Abstract
N-glycans are major components of many glycoproteins. These sugar moieties are frequently involved in important physiological and disease processes via their interactions with a variety of glycan-binding proteins (GBP). Clustering effect is an important feature in many glycan-lectin interactions. We describe in this paper a chemoenzymatic synthesis of novel N-glycan clusters using a tandem endoglycosidase-catalyzed transglycosylation. It was found that the internal β-1,2-linked GlcNAc moieties in the N-glycan core, once exposed in the non-reducing terminus, was able to serve as acceptors for transglycosylation catalyzed by Endo-A and EndoM-N175A. This efficient chemoenzymatic method allows a quick extension of the sugar chains to form a class of glycan clusters in which sugar residues are all connected by native glycosidic linkages found in natural N-glycans. In addition, a discriminative enzymatic reaction at the two GlcNAc residues could be fulfilled to afford novel hybrid clusters. Lectin microarray studies revealed unusual properties in glyco-epitope expression by this panel of structurally well-defined synthetic N-glycans. These new compounds are likely valuable for functional glycomics studies to unveil new functions of both glycans and carbohydrate-binding proteins.
Keywords: chemoenzymatic synthesis, N-glycans, clusters, transglycosylation, lectin microarray
Introduction
N-linked glycosylation is a predominant covalent modification of proteins in eukaryotes. It is well documented that N-glycans of glycoproteins are involved in many important biological events including protein folding, ER-associate protein degradation, cell differentiation, cell adhesion, host-pathogen interaction, cancer metastasis, and autoimmunity 1. Glycoproteins are often characterized by their structural micro-heterogeneity in terms of the components of the attached glycans. It becomes clear that distinct N-glycans can confer significantly different effects on the structure and function of a given glycoprotein. This was exemplified by recent discoveries that the attachment of subtly different N-glycans at the conserved N-glycosylation site of the Fc domain could result in dramatically different impacts on the ADCC function of monoclonal antibodies and on the anti-inflammatory activity of intravenous immunoglobulin (IVIG) 2. On the other hand, not only does the fine structure of the glycans provide the basis for molecular recognition, but the way of their presentation, e.g., the monovalent vs. multivalent format, is also of paramount importance in governing the specificity and strength in carbohydrate-protein interactions 3. Recent advances in glycan and glycan-binding protein microarray technology have offered exciting new opportunities to unveil the mysteries of glycans in various cellular processes and disease states 4. Nevertheless, functional glycomics studies are still limited by the availability of structurally well-defined N-glycans and related glycoconjugates, which are difficult to obtain in homogeneous forms from natural source 5.
We and others have previously demonstrated that a class of endo-β-N-acetylglucosaminidases (ENGases), including the Endo-A from Arthrobactor protophormiae and the Endo-M from Mucor hiemali, were able to transfer an oligosaccharide en bloc from either natural N-glycans or synthetic sugar oxazolines to a GlcNAc-containing moiety to form a new β-1,4-glycosidic linkage in a regio- and stereo-specific manner, leading to the synthesis of complex oligosaccharides, N-glycopeptides and N- glycoproteins 6. Endo-A is specific for high-mannose or hybrid type N-glycans and has been applied for the synthesis of high-mannose type oligosaccharides and N-glycopeptides 7, whereas Endo-M is able to work on three major types (high-mannose, hybrid, and complex type) of N-glycans and has been particularly useful for synthesizing complex type N-glycopeptides 8. In particular, the recent findings that synthetic oligosaccharide oxazolines (the mimics of the oxazolinium ion intermediate of the enzymatic reaction) could be used for Endo-A catalyzed transglycosylation have significantly expanded the scope of the chemoenzymatic method for glycopeptide and glycoprotein synthesis 9–11. It was found that the highly activated sugar oxazolines corresponding to the truncated or modified N-glycans could serve as substrates for the Endo-A catalyzed transglycosylation but the ground-state products formed were refractory to enzymatic hydrolysis due to the slight structural modification. Moreover, the discovery of several ENGase-based glycosynthases, including EndoM-N175A; EndoA-N171A, and EndoA-E173Q that could promote transglycosylation with sugar oxazolines of natural N-glycans but lack the ability to hydrolyze the product, has enabled the synthesis of homogeneous glycoproteins carrying full-size natural N-glycans 12–14. Subsequent studies indicate that Endo-A and Endo-M could accommodate diverse structures in the aglycon portions of GlcNAc- or Glc-tagged acceptors for transglycosylation, permitting the introduction of N-glycans into a wide range of natural products, unnatural peptides, and even polysaccharides 15, 16. The relaxed substrate specificity of Endo-A and Endo-M, together with the powerful transglycosylation potential of the glycosynthase mutants, prompted us to examine the possibility to glue multiple N-glycans to the complex-type GlcNAc2Man3GlcNAc2-Asn core through tandem enzymatic transglycosylation. We report in this paper the chemoenzymatic synthesis of a class of novel N-glycan clusters containing multiple N-glycan cores, in which all monosaccharide residues are connected via defined native glycosidic bonds found in natural N-glycans. Lectin microarray analysis of the synthetic N-glycan clusters has revealed unusual lectin-carbohydrate recognition patterns that were not observed before.
Results and Discussions
Enzymatic transglycosylation onto the β-1,2-linked GlcNAc residues in the Asn-linked GlcNAc2Man3GlcNAc2 core
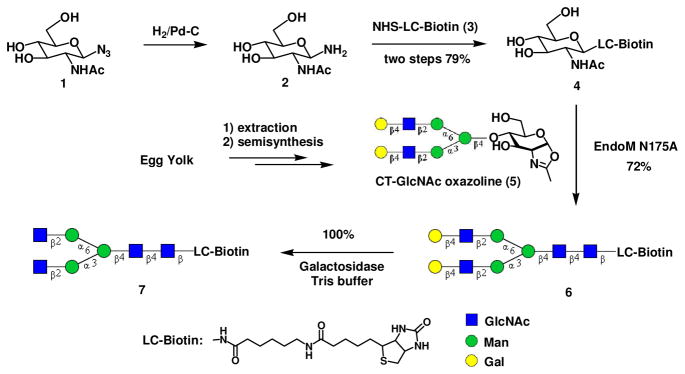
Construction of an array of N-glycan clusters started with the preparation of a biotinylated bi-antennary complex type N-glycan (Scheme 1). Coupling of glycosylamine 2 and an activated biotin tag (3) gave the GlcNAc-LC-biotin (4), which was then used as an acceptor for enzymatic transglycosylation. We have previously reported the synthesis of an asialoglycan oxazoline (5) and have shown that it was a good substrate of the glycosynthase mutant EndoM-N175A for glycoprotein synthesis 13. Thus the reaction of oxazoline (5) and acceptor (4) (donor/acceptor, 3:1) under the catalysis of EndoM-N175A gave the biotinylated N-glycan product (6) in 72% yield. The two terminal galactose residues were then selectively removed by treatment with β-1,4-galactosidase from Bacteroides fragilis to provide GlcNAc2Man3GlcNAc2-LC-biotin (7), in which the two internal GlcNAc residues were exposed at the non-reducing terminus. Enzymatic extension of the sugar chain from the two GlcNAc residues would enable the synthesis of novel glycan structures. The advantage of introducing a biotin tag in the aglycon portion of the glycan primer (7) was apparent: it would facilitate the detection of the synthetic N-glycans bound in a glycan-binding protein microarray platform, or the biotinylated glycans could be directly immobilized on a streptavidin plates to provide a glycan array platform.
Scheme 1.
