Abstract
The fidelity of poliovirus RNA-dependent RNA polymerase (3Dpol) was determined using a modified α-complementation assay. Several polymerases have been analyzed by this approach allowing comparisons to be drawn. Various conditions including high and low MgCl2, replacing MgCl2 with MnCl2, skewed nucleotide pools, and the presence of poliovirus protein 3AB were analyzed. The assay included RNA synthesis by 3Dpol on an RNA template that coded for a region of the alpha peptide of β-galactosidase (LacZ-α). The product of this reaction was used as a template for a second round of 3Dpol synthesis and the resulting RNA was reverse transcribed to DNA by reverse transcriptase. The DNA was amplified by PCR and inserted into a vector used to transform E. coli. The bacteria were screened for β-galactosidase activity by blue-white phenotype analysis with white or faint blue colonies scored as errors made during synthesis on lacZ-α. Although 3AB strongly stimulated 3Dpol synthesis as expected, no change in fidelity was detected. Changes in MgCl2 also showed little effect. Mutation rates of ~9 × 10−5 (~1 error per 11,000 incorporations) were estimated for these conditions. In contrast, MnCl2 or skewed nucleotide pools were highly mutagenic resulting in lowered fidelity.
Keywords: poliovirus, RNA-dependent RNA polymerase, 3Dpol, mutation rate, 3AB
1. Introduction
Several factors including recombination in plus sense viruses and the relatively low fidelity of RNA-dependent RNA polymerase in plus and minus sense viruses contribute to the ability of RNA viruses to rapidly generate genetic diversity (Domingo et al., 1996; Holland, 1993). Low polymerase fidelity is due at least in part to the lack of a 3′-5′ exonuclease proofreading activity (Domingo et al., 1996). With respect to replication fidelity, poliovirus has been the most heavily studied of those viruses using RNA-dependent RNA polymerase (RDRP). Polio’s RDRP (referred to as 3Dpol) can be easily expressed and purified and shows reproducible activity in vitro. Other viral polymerases, especially those of negative sense viruses have proven more difficult to work with.
Despite decades of work examining 3Dpol fidelity in vitro and replication fidelity in cells, both remain largely uncertain with the results highly dependent on the method employed. Several methods have been used to test the mutation frequency of poliovirus during cellular replication, however, results show values that span approximately 3 orders of magnitude from about 10−3 to 10−6 (Crotty et al., 2001; De la Torre et al., 1990; Jarvis and Kirkegaard, 1992; Parvin et al., 1986). The theoretically less complex and simpler problem of measuring the error rate of 3Dpol in vitro has nonetheless produced highly divergent apparently method-dependent results. Interestingly, reported results span approximately the same range (10−3 to 10−6) as cellular assays (Arias et al., 2008; Freistadt et al., 2007; Ward et al., 1988; Wells et al., 2001).
One of the most common in vitro assay for determining the fidelity of purified polymerases is the α-complementation assay which examines synthesis on templates that code for the alpha peptide of β-galactosidase (LacZ-α) (Boyer et al., 1996; DeStefano et al., 1998; Ji and Loeb, 1992; Kunkel, 1985; Stuke et al., 1997). However, this system requires production of a DNA strand for fidelity measurements and cannot be used to directly assess RDRP fidelity. We previously reported a method to adapt the α-complementation assay to RDRPs. An important strength of this approach is that many polymerases have been screened in the assay. This allows for direct comparisons with several polymerases. In this report we have used a modified version of this method to examine the fidelity of 3Dpol under several reaction conditions including different Mg2+ concentrations and the presence or absence of poliovirus protein 3AB, which is known to stimulate 3Dpol (Lama et al., 1995; Paul et al., 1994; Paul et al., 2000; Plotch and Palant, 1995; Rodriguez-Wells et al., 2001). A mutation frequency corresponding to ~1 mistake per 11,000 nucleotides copied was determined and was not perturbed by 3AB or changes in Mg2+, though synthesis at limiting nucleotide concentrations or in the presence of Mn2+ produced significant increases in the mutation frequency. The results are discussed in the context of previous mutation frequency measurements for poliovirus and other viruses.
2. Materials and methods
2.1. Materials
Calf intestinal alkaline phosphatase (CIP), RNase (ribonuclease)-DNase (deoxyribonuclease) free, T3 RNA polymerase, ribonucleotides, and deoxiribonucleotides were obtained from Roche. Rapid DNA ligation kit and RNasin RNase inhibitor were from Promega. Restriction enzymes, Moloney murine leukemia virus reverse transcriptase (MMLV-RT), and T4 polynucleotide kinase (PNK) were from New England Biolabs. Pfu DNA polymerase was from Stratagene. DNA and RNA oligonucleotides used as primers were purchased from Integrated DNA Technologies. G-25 spin columns were from Harvard Apparatus. RNeasy RNA purification kit and gel extraction kit were from Qiagen. X-gal was obtained from Denville Scientific, Inc. IPTG and media were obtained from Gibco, Life Technologies. All other chemicals were obtained from Fisher Scientific, VWR, or Sigma.
2.2. Polymerase (3Dpol) and 3AB purification
The polymerase of poliovirus type 1 (Mahoney strain) used for these studies was expressed in Escherichia coli (E. coli) using plasmid pT7pol and purified as described (Plotch et al., 1989). Protein 3AB was expressed as a Glutathione S-transferase (GST) conjugate in E. coli using plasmid pGEX-3AB and purified as described (Plotch and Palant, 1995). The GST was removed during purification using thrombin.
