldm(calculated, = Lsm1 × Lsm2. Experimental data are taken from Table 1. The R factors inform about additivity, cooperativity and anticooperativity (see Materials and Methods). Notice the slight anticooperative nature of duplex 16 (R = 0.2), that disappears when the cofactor circle is open (data not shown).
Altogether, the functional assays indicate specificity of histidylation because mutating putative identity positions modifies charging efficiencies. Thus resected pseudoknots obey similar identity rules than canonical tRNAs and the functional analogy of positions 117 and 4 in the duplexes with identity positions -1 and 73 in tRNAHis is demonstrated.
Evolutionary Considerations.
Considering the large architectural differences between RNA substrates of HisRS [canonical tRNAHis, tRNA-like structures (4, 24, 33, 37) and the derived minisubstrates (this work and refs. 10, 24, and 37)], it follows that the nature of identity elements and their correct presentation to the synthetase are more important than the RNA architecture in which they are embedded. The nonrequirement of a defined structural framework reflects a probable convergent evolution of several primitive RNA structures toward contemporary and more elaborate RNAs specific for HisRS. The functionality of minimalist RNAs derived from contemporary histidine acceptors reminisces this early history of tRNA and is corroborated by the location within duplexes of the major identity signals. This indicates in turn that function of contemporary HisRSs is primarily triggered by RNA contacts near the enzyme catalytic center, as also suggested by analysis of the three-dimensional structure of E. coli HisRS (38). Even if the propensity of HisRS to recognize diverse RNAs (39) is not fully understood, it is tempting to correlate it with ranking of this enzyme in class II of synthetases (40) that appear the most primitive because they aminoacylate well RNA minisubstrates (see refs. 10 and 11). Histidine systems are pivotal not only because of the variety of HisRS substrates but also because of the particular nature of the N-1 (or its mimic) identity element. In primitive aminoacylation systems this element may had a broader role that was lost during evolution except for specifying histidine identity. The fact that the arginine and aspartate synthetases charge well a tRNAAsp variant with a 5′-extension (41) may reminisce this role.
The moderate functional differences between duplexes mutated at identity positions (Table 1) but the strict necessity for histidylation of N-1 and N73 (13, 14) or their structural analogues in tRNA-like structures (7), suggest that many contemporary RNAs or RNA associations folded in helical conformations, either of canonical or pseudoknotted type, may interact with HisRS and, if terminating by a overhanging 3′-CCAOH, may become histidine acceptors. Therefore, one can expect direct or indirect participation of HisRS and its RNA substrates in metabolic processes other than genetic code translation or RNA replication, as for the tRNA-like molecules (e.g., refs. 42–45), that remain to be unraveled. From another viewpoint it can be anticipated that when targeting RNA structures with antisense RNAs (30), resected pseudoknots may form that could become HisRS competitors so that biological consequences of antisense hybridization could originate from HisRS inhibition. More work will be necessary to explore these possibilities.
Acknowledgments
We are indebted to C. Florentz, J. Rudinger, J. Ng, and E. Westhof for stimulating discussions and A. Hoeft for synthesis of DNA fragments. This work was supported by Centre National de la Recherche Scientifique, Ministère de l’Enseignement Supérieur et de la Recherche, and Université Louis Pasteur (Strasbourg). B.F. was partially supported by grants from MESR and Association pour la Recherche sur le Cancer.
ABBREVIATIONS
- BMV
brome mosaic virus
- HisRS
histidyl-tRNA synthetase
Footnotes
A commentary on this article begins on page 10351.
References
- 1.Giegé R, Puglisi J D, Florentz C. Prog Nucleic Acid Res Mol Biol. 1993;45:129–206. doi: 10.1016/s0079-6603(08)60869-7. [DOI] [PubMed] [Google Scholar]
- 2.Saks M E, Sampson J R, Abelson J N. Science. 1994;263:191–197. doi: 10.1126/science.7506844. [DOI] [PubMed] [Google Scholar]
- 3.McClain W H. In: tRNA: Structure, Biosynthesis and Function. Söll D, RajBhandary U L, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 335–347. [Google Scholar]
- 4.Florentz C, Giegé R. In: tRNA: Structure, Biosynthesis and Function. Söll D, RajBhandary U L, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 144–163. [Google Scholar]
- 5.Florentz C, Dreher T W, Rudinger J, Giegé R. Eur J Biochem. 1991;195:229–234. doi: 10.1111/j.1432-1033.1991.tb15698.x. [DOI] [PubMed] [Google Scholar]
- 6.Dreher T W, Tsai C-H, Florentz C, Giegé R. Biochemistry. 1992;31:9183–9189. doi: 10.1021/bi00153a010. [DOI] [PubMed] [Google Scholar]
- 7.Felden B, Florentz C, Westhof E, Giegé R. Biochem Biophys Res Commun. 1998;243:426–434. doi: 10.1006/bbrc.1997.7753. [DOI] [PubMed] [Google Scholar]
- 8.Graffe M, Dondon J, Caillet J, Romby P, Ehresmann C, Ehresmann B, Springer M. Science. 1992;255:994–996. doi: 10.1126/science.1372129. [DOI] [PubMed] [Google Scholar]