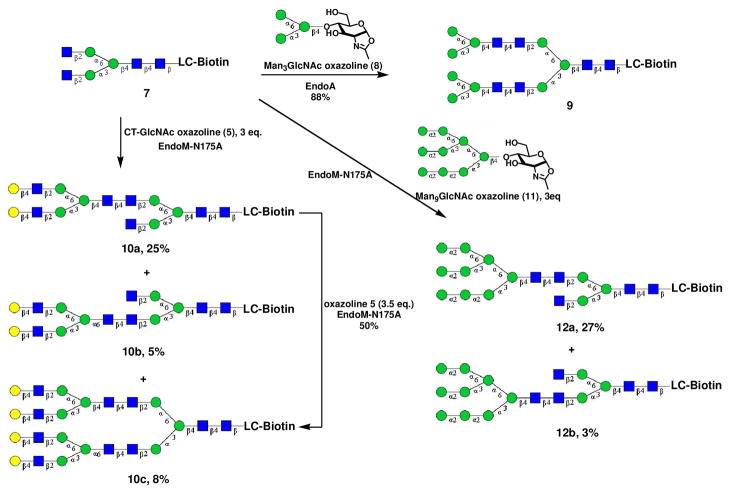
To test whether the two terminal β-1,2-linked GlcNAc residues of 7 are able to serve as acceptors for the next round enzymatic transglycosylation, we first examined the Endo-A catalyzed transglycosylation of 7 using Man3GlcNAc oxazoline (8) 10 as the donor substrate (Scheme 2). It was found that when oxazoline 8 and GlcNAc2Man3GlcNAc2-LC-biotin (7) (donor/acceptor, 6:1) were incubated with Endo-A in a phosphate buffer at 30°C, the transglycosylation reaction proceeded very efficiently and, after 2h, almost all the starting material 7 was converted to the doubly glycosylated product 9 (as revealed by HPLC). The product was readily isolated by HPLC in 88% yield. MALDI-TOF MS analysis showed a single m/z species at 3055.12, which matches well with the theoretical data of 9 that carries three Man3GlcNAc2 cores (calculated, M = 3032.14; found M+Na = 3055.12). The newly formed glycosidic bonds in the Man3GlcNAc2 cores were assumed to be in the natural β-1,4-glycosidic linkage between the two GlcNAc residues, on the basis of all previously reported stereo-specificity of Endo-A catalyzed transglycosylation 10, 11, 15. To further substantiate this assumption, compound 9 was treated with wild type Endo-M that specifically hydrolyzes the β-1,4-glycosidic bond in natural N-glycan. It was found that treatment of 9 with Endo-M gave GlcNAc-LC-biotin (1 equiv.), GlcNAc2Man3GlcNAc (1 equiv.) and Man3GlcNAc (2 equiv.) (data not shown), suggesting that the three Man3GlcNAc2 cores all are having the natural GlcNAc-β-1,4-GlcNAc linkage in the core. The successful simultaneous enzymatic transfer of two Man3GlcNAc moieties to the GlcNAc2Man3GlcNAc2-LC-biotin indicates that the two terminal β-1,2-linked GlcNAc residues, albeit in a rigid and crowded environment on the N-glycan core, are accessible to Endo-A catalyzed glycosylation.
Scheme 2.
Next, we examined the glycosylation of 7 by the glycosynthase mutant EndoM-N175A, using a large complex type glycan oxazoline (5) as the donor substrate. Interestingly, when a limited amount of oxazoline donor substrate was used, a selective glycosylation on the two terminal GlcNAc residues was observed. Thus, incubation of oxazoline 5 and acceptor 7 (donor/acceptor, 3:1) with EndoM-N175A at 30°C for 4h gave three products: the two mono-glycosylated products (10a and 10b) and the doubly transglycosylated product 10c in 25, 5, and 8% yields, respectively (Scheme 2). The GlcNAc linked to the mannose on the α-1,6-arm was much more favorable for transglycosylation than the GlcNAc linked to the mannose at the α-1,3-arm, with a selectivity of 5:1 (25% vs. 5%). Although both GlcNAc are β-1,2-linked to the mannose moiety, the regio-selectivity might result from the difference in steric hindrance, as the GlcNAc at the 6-arm would be spatially less hindered than the GlcNAc located on the α-1,3-arm. Nevertheless, double glycosylation could be achieved by further chemoenzymatic transglycosylation with excess amount of donor substrate for a longer incubation time. This was exemplified by further glycosylation of the purified mono-glycosylated glycan 10a and oxazoline 5 (3.5 molar equiv.) under the catalysis of EndoM-N175A, giving the doubly-glycosylated product (10c) in 50% yield. When Man9GlcNAc oxazoline (11) was used as the donor substrate, an even higher regio-selectivity in transglycosylation was observed. Incubation of 11 and 7 (3:1) with EndoA-N175A for 4h gave 27% of 12a, 3% of 12b, and trace amount of the doubly glycosylated product (12a:12b = 9:1) (Scheme 2). The higher selectivity might be explained by the bulky structure of tri-antennary glycan (11) in comparison with the bi-antennary complex glycan 5. It should be mentioned that the use of wild type Endo-M for the reaction between acceptor 7 and the oxazoline 5 or 11 failed to provide the transglycosylation products due to the enzymatic hydrolysis of the acceptor 7 as well as the products that were formed (data not shown).
It should be mentioned that in the synthesis of large complex N-glycan clusters, most of the transglycosylation reactions were carried out on a relatively small scale. Nevertheless, the specific enzymatic transglycosylation usually gave a very clear HPLC profile allowing easy quantification and isolation of the reaction products by HPLC. To characterize the transglycosylation products, the new compounds were purified by HPLC and were subjected to MALDI-TOF MS analysis, which confirmed that the products 10a and 10b were mono-transglycosylated product and the 10c was the doubly glycosylated product (see Experimental Section). To discriminate the two mono-transglycosylated products 10a and 10b, specific enzymatic transformation coupled with MS analysis was applied (Figure S1, Supporting Information). Treatment of 10a and 10b with β-N-acetylglucosaminidase from Xanthomonas manihotis resulted in the removal of the remaining terminal GlcNAc, which exposed the α-1,3-Man residue in 10a or the α-1,6-Man residue in 10b. When the two intermediates, which had the same molecular mass, were then treated further with an α-1,2/α-1,3-mannosidase from Xanthomonas manihotis, only the exposed α-1,3-man residue in 10a would be removed, leading to the formation of a species with a loss of 162 Da in molecular mass, while the α-1,6-Man residue exposed in 10b would not be hydrolyzed. Thus, MALDI-TOF MS analysis of the resulting products led to an unambiguous assignment of 10a and 10b as the clusters with the additional glycan attached at the α-1,6-arm and the α-1,3-arm positions, respectively. Discrimination between 12a and 12b was achieved by similar enzymatic transformations of 12a with β-N-acetylglucosaminidase and α-1,2/α-1,3-mannosidase coupled with MS analysis (Figure S2, Supporting Information).
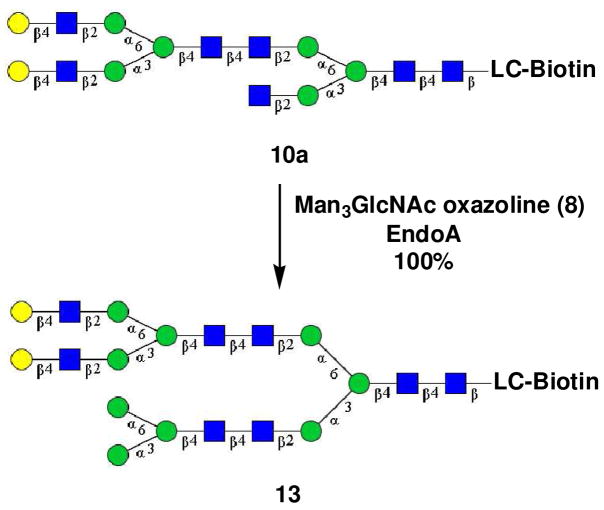
The regio-selectivity observed in the EndoM-N175A catalyzed transglycosylation also provided an opportunity to introduce distinct N-glycans at the α-1,6- and α-1,3-arms of the core, permitting the synthesis of novel hybrid N-glycan clusters. Thus, after the first selective transglycosylation of 7 with oxazoline (5) to produce compound 10a, another distinct glycan, Man3GlcNAc, was successfully introduced to the remaining terminal GlcNAc at the α-1,3-arm by Endo-A to give the hybrid N-glycan cluster 13 in essentially quantitative yield (Scheme 3). This result represents a remarkable example on the potential of the enzymatic transglycosylation for expanding the diversity of the glycan cluster structures.
Scheme 3.