2.3. Polyacrylamide gel electrophoresis
Denaturing polyacrylamide gels (6% or 8% w/v), native polyacrylamide gels (12% w/v), and 1% agarose gels were prepared and electrophoresed as described (Sambrook and Russell, 2001).
2.4. Primer labeling
Reactions for primer labeling were done in a 50 μl volume containing 50 mM Tris-HCl, pH (7.6), 10 mM MgCl2, 10 mM betamercaptoethanol, 10 μl of [γ-32P]ATP (3000 Ci/ mmol at 10 μCi/μl) and 5 units of PNK. The reaction mixture was incubated for 30 minutes at 37°C and the PNK was heat inactivated for 15 minutes at 65°C according to manufacturer’s recommendation. Reactions were run over G-25 spin columns to remove excess ATP.
2.5. Preparation of RNA for fidelity assay
Transcripts were prepared with T3 RNA polymerase as recommended by the enzyme manufacturer. The transcript used as a template for 3Dpol was derived from plasmid pBSΔPvuII1146 prepared as described (DeStefano et al., 1998). The plasmid was cleaved with NdeI and T3 polymerase was used to prepare run-off RNA transcripts of ~760 nucleotides in length. After synthesis 10 units of DNase I-RNase free were added and incubation was continued for 15 min. The RNA was purified using a Qiagen RNeasy kit and checked by gel electrophoresis. The RNA was quantified by spectrophotometric analysis. To calculate the molecular weight of the RNA the following equation was used: ([A x 328.2.2) + (G x 344.2) + (C x 304.2) + (U x 305)]). The molecular weight was used to determine the molar concentration of the RNA transcripts using the standard conversion of 1 OD260 equals 40 μg/ml for single strand RNA.
2.6. RNA-RNA and RNA-DNA hybridization
Primers were hybridized to template RNA by mixing primer-RNA transcript at a 2:1 ratio in 10 μl of buffer containing 50 mM HEPES, (pH 7), 80 mM KCl, and 5 mM DTT. The mixture was heated at 65°C for 5 minutes then slow cooled to room temperature and used immediately in assays.
2.7. RNA primer extension reactions with 3Dpol
The ~760 nucleotide RNA template was hybridized to a 5′ P-32 labeled 25 nucleotide RNA primer (as described above). The primer sequence was 5′-CCUCUUCGCUAUUACGCCAG-3′. Full extension of the primer produced a 199 nucleotide final product (see Fig. 1A). The primer-template complex was pre-incubated in 17 μl of buffer (see below) for 3 min at 30°C. Eight μl of 4 μM 3Dpol in 50 mM HEPES (pH 7), 5 mM DTT and 10 % glycerol was added to initiate the reaction and incubation was continued for 1 hour. The final concentration of reaction components were 1.28 μM 3Dpol, 100 nM template, 200 nM primer, 50 mM HEPES (pH 7), 10 mM KCl, 0.8 mM MgCl2, 5 mM DTT, 50 μM rNTPs, 0.4% glycerol and 0.4 units/μl RNasin. The conditions used were determined to be optimal conditions for primer extension (Rodriguez-Wells et al., 2001). Modifications including addition of 3AB, changes in MgCl2 and rNTP concentrations, and replacement of MgCl2 with MnCl2 are described in the figure legends and text. Reactions were stopped by adding 5 μl of Proteinase K solution (2 mg/ml in 1.25 % SDS, 15 mM EDTA (pH 8.0) and 10 mM Tris-HCl (pH 8.0)) and incubating for 15 min at 37°C. Twenty-five μl of 2X sample buffer (90% formamide, 10 mM EDTA (pH 8), 0.25% each bromophenol blue and xylene cyanol) was added and products were electrophoresed on 6% polyacrylamide gels (19:1 acrylamide:bis-acrylamide), as described above. Fully extended 199 nucleotide RNA was located by autoradiography, and recovered by the crush and soak method (Sambrook and Russell, 2001) in 500 μl of elution buffer containing 50 mM Tris-HCl (pH 7), 1 mM EDTA (pH 8), and 80 units of RNasin. After overnight elution, this material was passed through a 0.45 μm syringe filter and recovered by ethanol precipitation after addition of 1/10th volume 3M NaAc (pH 7) and 30 μg of glycogen. Typically material from two separate reactions was combined during recovery. The recovered RNA was hybridized to another 20 base 5′ P-32 labeled RNA primer (5′-AGGAUCCCCGGGUACCGAGC-3′) with 50–100 fold greater specific activity than the primer used for round 1, and a second round of synthesis by 3Dpol was performed as described above. Reactions were started, terminated, and fully extended RNA synthesis product was gel purified as describe above but on an 8% gel which was run for ~1 hr after the xylene cyanol dye had run off. This was done to fully separate the starting 199 nucleotide templates from the 162 nucleotide full extension product of round 2. The RNA recovered from the second round of synthesis was hybridized to a DNA primer and reverse transcribed as describe below.
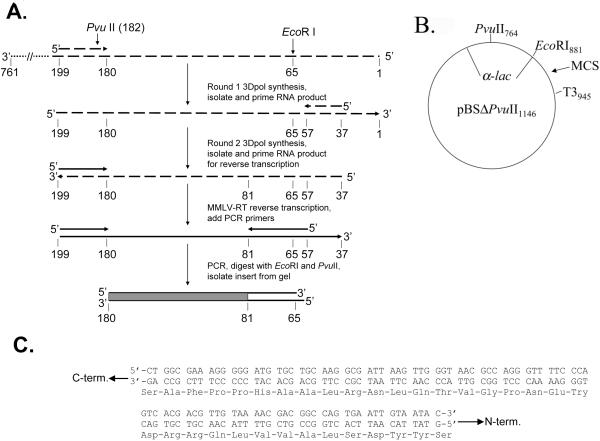
Fig. 1.
