Abstract
A novel recombinant human parainfluenza virus type 1 (rHPIV1), rHPIV1-C+P, in which the overlapping open reading frames of the C and P genes were separated in order to introduce mutations into the C gene without affecting P, was generated. Infectious rHPIV1-C+P was readily recovered and replicated as efficiently as HPIV1 wild-type (wt) in vitro and in African green monkeys (AGMs). rHPIV1-C+P expressed increased levels of C protein and, surprisingly, activated the type I IFN and apoptosis responses more strongly than HPIV1 wt. rHPIV1-C+P provided a useful backbone for recovering an attenuated P/C gene mutation, (Δ84–85), which was previously unrecoverable, likely due to detrimental effects of the deletion on the P protein. rHPIV1-CΔ84–85+P and an additional mutant, rHPIV1-CΔ169–170 +P, were found to replicate to similar titers in vitro and to activate the type I IFN and apoptosis responses to a similar degree as rHPIV1-C+P. rHPIV1-CΔ84–85+P was found to be highly attenuated in AGMs, and all viruses were immunogenic and effective in protecting AGMs against challenge with HPIV1 wt. rHPIV1-CΔ84–85+P will be investigated as a potential live-attenuated vaccine candidate for HPIV1.
Keywords: Human parainfluenza virus, C protein, vaccine candidates
Introduction
Human parainfluenza virus type 1 (HPIV1) is a clinically important pediatric respiratory pathogen. The virus belongs to the Paramyxoviridae family, which includes other important human pathogens such as HPIV2 and HPIV3, respiratory syncytial virus (RSV), measles virus, mumps virus, and human metapneumovirus [1]. The HPIVs are enveloped, nonsegmented, single-stranded, negative-sense RNA viruses, which can be further classified in the genera Respirovirus (HPIV1 and HPIV3) and Rubulavirus (HPIV2). HPIV1, -2, and -3 are significant respiratory pathogens for infants and young children and, collectively, they are second only to RSV as the leading cause of pediatric hospitalization for virally-induced respiratory disease [1–3]. HPIV1 is the most common cause of croup, and other clinical manifestations range from mild upper respiratory illness, including rhinitis and pharyngitis, to severe lower respiratory illness, including bronchiolitis and pneumonia [1, 4–7]. A licensed vaccine is currently not available for the prevention of HPIV disease, but experimental live attenuated candidate vaccines for HPIV1, -2, and -3 are currently in clinical trials [8–14]. The current experimental HPIV1 vaccine candidate, rHPIV1-CR84G/Δ170HNT553ALY942A, is a cDNA-derived recombinant HPIV1 (rHPIV1), which was generated using a reverse genetics system [12]. This virus contains a combination of temperature sensitive (ts) attenuating (att) mutations and non-ts att mutations in the P/C, HN and L genes, which include a non-ts att “set” of point mutations (P/CR84G and HNT553A), a deletion (Δ) involving codon 170 in P/C (P/CΔ170) that is non-ts att, and a codon optimized for genetic stability at codon 942 in L (LY942A) that is ts att [15, 16].
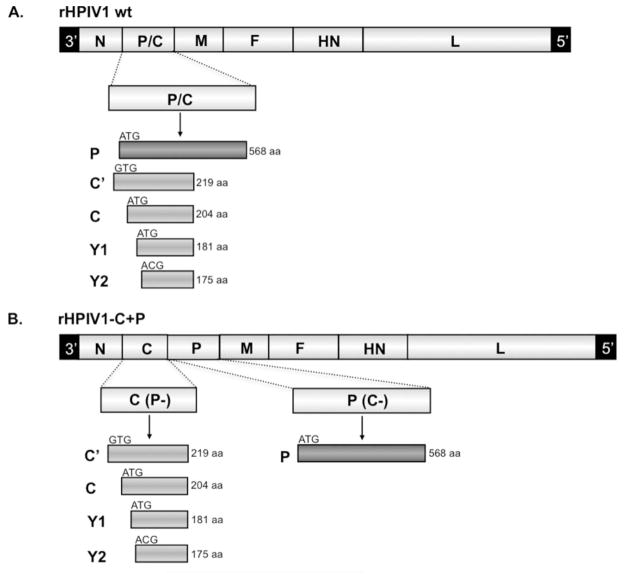
The HPIV1 genome contains six genes in the order 3′-N-P/C-M-F-HN-L-5′ [17]. Each gene encodes a single protein, with the exception of the P/C gene which contains two overlapping open reading frames (ORFs). One P/C gene ORF encodes the phosphoprotein, P, and the second encodes up to four accessory C proteins, C′, C, Y1, and Y2, which initiate at different, staggered initiation codons and are carboxy co-terminal (Figure 1A)[1]. C proteins are expressed by members of the Respirovirus, Morbillivirus, and Henipavirus genera, but not by viruses that belong to the Rubulavirus and Avulavirus genera. The paramyxovirus C proteins studied to date are nonessential accessory proteins that contribute significantly to virus replication and virulence in vivo [16, 18–21]. Paramyxovirus C proteins have been shown to play an important role in virus replication and pathogenesis through a number of different mechanisms including: inhibition of the host innate immune response via antagonism of type I interferon (IFN) induction and/or IFN signaling [21–26]; regulation of viral mRNA synthesis via binding to the L polymerase protein [20, 27–30]; involvement in virion assembly and budding via an interaction with host cell AIP1/Alix [31–33]; and regulation of apoptosis [21, 34–38]. Since the C proteins are multifunctional, it would be desirable to define the contribution of the individual C proteins to viral replication, host interactions, and pathogenesis as well as the domains within each protein that are responsible for each of these functions. Unfortunately, since there is extensive overlap between the C and P ORFs, mutations introduced into the C ORF frequently also affect the P protein.
Figure 1.
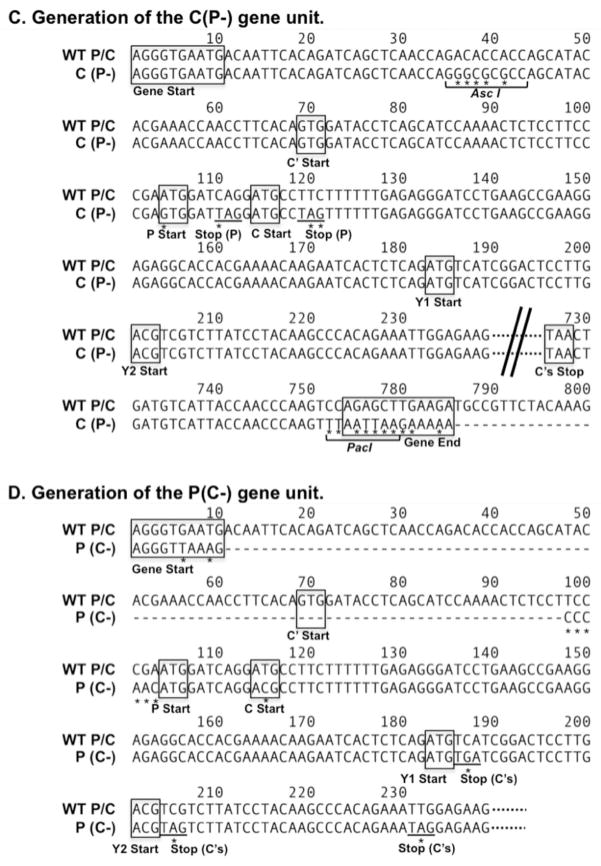
Designing the rHPIV1-C+P cDNA. (A) The rHPIV1 wt genome, shown 3′ to 5′, contains six genes including the P/C gene that translates the phosphoprotein P from one ORF and the four carboxy-coterminal C proteins, C′, C, Y1 and Y2, from a second, overlapping ORF. (B) A rHPIV1 wt genome, modified to contain unique AscI and NotI restriction enzyme sites (HPIV1 at nucleotide positions 1776 and 3609 of the genome, flanking the P and C ORFs), was used to generate the novel virus genome, rHPIV1-C+P. This novel genome contains seven genes, in which the P/C gene has been separated into two distinct genes, C and P (inserted in that order), where the C gene encodes only C′, C, Y1 and Y2 and the P gene encodes only P. In order to generate this new virus genome, the P/C gene was duplicated and one copy, C(P-), was modified to silence the P ORF without affecting C, and the other copy, P(C-), was modified to silence the C ORF without affecting P, as shown in panels (C) and (D), respectively; nucleotide substitutions are indicated (*). (C) To generate the C(P-) gene, expression of the P protein was silenced by changing the methionine P start codon (ATG) to valine (GTG) and introducing two stop codons at P codons 3 and 6. In addition, the downstream end of the gene was deleted beginning 23 nucleotides after the C stop codon, and a PacI restriction enzyme site was introduced for cloning purposes, along with the indicated (boxed) gene-end sequence. (D) For the P(C-) gene, the P gene start sequence was replaced with the N gene start sequence (an incidental change) and the 93 nt between the gene-start signal and P start codon were deleted, as indicated (−), and replaced with a 6-nt spacer CCCAAC, thus eliminating the first 11 codons of C′ including its start codon. The placement of this CCCAAC spacer immediately upstream of the P start codon (AACATG) was designed to optimize the Kozak sequence and to reduce translational initiation at the downstream start codons. The methionine start codon of the C protein (ATG) was converted to threonine (ACG). Since the Y1 and Y2 start codons could not be modified without affecting the P ORF, one stop codon was introduced immediately downstream of the Y1 start codon and two stop codons were introduced downstream of the Y2 start codon: these stop codons would affect all four C proteins. The cDNA incorporating these modifications was used to recover infectious rHPIV1-C+P.