Further extension of the sugar chains in the glycan clusters by tandem enzymatic transglycosylation
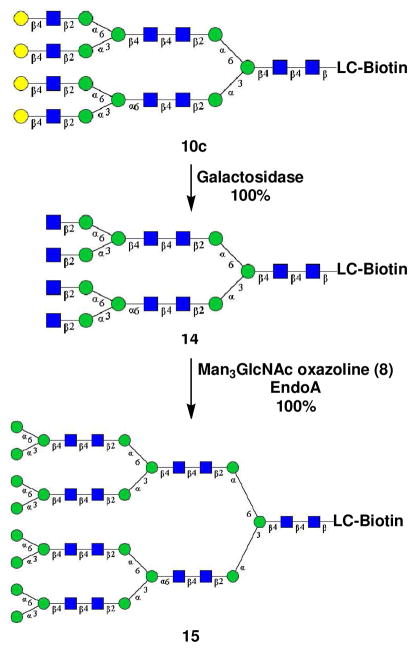
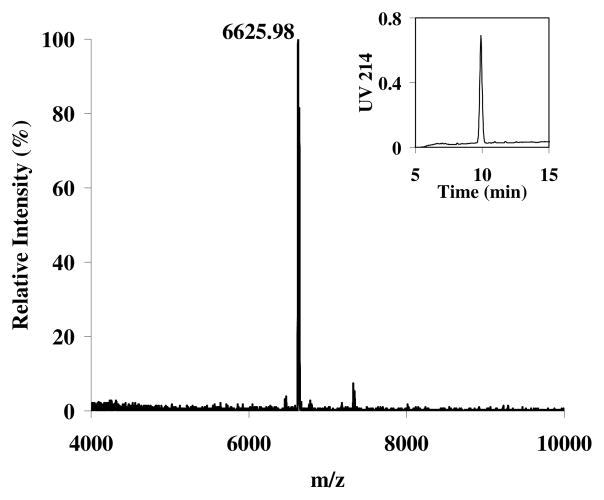
For those glycan clusters that contain terminal LacNAc moieties such as compounds 10a, 10b, 10c, and 13, further sugar chain extensions could be readily achieved by unmasking the terminal GlcNAc residues followed by repeat enzymatic transglycosylation. For example, treatment of 10c with β-1,4-galactosidase from Bacteroides fragilis resulted in selective removal of the four terminal galactose residues to provide compound 14, in which the four internal GlcNAc residues were exposed. Simultaneous glycosylation of these GlcNAc residues in 14 was achieved in a single step by its Endo-A catalyzed transglycosylation with an excess amount of Man3GlcNAc-oxazoline (8) (4 equiv. per terminal GlcNAc) to give N-glycan cluster 15 in essentially quantitative yield (Scheme 4). The HPLC and MALDI-TOF MS profiles of the purified N-glycan cluster 15 were shown in Figure 1. The m/z species observed in the MS spectrum is in good agreement with the calculated molecular mass (observed, m/z = 6625.98; calculated, [M + Na] = 6625.41). Glycan cluster 15 consists of four terminal Man3GlcNAc2 cores and three internal Man3GlcNAc2 cores. It would be worthwhile to emphasize that all the sugar residues in these enzymatically synthetic N-glycan clusters are connected to each other by the defined natural glycosidic bonds found in natural N-glycans, which usually demonstrate confined rotations of the chains than those linked through flexible spacers. Thus the N-glycan clusters described in this work represent a new class of N-glycan clusters that are different from other synthetic oligosaccharide clusters in which mono- or oligosaccharides are linked with various spacers.
Scheme 4.
Figure 1.
HPLC and MALDI-TOF MS profiles of N-glycan cluster 15.
Lectin array characterization of the synthetic N-glycan clusters
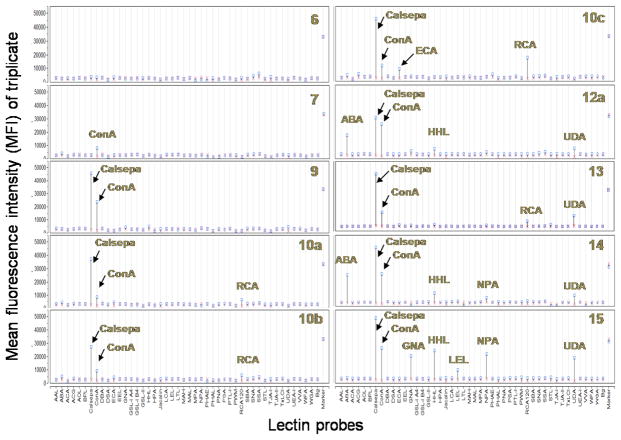
The availability of a class of structurally well-defined synthetic N-glycan clusters has now provided an opportunity to investigate how the unusual configuration of multiple N-glycan cores in a molecule would contribute to their recognition with various glycan-binding proteins such as lectins. For a preliminary study, a lectin microarray consisting of 45 lectins was used (Figure 2a), in which the lectins were immobilized on a glass slide as previously reported 17. All the biotinylated N-glycan cluster were assayed at a concentration of 100 nM, and the bound glycans were detected by a fluorescence(Cy3)-labeled streptavidin. Figure 2b demonstrated a typical fluorescent imaging of the microarray profiles for glycan clusters 9 and 15. The lectin-binding profiles of all the synthetic N-glycan clusters were summarized in Figure 3. For each compound, its responses to the respective lectins were quantitatively determined and expressed as mean fluorescence intensity (MFI) of triplicate detections.
Figure 2. A 45-lectin microarray platform for probing N-glycan clusters.
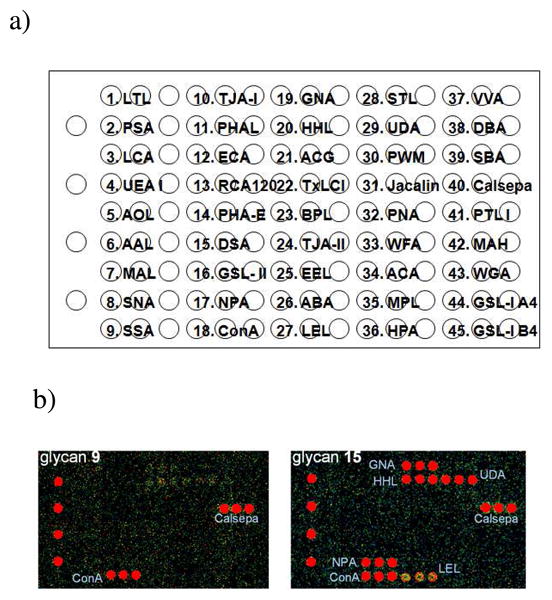
a) lectin printing pattern; b) Fluorescent imaging of the lectin-recognition profiles of glycan clusters 9 and 15.
Figure 3. Glyco-epitope profiles of novel synthetic N-glycans detected by lectin arrays.
overlay plots were produced using JMP-Genomics 4.0 software package (SAS Institute in Cary, North Carolina). Each plot was an overlay plot of GlcNAc-LC-biotin (4) that served as a control in the assay (marked with a symbol “X”) and the respective N-glycan cluster (marked with a symbol “□”).
For the simple biotinylated N-glycans, 6 and 7, no apparent lectin interactions were observed under the screening conditions (at 100 nM glycan concentration) except for a weak binding of the GlcNAc-terminated glycan 7 to ConA, a lectin that is specific for terminal GlcNAc, Man, and Glc residues. The known sugar-binding specificity of the 45 lectins used in the present microarray analysis was listed in Figure S3 (Supporting Information). The cluster 9, which contains two terminal Man3GlcNAc2 cores, showed significant signals on spots for lectins Calsepa and ConA. These results are consistent with the known specificity of these two lectins, which recognize terminal mannose/GlcNAc residues and particularly high-mannose glycan 18. Glycans 10a and 10b picked up three lectins, Calsepa, ConA, and RCA120. RCA120 is a lectin specific for terminal β-Gal residues 19. The strong interactions between lectin Calsepa and 10a or 10b is an interesting observation. Calsepa is a mannose-binding-type Jacalin-realated lectin (mJRL) with binding preference to small high-mannose N-glycan (Man2–6). It is also reported that Calsepa interacts with sialo/asialo complex-type N-glyans with terminal Gal or GlcNAc. Interestingly bisecting GlcNAc dramatically enhances binding affinity of complex-type N-glycans with Calsepa 18. The strong interaction between Calsepa and 10a or 10b suggests that both the terminal Man3GlcNAc core and the remaining terminal GlcNAc in 10a and 10b might bind to two distinct binding sites in the lectin simultaneously in a concerted manner, thus dramatically enhancing its affinity to the lectin. It should be noted that Calsepa did not show detectable affinity to the complex-type N-glycans 6 and 7 at 100 nM, despite the presence of terminal GalNAc and GlcNAc in the glycans.