A, B, and C. System used to determine the fidelity of 3Dpol. An overview of the procedure used to assess polymerase fidelity is presented. RNAs are broken lines and DNAs are solid. Primers have arrowheads at the 3′ end. The ~760 nucleotide template RNA used as the initial template for 3Dpol-directed RNA synthesis is shown at the top with the 3′ and 5′ ends indicated. The positions of PvuII and EcoRI restriction sites are indicated for reference to the vector. The filled box at the bottom of the figure is the 99 base region of the lacZ-α gene that was scored in the assay. The region between 65 and 81 was not included in the error rate calculations because it was derived from one of the PCR primers and not 3Dpol. Details for specific steps are provided under Materials and Methods. (B) Plasmid pBSM13ΔPvuII1146, is shown. Relevant sites on the plasmid are indicated and numbering is based on the parent plasmid (pBSM13+ (Stratagene)). (C), the nucleotide and amino acid sequence for the 99 base region of the lacZ-α gene that was scored in the assay is shown. Both strands of the DNA plasmid are shown since 3Dpol synthesis was performed in both directions (see Fig. 1A). Refer to (Bebenek and Kunkel, 1995) for a description of the types of mutations and specific nucleotide changes that can be detected in this region.
2.8. RNA-directed DNA synthesis with the MMLV reverse transcriptase
The RNA generated by two rounds of synthesis with 3Dpol was isolated and hybridized as described above but using 1 pm of a DNA copy of the RNA primer from round 1 (5′-CCTCTTCGCTATTACGCCAG-3′). For reactions used to estimate the assay background the round one 760 nucleotide template RNA (0.1 pm) was used instead of RNA produced by 3Dpol. Reverse transcription was performed essentially as described (Ji and Loeb, 1992). The primer-template complex (all recovered material, ~0.1 pm of template) was resuspended in a buffer containing 50 mM Tris-HCl (pH 8), 1 mM DTT, 6 mM MgCl2, 0.5 mM EDTA (pH 8), 50 mM KCl, 100 μM dNTPs, and 0.1 unit/μl RNasin. Five units of MMLV-RT were added to initiate reactions. The final reaction volume was 25 μl. Reactions were continued for 15 minutes at 37°C. RNase-DNase free (0.25 units) was added and incubated for 5 min to digest the remaining RNA. The reverse transcribed DNA was amplified using PCR as described below.
2.9. Polymerase chain reaction (PCR)
The DNA produced by reverse transcription was amplified by PCR using the following primers: 5′-CCTCTTCGCTATTACGCCAG-3′ and 5′-GCTCGAATTCGCCCTATAGTGAGTC-3′. The first primer is the one used for the RNA-directed DNA synthesis by MMLV-RT and the second primer overlapped a region of the EcoRI site (in bold italics) on the plasmid from which the RNA was originally transcribed (Fig. 1). Reactions were performed in 100 μl of Tris-HCl (pH 9), 50 mM KCl, 2 mM MgCl2, 0.1% Triton X-100, 200 μM dNTPs, 50 pm of each primer, and 5 units of Pfu polymerase. Five μl of material from the reverse transcription reaction was used as the DNA source. Twenty-five cycles at 94°C (1 min), 50°C (1 min) then 72°C (1 min), followed by a 5 min incubation at 72°C were performed. The aqueous phase from the PCR reactions was phenol-chloroform extracted and recovered by ethanol precipitation. PCR products were then digested with 30 units each of PvuII and EcoRI in 75 μl of EcoRI buffer for 1 hour at 37°C. After restriction digest, 15 μl of 6X loading buffer (40% sucrose, 0.25% (w/v) bromophenol blue, 0.25% (w/v) xylene cyanol) was added to each reaction and samples were electrophoresed on a 12% native polyacrylamide gel as described above. The DNA was located by UV shadowing and recovered by overnight elution as described above. Spectrophotometry was used to quantify the recovered DNA as described above.
2.10. Preparation of vector for fidelity assay
Ten μg of the plasmid pBSΔPvuII1146 was digested with 50 units each of EcoRI and PvuII in 100 μl using EcoRI buffer. After 2 hours, 6X loading buffer was added and the DNA was electrophoresed on a 1% agarose gel as described above. The DNA was located by minimal exposure to long wave UV and excised. DNA was recovered using a Qiagen gel extraction kit and treated with 5 U of CIP for 2 hours at 37°C. Dephosphorylated vector was recovered by phenol-chloroform extraction and ethanol precipitation and quantified with a spectrophotometer. The quality of the vectors for the fidelity assay was assessed by re-ligating the cleaved vector and PvuII-EcoRI vector fragment (recovered from agarose gels after cleavage of pBSΔPvuII1146) as described below. Vectors producing colony mutation frequencies of less than ~0.005 (1 white or faint blue colony in 200 total) were used in fidelity assays.
2.11. Ligation of PCR fragments into vectors
The cleaved vector and insert fragments were ligated using a rapid DNA ligation kit from Promega. Vector (100 ng) and insert (0.05 pm) were ligated at a ~1:1 molar ratio for 10 min as described by the manufacturer. The ligation mixture (0.5 μl) was used to transform E. coli GC5 cells according to the manufacturer’s protocol. Transformed cells were plated in the presence of IPTG and X-Gal and grown overnight. Colonies were scored for α-complementation by counting white or faint blue colonies (mutations in lacZ-α) and blue colonies (non-mutated). The colony mutation frequency was the ratio of white + faint blue colonies to total colonies. The nucleotide misincorporation frequency was estimated as described in the Discussion section.