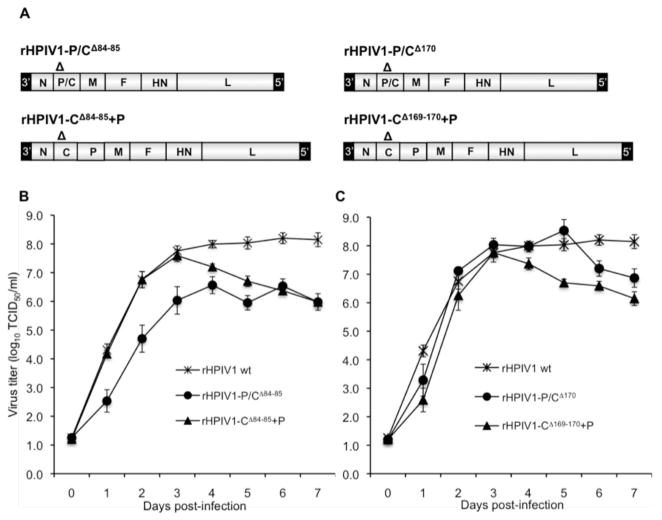
Since the C proteins are non-essential proteins that antagonize the IFN response and since mutations that abrogate the function of viral IFN antagonists are often attenuating in vivo [18], the HPIV1 C proteins are attractive targets for attenuating the virus through site-specific mutagenesis [16], although complete deletion of the C protein has been found to be over-attenuating [21]. Two P/C gene mutations, P/CR84G and P/CΔ170, have previously been found to modify the innate immune response and to restrict replication of HPIV1 in vivo [16, 26], and therefore can be used to develop live attenuated virus vaccines. The CR84G mutation attenuates HPIV1 for the upper respiratory tract (URT) of African Green monkeys (AGMs) only in combination with the HNT553A mutation [39]. However, HPIV1 mutants containing the P/CR84G mutation alone, in contrast to HPIV1 wild type (wt), permit the efficient induction of type I IFN but, like HPIV1 wt, inhibit type I IFN signaling [16, 26]. This suggests that residue 84 of the C proteins is involved in the suppression of IFN production but does not affect IFN signaling. However, this suggestion is offered with the caveat that the CR84G mutation also introduces a single amino acid substitution in P (E87G) [16], so it remained unclear whether the effect of the mutation in C or P was responsible for the effect on the induction of IFN. Either a deletion or point mutation involving codon 170 (P/CΔ170 and P/CF170S, respectively) enable efficient type I IFN production and signaling during infection, and both mutants are associated with attenuation in vivo [16, 26, 40]. Since the change in P associated with the P/CF170S mutation is translationally silent [16] the association of this phenotype with a mutation at codon 170 in C was unambiguous. The P/CΔ170 mutation was chosen for inclusion in the current HPIV1 vaccine candidate because reversion to wild type phenotype is much less likely with deletion mutations compared to point mutations. The P/CΔ170 mutation involves a six-nucleotide deletion affecting codons 168–170 (from the third nucleotide of codon 168 to the second nucleotide of codon 170) in C and results in an amino acid change of RDF to S; this deletion also affects P, deleting the 6 nucleotides encoding amino acids 172 and 173 [16] (Table 1). We have also previously attempted to generate a deletion of codons 84 and 85 in an effort to stabilize the P/CR84G mutation [16]. In that study, rHPIV1-P/CΔ84–85 proved difficult to recover using HEp-2 [16], and the viruses that were recovered contained multiple adventitious mutations and did not grow efficiently. Deletion of codons 84 and 85 in C also affect the P protein, resulting in codon changes that have the effect of deleting amino acids 87 and 88 in P (Table 1). We hypothesized that the difficulty in recovering the P/CΔ84–85 deletion mutation might be due to a negative effect of this deletion on P, a protein required for transcriptase/replicase activities. Nonetheless, for the present study, we finally were able to recover a rHPIV1-P/CΔ84–85 virus lacking adventitious mutations.
Table 1.
Summary of the effect of the deletion mutations on the P and C ORFs of the rHPIV1 mutant viruses.
| Virus a | P ORF Amino Acid/Nucleotide change (wild type sequence → mutant sequence) b | C ORF Amino Acid/Nucleotide change (wild type sequence → mutant sequence) b | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| codon #: | 87 | 88 | 89 | codon #: | 83 | 84 | 85 | 86 | ||||||||
| rHPIV1-P/CΔ84–85 | aa: | E | S | G | → | G | aa: | R | R | V | E | → | R | E | ||
| nt: | GAG | AGT | GGA | → | GGA | nt: | AGG | AGA | GTG | GAA | → | AGG | GAA | |||
| rHPIV1-CΔ84–85+P | No change | codon #: | 83 | 84 | 85 | 86 | ||||||||||
| aa: | R | R | V | E | → | R | E | |||||||||
| nt: | AGG | AGA | GTG | GAA | → | AGG | GAA | |||||||||
| codon #: | 171 | 172 | 173 | 174 | codon #: | 168 | 169 | 170 | ||||||||
| rHPIV1-P/CΔ170 | aa: | E | G | F | P | → | E | P | aa: | R | D | F | → | S | ||
| nt: | GAG | GGA | TTT | CCA | → | GAG | CCA | nt: | AGG | GAT | TTC | → | AGC | |||
| codon #: | 168 | 169 | 170 | |||||||||||||
| rHPIV1-CΔ169–170+P | No change | aa: | R | D | F | Q | → | R | Q | |||||||
| nt: | AGG | GAT | TTC | CAG | → | AGG | CAG | |||||||||
The virus nomenclature, as used throughout the study.
Nucleotide deletions are indicated by underlined text.
In the present study, we also generated a novel rHPIV1, designated rHPIV1-C+P, in which the P/C gene was separated into two distinct genes that express the C proteins and the P protein independently. Thus mutations, including deletion mutations, could be introduced into C without affecting P. Using the rHPIV1-C+P cDNA, we have been able to generate mutants that express wt P protein and that contain the Δ84–85 or Δ169–170 mutations in the C proteins only. These viruses were successfully recovered and evaluated in vitro and in vivo to determine: i) the effect of separating the C and P genes on HPIV1 replication in vitro and in vivo, on the IFN response to virus infection in vitro, on IFN signaling, and on apoptosis; ii) the effect of the deletion mutations on these same properties; and iii) the potential of rHPIV1-C+P containing the Δ84–85 or Δ169–170 deletion mutations in the C proteins for use as live attenuated HPIV1 vaccine candidates. Each of the novel viruses grew efficiently in vitro and, most notably, rHPIV1-CΔ84–85+P replicated to significantly higher titer than the newly generated rHPIV1-P/CΔ84–85. In addition, both the rHPIV1-C+P and the two C+P deletion mutants induced type 1 IFN to varying levels, and all three viruses induced a similar degree of apoptosis. Separation of the C and P genes did not have a significant attenuating effect in AGMs, but introduction of the Δ84–85 mutation into the C protein of rHPIV1-C+P restricted virus replication in AGMs significantly. rHPIV1-CΔ84–85+P is currently considered a HPIV1 vaccine candidate suitable for clinical development due to its high level of attenuation in both the URT and lower respiratory tract (LRT) of non-human primates and its efficacy in protecting against HPIV1 wt challenge.
Materials and Methods
Cells and Viruses
LLC– MK2 cells (ATCC CCL-7.1) and HEp–2 cells (ATCC CCL-23) were maintained in Opti– MEM I (Gibco–Invitrogen, Inc. Grand Island, NY) supplemented with 5% FBS and gentamicin sulfate (50 μg/ml). A549 cells (ATCC CCL -185) were maintained in F-12 nutrient mixture (HAM) (Gibco-Invitrogen, Inc.) supplemented with 10% FBS, gentamicin sulfate (50 μg/ml) and L-glutamine (4 mM). Vero cells (ATCC CCL-81) were maintained in MEM (Gibco-Invitrogen, Inc.) supplemented with 10% FBS, gentamicin sulfate (50 μg/ml) and L-glutamine (4 mM). BHK-T7 cells, which constitutively express T7 RNA polymerase [41], were kindly provided by Ursula Buchholz, NIAID, and were maintained in GMEM (Gibco-Invitrogen, Inc.) supplemented with 10% FBS, geneticin (1 mg/ml), MEM amino acids, and L-glutamine (2 mM). Cells were maintained at 37°C.
Biologically-derived wt HPIV1 Washington/20993/1964 (HPIV1 wt), the parent of rHPIV1 wt, was originally isolated from a clinical sample in primary AGM kidney (AGMK) cells, passaged two more times in primary AGMK cells [42] and once in LLC-MK2 cells, and is also known as HPIV1LLC1 [39]. The rHPIV1 wt referred to in this study was identical to this biologically-derived virus except for two translationally silent point mutations in the L gene [17] and a mutation in the HN gene, HNT553A, which does not affect virus replication [16]. This rHPIV1 wt was used in all experiments, with the exception that the biological HPIV1 wt was used for the AGM studies, as indicated. In addition to the HPIV1 wt viruses and novel viruses constructed in this study, several viruses which were previously described were used, including the C gene mutants rHPIV1-CF170S and rHPIV1-P/CΔ170 [16, 39], and the C gene deletion mutant, rHPIV1-P(C-) [21]. All virus infections were performed at 32°C in serum-free media containing 1.2% TrypLE Select, a recombinant trypsin-like enzyme (Gibco-Invitrogen, Inc.), in order to cleave and activate the HPIV1 fusion (F) protein, as described previously [17]. For IFN assays, virus stocks were sucrose-purified, as previously described, to minimize contamination with cellular factors, especially IFN [26]. Recombinant vesicular stomatitis virus expressing the green fluorescent protein (VSV-GFP) was originally obtained from John Hiscott [43]. Stocks of VSV-GFP were propagated in Vero cells and sucrose purified as indicated above.
Virus titers in samples were determined by 10-fold serial dilution of virus in 96-well LLC-MK2 monolayer cultures, using two or four wells per dilution, as described previously [44]. Virus titers are expressed as log10 50% tissue culture infectious dose per ml (log10 TCID50/ml). VSV stock titers were determined by plaque assay on Vero cells under a 0.8% methylcellulose overlay.
Antibodies
Polyclonal antisera directed against the HPIV1 N, C, P and F proteins were generated by repeated immunization of rabbits with the following KLH-conjugated peptides: (i) RRLADRKQRLSQANN (amino acid (aa) residues 485–499 of N), for HPIV1-N-485; (ii) LKPERHHENKNHSQMSS (aa residues 24–43 of HPIV1 C′; directed against the amino terminal region of C′ and C), for HPIV1-C-36; (iii) RDPEAEGEAPRKQES (aa residues 10–24 of P), for HPIV1-P-2; and (iv) CYTLESRMRNPYMGNNSN (aa residues 538–555 of F), for HPIV1-F-15. Antisera and peptides were generated at Spring Valley Labs (Woodbine, MD) and ProSci Incorporated (Poway, CA). Two murine monoclonal antibodies directed against the HPIV1 HN protein, designated 8.2.2.A and 4.5, were kindly provided by Dr. Yasuhiko Ito [45]. Other antibodies used included FITC-labeled rabbit anti-human activated caspase 3 (BD Pharmingen, San Jose, CA), peroxidase-labeled goat anti-rabbit IgG (KPL, Gaithersburg, MD) and allophycocyanin-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA).