The lectin microarray also revealed interesting properties for glycan cluster 10c, which bears four terminal Galβ1-4GlcNAc moieties. First, it showed significantly enhanced affinity to the LacNAc-specific galectins RCA120 and ECA 20 as compared with the glycans 6, 10a, and 10b. The dramatic enhancement of affinity could be explained by the clustering effect of the Galβ1-4GlcNAc ligands in 10c. Secondly, the Calsepa binding signals were also enhanced. More surprisingly, glycan cluster 10c also demonstrated significant affinity to ConA, although it does not contain terminal GlcNAc/Man residues. These results suggest that the appropriate clustering arrangement of the internal Man3GlcNAc2 cores in 10c may generate novel new binding sites or interfaces for lectin ConA that hitherto favors terminal mannose/GlcNAc residues. As to the Man9GlcNAc2 containing cluster 12a, it was recognized by 5 lectins in the microarray. In addition to its expected affinity to the high-mannose recognizing lectins, including Calsepa, ConA, and HHL, it also picked up two unexpected lectins, ABA and UDA. Lectin ABA has a known specificity for terminal Galβ1-3GalNAc structure and a very weak affinity to terminal GlcNAc residue on complex-type N-glycans (Kd at 100 μM level) 21. The reason why 12a exhibits such a high-affinity to lectin ABA is yet to be elucidated, but perhaps both the Man9 structure and the terminal GlcNAc at the other arm are involved in a simultaneous interaction with two binding sites in the lectin. The glycan clusters 10a and 10b, which contain a terminal GlcNAc but carry a complex glycan on the other arm, were not recognized by ABA. Lectin UDA (Urtica dioica agglutinin) is known to be specific for chitin oligomers (N,N′,N″-triacetylchitotriose and higher chitin oligosaccharides) 22, 23. The significant interaction between UDA and compound 12a may suggest that the three internal GlcNAc residues may form a discontinuous glyco-epitope that would fit to the chitotriose-binding domain in the lectin 23. Alternatively, the Man9GlcNAc2 moiety itself may also contributes directly to the binding, as compound 10a, which contains a similar three internal GlcNAc motif but carries a complex N-glycan at the 6-arm, clearly did not bind to UDA. The glycan cluster 13, which possesses both terminal Galβ1-4GalNAc and high-mannose moieties, demonstrated the expected binding specificity for lectins Calpesa, ConA, and RCA120. But again, it also showed significant binding affinity to UDA. The glycan cluster 14, which possesses multiple terminal GlcNAcβ1,2-Man structures, demonstrated strong affinity to lectins Calsepa, ConA, and ABA, and moderate binding capacity to HHL and UDA. While its strong interactions with Calsepa, ConA and ABA are expected because of the clustering effect of multiple terminal GlcNAc residues, the interaction between glycan 14 and lectin HHL that has a known specificity for mannose is a new observation. Compound 15, which possesses 4 terminal Man3GlcNAc2 cores and 3 internal Man3GlcNAc2 cores, is another novel N-glycan cluster that demonstrated unusual lectin recognition properties. A comparison of this glycan cluster with the simpler glycan cluster 9 (2 terminal Man3GlcNAc2 cores and 1 internal Man3GlcNAc core), revealed a clear cluster effects in lectin recognition (Figure 3). Under the same assay conditions, glycan 9 recognized only two lectins, Calsepa and ConA that are specific for terminal mannose residues and high-mannose type N-glycans. However, glycan cluster 15 was recognized by 7 different lectins in the microarray. In addition to Calsepa and ConA, compound 15 showed high affinity to the other three high-mannose specific lectins, GNA 24, HHL, and NPA 25. The fact that glycan cluster 9 did not show sufficient affinity to these three lectins (negative at 100 nM) strongly suggest that these three lectins recognize particularly a high-density cluster of high-mannose type glycans, as demonstrated by the glycan cluster 15. Moreover, glycan cluster 15 also picked up two chitin oligosaccharide-specific lectins, UDA 23 and LEL 26 that recognizes chitin oligosaccharides with 3 or more GlcNAc moieties. A plausible explanation is that the compact cluster of the multiple GlcNAcβ1-4GlcNAc moieties in 15 forms a novel discontinuous epitope that mimics the chitin oligosaccharide to provide a high-affinity structure to interact with the binding domain in the lectin. Notably, among the synthetic glycan clusters, only compound 15 showed significant binding to lectin LEL that is known to be specific for N-acetyl-chitooligosaccharide moieties (Figure 3). These experimental data suggest that the unusual configuration of the N-glycan cores in the defined N-glycan clusters create novel lectin recognition motifs or glyco-epitopes that may implicate special roles of unusual N-glycans in a biological system. Recently, Dennis and co-workers has reported that the number and degree of branching (e.g., tetra-antennary vs. bi-antennary) of complex type N-glycans in cell-surface receptors are critical factors to regulate cell proliferation and differentiation, due to the distinct affinities of the differentially branched N-glycans to galectins 27. On the other hand, the highly branched and compactly packed N-glycans clusters described in this work, which show unusually high-affinity to some specific lectins, may be used as specific inhibitors to decipher the functional roles of N-glycans and lectins in a given biological process.
Conclusion
A facile synthesis of a class of novel N-glycan clusters was achieved via tandem chemoenzymatic tranglycosylation. The unusual configurations and highly branched packing of the subunit N-glycans in the clusters create new structural motifs, demonstrating novel carbohydrate-lectin recognition patterns. These synthetic N-glycan clusters should be valuable for functional glycomics studies to unveil new functions for both glycans and carbohydrate-binding proteins.
Experimental
Materials and methods
1-β-Azido-GlcNAc (1) was prepared by reported method 28. Succinimidyl-6-(biotinamido)hexanoate (NHS-LC-biotin, 3) was purchased from Pierce Biotechnology, Inc. CT-GlcNAc oxazoline (5) 13, Man3GlcNAc oxazoline 10, and Man9GlcNAc oxazoline 12 were synthesized following our previously reported procedures. The β-1,4-galactosidase, β-N-acetylglucosaminidase and α-1,2/1,3-mannosidase were purchased from New England Biolabs, Inc. Endo-A was overproduced following the literature 29. EndoM-N175A was overproduced according to the previously reported method 12. The activity unit of Endo-A was defined as following: 1 unit of Endo-A is the amount of enzyme required to hydrolyze 1 μmol Man9GlcNAc2Asn (substrate concentration, 10 mM) in one minute at 30 °C in a phosphate buffer (50 mM, pH 6.5). The unit of glycosynthase activity of EndoM-N175A was defined as following: 1 unit of EndoM-N175A is the amount of enzyme required to transfer 1μmol Man9GlcNAc oxazoline to GlcNAc-pNP at 1:2 donor/acceptor ratio in one minute. All the other reagents were purchased from Sigma/Aldrich and used as received.
Analytical RP-HPLC was performed on a Waters 626 HPLC instrument with a Symmetry300™ C18 column (5.0 μm, 4.6 × 250 mm) at 40°C. The column was eluted at a flow rate of 1.0 mL/min using a linear gradient of 0–90% MeCN containing 0.1% trifluoroacetic acid (TFA) for 30 min. The yields were calculated based on the HPLC quantification of both starting materials and products [absorbance (abs) at 214 nm], using the following formula: yield (%) = [product abs/(starting material Abs + product abs)] × 100. Preparative HPLC was performed on a Waters 600 HPLC instrument with a preparative C18 column (Symmetry300™, 7.0 μm, 19 × 300 mm). The column was eluted with a suitable gradient of water/acetonitrile containing 0.1% TFA. NMR spectra were measured with JEOL ECX 400 MHz and/or Inova 500 MHz NMR spectrometers. All chemical shifts were assigned in ppm. The ESI-MS Spectra were measured on a Waters Micromass ZQ-4000 single quadruple mass spectrometer. MALDI-TOF MS measurement was performed on an Autoflex II MALDI-TOF mass spectrometer (Bruker Daltonics). The instrument was calibrated by using ProteoMass Peptide MALDI-MS calibration kit (MSCAL2, Sigma/Aldrich). The matrix used for glycans is 2,5-dihydroxybenzoic acid (DHB) (10.0 mg/mL in 50% acetonitrile containing 0.1% trifluoroacetic acid). The measuring conditions in detail: 337 nm nitrogen laser with 100 μJ output; laser frequency 50.0 Hz; laser power 30–45%; linear mode; positive polarity; detection range 1000–10000; pulsed ion extraction: 70ns; high voltage: on; realtime smooth: high; shots: 500–2000.