3. Results
3.1. Determination of fidelity assay background
Conversion of 3Dpol RNA synthesis products to DNA for the α-complementation assay necessitates an additional reverse transcription step which is not required for DNA polymerases (Fig. 1). In addition, due to the low yield of RNA synthesis products after two rounds of 3Dpol synthesis, the level of reverse transcription products is too low to be used directly and PCR is required to amplify the products. These amplified products are then cleaved with restriction enzymes and ligated into a linearized vector that is missing the portion of the lacZ-α gene contained in the amplified products. The 99 nucleotide region that is screened for mutations in the current assay is shown in Fig. 1C. All frameshift mutations and several substitutions in this region can be detected in the assay (see (Bebenek and Kunkel, 1995)). This approach has several steps where mutations can be introduced in addition to those derived from 3Dpol. These include mutations generated by T3 RNA polymerase, reverse transcriptase (RT), Taq polymerase, and errors resulting from inaccurate restriction digestion and subsequent ligation. The latter group contributed significantly to background error in previous experiments using this basic approach (Wells et al., 2001). The current approach used a much longer starting template (~760 vs 230 nucleotides) to make it easier to separate synthesis product from the starting material on a gel. Larger amounts of starting RNA template (100 nM vs. 5 nM) and enzyme (1.28 μM vs. 26 nM) were used because we found this yielded more fully synthesized products from round 1. Finally a different, less complicated dephosphorylation scheme was used in which the vector DNA was dephosphorylated at both the EcoRI and PvuII sites and the inserts were left phosphorylated at both sites. The assay background was determined by performing reverse transcription with MMLV-RT on the starting T3 transcript material (see Methods). The generated cDNA was then amplified and used to make inserts as was done for cDNAs from 3Dpol-derived RNA. Therefore the control should have errors from all steps except the two rounds of 3Dpol synthesis. A control was performed along with each set of experiments (Table 1). The ratio of white and faint blue colonies to total colonies (colony mutation frequency) for the control was 0.011 +/− 0.003 (ave. 6 exp. +/− S.D.) which corresponds to ~1 white or faint blue colony in 90 total.
TABLE 1. Colony mutation frequencies for experiments with 3Dpol.
|
aExp. # |
bCondition | White/Faint Blue Colonies |
Total Colonies |
cColony Mutation Frequency (CMF) |
CMF- BKG. |
|---|---|---|---|---|---|
| 1 | Background | 49 | 3609 | 0.014 | 0 |
| 0.8 mM MgCl2, 50 μM rNTPs | 71 | 2712 | 0.026 | 0.012 | |
| 4 mM MgCl2, 500 μM rNTPs | 87 | 2529 | 0.034 | 0.020 | |
| 0.8 mM MgCl2, 50 μM G, U, CTP, 5 | 51 | 1900 | 0.027 | 0.013 | |
| μM ATP | |||||
| 0.8 mM MgCl2, 50 μM G, U, CTP, 0.5 | 111 | 2526 | 0.044 | 0.030 | |
| μM ATP | |||||
| 2 | Background | 61 | 3705 | 0.016 | 0 |
| 0.8 mM MgCl2, 50 μM rNTPs | 63 | 2487 | 0.025 | 0.009 | |
| 4 mM MgCl2, 500 μM rNTPs | 75 | 3023 | 0.025 | 0.009 | |
| 4 mM MnCl2, 500 μM rNTPs | 162 | 1734 | 0.093 | 0.077 | |
| 3 | Background | 26 | 2633 | 0.010 | 0 |
| 4 mM MgCl2, 500 μM rNTPs | 45 | 3067 | 0.015 | 0.005 | |
| 4 mM MgCl2, 500 μM rNTPs +3AB | 44 | 2648 | 0.017 | 0.007 | |
| 4 | Background | 10 | 1261 | 0.008 | 0 |
| 4 mM MgCl2, 500 μM rNTPs | 16 | 1070 | 0.015 | 0.007 | |
| 4 mM MgCl2, 500 μM rNTPs +3AB | 31 | 1874 | 0.014 | 0.006 | |
| 5 | Background | 23 | 2587 | 0.009 | 0 |
| 0.8 mM MgCl2, 50 μM rNTPs | 50 | 2784 | 0.018 | 0.009 | |
| 4 mM MgCl2, 500 μM rNTPs | 50 | 3467 | 0.014 | 0.005 | |
| 6 | Background | 11 | 1427 | 0.008 | 0 |
| 0.8 mM MgCl2, 50 μM rNTPs | 28 | 1971 | 0.014 | 0.006 | |
| 4 mM MgCl2, 500 μM rNTPs | 20 | 1365 | 0.015 | 0.007 | |
| Average values +/− standard deviation for experiments with multiple repeats | |||||
| Ave. | Ave. -BKG. | ||||
| Background | 0.011 +/− 0.003 | 0 | |||
| 0.8 mM MgCl2, 50 μM rNTPs | 0.021 +/− 0.006 | 0.009 +/− 0.002 | |||
| 4 mM MgCl2, 500 μM rNTPs | 0.020 +/− 0.008 | 0.009 +/− 0.006 | |||
| 4 mM MgCl2, 500 μM rNTPs +3AB | 0.016 +/− 0.002 | 0.007 +/− 0.001 | |||
Independent experiments performed at different times.