Construction of novel rHPIV1s containing separated C and P genes
rHPIV1 wt contains a single P/C gene, which expresses the P and C proteins from overlapping ORFs (Figure 1A) [17]. A novel virus, rHPIV1-C+P, was generated in which the antigenomic cDNA of the rHPIV1 wt genome was modified so that the C and P proteins could be expressed from two distinct genes (Figure 1B). A recombinant version of the wt HPIV1 Washington/20993/1964 (Genbank accession number NC_003461), which was modified to contain unique AscI and NotI restriction enzyme sites (rHPIV1 genome positions 1776 and 3609, respectively, flanking the P and C ORFs), was used. The wt P/C gene was excised from the genome by digestion with these two enzymes, and two novel genes were PCR-amplified using the wt P/C gene as a template. In the first gene, C(P-) (Figure 1C), which was modified to express C proteins only, P protein expression was silenced by introducing mutations into the P ORF which did not affect the C ORF (refer to Figure legend 1C for further detail). In the second gene, P(C-) (Figure 1D), which was modified to express only the P protein, silencing of the expression of the four C proteins was achieved by the introduction of deletions and nucleotide substitutions (refer to Figure legend 1D for further detail). The gene order for the new rHPIV1-C+P is: 3′-N-C-P-M-F-HN-L-5′. In addition, rHPIV1-C+P contains the HNT553A mutation. Mutant versions of rHPIV1-C+P were generated to contain codon deletions in the C(P-) gene. These included rHPIV1-CΔ84–85+P and rHPIV1-CΔ169–170+P, which contain deletions of codons 84–85 (both with and without the HNT553A mutation) and 169–170 (without the HNT553A mutation) in the C proteins of the C(P-) gene, respectively. Thus, the deletion in rHPIV1-CΔ169–170+P is slightly different from the deletion mutation present in rHPIV1-P/CΔ170 [16]: the rHPIV1-P/CΔ170 mutation affects codons 168–170 in C, resulting in an amino acid change of RDF to S, whereas rHPIV1-CΔ169–170+P contains a six nucleotide deletion resulting in the complete deletion of codons 169 and 170 (DF; Table 1).
The changes described were achieved with a modified PCR mutagenesis protocol [46], using the Advantage– HF PCR Kit (Clontech Laboratories, Palo Alto, CA). Full– length antigenomic cDNA clones (FLCs) of HPIV1 containing the desired mutations were assembled in T7 polymerase-driven plasmids using standard molecular cloning techniques [17]. Each FLC was sequenced using a Perkin– Elmer ABI 3100 sequencer with the Big Dye sequencing kit (Perkin– Elmer Applied Biosystems, Warrington, UK) to confirm the presence of the introduced mutations and absence of adventitious changes. Each virus genome was designed to conform to correct phasing [17] and to the rule of six, i.e., the nucleotide length of each genome was designed to be an even multiple of six, a requirement for efficient replication of HPIV1 [47].
Recovery of infectious rHPIV1-C+P viruses
All rHPIV1s were generated using a reverse genetics system, in HEp-2 or BHK-T7 cells, as previously described [17, 21]. Upon recovery of each infectious virus, the viral genome was sequenced in its entirety to confirm its sequence and the absence of adventitious mutations, as previously described [12]. Studies presented here include viruses that have been described previously and are included here for comparative purposes, namely rHPIV1 wt, rHPIV1-P/CF170S, rHPIV1-P/CΔ170 and rHPIV1-P(C-) [17].
Western Blot
LLC-MK2 monolayer cultures grown in 6 well plates (Costar) were mock-infected or infected at a multiplicity of infection (MOI) of 5 TCID50 per cell with sucrose-purified rHPIV1 wt or rHPIV1-C+P. Cell lysates were harvested 48 h post infection (p.i.) with 100 μl of 1× Loading Dye Solution sample buffer (Qiagen, Inc) and purified on QIAshredder spin columns (Qiagen, Inc.). Fifteen μl (for C protein probing), 5 μl (for P and F protein probing) or 2.5 μl (for N protein probing) of each sample were reduced, denatured and loaded onto 10% Bis-Tris gels (Invitrogen, Inc.). Gels were run in MOPS buffer (Invitrogen, Inc.), and protein was transferred onto PVDF membranes (Invitrogen, Inc.) and detected as previously described [21].
Kinetics of replication of rHPIV1-P(C-)
The rHPIV1 wt and rHPIV1-C+P viruses were compared in multiple cycle growth curves at a MOI=0.01 TCID50/cell as previously described [12, 21]. Virus titers in the supernatant were determined as described previously [44].
Quantitation of type I IFN production
The amount of type I IFN produced by HPIV1-infected A549 cell cultures was determined by an IFN bioassay and IFN-β ELISA. Confluent A549 cells in 6-well plates were infected with sucrose-purified rHPIV1s at a MOI=5.0 for 1 h at room temperature (RT), in media containing 1.2% TrypLE Select. The cells were then washed and overlaid with media containing 10% FBS without TrypLE Select. Supernatants were harvested at 24, 48, 72 and 96 h p.i. Type I IFN in pH 2.0-treated supernatants was quantitated by type I IFN bioassay on HEp-2 cells and expressed as mean ± SE (pg/ml), as previously described [26]. For the IFN-β ELISA, samples were not pH 2.0-treated, and the procedure was performed according to manufacturer’s instructions (Fujirebio Inc., Tokyo, Japan). IFN-β concentrations determined by ELISA were expressed as mean international units (IU)/ml ± SE.
VSV plaque inhibition assay
The VSV plaque inhibition assay measures the effectiveness of the IFN signaling pathway in establishing an antiviral state with or without prior HPIV1 infection, as previously described [26]. Briefly, Vero cells in 6-well plates were mock-infected or infected with sucrose-purified rHPIV1s at a MOI=5.0 TCID50/cell for 24 h. Cells were then treated with 0, 100 or 1000 IU/ml IFN-β for 24 h prior to infection with VSV-GFP at 500 pfu/well, and the cultures were overlaid with 0.8% methylcellulose in MEM. GFP plaques were determined, as previously described [26]. The data presented are representative of three experiments.
FACS analysis
Fluorescence-activated cell sorting (FACS) analysis was performed for HPIV1 HN antigen and activated caspase 3 as previously described [21]. Briefly, LLC-MK2 cells in six-well plates were mock-infected or infected with rHPIV1 wt or various mutants at a MOI=5 TCID50/cell. Cells were harvested at 72 h p.i., fixed, stained, and analyzed by FACS.
Evaluation of virus replication in AGMs and determination of protective efficacy against HPIV1 wt challenge
Replication of HPIV1 wt and rHPIV1 mutants was determined in AGMs, as previously described [48]. AGMs were inoculated intranasally (i.n.) and intratracheally (i.t.) with 106 TCID50 of HPIV1 in a 1ml inoculum, diluted in L-15. Briefly, nasopharyngeal (NP) swab samples were collected daily for virus quantitation from days 0 to 10 p.i., and tracheal lavage (TL) fluid samples were collected on days 2, 4, 6, 8 and 10 p.i. On day 28 p.i., the AGMs were challenged i.n. and i.t. with 106 TCID50 per site of HPIV1 wt, as described above. NP swabs and TL samples were collected on days 2, 4, 6 and 8 post-challenge. The specimens were collected and stored and virus titers were determined, as previously described [44, 48]. The limit of detection was 0.5 log10 TCID50/ml. The mean sum of daily virus titers for each group was the sum of the titers for all of the days of sampling, excluding day 0.
All animal studies were performed under protocols approved by the National Institute of Allergy and Infectious Disease (NIAID) Animal Care and Use Committee (ACUC).
Evaluation of immune responses in AGMs
Serum was collected from each monkey on days 0 and 28 post-immunization and on day 28 post-challenge (day 56 post-first inoculation), and HPIV1 hemagglutination inhibiting (HAI) antibody titers were determined, as described previously [49], using 0.5% v/v guinea pig erythrocytes and HPIV1 wt as the antigen. The antibody titer was defined as the end-point serum dilution that inhibited hemagglutination and is expressed as the mean reciprocal log2 based titer ± SE.
Statistical analysis
The one-way ANOVA test (Student-Newman-Keuls multiple comparison test) was used for determination of statistically significant differences between data groups (P < 0.05) using Prism 5 (GraphPad Software Inc., San Diego, CA).
Results
Recovery of rHPIV1-C+P
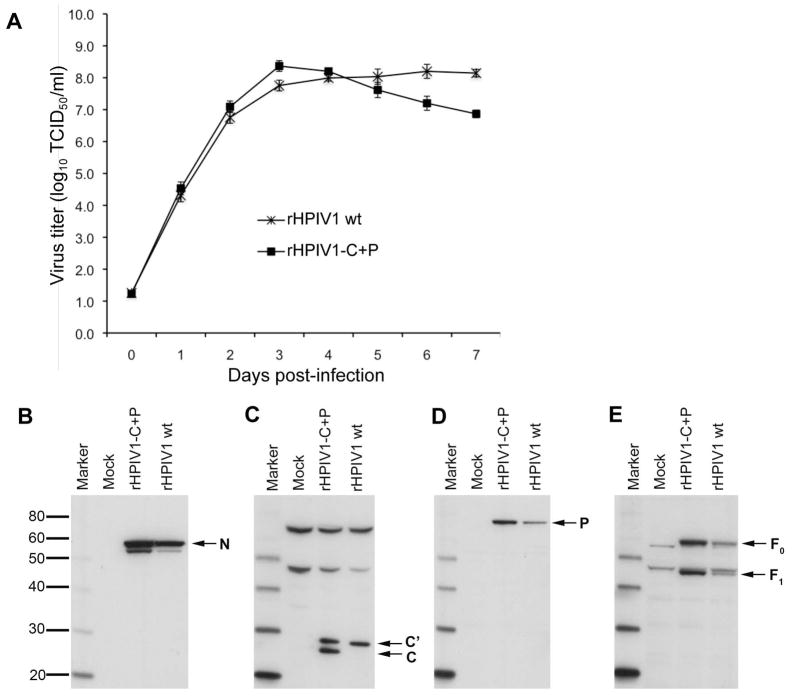
The P/C gene of HPIV1 wt encodes the P protein in one ORF and four carboxy-coterminal C proteins, C′, C, Y1, and Y2, in a second, overlapping ORF (Figure 1A). We engineered a novel virus, rHPIV1-C+P, to contain two distinct genes, designated C(P-) and P(C-), which would express the C proteins only and the P protein only, respectively. The specific changes made to access only one ORF from each unit are described in the materials and methods and in the legend for Figure 1. rHPIV1-C+P was recovered from cDNA in BHK-T7 cells, and sequence analysis of the entire virus genome revealed that rHPIV1-C+P contained all the intended mutations and no unintended changes (data not shown). The replication kinetics of rHPIV1-C+P in LLC-MK2 cells was evaluated in a multiple cycle replication assay. rHPIV1-C+P grew as efficiently as rHPIV1 wt, reaching a peak titer of 8.4 log10 TCID50/ml early in infection (Figure 2A), indicating that separating the P and C genes and thereby adding an extra gene did not significantly affect virus replication in vitro. However, starting on day 4 p.i., rHPIV1-C+P titers declined, likely as a result of cytopathic effect (cpe) observed in the cultures.
Figure 2.
Virus replication and identification of viral proteins during infection with rHPIV1 wt and rHPIV1-C+P in LLC-MK2 cells. (A) LLC-MK2 cells were infected with rHPIV1 wt and rHPIV1-C+P at a MOI of 0.01 TCID50/cell and the media was harvested for virus titration on days 0–7 p.i. Virus titers are shown as the means of 3 replicate cultures ± SEM. (B) LLC-MK2 cell lysates were prepared 48 h following mock-infection or infection with sucrose-purified rHPIV1 wt or rHPIV1-C+P at an input MOI of 5 TCID50/cell. Reduced, denatured cell lysates were resolved by SDS-PAGE electrophoresis and Western blots were prepared and analyzed using rabbit anti-peptide antisera against the (B) HPIV1 N protein, (C) HPIV1 C proteins (C′ and C), (D) HPIV1 P protein, and (E) HPIV1 F protein (uncleaved, F0, and cleaved, F1, forms).