Synthesis of 1-(6-biotinamido-hexanoylamino)-1-deoxy-2-acetamido-2-deoxy-β-D-glucopyranose (4)
1-β-Azido-GlcNAc (1) (10 mg, 38 μmol) was dissolved in MeOH (1.0 mL) containing 5% palladium on carbon (5.0 mg). The mixture was hydrogenated at r.t. under atmospheric pressure overnight. The residue was filtered via a Celite pad and the filtrate was concentrated. The obtained crude syrup of 1-β-amino-GlcNAc (2) was directly used without additional purification. A solution of the crude compound 2 and NHS-LC-biotin (3) (20 mg, 44 μmol) in a mixed solvent of phosphate buffer (50 mM, pH 7.5, 1.0 mL), MeCN (1.0 mL) and DMSO (1.0 mL) was shaken at r.t. overnight. The residue was subject to preparative HPLC for purification. The fractions containing product were combined and lyophilized to give GlcNAc-LC-biotin (4) as white powder (17 mg, 79% for two steps). 1H NMR (D2O, 400 MHz): δ 4.93 (d, 1H, J = 9.6 Hz, H-1 of GlcNAc), 4.47 (dd, 1H, J = 4.8, 8.0 Hz, H-7 of biotin), 4.29 (dd, 1H, J = 4.4, 8.0 Hz, H-8 of biotin), 3.74 (dd, 1H, J = 2.0, 12.4 Hz, H-6a of GlcNAc), 3.67 (t, 1H, J = 10.0 Hz, H-2 of GlcNAc), 3.61 (dd, 1H, J = 4.6, 12.4 Hz, H-6b of GlcNAc), 3.47 (t, 1H, J = 10.0 Hz, H-3 of GlcNAc), 3.38 (m, 2H, H-4 and H-5 of GlcNAc), 3.21 (m, 1H, H-4 of biotin), 3.03 (t, 2H, J = 6.8 Hz, CH2NHCO), 2.87 (dd, 1H, J = 4.8, 13.0 Hz, H-6a of biotin), 2.63 (d, 1H, J = 13.0 Hz, H-6b of biotin), 2.14 (m, 4H, CH2CONH), 1.86 (s, 3H, Ac of GlcNAc), 1.61-1.34 (m, 8H, CH2), 1.26 (m, 2H, CH2), 1.16 (m, 2H, CH2); 13C NMR (D2O, 100 MHz) δ 177.7, 176.6, 174.5, 165.3, 78.3, 77.6, 74.1, 71.2, 69.4, 62.1, 60.5, 60.2, 55.4, 54.3, 39.7, 39.0, 35.7, 35.5, 28.0, 27.8, 27.7, 25.5, 25.2., 25.0, 22.0; analytical HPLC: tR = 12.2 min; ESI-MS: calculated for C24H41N5O8S, M = 559.27 Da, found, 560.67 [M + H]+.
Synthesis of biotinylated complex type N-glycan (6)
A solution of GlcNAc-LC-biotin (4) (1.0 mg, 1.8 μmol) and CT-GlcNAc oxazoline (5) (7.5 mg, 5.3 μmol) in a phosphate buffer (50 mM, pH 7.5, 100 μL) was incubated with EndoM-N175A (125 mU) at 30°C for 8h. The residue was subject to preparative HPLC for purification. The fractions containing product were combined and lyophilized to give CT-GlcNAc2-LC-biotin (6) as white powder (2.5 mg, 72%). 1H NMR (D2O, 400 MHz): δ 4.97 (s, 1H, H-1 of Man4), 4.92 (d, 1H, J = 9.6 Hz, H-1 of GlcNAc1), 4.78 (s, 1H, H-1 of Man4′), 4.62 (s, 1H, H-1 of Man3), 4.46 (m, 4H, H-1 of GlcNAc2, H-1 of GlcNAc5, H-1 of GlcNAc5′, H-7 of biotin), 4.33 (dd, 2H, J = 2.4, 7.8 Hz, H-1 of Gal6 and Gal6′), 4.28 (dd, 1H, J = 4.6, 7.8 Hz, H-8 of biotin), 4.11 (m, 1H), 4.06 (m, 1H), 3.97 (m, 1H), 3.19 (quintet, 1H, J = 4.8 Hz, H-4 of biotin), 3.03 (t, 2H, J = 6.8 Hz, CH2NHCO), 2.86 (dd, 1H, J = 4.8, 12.8 Hz, H-6a of biotin), 2.65 (d, 1H, J = 12.8 Hz, H-6b of biotin), 2.12 (m, 4H, CH2CONH), 1.94 (s, 3H, Ac), 1.91 (s, 3H, Ac), 1.90 (s, 3H, Ac), 1.85 (s, 3H, Ac), 1.61-1.34 (m, 8H, CH2), 1.28 (m, 2H, CH2), 1.16 (m, 2H, CH2); 13C NMR (D2O, 100 MHz) δ 177.7, 176.6, 174.7, 174.6, 174.5, 165.3 (7 × CO), 102.9 (C-1 of Gal6 and Gal6′), 101.5 (C-1 of GlcNAc2), 100.4 (C-1 of Man3), 99.7 (C-1 of Man4), 99,6 (C-1 of GlcNAc5 and GlcNAc5′), 97.0 (C-1 of Man4′), 78.6 (C-1 of GlcNAc1), 62.8 (C-8 of biotin), 60.5 (C-7 of biotin), 55.8 (C-4 of biotin), 39.7 (C-6 of biotin), 39.0 (2 × CH2NHCO), 35.7, 35.5 (2 × CH2CONH), 27.9, 27.8, 27.6, 25.4, 25.2., 24.8 (6 × CH2), 22.3, 22.2, 22.0 (4 × Ac); analytical HPLC: tR = 11.2 min; MALDI-TOF-MS: calculated for C78H130N8O48S, M = 1978.77 Da, found, 2002.68 [M + Na]+.
Synthesis of the biotinylated complex type N-glycan (7)
A solution of CT-GlcNAc2-LC-Biotin (6) (2.5 mg, 1.3 μmol) in a phosphate buffer (50 mM, pH 5.5, 300 μL) was incubated with β-1,4-galactosidase (100 U) from Bacteroides fragilis (New England Biolabs) at 37 °C for 24h. The reaction was monitored by analytical HPLC until complete removal of terminal galactose. The residue was subject to preparative HPLC. The fractions containing product were combined and lyophilized to give GlcNAc2Man3GlcNAc2-LC-biotin (7) as white powder (2.1 mg, quantitative yield). 1H NMR (D2O, 400 MHz): δ 4.98 (s, 1H, H-1 of Man4), 4.93 (d, 1H, J = 9.6 Hz, H-1 of GlcNAc1), 4.78 (s, 1H, H-1 of Man4′), 4.64 (s, 1H, H-1 of Man3), 4.48 (m, 2H, H-1 of GlcNAc2, H-7 of biotin), 4.43 (d, 2H, J = 8.6 Hz, H-1 of GlcNAc5 and GlcNAc5′), 4.29 (dd, 1H, J = 4.4, 8.0 Hz, H-8 of biotin), 4.12 (m, 1H), 4.05 (m, 1H), 3.97 (m, 1H), 3.20 (quintet, 1H, J = 4.8 Hz, H-4 of biotin), 3.03 (t, 2H, J = 6.8 Hz, CH2NHCO), 2.87 (dd, 1H, J = 4.8, 13.2 Hz, H-6a of biotin), 2.63 (d, 1H, J = 12.8 Hz, H-6b of biotin), 2.13 (m, 4H, CH2CONH), 1.94 (s, 3H, Ac), 1.91 (s, 6H, 2 × Ac), 1.86 (s, 3H, Ac), 1.62-1.34 (m, 8H, CH2), 1.27 (m, 2H, CH2), 1.15 (m, 2H, CH2); 13C NMR (D2O, 100 MHz) δ 177.7, 176.6, 174.7, 174.6, 174.5, 165.3 (7 × CO), 101.3 (C-1 of GlcNAc2), 100.4 (C-1 of Man3), 99.6 (C-1 of Man4, C-1 of GlcNAc5 and GlcNAc5′), 97.0 (C-1 of Man4′), 80.4, 79.5, 78.6 (C-1 of GlcNAc1), 78.2, 76.4, 76.2, 75.8, 74.4, 74.3, 73.5, 73.3, 73.2, 72.8, 72.7, 72.0, 70.2, 69.9, 69.5, 69.4, 67.3, 65.8, 65.7, 62.1 (C-8 of biotin), 61.7, 61.6, 60.6 (C-7 of biotin), 60.2, 59.8, 55.4, 55.3 (C-4 of biotin), 39.7 (C-6 of biotin), 39.0 (2 × CH2NHCO), 35.7, 35.5 (2 × CH2CONH), 27.9, 27.8, 27.6, 25.4, 25.2., 24.8 (6 × CH2), 22.3, 22.2, 22.0 (4 × Ac); analytical HPLC: tR = 11.4 min; MALDI-TOF-MS: calculated for C66H110N8O38S, M = 1654.66 Da, found, 1678.07 [M + Na]+.