In background assays the T3 RNA polymerase template RNA was directly reverse transcribed by RT and this cDNA product was used in PCR reactions. Assays to test 3Dpol fidelity had two additional 3Dpol synthesis steps prior to PCR. These included extension of a primer on the original T3 template, recovery of the synthesized product, and a second round of 3Dpol synthesis using this product as the template.
Numbers shown are the “colony mutation frequency” (CMF) defined as white + faint blue colonies divided by total colonies. Refer to the Results and Methods sections for details.
3.2. RNA synthesis products produced by 3Dpol under various reaction conditions
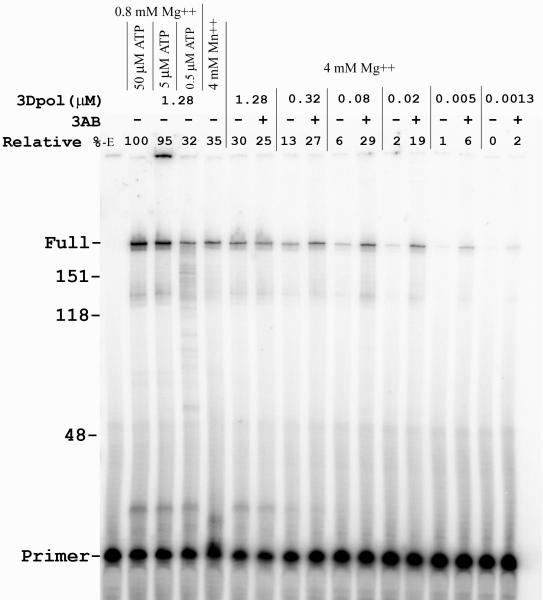
Synthesis products produced in the first round of synthesis with 3Dpol under several conditions are shown in Fig. 2. Consistent with previous observations (Rodriguez-Wells et al., 2001), optimal extension occurred with 0.8 mM MgCl2 and 50 μM rNTPs. Increasing MgCl2 to 4 mM and rNTPs to 500 μM (optimal [rNTP] for this divalent cation concentration) resulted in a ~3 fold decrease in fully extended products. Reductions were also observed when the concentration of ATP was deceased to 0.5 μM (C, G, and UTP held constant at 50 μM) while replacing 4 mM MgCl2 with the same level of MnCl2 did not significantly effect the yield. Poliovirus 3AB protein is known to stimulate 3Dpol synthesis, especially at low 3Dpol concentrations (see Introduction). In order to test the effect of 3AB on 3Dpol fidelity, conditions where stimulation was observed were determined. To do this, a fixed concentration of 4 μM 3AB was used while the concentration of 3Dpol was lowered in reactions containing 4 mM MgCl2 and 500 μM rNTPs. No stimulation was observed with the standard 1.28 μM 3Dpol concentration (compare with and without 3AB) while stimulation by 3AB was evident when 0.32, 0.08, 0.02, 0.005, and 0.00125 μM 3Dpol was used. The 0.08 μM 3Dpol level was used for fidelity assays with 3AB because the yield of total product from these reactions was about the same as standard conditions (1.28 μM 3Dpol) without 3AB.
Fig. 2.
3Dpol synthesis on the ~760 nucleotide RNA template under different conditions. Shown is an autoradiogram with extension of a 20 nucleotide 5′ P-32 end-labeled RNA primer on the round 1 RNA template. Full extension of the primer resulted in a 199 nucleotide product (see Fig. 1A). Nucleotide size positions on the left were bases on a DNA ladder. “Relative %” indicates the amount of fully extended product (determined using a phosphoimager) in each lane compared to the 100% value in lane 2. The presence or absence of 3AB (4 μM final concentration) is indicated above each lane as is the amount of 3Dpol used and the divalent cation conditions. In reactions with 0.8 mM MgCl2, the concentration of ATP is indicated above the lane while all other nucleotides were at 50 μM. In reactions with 4 mM MnCl2 or 4 mM MgCl2 all nucleotides were present at 500 μM. -E: control reaction in the absence of 3Dpol. Refer to Materials and Methods for details.
Products from first round synthesis were used as a template for a second round of 3Dpol synthesis (see Methods and Fig. 1A). This was required to increase the proportion of errors in the assay that resulted from 3Dpol, thus increasing the colony mutation frequency sufficiently above background. The primer for the second strand reaction was labeled to ~100 fold greater specific activity than the one used for first strand synthesis so the second strand products could be more easily located. The level of second strand products varied depending on conditions and from assay to assay.
3.3. Determination of the fidelity of 3Dpol under various conditions
The colony mutation frequency for 3Dpol was determined by subtracting the MMLV-RT results (background) from the colony mutation frequencies obtained with 3Dpol (see Methods). Results for the various conditions are reported in Table 1. Individual experiments and average results are reported in the Table. The standard condition (0.8 mM MgCl2, 50 μM rNTPs) was repeated in 4 separate experiments yielding an average colony mutation frequency value after background subtraction of 0.009 +/− 0.002. Based on the average background value of 0.011 (see above) this indicates that 45% (0.009/(0.009 + 0.011)) of the total white or faint blue colonies resulted from errors made by 3Dpol while ~55% were due to assay background. No change was observed in the colony mutation frequency with 4 mM MgCl2 and 500 μM rNTPs (0.009 +/− 0.006 (6 exp.). Inclusion of 3AB under these conditions did not significantly alter this result (0.007 +/− 0.001 (2 exp.)). In contrast, replacing 4 mM MgCl2 with the same concentration of MnCl2 increased the colony mutation frequency ~9 fold to 0.077 (1 exp.), a result consistent with the mutagenic nature of this divalent cation in 3Dpol reactions (Arnold et al., 1999). A single experiment was also conducted using 0.8 mM MgCl2 and 50 μM of C, G, and UTP along with 50, 5, or 0.5 μM ATP. Results with 50 or 5 μM ATP were essentially the same at 0.012 and 0.009 (results - BKG), respectively, while the colony mutation frequency approximately doubled with 0.5 μM ATP (0.026).