Viral protein expression in rHPIV1 wt and rHPIV1-C+P infected cells
Western blots of infected LLC-MK2 cell lysates using antibodies against HPIV1 N, C, P and F proteins were performed to determine whether viral protein expression was affected by separating the C and P ORFs into two separate monocistronic genes (Figure 2B–D). A band corresponding to the predicted size of the N protein, 57.6kDa, and an additional smaller unidentified band were observed in lysates from cells infected with HPIV1 wt and rHPIV1-C+P using an antiserum specific to the N protein (HPIV1-N-485; Figure 2B). Bands corresponding to the C′ and C proteins, with a predicted mass of 25.6 kDa and 24.1 kDa, respectively, were seen in lysates from cells infected with HPIV1 wt and rHPIV1-C+P using an antiserum (HPIV1-C-36) directed against the amino terminus of the C proteins (Figure 2C). C′ is the most abundant C protein in rHPIV1 wt-infected cells, and although C is not visible in this particular Western blot, it was detected in other gels at low levels (relative to C′) in rHPIV1 wt-infected cells (data not shown). In rHPIV1-C+P-infected cells it appears that C′ and C are expressed at approximately equivalent levels. The basis for the apparent increased expression of C by the rHPIV1-C+P virus is not known, but conceivably could be a consequence of the introduction of a stop codon in the P ORF immediately upstream of the C translational initiation start site (Figure 1C): ribosomes that prematurely exit the P ORF at this introduced stop site might re-initiate at the translational start site of C. Y1 and Y2 were not detected in either lysate using antiserum directed against the carboxy-terminal end of the C proteins (data not shown). A band corresponding in size to the HPIV1 P protein, predicted to be 64.5 kDa, was detected with P specific antiserum (HPIV1-P-2) in both rHPIV1 wt- and rHPIV1-C+P-infected cells (Figure 2D). Using F-specific antiserum (HPIV1-F-15), bands corresponding in size to the uncleaved HPIV1 F0 protein (60.8 kDa) and one of the cleavage products, F1 (48.5kDa), were detected in both rHPIV1 wt- and rHPIV1-C+P-infected cells (Figure 2E). Overall, viral protein expression appeared slightly higher during infection with rHPIV1-C+P compared to rHPIV1 wt in LLC-MK2 cells. Since these Western blots were not quantitative, viral protein expression between protein species could not be compared.
Introduction of the CΔ84–85 or CΔ169–170 mutations into rHPIV1-C+P
Previously, we generated a rHPIV1 mutant containing a deletion affecting codons 168–170 in C, designated as rHPIV1-P/CΔ170, as a means of developing vaccine strains with enhanced stability [16]. In that study, we also attempted to recover a virus with deletion of codons 84 and 85 in C, designated as rHPIV1-P/CΔ84–85. Note that, in each case, six nucleotides were deleted so that the genome length complied with the rule of six [47] (Table 1). Whereas rHPIV1-P/CΔ170 was readily recovered from cDNA and grew efficiently, rHPIV1-P/CΔ84–85 was difficult to recover and all of the recovered viruses contained multiple adventitious mutations throughout the genome [16]. However, in the current study, we succeeded in recovering an isolate of rHPIV1-P/CΔ84–85 that was free of adventitious mutations (Figure 3A). In addition, we engineered the CΔ84–85 and CΔ169–170 mutations into the new rHPIV1-C+P virus and recovered viruses (rHPIV1-CΔ84–85+P and rHPIV1- CΔ169–170+P, respectively) that were free of adventitious mutations (Figure 3A). This made it possible to directly compare the effect of the CΔ84–85 and CΔ169–170 mutations, in either the wt or C+P backbone, on replication in vitro. The level of replication of rHPIV1-CΔ84–85+P was compared with that of rHPIV1-P/CΔ84–85 and HPIV1 wt virus in a multiple cycle growth curve in LLC-MK2 cells (Figure 3B). The peak virus titer of rHPIV1-P/CΔ84–85 in LLC-MK2 cells was 50- to 100-fold lower than that of rHPIV1 wt (Figure 3B) or rHPIV1-C+P (data not shown) and 10-fold lower than that of rHPIV1-CΔ84–85+P. In contrast, the deletion affecting codon 170 of C did not affect peak virus replication in either the rHPIV1 wt or rHPIV1-C+P backbone (Figure 3C). However, the titers of both rHPIV1-CΔ169–170+P and rHPIV1-CΔ84–85+P viruses decreased after day 3, as previously observed for HPIV1 mutants that induce apoptosis in vitro [21]. Importantly, in Vero cells, the cell substrate used for vaccine manufacture, rHPIV1-CΔ84–85+P replicated to peak titers 1000 times higher than those reached by rHPIV1-P/CΔ84–85 (data not shown). This data demonstrates that the rHPIV1-C+P is a useful backbone for inserting attenuating mutations that would otherwise restrict viral replication in vitro to a degree that impedes vaccine manufacture. In addition, the effect of the CΔ84–85 and CΔ169–170 mutations on C function, e.g., on IFN induction/signaling, could now be studied independent of mutations in P.
Figure 3.
Comparison of the multiple cycle replication of rHPIV1 wt with novel rHPIV1-C+Ps. (A) HPIV1 C gene mutants, containing deletion mutations (Δ) at codons 84–85 and 169–170, were prepared on the HPIV1 wt or C+P backbone. (B-C) LLC-MK2 cells were infected with rHPIV1s at a MOI of 0.01 TCID50/cell, and the media was harvested for virus titration on days 0–7 p.i. Virus titers are shown as the mean of 3 replicate cultures ± SEM. Viruses are grouped for direct comparison: (B) rHPIV1 wt, rHPIV1-P/CΔ84–85 and rHPIV1-CΔ84–85+P, and (C) rHPIV1 wt, rHPIV1-P/CΔ170 and rHPIV1-CΔ169–170+P.
IFN production in cells infected with rHPIV1-C+P, rHPIV1-CΔ84–85+P, or rHPIV1-CΔ169–170+P
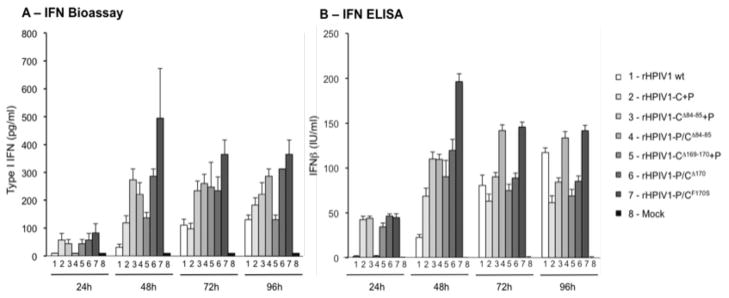
The C proteins of HPIV1 wt inhibit the type I IFN response at the level of IFN production and signaling [21, 25, 26]. Here, we evaluated type I IFN production in human respiratory epithelial A549 cells infected with rHPIV1 wt, rHPIV1-C+P, rHPIV1-CΔ84–85+P, rHPIV1- CΔ169–170+P, and a previously isolated C mutant virus, rHPIV1-P/CF170S, which is a strong inducer of type I IFN and serves as a positive control virus [26] (Figure 4). IFN production was measured with a bioassay using VSV-GFP as an indicator (Figure 4A) and by an IFN-β ELISA (Figure 4B). As expected, type I IFN was not detected in mock-infected cells. IFN production was delayed and of low magnitude in rHPIV1 wt infected cells compared to that in rHPIV1-P/CF170S virus-infected cells (groups 1 and 7, Figure 4). In rHPIV1-C+P-infected cells (group 2, Figure 4), type I IFN was present at 24 and 48 h p.i. at significantly higher concentrations than in rHPIV1 wt-infected cells (P<0.05, compare groups 1 and 2, Figure 4), although this difference disappeared at later time points. We were surprised by the observation that separating the C and P ORFs resulted in early induction of IFN in A549 cells since this virus, like rHPIV1 wt, expresses wt C proteins, albeit in different ratios and concentrations.
Figure 4.
rHPIV1 C gene mutants induce type I IFN production. A549 cell monolayers were either mock-infected or infected with rHPIV1 wt, rHPIV1-C+P or various mutants, at a MOI of 5 TCID50/cell. Aliquots of the overlying medium were taken at 24, 48, 72 and 96 h p.i. and type I IFN proteins were quantified by (A) type I IFN bioassay or (B) IFN-β ELISA. (A) For the type I IFN bioassay, supernatants were pH 2.0-treated and assayed on HEp-2 cells in comparison to an AVONEX® IFN-β standard and data are expressed in pg/ml ± SE based on triplicate samples. (B) For the IFN-β ELISA, supernatants were tested against IFN-β standards, as per the manufacturer’s protocol, and IFN concentrations are expressed in IU/ml ± SE based on triplicate samples.
IFN expression was modestly increased at 48 and 72h p.i. in rHPIV1-CΔ84–85+P-infected cells compared to cells infected with the parental rHPIV1-C+P (compare groups 3 and 2, respectively, Figure 4), suggesting that deletion of codons 84–85 and separation of C and P exert a modest additive effect on the induction of IFN. rHPIV1-CΔ169–170+P and its parent, rHPIV1-C+P, had similar IFN induction profiles (groups 5 and 2, respectively, Figure 4). The effect of the CΔ169–170 and CΔ84–85 mutations on IFN induction was comparable when the mutations were present in either the wt or C+P backbone (Figure 4, compare rHPIV1-CΔ170 versus rHPIV1-CΔ169–170+P, groups 6 and 5, respectively, and rHPIV1-P/CΔ84–85 versus rHPIV1-CΔ84–85+P, groups 4 and 3, respectively).