Synthesis of the N-glycan cluster (9)
A solution of GlcNAc2Man3GlcNAc2-LC-Biotin (7) (0.20 mg, 0.12 μmol) and Man3GlcNAc oxazoline (8) (0.50 mg, 0.72 μmol) in phosphate buffer (50 mM, pH 7.5, 15 μL) was incubated with Endo-A (2.0 mU) at 30 °C for 1h. The reaction was monitored by analytical HPLC until the completion of the reaction. The residue was subject to preparative HPLC purification. The fractions containing product were combined and lyophilized to give Man3GlcNAc2Man(1,6)- [Man3GlcNAc2Man(1,3)]-ManGlcNAc2-LC-biotin (9) as a white powder (88% yield). Analytical HPLC: tR = 11.1 min; MALDI-TOF-MS: calculated for C118H196N10O78S, M = 3033.14 Da, found, 3055.12 [M + Na]+.
Transglycosylation of glycan 7 with the complex type glycan oxazoline 5
Synthesis of glycan clusters 10a and 10b
A solution of GlcNAc2Man3GlcNAc2-LC-biotin (7) (0.50 mg, 0.30 μmol) and CT-GlcNAc oxazoline (5) (1.3 mg, 0.91 μmol) in a phosphate buffer (50 mM, pH 7.5, 20 μL) was incubated with EndoM-N175A (30 mU) at 30°C. The reaction was monitored by analytical HPLC. After 4h, HPLC showed the formation of three new products, which were readily purified by HPLC to give the 6-arm glycosylated product CT-GlcNAc2Man(1,6)-[GlcNAcMan(1,3)]-LC-biotin (10a) in 25% yield; the 3-arm glycosylated product CT-GlcNAc2Man(1,3)-[GlcNAcMan(1,6)]-LC-biotin (10b) in 5% yield, and the doubly glycosylated product 10c in 8% yield.
CT-GlcNAc2Man(1,6)-[GlcNAcMan(1,3)]-ManGlcNAc2-LC-biotin (10a): white powder, analytical HPLC: tR = 10.8 min; MALDI-TOF-MS: calculated for C120H199N11O78S, M = 3074.17 Da, found, 3097.78 [M + Na]+.
CT-GlcNAc2Man(1,3)-[GlcNAcMan(1,6)]-ManGlcNAc2-LC-biotin (10b): white powder, analytical HPLC: tR = 11.1 min; MALDI-TOF-MS: calculated for C120H199N11O78S, M = 3074.17 Da, found, 3098.36 [M + Na]+.
CT-GlcNAc2Man(1,6)-[CT-GlcNAc2Man(1,3)]-ManGlcNAc2-LC-biotin (10c): white powder, analytical HPLC: tR = 10.2 min; MALDI-TOF-MS: calculated for C174H288N14O118S, M = 4496.24 Da, found, 4519.69 [M + Na]+.
Synthesis of N-glycan cluster 10c from the mono-transglycosylated compound 10a
A solution of 10a (0.30 mg, 0.10 μmol) and CT-GlcNAc oxazoline (8) (0.50 mg, 0.35 μmol) in a phosphate buffer (50 mM, pH 7.5, 5.0 μL) was incubated with EndoM-N175A (30 mU) at 30 °C for 8h. The reaction mixture was subject to HPLC purification to give the doubly glycosylated product 10c (50% yield).
Transglycosylation with the Man9GlcNAc oxazoline (9) by EndoM-N175A
A solution of GlcNAc2Man3GlcNAc2-LC-biotin (7) (0.20 mg, 0.12 μmol) and Man9GlcNAc oxazoline (11) (0.60 mg, 0.36 μmol) in a phosphate buffer (50 mM, pH 7.5, 15 μL) was incubated with EndoM-N175A (10 mU) at 30°C. The reaction was monitored by analytical HPLC. After 4h, HPLC indicated the formation of the 6-arm glycosylated product 12a in 27% yield and the 3-arm glycosylated product 12b in 3% yield. The products were purified by HPLC. The doubly glycosylated product was formed in only trace amount (as indicated by MS analysis), which was not isolated for further analysis.
Man9GlcNAc2Man(1,6)-[GlcNAcMan(1,3)]-ManGlcNAc2-LC-biotin (12a): white powder, analytical HPLC: tR = 10.7 min; MALDI-TOF-MS: calculated for C128H213N9O88S, M = 3318.13 Da, found, 3341.78 [M + Na]+.
Man9GlcNAc2Man(1,3)-[GlcNAcMan(1,3)]-ManGlcNAc2-LC-biotin (12b): white powder, analytical HPLC: tR = 11.0 min; MALDI-TOF-MS: calculated for C128H213N9O88S, M = 3318.13 Da, found, 3341.57 [M + Na]+.
Synthesis of N-glycan cluster 13
A solution of 10a (0.10 mg, 33 nmol) and Man3GlcNAc oxazoline (5) (0.10 mg, 0.14 μmol) in a phosphate buffer (50 mM, pH 7.5, 5.0 μL) was incubated with Endo-A (1.0 mU) at 30°C for 2h. Analytical HPLC monitoring indicated the formation of the hybrid glycosylated product 13 in a quantitative yield. The product was purified by HPLC to give CT-GlcNAc2Man(1,6)-[Man3GlcNAc2Man(1,3)]-ManGlcNAc2-LC-biotin (13). Analytical HPLC: tR = 10.4 min; MALDI-TOF-MS: calculated for C146H242N12O98S, M = 3763.40 Da, found, 3786.87 [M + Na]+.
Synthesis of N-glycan cluster 14
A solution of 10c (0.15 mg, 33 nmol) in a phosphate buffer (50 mM, pH 5.5, 30 μL) was incubated with β-1,4-galactosidase (10 U) at 37°C for overnight. The reaction was monitored by analytical HPLC until the complete removal of all the terminal galactose moieties. The residue was subject to preparative HPLC purification. The fractions containing product were combined and lyophilized to give GlcNAc2Man3GlcNAc2Man(1,6)-[GlcNAc2Man3GlcNAc2Man(1,3)]-ManGlcNAc2-LC-biotin (14) (quantitative yield) as a white powder. Analytical HPLC: tR = 10.7 min; MALDI-TOF-MS: calculated for C150H248N14O98S, M = 3847.67 Da, found, 3870.97 [M + Na]+.
Synthesis of N-glycan cluster 15
A solution of 14 (0.10 mg, 26 nmol) and Man3GlcNAc oxazoline (5) (0.30 mg, 0.44 μmol) in a phosphate buffer (50 mM, pH 7.5, 15 μL) was incubated with Endo-A (2.0 mU) at 30°C. HPLC monitoring indicated the complete transglycosylation after 2h incubation. The reaction mixture was subject to preparative HPLC. The fractions containing product were combined and lyophilized to give the N-glycan cluster (15) (quantitative yield). Analytical HPLC: tR = 9.6 min; MALDI-TOF-MS: calculated for C254H420N18O178S, M = 6602.41 Da, found, 6625.98 [M + Na]+.