In the previous report using a α-complementation assay to estimate 3Dpol fidelity, an approximately 5-fold higher colony mutation frequency was observed using conditions similar to the low Mg2+ conditions used here (Rodriguez-Wells et al., 2001). The reason for the disparity between the results is unclear although it may stem, in part, from the different protocols used in the assays. In the current work much larger amounts of a longer template RNA were used and a different dephosphorylation scheme was employed (see above). A different enzyme preparation was also used in the earlier work although no significant difference in results in the current work was observed between two independently prepared enzyme preparations.
4. Discussion
Analysis of the fidelity of RDRPs in vitro has typically been accomplished using biased nucleotide pools in primer extension assays (Arias et al., 2008; Arnold et al., 1999; Freistadt et al., 2007). This approach has the advantage of being simple and rapid and allows conditions to be set up to accurately test specific types of mutations. A clear disadvantage is that only a very small subset of the possible sequence contexts are typically examined in these assays. It is also difficult to directly compare different polymerases unless the same sets of primers and templates are used. Direct sequencing of reverse transcribed and PCR amplified RNA products has also been used to determine replication fidelity (Crotty et al., 2001; Ji and Loeb, 1994). This approach has the clear advantage of examining fidelity over the native genome sequence. Drawbacks include a potentially high background from reverse transcription and PCR and relatively high cost, despite declining costs for DNA sequencing. As the cost of sequencing declines further this approach will likely become more common. The α-complementation assay examines fidelity over a larger set of sequences than other in vitro assays (though less than direct sequencing) and allows direct comparisons with other polymerases. Thus far, with the exception of 3Dpol, only DNA polymerases have been tested in the assay. The major drawback of this assay for RDRPs vs. DNA polymerases is that several additional steps are required (sec. 3.1 and Fig. 1) which are time consuming and lead to a high background. Because of the high background the assay is not particularly useful for determining the types of mutations made except when mutagenic conditions (Mn2+ or biased rNTP pools for example) are employed. Despite this drawback the method yields an estimate of fidelity for RDRP in one of the best developed and most heavily employed in vitro fidelity assays.
For α-complementation assays, an estimate of the base misincorporaton frequency can be made from the colony mutation frequency. How this number translates into a misincorporation frequency depends on several factors including the proportion of error that were deletions or insertions vs. substitutions and whether a particular substitution error produces the white or faint blue phenotype that is scored in the assay. Deletions and insertions result in frameshift mutations that are nearly always detected as white or faint blue colonies. In contrast, less than half of the substitutions made in the assay result in a mutant phenotype. This is due to wobble substitutions and others that produce amino acid changes that do not significantly affect complementation activity (Bebenek and Kunkel, 1995). A substitution detection rate of 35% was estimated using a region of lacZ-α similar to the one used in this work (Ji and Loeb, 1992). Sequencing of plasmid DNA from the current work (data not shown) revealed a spectrum of mutations similar to that found in our previous work (Wells et al., 2001), however, it was not possible to determine the proportion of deletions/insertions vs. substitutions in the current work as the background was too high to differentiate between mutations made by 3Dpol and those due to background sources. In α-complementation assays with HIV-RT from this lab ~30% of recovered mutations were insertions/deletions (DeStefano et al., 1998) and ~70% substitutions, although there has been significant variation in this proportion depending on the particular lab and version of the α-complementation assay used (Bebenek et al., 1989; Boyer et al., 1992; Ji and Loeb, 1994; Ji and Loeb, 1992; Roberts et al., 1988; Weber and Grosse, 1989). The assay used here included 99 nts of the lacZ-α gene that were derived from 3Dpol synthesis in each of two rounds or a total of 198 nts (see Fig. 1A). Based on this, and using the 35% detection rate for substitutions in this region, the 3Dpol mutation frequency can be estimated. For example, taking the extreme cases, if 100% of the mutations made by 3Dpol were deletions/insertions, then the mutation frequency for the 0.8 mM MgCl2 50 μM rNTPs condition would be 4,6 × 10−5 or 1 error per ~20,000 incorporations (0.009 colony mutation frequency/198 nt= 4.6 × 10−5); and if 100% were substitutions it would be 1.3 × 10−4, or ~1 error per 8,000 incorporations (4.6 × 10−5/0.35= 1.3 × 10−4). Taking a more likely scenario of about 50% deletions/insertions and 50% substitutions yields a mutation frequency of 8.8 × 10−5, or ~1 error per 11,000 incorporations ((0.009 × 0.5)/198= 2.3 × 10−5 for deletions/insertions, and ((0.009 × 0.5)/198)/0.35 = 6.5 × 10−5 for substitutions, total is 8.8 × 10−5 for both).