IFN signaling in response to HPIV1 infection is modified in cells infected with rHPIV1-C+P viruses
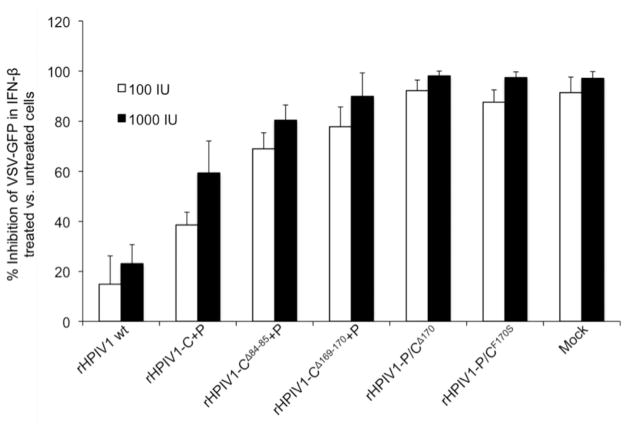
The C proteins of HPIV1 wt have previously been shown to inhibit type I IFN signaling [26]. Here, we evaluated the ability of newly generated rHPIV1 mutants to inhibit the induction of an antiviral state by extrinsic IFN-β using a VSV plaque inhibition assay in Vero cells. Cells were infected with a rHPIV1 for 24 h before treatment with 100 or 1000 IU/ml IFN-β for 24 h. Cells were then challenged 24 h later with VSV-GFP and plaque numbers were determined at 48 h p.i. to evaluate the effect of prior infection with rHPIV1s on the ability of type I IFN to establish an antiviral state and to protect Vero cells from productive VSV infection. In mock-infected cells, an antiviral state was established using 100 and 1000 IU of IFN-β per ml, as evidenced by an almost complete inhibition of VSV-GFP replication (Figure 5, mock group). As previously observed, rHPIV1 wt inhibited this activity of IFN whereas rHPIV1-P/CΔ170 and rHPIV1P/CF170S did not (Figure 5) [26]. rHPIV1-P/CΔ84–85 did not grow to a high enough titer for use in this assay; therefore this virus could not be included here. Infection with rHPIV1-C+P had an intermediate inhibitory effect on IFN signaling, i.e., there was a significant difference in the number of VSV plaques between rHPIV1-C+P and either rHPIV1 wt or mock-infected cells at both IFN concentrations (p<0.05). The deletion mutants, rHPIV1-CΔ84–85+P and rHPIV1-CΔ169–170+P, like the rHPIV1P/CF170S control, did not significantly inhibit IFN signaling. Two observations can be made from these findings: (1) rHPIV1-C+P-is less able to inhibit IFN signaling than rHPIV1 wt although both viruses express wt C protein, and (2) the mutant C proteins expressed in cells infected with rHPIV1-CΔ84–85+P and rHPIV1-CΔ169–170+P were unable to inhibit IFN signaling.
Figure 5.
Type I IFN signaling in rHPIV1 wt or mutant infected cells. Vero cells in 6-well plates were infected with the indicated rHPIV1s at a MOI of 5 TCID50/cell and incubated for 24 h. Cells were then left untreated or were treated with 100 or 1000 IU/ml IFN-β (1 well per treatment per virus) for 24 h. The cells were then infected with VSV-GFP and incubated for 48 h. The VSV-GFP foci were visualized using a phosphorimager and counted. The graph represents the percent inhibition of VSV-GFP replication ± S.E. in IFN-β treated versus untreated cells based on three independent experiments.
Infection with rHPIV1-C+P and mutants induces apoptosis
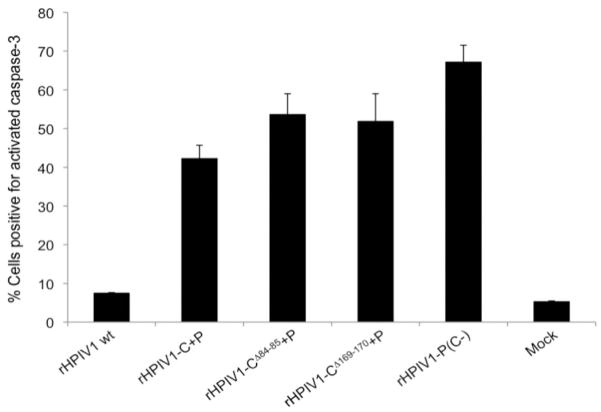
We previously observed enhanced cpe associated with apoptosis during infection with a C gene deletion mutant, rHPIV1-P(C-), compared to rHPIV1 wt [21]. This identified a role of the HPIV1 C proteins in inhibiting apoptosis in vitro. Here, enhanced cpe compared to rHPIV1 wt was observed during infection with rHPIV1-C+P, rHPIV1-CΔ84–85+P and rHPIV1-CΔ169–170+P. To explore the basis of the enhanced cpe, we examined the activation of caspase 3, the major effector caspase in the apoptotic pathway, in infected LLC-MK2 cells. Replicate cultures of LLC-MK2 cells were mock-infected or infected with rHPIV1 wt, rHPIV1-C+P, rHPIV1-CΔ84–85+P, rHPIV1-CΔ169–170+P, or rHPIV1-P(C-) at an MOI of 5, and cells were prepared for FACS analysis at 72 h p.i. Staining for HPIV1 HN antigen demonstrated that all of the viruses analyzed infected LLC-MK2 cells at high and comparable frequency (data not shown). Little to no caspase 3 activation was detected in mock and rHPIV1 wt-infected cells, i.e., 5.3% and 7.5% of cells, respectively, stained positive for activated caspase 3 (Figure 6). Cells infected with rHPIV1-P(C-) were used as a positive control for activation of caspase 3, and 67.2% of cells in this group stained positive at 72 h. The C+P viruses also induced activation of caspase 3, although at a lower frequency; 42.3% for rHPIV1-C+P, 53.6% for rHPIV1-CΔ84–85+P and 51.9% for rHPIV1-CΔ169–170+P. There was no significant difference in caspase 3 activation between these three viruses, indicating that the presence of the deletion mutations in the rHPIV1-C+P backbone did not have an additive effect on the induction of apoptosis over that seen in rHPIV1-C+P-infected cells. We were surprised to see apoptosis in cells infected with rHPIV1-C+P since this virus expresses wt C proteins and since HPIV1 wt infection of LLC-MK2 cells does not cause apoptosis.
Figure 6.
Infection with rHPIV1-C+P containing wt or mutant C genes, induces activation of caspase 3, which is indicative of apoptosis. Caspase 3 activation was evaluated by FACS analysis. LLC-MK2 cells were mock-infected or infected with rHPIV1s at a MOI of 5 TCID50/cell. At 72 h p.i., cells were harvested, fixed, permeabilized and immunostained for HPIV1 HN and activated caspase 3 in FACS buffer prior to analysis. Sample analysis was carried out using a FACSCalibur flow cytometer and FlowJo software. The percentage of cells positive for activated caspase 3 at 72 h p.i., as determined by FACS analysis ± S.E., is shown and is representative of two independent experiments.
C+P deletion mutants are attenuated in AGMs
In order to evaluate whether rHPIV1-C+P or its derivatives have potential for use as live attenuated HPIV1 vaccines, the magnitude and duration of replication was assessed in the URT and LRT of AGMs. As expected, HPIV1 wt replicated efficiently in the URT and the LRT of AGMs (Table 2). rHPIV1-C+P, containing the non-attenuating HNT553A mutation, replicated as well as HPIV1 wt, i.e. there was no statistically significant difference in mean peak virus titer or mean sum of daily virus titer (compare groups 1 and 3 in Table 2). As previously demonstrated, rHPIV1-P/CΔ170 replication was significantly restricted compared to HPIV1 wt in both the URT and the LRT (compare groups 1 and 2 in Table 2) [16]. In contrast, replication of rHPIV1-CΔ169–170+P was not restricted with regard to mean peak virus titer or with regard to the mean sum of daily virus titer in the LRT. Only the mean sum of daily virus titer in the URT was significantly lower than in rHPIV1 wt-infected animals (compare groups 1 and 4 in Table 2). In sum, these data indicate that for rHPIV1-P/CΔ170 the effect of the deletion in C only partially accounts for the restriction of virus replication in AGMs and that the effect of the deletion on the P protein likely contributes to its attenuation in AGMs. The titer of rHPIV1-P/CΔ84–85 was too low for this virus to be included in the AGM study. However, replication of rHPIV1-CΔ84–85+P, with (*) and without the HNT553A mutation (groups 5 and 6, in Table 2), was highly restricted in both the URT and LRT of AGMs, as indicated by a 400- to 500-fold reduction in mean peak titer in the URT and a 200–250- fold reduction of mean peak virus titer in the LRT, compared to HPIV1 wt (compare groups 1 and 5 or 6 in Table 2). Therefore, deletion of codons 84–85 in the HPIV1 C protein severely restricted virus replication in vivo, and this effect was independent of any contribution of the mutation in HN. Thus the CΔ84–85 mutation was up to 100-fold more restrictive in replication than the CΔ169–170 mutation in the rHPIV1-C+P backbone.
Table 2.
Replication of rHPIV1-C+P mutants in AGMs.
| Group | Virusa | No. animals | Mean peak virus titer (log10 TCID50/ml) b |
Mean sum of the daily virus titers (log10 TCID50/ml)c |
||
|---|---|---|---|---|---|---|
| NP swab | TL | NP swab | TL | |||
| 1 | HPIV1 wt | 18 | 4.3±0.1 | 4.1±0.3 | 27.4±1.4 | 13.6±1.4 |
| 2 | rHPIV1-P/CΔ170 | 6 | 3.4±0.5 | 2.3±0.5d | 14.8±1.9 | 5.1±0.8 |
| 3 | rHPIV1-C+P * | 6 | 3.4±0.4 | 4.7±0.4 | 22.3±2.3 | 15.3±1.5 |
| 4 | rHPIV1-CΔ169–170+P | 4 | 3.4±0.4 | 3.7±0.3 | 19.4±1.2 | 10.2±1.4 |
| 5 | rHPIV1-CΔ84–85+P | 4 | 1.6±0.3 | 1.7±0.1 | 7.9±1.3 | 7.8±0.2 |
| 6 | rHPIV1-CΔ84–85+P * | 4 | 1.7±0.2 | 1.8±0.3 | 9.4±0.9 | 7.1±0.2 |
Monkeys were inoculated i.n. and i.t. with 106 TCID50 of the indicated virus in a 1ml inoculum at each site. Monkeys lacked HAI antibody to HPIV1 wt at the start of the study.
Mean peak titer: mean ± S.E of the individual peak virus titers for the animals in each group, irrespective of day. Virus titrations were performed on LLC-MK2 cells at 32°C. The limit of detection was 0.5 log10 TCID50/ml. NP samples were collected on days 0 to 10 post-infection. TL samples were collected on days 2, 4, 6, 8, and 10 post-infection.
Mean sum of the daily virus titers: the sum of the titers for all of the days of sampling, excluding day 0, was determined for each animal individually, and the mean was calculated for each group. On days when no virus was detected, a value of 0.5 log10 TCID50/ml was assigned for the purpose of calculation. The mean sum of the lower limit of detection was 5.0 log10 TCID50/ml for NP swabs and 2.5 log10 TCID50/ml for TL samples.
The underlined values indicate a significant reduction in virus replication compared to the corresponding data point for the HPIV1 wt group, as measured by the Newman-Keuls multiple comparison test (p<0.05).
These viruses contain the HNT553A mutation.