Lectin microarray analysis
Lectins were immobilized on glass slides according to the previously reported method 17. The lectin microarray profilings of the glycan clusters were carried out following the literature method 17, with some modifications. Briefly, the biotinylated N-glycan clusters were applied on lectin arrays at a concentration of 100 nM and incubated at 4°C for 5h. After washing to remove the unbound N-glycans, the lectin arrays were incubated with a fluorescence (Cy3)-labeled Streptavidin at a concentration of 1 μg/ml at r.t. for 30 min. The array-captured N-glycans were then visualized and quantified by scanning the arrays with a specialized GlycoStation (GP Biosciences Ltd, Yokohama, Japan). For each compound, the binding responses with the lectin arrays were quantitatively determined and expressed as mean fluorescence intensity (MFI) of triplicate detections. In all the assays, GlcNAc-LC-biotin (4) was used as the control. Overlay plots were produced using JMP-Genomics 4.0 software package (SAS Institute in Cary, North Carolina).
Supplementary Material
Acknowledgments
We thank Prof. Kaoru Takegawa for providing the pGEX-2T/Endo-A plasmid; Prof. Kenji Yamamoto and Ms. Midori Umekawa for providing the pET23b-Endo-M plasmid; and Dr. Cishan Li for expressing the endo-enzymes. This work was supported by the National Institutes of Health (NIH) grants R01 GM080374 (to LXW) and U01 CA128416 (To DW).
Footnotes
Supporting Information available: Complete Ref. 4a; the enzymatic transformation coupled with MS analysis for the characterization of the regio-selective transglycosylation products 10a, 10b, and 12a; the 1H-NMR, 13C-NMR, MS spectra, and HPLC profiles of key compounds. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.(a) Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dwek RA. Chem Rev. 1996;96:683–720. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]; (b) Helenius A, Aebi M. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]; (c) Haltiwanger RS, Lowe JB. Annu Rev Biochem. 2004;73:491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]; (d) Dube DH, Bertozzi CR. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]; (e) Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. Annu Rev Immunol. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]; (f) Gamblin DP, Scanlan EM, Davis BG. Chem Rev. 2009;109:131–163. doi: 10.1021/cr078291i. [DOI] [PubMed] [Google Scholar]
- 2.(a) Jefferis R. Biotechnol Prog. 2005;21:11–16. doi: 10.1021/bp040016j. [DOI] [PubMed] [Google Scholar]; (b) Kaneko Y, Nimmerjahn F, Ravetch JV. Science. 2006;313:670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]; (c) Anthony RM, Nimmerjahn F, Ashline DJ, Reinhold VN, Paulson JC, Ravetch JV. Science. 2008;320:373–376. doi: 10.1126/science.1154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Lee RT, Lee YC. Glycoconj J. 2000;17:543–551. doi: 10.1023/a:1011070425430. [DOI] [PubMed] [Google Scholar]; (b) Lundquist JJ, Toone EJ. Chem Rev. 2002;102:555–578. doi: 10.1021/cr000418f. [DOI] [PubMed] [Google Scholar]; (c) Brewer CF, Miceli MC, Baum LG. Curr Opin Struct Biol. 2002;12:616–623. doi: 10.1016/s0959-440x(02)00364-0. [DOI] [PubMed] [Google Scholar]
- 4.(a) Blixt O, et al. Proc Natl Acad Sci USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Feizi T, Chai W. Nat Rev Mol Cell Biol. 2004;5:582–588. doi: 10.1038/nrm1428. [DOI] [PubMed] [Google Scholar]; (c) Xia B, Kawar ZS, Ju T, Alvarez RA, Sachdev GP, Cummings RD. Nat Methods. 2005;2:845–850. doi: 10.1038/nmeth808. [DOI] [PubMed] [Google Scholar]; (d) Oyelaran O, Gildersleeve JC. Expert Rev Vaccines. 2007;6:957–969. doi: 10.1586/14760584.6.6.957. [DOI] [PubMed] [Google Scholar]; (e) Pilobello KT, Mahal LK. Curr Opin Chem Biol. 2007;11:300–305. doi: 10.1016/j.cbpa.2007.05.002. [DOI] [PubMed] [Google Scholar]; (f) Hirabayashi J. J Biochem. 2008;144:139–147. doi: 10.1093/jb/mvn043. [DOI] [PubMed] [Google Scholar]; (g) Zhao J, Patwa TH, Lubman DM, Simeone DM. Curr Opin Mol Ther. 2008;10:602–610. [PMC free article] [PubMed] [Google Scholar]; (h) Liang PH, Wu CY, Greenberg WA, Wong CH. Curr Opin Chem Biol. 2008;12:86–92. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Hsu KL, Mahal LK. Curr Opin Chem Biol. 2009;13:427–432. doi: 10.1016/j.cbpa.2009.07.013. [DOI] [PubMed] [Google Scholar]; (j) Oyelaran O, Gildersleeve JC. Curr Opin Chem Biol. 2009;13:406–413. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF, Cummings RD. Chem Biol. 2009;16:36–47. doi: 10.1016/j.chembiol.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Raman R, Raguram S, Venkataraman G, Paulson JC, Sasisekharan R. Nat Methods. 2005;2:817–824. doi: 10.1038/nmeth807. [DOI] [PubMed] [Google Scholar]; (b) Paulson JC, Blixt O, Collins BE. Nat Chem Biol. 2006;2:238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 6.(a) Yamamoto K. J Biosci Bioeng. 2001;92:493–501. doi: 10.1263/jbb.92.493. [DOI] [PubMed] [Google Scholar]; (b) Wang LX. Carbohydr Res. 2008;343:1509–1522. doi: 10.1016/j.carres.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Takegawa K, Yamaguchi S, Kondo A, Kato I, Iwahara S. Biochem Int. 1991;25:829–835. [PubMed] [Google Scholar]; (b) Takegawa K, Yamaguchi S, Kondo A, Iwamoto H, Nakoshi M, Kato I, Iwahara S. Biochem Int. 1991;24:849–855. [PubMed] [Google Scholar]; (c) Fan JQ, Takegawa K, Iwahara S, Kondo A, Kato I, Abeygunawardana C, Lee YC. J Biol Chem. 1995;270:17723–17729. doi: 10.1074/jbc.270.30.17723. [DOI] [PubMed] [Google Scholar]; (d) Takegawa K, Tabuchi M, Yamaguchi S, Kondo A, Kato I, Iwahara S. J Biol Chem. 1995;270:3094–3099. doi: 10.1074/jbc.270.7.3094. [DOI] [PubMed] [Google Scholar]; (e) Wang LX, Fan JQ, Lee YC. Tetrahedron Lett. 1996;37:1975–1978. [Google Scholar]; (f) Wang LX, Tang M, Suzuki T, Kitajima K, Inoue Y, Inoue S, Fan JQ, Lee YC. J Am Chem Soc. 1997;119:11137–11146. [Google Scholar]; (g) Deras IL, Takegawa K, Kondo A, Kato I, Lee YC. Bioorg Med Chem Lett. 1998;8:1763–1766. doi: 10.1016/s0960-894x(98)00306-0. [DOI] [PubMed] [Google Scholar]; (h) Fujita K, Takegawa K. Biochem Biophys Res Commun. 2001;282:678–682. doi: 10.1006/bbrc.2001.4631. [DOI] [PubMed] [Google Scholar]; (i) Fujita K, Miyamura T, Sano M, Kato I, Takegawa K. Journal of Biosci Bioeng. 2002;93:614–617. [PubMed] [Google Scholar]; (j) Singh S, Ni J, Wang LX. Bioorg Med Chem Lett. 2003;13:327–330. doi: 10.1016/s0960-894x(02)01025-9. [DOI] [PubMed] [Google Scholar]; (k) Li H, Singh S, Zeng Y, Song H, Wang LX. Bioorg Med Chem Lett. 2005;15:895–898. doi: 10.1016/j.bmcl.2004.12.066. [DOI] [PubMed] [Google Scholar]; (l) Wang LX, Song H, Liu S, Lu H, Jiang S, Ni J, Li H. ChemBioChem. 2005;6:1068–1074. doi: 10.1002/cbic.200400440. [DOI] [PubMed] [Google Scholar]