The overall mutation frequency of 3Dpol as determined above is comparable to most in vitro results for HIV-RT using α-complementation assays which range from ~5 × 10−4 – ~5 × 10−5 (1 error in 2000–20,000 incorporated bases). Other RTs tested have modestly lower error rates than 3Dpol while DNA-dependent DNA polymerases with proof-reading activity are in general significantly more accurate (Bebenek et al., 1989; Bebenek and Kunkel, 1995; Boyer et al., 1992; Ji and Loeb, 1994; Ji and Loeb, 1992; Roberts et al., 1988; Weber and Grosse, 1989). Most other in vitro estimations of 3Dpol fidelity have used nucleotide biased primer extension assays on heteropolymeric templates. Results vary with some reports suggesting misincorporation rates similar to those reported here (Arias et al., 2008) while other reported much lower error frequencies (Freistadt et al., 2007). Differences in the methods and conditions or enzyme preparations probably account for some of the discrepancies. However, these inconsistencies highlight the difficulties involved with accurately determining the fidelity of viral polymerases in general. Different approaches, different conditions and variations between enzyme preparations, coupled with the apparent high variability of these assays have resulted in widely variable results.
The ~ 1 error per 11,000 incorporation (using the 50/50 estimate of deletion/insertion vs. substitutions from above) determined here is also consistent with estimates of the mutation rate per genome per replication event (μg), a value determined from results in cells. Estimates from several experiments with various RNA viruses lead to a μg value of 0.76 (Drake and Holland, 1999). For poliovirus with a genome length of approximately 7,500, this value suggests a mutation rate of approximately 1 × 10−4, or about 1 error per 10,000 bases synthesized. Since μg is based on results in cells where host factors and selection can potentially influence fidelity, a direct correlation with polymerase fidelity in vitro would not necessarily be expected. A close correlation would imply minimal impact of cellular or other viral factors on polymerase fidelity.
The fidelity of 3Dpol was not significantly different using high and low MgCl2 conditions. We had previously determined that low Mg2+ (0.8 mM), though not commonly used by others in previous reports, is actually optimal for primer extension providing that lower rNTP (50 μM) concentrations are used (Rodriguez-Wells et al., 2001). Despite significantly decreasing primer extension (Fig. 2), 4 mM MgCl2 with 500 μM rNTPs did not affect the mutation frequency (Table 1). This was not due to the assay not being sensitive enough to detect a change in the mutation rate as mutagenic conditions (biased nucleotide pools (0.5 μM ATP) or replacing Mg2+ with Mn2+) significantly increased the mutations frequency (Table 1). The results with lowered ATP suggest that a 2-fold change in the error rate could have been easily detected while it is unlikely that small changes in fidelity would be detected in the assay.
Despite strongly stimulating 3Dpol synthesis under low enzyme conditions (Fig. 2), 3AB did not alter 3Dpol’s fidelity (Table 1). This lab and others have shown that 3AB is highly stimulatory in primer-dependent in vitro 3Dpol reactions (see Introduction). Others have also shown that these proteins can bind to each other and the domains responsible for binding have been mapped (Hope et al., 1997; Plotch and Palant, 1995; Strauss et al., 2003; Xiang et al., 1998; Xiang et al., 1995). Still, the mechanism of stimulation is unknown and whether it can affect fidelity has not previously been tested. Again a small effect beyond the detection limits of the assay cannot be ruled out. Interestingly, we have also shown that HIV NC protein which can enhance the processivity of DNA synthesis by HIV-RT, also does not influence fidelity (DeStefano et al., 1998). Like NC, 3AB possesses nucleic acid chaperone activity (Levin et al., 2005; DeStefano and Titilope, 2006). Both NC and 3AB can bind and “coat” nucleic acid strands, consistent with the chaperone activity. The current results with 3AB and previous results with NC indicate that chaperone activity, and in these cases stimulation of nucleic acid synthesis, do not influence the fidelity of nucleotide incorporation.
Acknowledgements
This work was supported by a National Institutes of General Medicine grant number GM051140. We would like to thank Dr. Stephen Plotch (formerly of Wyeth-Ayerst Research) for the plasmids for 3AB and 3Dpol production and for his assistance in purifying the proteins.
Abbreviations
- RDRP
RNA dependent RNA polymerase
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- X-Gal
bromo-chloro-indolyl-galactopyranoside
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- GST
glutathione S-transferase
- CIP
calf intestinal alkaline phosphatase
- PNK
T4 polynucleotide kinase
- MMLV-RT
Maloney murine leukemia virus reverse transcriptase
- NC
HIV nucleocapsid protein
- PCR
polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Arias A, Arnold JJ, Sierra M, Smidansky ED, Domingo E, Cameron CE. Determinants of RNA-dependent RNA polymerase (in)fidelity revealed by kinetic analysis of the polymerase encoded by a foot-and-mouth disease virus mutant with reduced sensitivity to ribavirin. J. Virol. 2008;82(24):12346–12355. doi: 10.1128/JVI.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JJ, Ghosh SK, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Divalent cation modulation of primer, template, and nucleotide selection. J. Biol. Chem. 1999;274(52):37060–37069. doi: 10.1074/jbc.274.52.37060. [DOI] [PubMed] [Google Scholar]