Immunization of AGMs with a C+P deletion mutant protects against challenge with HPIV1 wt
The potential of the rHPIV1-C+P deletion mutants for use as vaccines was determined by evaluating the ability of immunization with mutant rHPIV1s to induce serum HAI antibodies and to protect against challenge with HPIV1 wt. Serum HAI antibody titers were determined prior to and following challenge with HPIV1 wt in order to assess the immunogenicity of the rHPIV1 vaccine candidates in AGMs (Table 3). All of the HPIV1s elicited a detectable HAI antibody response following immunization; however, there was considerable variation between viruses with respect to antibody titers. As expected, HPIV1 wt and rHPIV1-C+P induced high titers of HAI antibodies, as did rHPIV1-P/CΔ170, as described previously [16]. rHPIV1-CΔ169–170+P elicited moderate levels of HAI antibody and, despite its high levels of attenuation, rHPIV1-CΔ84–85+P* also elicited a moderate level of HAI antibody, while rHPIV1-CΔ84–85+P elicited a very modest antibody response. Corresponding with the induction of antibody, immunization with each of the viruses protected AGMs against replication of the HPIV1 wt challenge virus four weeks after a single mucosal vaccination (Table 3). Notably, rHPIV1-CΔ84–85+P and rHPIV1-CΔ84–85+P*, despite their highly restricted replication, were able to significantly restrict HPIV1 wt challenge virus replication in the LRT. Challenge virus titers in the URT were more variable (Table 3). However, their ability to restrict challenge virus replication corresponded to the level of serum HAI antibody following immunization, with rHPIV1-CΔ84–85+P demonstrating a weak antibody response and less protection against HPIV1 wt. Following challenge, HAI antibody titers increased in animals that had low to moderate pre-challenge HAI antibody titers (groups 4, 5 and 6). In summary, for most of the virus mutants examined, a correlation exists between the level of vaccine virus replication and its immunogenicity and efficacy against HPIV1 wt challenge virus replication. For example, the level of replication and the efficacy of rHPIV1-CΔ169–170+P and rHPIV1-P/CΔ170 are greater that that of rHPIV1-CΔ84–85+P*. In the authors judgment each of these viruses could be considered as a vaccine candidate against HPIV1
Table 3.
Immunization with rHPIV1-C+P mutants protects AGMs against challenge with HPIV1 wt.
| Group | Virus a | No. animals | Serum HAI antibody titer (mean reciprocal log2 ± S.E.) b |
Mean peak challenge virus titer (log10 TCID50/ml)c |
||
|---|---|---|---|---|---|---|
| Pre-challenge | Post-challenge | NP swab | TL | |||
| 1 | HPIV1 wt | 16 | 7.0±0.4 | 6.2±0.4 | 1.0±0.2 | 0.7±0.1 |
| 2 | rHPIV1-P/CΔ170 | 6 | 5.9±0.2 | 6.4±0.4 | ≤0.5±0.0 | ≤0.5±0.0 |
| 3 | rHPIV1-C+P* | 6 | 6.5±0.4 | 4.2±0.2 | 1.2±0.2 | 1.5±0.3 |
| 4 | rHPIV1-CΔ169–170+P | 4 | 4.2±1.6 | 3.8±0.9 | 1.5±0.4 | ≤0.5±0.0 |
| 5 | rHPIV1-CΔ84–85+P | 4 | 1.8±0.8 | 3.3±1.3 | 4.3±0.3 | 3.5±0.4 |
| 6 | rHPIV1-CΔ84–85+P * | 4 | 3.3±0.5 | 5.0±0.0 | 2.7±0.7 | 2.1±0.8 |
| 7 | Non-Immunized Controls | 16 | 1.0±0.0 | 5.8±0.4 | 4.7±0.3 | 4.6±0.3 |
AGMs previously immunized with the indicated viruses were inoculated i.n. and i.t. with 106 TCID50 of HPIV1 wt, 1ml inoculum per site, at day 28 post-immunization.
HAI titers to HPIV1 were determined by HAI assay of sera collected at day 28 (pre-challenge) and day 56 (post-challenge) following initial inoculation. Titers are expressed as mean reciprocal log2 ± S.E., the limit of detection is 1.0 ± 0.0.
Mean peak challenge virus titer: mean ± S.E of the individual peak challenge virus titers for the animals in each group, irrespective of day. Virus titrations were performed on LLC-MK2 cells at 32°C. The limit of detection was 0.5 log10 TCID50/ml. NP samples were collected on days 0, 2, 4, 6, and 8 post-infection. TL samples were collected on days 2, 4, 6, and 8.
These viruses contain the HNT553A mutation.
Discussion
The recovery of live infectious HPIV1 from cDNA has been an important step towards the rational development of a HPIV1 vaccine, and clinical trials are currently in progress to evaluate the first cDNA-derived live attenuated HPIV1 vaccine, rHPIV1-CR84G/Δ170HNT553ALY942A [12]. However, development of additional live attenuated HPIV1 vaccine candidates with varying degrees of attenuation is needed in case the rHPIV1-CR84G/Δ170HNT553ALY942A vaccine encounters problems during clinical development with regard to safety, attenuation, genetic stability, immunogenicity, or efficacy. In this study, rHPIV1-C+P, a novel rHPIV1 that expresses the P and C proteins from two separate genes, was generated as a backbone for developing additional HPIV1 vaccines since it permits the introduction of mutations into C without affecting P.
The CΔ84–85 mutation was developed as an alternative to the CR84G point mutation in order to enhance both genetic stability and attenuation and was initially studied on the rHPIV1 wt backbone containing overlapping P and C ORFs. This mutant, rHPIV1-P/CΔ84–85, was highly restricted for replication in vitro, precluding its use as a live attenuated virus vaccine due to its poor yield in Vero cells, the substrate used for vaccine manufacture. We hypothesized that the highly restricted replication of rHPIV1-P/CΔ84–85 might be due to impaired P protein function because the CΔ84–85 deletion affects not only the C ORF but also the P ORF. Paramyxovirus P proteins are essential for virus replication and play a role in both mRNA synthesis and genome replication. The P protein binds to the L protein to form the viral RNA-dependent RNA polymerase (vRdRp), which is required for viral RNA synthesis [27]. The L-binding domain has been identified for Sendai virus P and is located in the carboxy-terminal half of the protein (aa 413–435) [50, 51]. In addition, the P protein acts as a chaperone, forming a complex with unassembled nucleoprotein during the nascent chain assembly step of genome replication [52]. For Sendai virus, the amino-terminal half of P has been shown to contain not only the N binding domain (aa 33–41) [52] but also a region from aa 1–144 that is required for synthesis of both mRNA and vRNA [52]. Sendai virus P binds to the N-RNA complex via a region in the C-terminal domain of P during RNA synthesis and vRdRp scanning [53]. In generating rHPIV1-P/CΔ84–85, we deleted codons 84 and 85 in the C ORF, which corresponded to a complete deletion of codon 88 and partial deletions of codons 87 and 89 in the P ORF (nt: GAG AGT GGA → GGA; aa: ESG → G; Table 1). Since codons 87 and 88 of the P ORF are i) within the region required for mRNA and vRNA synthesis for Sendai virus and ii) conserved in HPIV1 and Sendai virus, it seemed plausible that the effect of the P/CΔ84–85 mutation on the P protein could be responsible for the restricted replication of the rHPIV1-P/CΔ84–85 mutant in Vero cells. Therefore, rHPIV1-C+P was designed such that mutations could be introduced into C without affecting P, thereby permitting evaluation of the effect of a mutation in C independent of its effect on P.
rHPIV1-C+P was recovered and found to replicate efficiently both in vitro and in vivo in AGMs and, therefore, it could serve as a suitable virus for introduction of attenuating mutations in C. Since the C proteins of HPIV1 wt inhibit a) the induction of type 1 IFN, b) the signaling of type 1 IFN through its receptor, and c) development of apoptosis, it was expected that these three properties of HPIV1 wt would be shared by rHPIV1-C+P since both viruses express wt C and P proteins. However, this was not the case. For reasons that remain undefined, rHPIV1-C+P induced more IFN than HPIV1 wt, inhibited IFN signaling less than HPIV1 wt, and induced more apoptosis than HPIV1 wt. Compared to rHPIV1 wt, rHPIV1-C+P manifests a clear difference in the ratio of C to C protein expression (Figure 2) and a slight increase in the level of N, P, and F expression. These differences in protein expression likely contribute to the phenotypic differences between rHPIV1-C+P and rHPIV1 wt. The higher level of C versus C′ protein expression in rHPIV1-C+P is not surprising since translation is initiated most efficiently at the most 5′-proximal AUG start codon in the mRNA [54]. In the rHPIV1 wt P/C mRNA the P and C start codons are separated by only 7 nucleotides and the P start codon is the first AUG of this mRNA (C′ is translated from an unconventional start codon). In rHPIV1-C+P, two separate mRNAs encode the C proteins and the P protein, respectively, and there is no P start codon in the C gene mRNA immediately preceding the C start. This would result in a higher level of C protein expression from a C(P-) gene than a wt P/C gene. It is possible that the resulting over-expression of C proteins could lead to endoplasmic reticulum stress resulting in apoptosis and induction of IFN-β during infection with rHPIV1-C+P [55–57]. Another mechanism to consider as an alternative, or in addition to this, is that the individual C proteins may have unique functions which are as yet unidentified. The greater level of C protein expression observed during infection with rHPIV1-C+P infected cells may result in a dominance of C function over C′ function. It is difficult to propose a hypothesis which adequately explains the set of phenotypes (increased type I IFN production, increased type I IFN signaling, and apoptosis) induced by separating the C and P genes of HPIV1 without knowing the individual functions of the C proteins. Studies to determine the mechanisms responsible for these phenotypic differences are underway. Since rHPIV1-C+P was less able than rHPIV1 wt to inhibit IFN production, IFN signaling and apoptosis, these phenotypes of the rHPIV1-C+P mutants need to be interpreted as the sum total of two effects: the effect of separating C and P and the effect of disabling the function of C by the introduced mutation. Fortunately, the level of replication of rHPIV1-C+P and rHPIV1 wt in vitro and in AGMs was similar so the effect of mutations introduced into C on replication of HPIV1 in vitro and on attenuation in vivo could be accurately assessed.
The Δ84–85 mutation, which, on the rHPIV1 wt backbone, rHPIV1-P/CΔ84–85, restricted replication in vitro, did not affect the replication of rHPIV1-C+P. The novel virus, rHPIV1-CΔ84–85+P, replicated like rHPIV1 wt in LLC-MK2 cells until extensive cpe became apparent. rHPIV1-CΔ84–85+P also grew more efficiently than rHPIV1-P/CΔ84–85 in all cell types studied, which would permit its manufacture as a vaccine virus in Vero cells. This difference in replication in vitro between rHPIV1-P/C 84–85 and rHPIV1-CΔ84–85+P suggests that an alteration of P protein function by the CΔ84–85 mutation in rHPIV1-P/CΔ84–85 was responsible, at least in part, for its restricted replication in vitro. The CΔ169–170 mutation did not affect replication in vitro in either the rHPIV1 wt or rHPIV1-C+P backbone, but the onset of apoptosis associated with rHPIV1-C+P, rHPIV1-CΔ84–85+P, and rHPIV1-CΔ169–170+P viruses did correlate with a reduction in replication, suggesting that apoptosis might be responsible for the decreased virus yield. In addition to apoptosis, rHPIV1-C+P and its rHPIV1-CΔ84–85+P and rHPIV1-CΔ169–170+P derivatives induced IFN early in infection and failed to inhibit IFN signaling. The two deletion mutants differed only modestly from the rHPIV1-C+P parent in each of these phenotypes. A similar phenomenon was previously observed for a HPIV1 C deletion mutant, rHPIV1-P(C-), but the decline in virus yield was steeper than that observed in the present study, consistent with more widespread apoptosis observed during infection with rHPIV1-P(C-) (Figure 6) [21]. Apoptosis has been observed in vivo with Sendai virus C mutant viruses, and this was associated with restricted replication in vivo [34, 58]. However, it remains unknown whether restriction of virus replication is caused by apoptosis or whether an antiviral pathway is activated in parallel with the induction of apoptosis.