- 8.(a) Yamamoto K, Kadowaki S, Watanabe J, Kumagai H. Biochem Biophys Res Commun. 1994;203:244–252. doi: 10.1006/bbrc.1994.2174. [DOI] [PubMed] [Google Scholar]; (b) Haneda K, Inazu T, Yamamoto K, Kumagai H, Nakahara Y, Kobata A. Carbohydr Res. 1996;292:61–70. doi: 10.1016/s0008-6215(96)91025-3. [DOI] [PubMed] [Google Scholar]; (c) Haneda K, Inazu T, Mizuno M, Iguchi R, Yamamoto K, Kumagai H, Aimoto S, Suzuki H, Noda T. Bioorg Med Chem Lett. 1998;8:1303–1306. doi: 10.1016/s0960-894x(98)00209-1. [DOI] [PubMed] [Google Scholar]; (d) Mizuno M, Haneda K, Iguchi R, Muramoto I, Kawakami T, Aimoto S, Yamamoto K, Inazu T. J Am Chem Soc. 1999;121:284–290. [Google Scholar]; (e) Mori T, Sekine Y, Yamamoto K, Okahata Y. Chem Commun (Camb) 2004:2692–2693. doi: 10.1039/b411082j. [DOI] [PubMed] [Google Scholar]; (f) Yamanoi T, Yoshida N, Oda Y, Akaike E, Tsutsumida M, Kobayashi N, Osumi K, Yamamoto K, Fujita K, Takahashi K, Hattori K. Bioorg Med Chem Lett. 2005;15:1009–1013. doi: 10.1016/j.bmcl.2004.12.040. [DOI] [PubMed] [Google Scholar]; (g) Fujita K, Yamamoto K. Biochim Biophys Acta. 2006;1760:1631–1635. doi: 10.1016/j.bbagen.2006.09.003. [DOI] [PubMed] [Google Scholar]; (h) Makimura Y, Watanabe S, Suzuki T, Suzuki Y, Ishida H, Kiso M, Katayama T, Kumagai H, Yamamoto K. Carbohydr Res. 2006;341:1803–1808. doi: 10.1016/j.carres.2006.04.024. [DOI] [PubMed] [Google Scholar]; (i) Haneda K, Takeuchi M, Tagashira M, Inazu T, Toma K, Isogai Y, Hori M, Kobayashi K, Takegawa K, Yamamoto K. Carbohydr Res. 2006;341:181–190. doi: 10.1016/j.carres.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 9.(a) Fujita M, Shoda S, Haneda K, Inazu T, Takegawa K, Yamamoto K. Biochim Biophys Acta. 2001;1528:9–14. doi: 10.1016/s0304-4165(01)00164-7. [DOI] [PubMed] [Google Scholar]; (b) Li H, Li B, Song H, Breydo L, Baskakov IV, Wang LX. J Org Chem. 2005;70:9990–9996. doi: 10.1021/jo051729z. [DOI] [PubMed] [Google Scholar]; (c) Wei Y, Li C, Huang W, Li B, Strome S, Wang LX. Biochemistry. 2008;47:10294–10304. doi: 10.1021/bi800874y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Rising TW, Claridge TD, Davies N, Gamblin DP, Moir JW, Fairbanks AJ. Carbohydr Res. 2006;341:1574–1596. doi: 10.1016/j.carres.2006.03.007. [DOI] [PubMed] [Google Scholar]; (e) Rising TW, Claridge TD, Moir JW, Fairbanks AJ. ChemBioChem. 2006;7:1177–1180. doi: 10.1002/cbic.200600183. [DOI] [PubMed] [Google Scholar]; (f) Rising TW, Heidecke CD, Moir JW, Ling Z, Fairbanks AJ. Chem Eur J. 2008;14:6444–6464. doi: 10.1002/chem.200800365. [DOI] [PubMed] [Google Scholar]
- 10.Li B, Zeng Y, Hauser S, Song H, Wang LX. J Am Chem Soc. 2005;127:9692–9693. doi: 10.1021/ja051715a. [DOI] [PubMed] [Google Scholar]
- 11.(a) Li B, Song H, Hauser S, Wang LX. Org Lett. 2006;8:3081–3084. doi: 10.1021/ol061056m. [DOI] [PubMed] [Google Scholar]; (b) Zeng Y, Wang J, Li B, Hauser S, Li H, Wang LX. Chem Eur J. 2006;12:3355–3364. doi: 10.1002/chem.200501196. [DOI] [PubMed] [Google Scholar]; (c) Ochiai H, Huang W, Wang LX. J Am Chem Soc. 2008;130:13790–13803. doi: 10.1021/ja805044x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umekawa M, Huang W, Li B, Fujita K, Ashida H, Wang LX, Yamamoto K. J Biol Chem. 2008;283:4469–4479. doi: 10.1074/jbc.M707137200. [DOI] [PubMed] [Google Scholar]
- 13.Huang W, Li C, Li B, Umekawa M, Yamamoto K, Zhang X, Wang LX. J Am Chem Soc. 2009;131:2214–2223. doi: 10.1021/ja8074677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heidecke CD, Ling Z, Bruce NC, Moir JW, Parsons TB, Fairbanks AJ. ChemBioChem. 2008;9:2045–2051. doi: 10.1002/cbic.200800214. [DOI] [PubMed] [Google Scholar]
- 15.Huang W, Ochiai H, Zhang X, Wang LX. Carbohydr Res. 2008;343:2903–2913. doi: 10.1016/j.carres.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Huang W, Groothuys S, Heredia A, Kuijpers BH, Rutjes FP, van Delft FL, Wang LX. ChemBioChem. 2009;10:1234–1242. doi: 10.1002/cbic.200800741. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ochiai H, Huang W, Wang LX. Carbohydr Res. 2009;344:592–598. doi: 10.1016/j.carres.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuno A, Uchiyama N, Koseki-Kuno S, Ebe Y, Takashima S, Yamada M, Hirabayashi J. Nat Methods. 2005;2:851–856. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura-Tsuruta S, Uchiyama N, Peumans WJ, Van Damme EJ, Totani K, Ito Y, Hirabayashi J. FEBS J. 2008;275:1227–1239. doi: 10.1111/j.1742-4658.2008.06282.x. [DOI] [PubMed] [Google Scholar]
- 19.Baenziger JU, Fiete D. J Biol Chem. 1979;254:9795–9799. [PubMed] [Google Scholar]
- 20.De Boeck H, Loontiens FG, Lis H, Sharon N. Arch Biochem Biophys. 1984;234:297–304. doi: 10.1016/0003-9861(84)90352-7. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura-Tsuruta S, Kominami J, Kuno A, Hirabayashi J. Biochem Biophys Res Commun. 2006;347:215–220. doi: 10.1016/j.bbrc.2006.06.073. [DOI] [PubMed] [Google Scholar]
- 22.(a) Shibuya N, Goldstein IJ, Shafer JA, Peumans WJ, Broekaert WF. Arch Biochem Biophys. 1986;249:215–224. doi: 10.1016/0003-9861(86)90577-1. [DOI] [PubMed] [Google Scholar]; (b) Broekaert WF, JVANP, Leyns F, Joos H, Peumans WJ. Science. 1989;245:1100–1102. doi: 10.1126/science.245.4922.1100. [DOI] [PubMed] [Google Scholar]
- 23.Harata K, Muraki M. J Mol Biol. 2000;297:673–681. doi: 10.1006/jmbi.2000.3594. [DOI] [PubMed] [Google Scholar]
- 24.(a) Hester G, Wright CS. J Mol Biol. 1996;262:516–531. doi: 10.1006/jmbi.1996.0532. [DOI] [PubMed] [Google Scholar]; (b) Wright CS, Hester G. Structure. 1996;4:1339–1352. doi: 10.1016/s0969-2126(96)00141-4. [DOI] [PubMed] [Google Scholar]
- 25.Kaku H, Van Damme EJ, Peumans WJ, Goldstein IJ. Arch Biochem Biophys. 1990;279:298–304. doi: 10.1016/0003-9861(90)90495-k. [DOI] [PubMed] [Google Scholar]
- 26.Kilpatrick DC. Biochem J. 1980;185:269–272. doi: 10.1042/bj1850269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 28.O’Reilly MK, Collins BE, Han S, Liao L, Rillahan C, Kitov PI, Bundle DR, Paulson JC. J Am Chem Soc. 2008;130:7736–7745. doi: 10.1021/ja802008q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujita K, Tanaka N, Sano M, Kato I, Asada Y, Takegawa K. Biochem Biophys Res Commun. 2000;267:134–138. doi: 10.1006/bbrc.1999.1963. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.