- 9.Felden B, Himeno H, Muto A, McCutcheon J P, Atkins J F, Gesteland R F. RNA. 1997;3:89–103. [PMC free article] [PubMed] [Google Scholar]
- 10.Martinis S A, Schimmel P. In: tRNA: Structure, Biosynthesis and Function. Söll D, RajBhandary U L, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 349–370. [Google Scholar]
- 11.Schimmel P, Giegé R, Moras D, Yokoyama S. Proc Natl Acad Sci USA. 1993;90:8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felden B, Florentz C, Giegé R, Westhof E. J Mol Biol. 1994;235:508–531. doi: 10.1006/jmbi.1994.1010. [DOI] [PubMed] [Google Scholar]
- 13.Himeno H, Hasegawa T, Ueda T, Watanabe K, Miura K-I, Shimizu M. Nucleic Acids Res. 1989;17:7855–7863. doi: 10.1093/nar/17.19.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklyn C, Schimmel P. Proc Natl Acad Sci USA. 1990;87:8655–8659. doi: 10.1073/pnas.87.21.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rietveld K, Pleij C W A, Bosch L. EMBO J. 1983;2:1079–1085. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joshi R L, Joshi S, Chapeville F, Haenni A L. EMBO J. 1983;2:1123–1127. doi: 10.1002/j.1460-2075.1983.tb01556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall T C, Shih D S, Kaesberg P. Biochem J. 1972;129:969–976. doi: 10.1042/bj1290969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perret V, Garcia A, Puglisi J D, Grosjean H, Ebel J-P, Florentz C, Giegé R. Biochimie. 1990;72:735–744. doi: 10.1016/0300-9084(90)90158-d. [DOI] [PubMed] [Google Scholar]
- 19.Uhlenbeck O C, Gumport R I. In: The Enzymes. Boyer P, editor. New York: Academic; 1982. pp. 31–57. [Google Scholar]
- 20.Favorova O O, Fasiolo F, Keith G, Vassilenko S K, Ebel J-P. Biochemistry. 1981;20:1006–1010. doi: 10.1021/bi00507a055. [DOI] [PubMed] [Google Scholar]
- 21.Mengual R. Thesis. Strasbourg, France: Université Louis Pasteur; 1977. [Google Scholar]
- 22.Pütz J, Puglisi J D, Florentz C, Giegé R. EMBO J. 1993;12:2949–2957. doi: 10.1002/j.1460-2075.1993.tb05957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolk M H, van der Graaf M, Wijmenga S S, Pleij C W, Heus H A, Hilbers C W. Science. 1998;280:434–438. doi: 10.1126/science.280.5362.434. [DOI] [PubMed] [Google Scholar]
- 24.Rudinger J, Florentz C, Giegé R. Nucleic Acids Res. 1994;22:5031–5037. doi: 10.1093/nar/22.23.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ten Dam E, Pleij K, Draper D. Biochemistry. 1992;31:11665–11676. doi: 10.1021/bi00162a001. [DOI] [PubMed] [Google Scholar]
- 26.Westhof E, Jaeger L. Curr Opin Struct Biol. 1992;2:327–333. [Google Scholar]
- 27.Massire C, Gaspin C, Westhof E. J Mol Graphics. 1994;12:201–206. doi: 10.1016/0263-7855(94)80088-x. [DOI] [PubMed] [Google Scholar]
- 28.Cheong C, Varani G, Tinoco I., Jr Nature (London) 1990;346:680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- 29.Musier-Forsyth K, Scaringe S, Usman N, Schimmel P. Proc Natl Acad Sci USA. 1991;88:209–213. doi: 10.1073/pnas.88.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ecker D J, Vickers T A, Bruice T W, Freier S M, Jenison R D, Manoharan M, Zounes M. Science. 1992;257:958–961. doi: 10.1126/science.1502560. [DOI] [PubMed] [Google Scholar]
- 31.Ahlquist P, Dasgupta R, Kaesberg P. Cell. 1981;23:183–189. doi: 10.1016/0092-8674(81)90283-x. [DOI] [PubMed] [Google Scholar]
- 32.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felden B, Florentz C, McPherson A, Giegé R. Nucleic Acids Res. 1994;22:2882–2286. doi: 10.1093/nar/22.15.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sissler, M., Giegé, R. & Florentz, C. (1998) RNA 4, in press. [DOI] [PMC free article] [PubMed]
- 35.Dietrich A, Kern D, Bonnet J, Giegé R, Ebel J-P. Eur J Biochem. 1976;70:147–158. doi: 10.1111/j.1432-1033.1976.tb10965.x. [DOI] [PubMed] [Google Scholar]
- 36.Frugier M, Florentz C, Giegé R. EMBO J. 1994;13:2218–2226. doi: 10.1002/j.1460-2075.1994.tb06499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Felden B, Florentz C, Giegé R, Westhof E. RNA. 1996;2:201–212. [PMC free article] [PubMed] [Google Scholar]
- 38.Arnez J G, Harris D C, Mitschler A, Rees B, Francklyn C S, Moras D. EMBO J. 1995;14:4143–4155. doi: 10.1002/j.1460-2075.1995.tb00088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rudinger J, Felden B, Florentz C, Giegé R. Bioorg Med Chem. 1997;5/6:1001–1009. doi: 10.1016/s0968-0896(97)00061-8. [DOI] [PubMed] [Google Scholar]
- 40.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Nature (London) 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 41.Perret V, Florentz C, Giegé R. FEBS Lett. 1990;270:4–8. doi: 10.1016/0014-5793(90)81221-9. [DOI] [PubMed] [Google Scholar]
- 42.Dreher T W, Rao A L N, Hall T C. J Mol Biol. 1989;206:425–438. doi: 10.1016/0022-2836(89)90491-9. [DOI] [PubMed] [Google Scholar]
- 43.Maizels N, Weiner A M. In: The RNA World. Gesteland R F, Atkins J F, editors. New York: Cold Spring Harbor; 1993. pp. 577–602. [Google Scholar]
- 44.Noller H F. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 137–156. [Google Scholar]
- 45.Giegé R. Proc Natl Acad Sci USA. 1996;93:12078–12081. doi: 10.1073/pnas.93.22.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]