The ability of the CΔ84–85 mutation to attenuate rHPIV1-C+P in AGMs was determined and compared to that of the previously characterized CΔ169–170 mutation [16]. The CΔ169–170 mutation was less attenuating in the rHPIV1-C+P backbone than in the HPIV1 wt backbone, suggesting that there was an effect of the CΔ169–170 mutation on P function in rHPIV1-P/CΔ170 that manifested itself in vivo. It is also possible that the difference in the sequence of the C proteins in the rHPIV1-P/CΔ170 and rHPIV1- CΔ169–170+P could contribute to the difference in replication in AGMs. Whereas replication of rHPIV1-CΔ169–170+P and rHPIV1-C+P was similar in the respiratory tract of AGMs, the level of attenuation specified by the CΔ84–85 mutation in rHPIV1-C+P was highly significant in both the URT and LRT. However, since rHPIV1-CΔ84–85+P and rHPIV1-CΔ169–170+P had similar profiles for IFN induction, IFN signaling, and induction of apoptosis, the mechanisms underlying the highly restricted replication of rHPIV1-CΔ84–85+P remain undefined. Given the previously observed attenuation phenotype specified by the point mutant, rHPIV1-CR84GHNT553A, which was att only in the URT of AGMs, and only in combination with the HNT553A mutation [39], we were surprised by the high degree of growth restriction demonstrated by the novel Δ84–85 mutants compared to the Δ169–170 mutants. Thus, conversion of a point mutation at codon 84 to a deletion mutation was accompanied by a greatly increased level of attenuation. Furthermore, the attenuation specified by the CΔ84–85 mutation in rHPIV1-C+P was not altered by the mutation in HN that was required for the attenuation effect previously observed for the CR84G mutation [16]. This indicates that the CΔ84–85 mutation is a strongly attenuating mutation that acts alone in attenuating rHPIV1-C+P for AGMs. However, in the current study the rHPIV1-CΔ84–85+P virus containing the HN mutation was found to be slightly more immunogenic and efficacious in AGMs. We cannot completely rule out an effect of the HN mutation, although this effect seems unlikely because the finding that the HN mutation conferred attenuation in combination with the CΔ84–85 mutation seems to be in contrast to the evidence suggesting that it augments the immunogenicity and efficacy of rHPIV1-CΔ84–85+P. Regarding the highly attenuated nature of the rHPIV1-CΔ84–85+P virus, it is possible that it will replicate more efficiently and subsequently be more immunogenic and efficacious in humans than in AGMs due to species specificity. Furthermore, this virus is not as defective as the C deletion mutant, rHPIV1-P(C-), that did not replicate at all in the URT and was barely detectable in the LRT, indicating that C deletion mutants are over attenuated and therefore not suitable as live-attenuated virus vaccines [21].
In summary, we have demonstrated that rHPIV1-C+P is a useful backbone for developing live attenuated HPIV1 vaccine candidates with genetically stable deletion mutations in the C gene. Specific C protein deletion mutations that render HPIV1 wt overly restricted in replication in vitro for vaccine manufacturing purposes due to ancillary effects on P can yield promising vaccine candidates when the mutation is introduced into rHPIV1-C+P. We identified two genetically stable attenuating mutations, CΔ84–85 and CΔ169–170, which specify a range in the level of attenuation conferred upon rHPIV1-C+P in AGMs. However, it remains to be determined how effective a highly restricted virus such as rHPIV1-CΔ84–85+P will be in humans, and the usefulness of this virus as a vaccine will need to be determined in clinical trials.
Acknowledgments
We thank Brad Finneyfrock, Marisa St. Claire, and Anthony Cook at Bioqual Inc., for carrying out the primate studies. We are also grateful to John Hiscott, McGill University, who provided the VSV-GFP, Ursula Buchholz, NIAID, NIH, who provided the BHK-T7 cells, and Yasuhiko Ito, Mie University School of Medicine, who provided the mouse anti-HPIV1 HN (8.2.2A and 4.5) antibodies.
This project was funded and supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. This project was partly performed under a cooperative research and development agreement (CRADA) between NIAID and MedImmune (CRADA #AI-5114) for the development of live attenuated viruses for respiratory syncytial viruses, parainfluenza viruses and human metapneumovirus.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Karron RA, Collins PL. Chapter 42. Parainfluenza Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1497–526. [Google Scholar]
- 2.Murphy BR, Prince GA, Collins PL, Van Wyke Coelingh K, Olmsted RA, Spriggs MK, et al. Current approaches to the development of vaccines effective against parainfluenza and respiratory syncytial viruses. Virus Res. 1988;11(1):1–15. doi: 10.1016/0168-1702(88)90063-9. [DOI] [PubMed] [Google Scholar]
- 3.Iwane MK, Edwards KM, Szilagyi PG, Walker FJ, Griffin MR, Weinberg GA, et al. Population-based surveillance for hospitalizations associated with respiratory syncytial virus, influenza virus, and parainfluenza viruses among young children. Pediatrics. 2004 Jun;113(6):1758–64. doi: 10.1542/peds.113.6.1758. [DOI] [PubMed] [Google Scholar]
- 4.Marx A, Torok TJ, Holman RC, Clarke MJ, Anderson LJ. Pediatric hospitalizations for croup (laryngotracheobronchitis): biennial increases associated with human parainfluenza virus 1 epidemics. J Infect Dis. 1997;176:1423–7. doi: 10.1086/514137. [DOI] [PubMed] [Google Scholar]
- 5.Reed G, Jewett PH, Thompson J, Tollefson S, Wright PF. Epidemiology and clinical impact of parainfluenza virus infections in otherwise healthy infants and young children < 5 years old. J Infect Dis. 1997;175(4):807–13. doi: 10.1086/513975. [DOI] [PubMed] [Google Scholar]
- 6.Heikkinen T, Thint M, Chonmaitree T. Prevalence of various respiratory viruses in the middle ear during acute otitis media. N Engl J Med. 1999;340(4):260–4. doi: 10.1056/NEJM199901283400402. [DOI] [PubMed] [Google Scholar]
- 7.Counihan ME, Shay DK, Holman RC, Lowther SA, Anderson LJ. Human parainfluenza virus-associated hospitalizations among children less than five years of age in the United States. Pediatr Infect Dis J. 2001;20(7):646–53. doi: 10.1097/00006454-200107000-00003. [DOI] [PubMed] [Google Scholar]
- 8.Karron RA, Makhene M, Gay K, Wilson MH, Clements ML, Murphy BR. Evaluation of a live attenuated bovine parainfluenza type 3 vaccine in two- to six-month-old infants. Pediatr Infect Dis J. 1996;15(8):650–4. doi: 10.1097/00006454-199608000-00003. [DOI] [PubMed] [Google Scholar]
- 9.Karron RA, Belshe RB, Wright PF, Thumar B, Burns B, Newman F, et al. A live human parainfluenza type 3 virus vaccine is attenuated and immunogenic in young infants. Pediatr Infect Dis J. 2003 May;22(5):394–405. doi: 10.1097/01.inf.0000066244.31769.83. [DOI] [PubMed] [Google Scholar]
- 10.Belshe RB, Newman FK, Anderson EL, Wright PF, Karron RA, Tollefson S, et al. Evaluation of combined live, attenuated respiratory syncytial virus and parainfluenza 3 virus vaccines in infants and young children. J Infect Dis. 2004 Dec 15;190(12):2096–103. doi: 10.1086/425981. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg DP, Walker RE, Lee MS, Reisinger KS, Ward JI, Yogev R, et al. A bovine parainfluenza virus type 3 vaccine is safe and immunogenic in early infancy. J Infect Dis. 2005 Apr 1;191(7):1116–22. doi: 10.1086/428092. [DOI] [PubMed] [Google Scholar]
- 12.Bartlett EJ, Castano A, Surman SR, Collins PL, Skiadopoulos MH, Murphy BR. Attenuation and efficacy of human parainfluenza virus type 1 (HPIV1) vaccine candidates containing stabilized mutations in the P/C and L genes. Virol J. 2007 Jul 2;4(1):67. doi: 10.1186/1743-422X-4-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nolan SM, Skiadopoulos MH, Bradley K, Kim OS, Bier S, Amaro-Carambot E, et al. Recombinant human parainfluenza virus type 2 vaccine candidates containing a 3′ genomic promoter mutation and L polymerase mutations are attenuated and protective in non-human primates. Vaccine. 2007 Aug 21;25(34):6409–22. doi: 10.1016/j.vaccine.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slobod KS, Shenep JL, Lujan-Zilbermann J, Allison K, Brown B, Scroggs RA, et al. Safety and immunogenicity of intranasal murine parainfluenza virus type 1 (Sendai virus) in healthy human adults. Vaccine. 2004 Aug 13;22(23–24):3182–6. doi: 10.1016/j.vaccine.2004.01.053. [DOI] [PubMed] [Google Scholar]
- 15.McAuliffe JM, Surman SR, Newman JT, Riggs JM, Collins PL, Murphy BR, et al. Codon substitution mutations at two positions in the L polymerase protein of human parainfluenza virus type 1 yield viruses with a spectrum of attenuation in vivo and increased phenotypic stability in vitro. J Virol. 2004 Feb;78(4):2029–36. doi: 10.1128/JVI.78.4.2029-2036.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartlett EJ, Amaro-Carambot E, Surman SR, Collins PL, Murphy BR, Skiadopoulos MH. Introducing point and deletion mutations into the P/C gene of human parainfluenza virus type 1 (HPIV1) by reverse genetics generates attenuated and efficacious vaccine candidates. Vaccine. 2006 Mar 24;24(14):2674–84. doi: 10.1016/j.vaccine.2005.10.047. [DOI] [PubMed] [Google Scholar]
- 17.Newman JT, Surman SR, Riggs JM, Hansen CT, Collins PL, Murphy BR, et al. Sequence analysis of the Washington/1964 strain of human parainfluenza virus type 1 (HPIV1) and recovery and characterization of wild-type recombinant HPIV1 produced by reverse genetics. Virus Genes. 2002;24(1):77–92. doi: 10.1023/a:1014042221888. [DOI] [PubMed] [Google Scholar]
- 18.Itoh M, Isegawa Y, Hotta H, Homma M. Isolation of an avirulent mutant of Sendai virus with two amino acid mutations from a highly virulent field strain through adaptation to LLC- MK2 cells. J Gen Virol. 1997;78(Pt 12):3207–15. doi: 10.1099/0022-1317-78-12-3207. [DOI] [PubMed] [Google Scholar]
- 19.Kurotani A, Kiyotani K, Kato A, Shioda T, Sakai Y, Mizumoto K, et al. Sendai virus C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes Cells. 1998;3(2):111–24. doi: 10.1046/j.1365-2443.1998.00170.x. [DOI] [PubMed] [Google Scholar]