- Bebenek K, Abbotts J, Roberts JD, Wilson SH, Kunkel TA. Specificity and mechanism of error prone replication by human immunodeficiency virus 1 reverse transcriptase. J. Biol. Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- Boyer JC, Bebenek K, Kunkel T,A. Analyzing the fidelity of reverse transcription and transcription. Methods Enzymol. 1996;275:523–537. doi: 10.1016/s0076-6879(96)75029-2. [DOI] [PubMed] [Google Scholar]
- Boyer JC, Bebenek K, Kunkel TA. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc. Natl. Acad. Sci. U. S. A. 1992;89(15):6919–6923. doi: 10.1073/pnas.89.15.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. U. S. A. 2001;98(12):6895–6900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Torre JC, Wimmer E, Holland JJ. Very high frequency of reversion to guanidine resistant in clonal pools of guanidine-dependent type 1 Poliovirus. J. Virol. 1990;64(2):664–671. doi: 10.1128/jvi.64.2.664-671.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano J, Ghosh J, Prasad B, Raja A. High fidelity of internal strand transfer catalyzed by human immunodeficiency virus reverse transcriptase. J. Biol. Chem. 1998;273(3):1483–1489. doi: 10.1074/jbc.273.3.1483. [DOI] [PubMed] [Google Scholar]
- Domingo E, Escarmis C, Sevilla N, Moya A, Elena SF, Quer J, Novella IS, Holland JJ. Basic concepts in RNA virus evolution. FASEB J. 1996;10:859–864. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 1999;96(24):13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistadt MS, Vaccaro JA, Eberle KE. Biochemical characterization of the fidelity of poliovirus RNA-dependent RNA polymerase. Virol. J. 2007;4:44. doi: 10.1186/1743-422X-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JJ. Replication error, quasispecies populations, and extreme evolution rates of RNA viruses. In: Morse S, editor. Emerging Viruses. Oxford University Press; New York: 1993. pp. 203–218. S. [Google Scholar]
- Hope DA, Diamond SE, Kirkegaard K. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J. Virol. 1997;71(12):9490–9498. doi: 10.1128/jvi.71.12.9490-9498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis TC, Kirkegaard K. Poliovirus RNA recombination: mechanistic studies in the absence of selection. Embo J. 1992;11(8):3135–3145. doi: 10.1002/j.1460-2075.1992.tb05386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Loeb LA. Fidelity of HIV-1 reverse transcriptase copying a hypervariable region of the HIV-1 env gene. Virology. 1994;199(2):323–30. doi: 10.1006/viro.1994.1130. [DOI] [PubMed] [Google Scholar]
- Ji JP, Loeb LA. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry. 1992;31(4):954–958. doi: 10.1021/bi00119a002. [DOI] [PubMed] [Google Scholar]
- Kunkel TA. The mutational specificity of DNA polymerase-beta during in vitro DNA synthesis. Production of frameshift, base substitution, and deletion mutations. J. Biol. Chem. 1985;260(9):5787–5796. [PubMed] [Google Scholar]
- Lama J, Sanz MA, Rodriguez PL. Role of 3AB protein in poliovirus genome replication. J. Biol. Chem. 1995;270(24):14430–14438. doi: 10.1074/jbc.270.24.14430. [DOI] [PubMed] [Google Scholar]
- Parvin JD, Moscona A, Pan WT, Leider JM, Palese P. Measurement of the mutation rates in animal viruses: Influenza A virus and Poliovirus type 1. J. Virol. 1986;59(2):377–383. doi: 10.1128/jvi.59.2.377-383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AV, Cao X, Harris KS, Lama J, Wimmer E. Studies with poliovirus polymerase 3Dpol. Stimulation of poly(U) synthesis in vitro by purified poliovirus protein 3AB. J. Biol. Chem. 1994;269(46):29173–29181. [PubMed] [Google Scholar]
- Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 2000;74(22):10359–10370. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch SJ, Palant O. Poliovirus protein 3AB form a complex with and stimulates the activity of the viral RNA polymerase, 3Dpol. J. Virol. 1995;69(11):7169–7179. doi: 10.1128/jvi.69.11.7169-7179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch SJ, Palant O, Gluzman Y. Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J. Virol. 1989;63(1):216–225. doi: 10.1128/jvi.63.1.216-225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JD, Bebenek K, Kunkel TA. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242(4882):1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Wells V, Plotch SJ, DeStefano JJ. Primer-dependent synthesis by poliovirus RNA-dependent RNA polymerase (3D(pol)) Nucleic Acids Res. 2001;29(13):2715–2724. doi: 10.1093/nar/29.13.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Strauss DM, Glustrom LW, Wuttke DS. Towards an understanding of the poliovirus replication complex: the solution structure of the soluble domain of the poliovirus 3A protein. J. Mol. Biol. 2003;330(2):225–234. doi: 10.1016/s0022-2836(03)00577-1. [DOI] [PubMed] [Google Scholar]
- Stuke AW, Ahmad-Omar O, Hoefer K, Hundmann G, Jentsch KD. Mutations in the SIV env and the M13 lacZa gene generated in vitro by reverse trancriptases and DNA polymerases. Arch. Virol. 1997;142:1139–1154. doi: 10.1007/s007050050148. [DOI] [PubMed] [Google Scholar]
- Ward CD, Stokes MA, Flanegan JB. Direct measurement of the Poliovirus polymerase error frequency in vitro. J. Virol. 1988;66(2):558–562. doi: 10.1128/jvi.62.2.558-562.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J, Grosse F. Fidelity of human immunodeficiency virus type I reverse transcriptase in copying natural DNA. Nucleic Acids Res. 1989;17(4):1379–1393. doi: 10.1093/nar/17.4.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells VR, Plotch SJ, DeStefano JJ. Determination of the mutation rate of poliovirus RNA-dependent RNA polymerase. Virus Res. 2001;74(1–2):119–132. doi: 10.1016/s0168-1702(00)00256-2. [DOI] [PubMed] [Google Scholar]
- Xiang W, Cuconati A, Hope D, Kirkegaard K, Wimmer E. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J. Virol. 1998;72(8):6732–6741. doi: 10.1128/jvi.72.8.6732-6741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang W, Cuconati A, Paul AV, Cao X, Wimmer E. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. Rna. 1995;1(9):892–904. [PMC free article] [PubMed] [Google Scholar]