- 20.Latorre P, Cadd T, Itoh M, Curran J, Kolakofsky D. The various Sendai virus C proteins are not functionally equivalent and exert both positive and negative effects on viral RNA accumulation during the course of infection. J Virol. 1998;72(7):5984–93. doi: 10.1128/jvi.72.7.5984-5993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartlett EJ, Cruz AM, Esker J, Castano A, Schomacker H, Surman SR, et al. Human parainfluenza virus type 1 C proteins are nonessential proteins that inhibit the host interferon and apoptotic responses and are required for efficient replication in nonhuman primates. J Virol. 2008 Sep;82(18):8965–77. doi: 10.1128/JVI.00853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcin D, Marq JB, Goodbourn S, Kolakofsky D. The amino-terminal extensions of the longer Sendai virus C proteins modulate pY701-Stat1 and bulk Stat1 levels independently of interferon signaling. J Virol. 2003 Feb;77(4):2321–9. doi: 10.1128/JVI.77.4.2321-2329.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gotoh B, Takeuchi K, Komatsu T, Yokoo J. The STAT2 activation process is a crucial target of Sendai virus C protein for the blockade of alpha interferon signaling. J Virol. 2003 Mar;77(6):3360–70. doi: 10.1128/JVI.77.6.3360-3370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komatsu T, Takeuchi K, Yokoo J, Gotoh B. C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-beta production. Virology. 2004 Jul 20;325(1):137–48. doi: 10.1016/j.virol.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 25.Bousse T, Chambers RL, Scroggs RA, Portner A, Takimoto T. Human parainfluenza virus type 1 but not Sendai virus replicates in human respiratory cells despite IFN treatment. Virus Res. 2006 Oct;121(1):23–32. doi: 10.1016/j.virusres.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 26.Van Cleve W, Amaro-Carambot E, Surman SR, Bekisz J, Collins PL, Zoon KC, et al. Attenuating mutations in the P/C gene of human parainfluenza virus type 1 (HPIV1) vaccine candidates abrogate the inhibition of both induction and signaling of type I interferon (IFN) by wild-type HPIV1. Virology. 2006 Aug 15;352(1):61–73. doi: 10.1016/j.virol.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 27.Curran J, Marq JB, Kolakofsky D. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology. 1992;189(2):647–56. doi: 10.1016/0042-6822(92)90588-g. [DOI] [PubMed] [Google Scholar]
- 28.Cadd T, Garcin D, Tapparel C, Itoh M, Homma M, Roux L, et al. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70(8):5067–74. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horikami SM, Hector RE, Smallwood S, Moyer SA. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology. 1997;235(2):261–70. doi: 10.1006/viro.1997.8702. [DOI] [PubMed] [Google Scholar]
- 30.Tapparel C, Hausmann S, Pelet T, Curran J, Kolakofsky D, Roux L. Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J Virol. 1997;71(12):9588–99. doi: 10.1128/jvi.71.12.9588-9599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hasan MK, Kato A, Muranaka M, Yamaguchi R, Sakai Y, Hatano I, et al. Versatility of the accessory C proteins of Sendai virus: contribution to virus assembly as an additional role. J Virol. 2000;74(12):5619–28. doi: 10.1128/jvi.74.12.5619-5628.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakaguchi T, Kato A, Sugahara F, Shimazu Y, Inoue M, Kiyotani K, et al. AIP1/Alix is a binding partner of Sendai virus C protein and facilitates virus budding. J Virol. 2005 Jul;79(14):8933–41. doi: 10.1128/JVI.79.14.8933-8941.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irie T, Nagata N, Yoshida T, Sakaguchi T. Recruitment of Alix/AIP1 to the plasma membrane by Sendai virus C protein facilitates budding of virus-like particles. Virology. 2008 Feb 5;371(1):108–20. doi: 10.1016/j.virol.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 34.Itoh M, Hotta H, Homma M. Increased induction of apoptosis by a Sendai virus mutant is associated with attenuation of mouse pathogenicity. J Virol. 1998;72(4):2927–34. doi: 10.1128/jvi.72.4.2927-2934.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heylbroeck C, Balachandran S, Servant MJ, DeLuca C, Barber GN, Lin R, et al. The IRF-3 transcription factor mediates Sendai virus-induced apoptosis. J Virol. 2000;74(8):3781–92. doi: 10.1128/jvi.74.8.3781-3792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bitzer M, Armeanu S, Prinz F, Ungerechts G, Wybranietz W, Spiegel M, et al. Caspase-8 and Apaf-1-independent caspase-9 activation in Sendai virus-infected cells. J Biol Chem. 2002 Aug 16;277(33):29817–24. doi: 10.1074/jbc.M111898200. [DOI] [PubMed] [Google Scholar]
- 37.Koyama AH, Irie H, Kato A, Nagai Y, Adachi A. Virus multiplication and induction of apoptosis by Sendai virus: role of the C proteins. Microbes Infect. 2003 Apr;5(5):373–8. doi: 10.1016/s1286-4579(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 38.Peters K, Chattopadhyay S, Sen GC. IRF-3 activation by Sendai virus infection is required for cellular apoptosis and avoidance of persistence. J Virol. 2008 Apr;82(7):3500–8. doi: 10.1128/JVI.02536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartlett EJ, Amaro-Carambot E, Surman SR, Newman JT, Collins PL, Murphy BR, et al. Human parainfluenza virus type I (HPIV1) vaccine candidates designed by reverse genetics are attenuated and efficacious in African green monkeys. Vaccine. 2005 Sep 7;23(38):4631–46. doi: 10.1016/j.vaccine.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 40.Newman JT, Riggs JM, Surman SR, McAuliffe JM, Mulaikal TA, Collins PL, et al. Generation of recombinant human parainfluenza virus type 1 vaccine candidates by importation of temperature-sensitive and attenuating mutations from heterologous paramyxoviruses. J Virol. 2004 Feb;78(4):2017–28. doi: 10.1128/JVI.78.4.2017-2028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73(1):251–9. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy BR, Richman DD, Chalhub EG, Uhlendorf CP, Baron S, Chanock RM. Failure of attenuated temperature-sensitive influenza A (H3N2) virus to induce heterologous interference in humans to parainfluenza type 1 virus. Infect Immun. 1975;12(1):62–8. doi: 10.1128/iai.12.1.62-68.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stojdl DF, Lichty BD, tenOever BR, Paterson JM, Power AT, Knowles S, et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003 Oct;4(4):263–75. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 44.Tao T, Durbin AP, Whitehead SS, Davoodi F, Collins PL, Murphy BR. Recovery of a fully viable chimeric human parainfluenza virus (PIV) type 3 in which the hemagglutinin-neuraminidase and fusion glycoproteins have been replaced by those of PIV type 1. J Virol. 1998;72(4):2955–61. doi: 10.1128/jvi.72.4.2955-2961.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komada H, Kusagawa S, Orvell C, Tsurudome M, Nishio M, Bando H, et al. Antigenic diversity of human parainfluenza virus type 1 isolates and their immunological relationship with Sendai virus revealed by using monoclonal antibodies. J Gen Virol. 1992;73(Pt 4):875–84. doi: 10.1099/0022-1317-73-4-875. [DOI] [PubMed] [Google Scholar]
- 46.Moeller K, Duffy I, Duprex P, Rima B, Beschorner R, Fauser S, et al. Recombinant measles viruses expressing altered hemagglutinin (H) genes: functional separation of mutations determining H antibody escape from neurovirulence. J Virol. 2001;75(16):7612–20. doi: 10.1128/JVI.75.16.7612-7620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72(2):891–9. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skiadopoulos MH, Tatem JM, Surman SR, Mitcho Y, Wu SL, Elkins WR, et al. The recombinant chimeric human parainfluenza virus type 1 vaccine candidate, rHPIV3-1cp45, is attenuated, immunogenic, and protective in African green monkeys. Vaccine. 2002;20(13):1846–52. doi: 10.1016/s0264-410x(02)00038-5. [DOI] [PubMed] [Google Scholar]
- 49.Clements ML, Belshe RB, King J, Newman F, Westblom TU, Tierney EL, et al. Evaluation of bovine, cold-adapted human, and wild-type human parainfluenza type 3 viruses in adult volunteers and in chimpanzees. J Clin Microbiol. 1991;29(6):1175–82. doi: 10.1128/jcm.29.6.1175-1182.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Curran J, Pelet T, Kolakofsky D. An acidic activation-like domain of the Sendai virus P protein is required for RNA synthesis and encapsidation. Virology. 1994;202(2):875–84. doi: 10.1006/viro.1994.1409. [DOI] [PubMed] [Google Scholar]
- 51.Smallwood S, Ryan KW, Moyer SA. Deletion analysis defines a carboxyl-proximal region of Sendai virus P protein that binds to the polymerase L protein. Virology. 1994;202(1):154–63. doi: 10.1006/viro.1994.1331. [DOI] [PubMed] [Google Scholar]
- 52.Curran J, Marq JB, Kolakofsky D. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J Virol. 1995;69(2):849–55. doi: 10.1128/jvi.69.2.849-855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryan KW, Murti KG, Portner A. Localization of P protein binding sites on the Sendai virus nucleocapsid. J Gen Virol. 1990;71(Pt 4):997–1000. doi: 10.1099/0022-1317-71-4-997. [DOI] [PubMed] [Google Scholar]
- 54.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999 Jul 8;234(2):187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 55.He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006 Mar;13(3):393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- 56.Medigeshi GR, Lancaster AM, Hirsch AJ, Briese T, Lipkin WI, Defilippis V, et al. West Nile virus infection activates the unfolded protein response, leading to CHOP induction and apoptosis. J Virol. 2007 Oct;81(20):10849–60. doi: 10.1128/JVI.01151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith JA, Turner MJ, DeLay ML, Klenk EI, Sowders DP, Colbert RA. Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-beta induction via X-box binding protein 1. Eur J Immunol. 2008 May;38(5):1194–203. doi: 10.1002/eji.200737882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcin D, Itoh M, Kolakofsky D. A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice, but not virus growth in cell culture. Virology. 1997;238(2):424–31. doi: 10.1006/viro.1997.8836. [DOI] [PubMed] [Google Scholar]