Abstract
Apoptosis through the TRAIL receptor pathway can be induced via agonistic IgG to either TRAIL-R1 or TRAIL-R2. Here we describe the use of phage display to isolate a substantive panel of fully human anti-TRAIL receptor single chain Fv fragments (scFvs); 234 and 269 different scFvs specific for TRAIL-R1 and TRAIL-R2 respectively. In addition, 134 different scFvs that were cross-reactive for both receptors were isolated. To facilitate screening of all 637 scFvs for potential agonistic activity in vitro, a novel high-throughput surrogate apoptosis assay was developed. Ten TRAIL-R1 specific scFv and 6 TRAIL-R2 specific scFv were shown to inhibit growth of tumor cells in vitro in the absence of any cross-linking agents. These scFv were all highly specific for either TRAIL-R1 or TRAIL-R2, potently inhibited tumor cell proliferation, and were antagonists of TRAIL binding. Moreover, further characterization of TRAIL-R1 agonistic scFv demonstrated significant anti-tumor activity when expressed and purified as a monomeric Fab fragment. Thus, scFv and Fab fragments, in addition to whole IgG, can be agonistic and induce tumor cell death through specific binding to either TRAIL-R1 or TRAIL-R2. These potent agonistic scFv were all isolated directly from the starting phage antibody library and demonstrated significant tumor cell killing properties without any requirement for affinity maturation. Some of these selected scFv have been converted to IgG format and are being studied extensively in clinical trials to investigate their potential utility as human monoclonal antibody therapeutics for the treatment of human cancer.
Key words: TRAIL-R1, TRAIL-R2, human antibody, scFv, phage display
Introduction
TNF-related apoptosis-inducing ligand (TRAIL/Apo2L/TNFSF10)1 is a member of the TNF family that promotes apoptosis in a broad range of cancer cells, but not in most normal cells.2,3 TRAIL exerts its apoptotic activity through binding to two receptors, TRAIL-R1 (also known as TNFRSF10A or DR4)4 and TRAIL-R2 (also known as TNFRSF10B, DR5 or TRICK2)5–8 that engage the cells' apoptotic machinery through conserved cytoplasmic regions known as death domains. TRAIL also binds to three other receptors, TRAIL-R3 (TNFRSF10C, DcR1)7,9 TRAIL-R4 (TNFRSF10D, DcR2)10 and osteoprotegerin (TNFRSF10D, OPG),11 but these lack functional death domains and therefore do not transmit an apoptotic signal. It has been suggested that these other TRAIL receptors act as decoy receptors to regulate the activities of TRAIL in vivo, either by directly competing for TRAIL binding or by forming complexes with TRAIL-R1 and TRAIL-R2.7,12,13
TRAIL is a homotrimeric protein that triggers oligomerization of TRAIL-R1 and/or TRAIL-R2, and subsequently the formation of a death-inducing signalling complex (DISC). This consists of death receptors, adaptor proteins, and procaspase 8, which lead to processing and activation of procaspase 8 by an autocatalytic mechanism.14 In some cell types (type I), such as SW480 (colon carcinoma) and H460 (human non small cell lung carcinoma) cells, activation of caspase 8 is sufficient for subsequent activation of the effector caspase 3 to execute cellular apoptosis (extrinsic pathway). In other cell types (type II), for example Jurkat (acute T cell leukaemia) and HCT116 (colon carcinoma) cells, amplification through the mitochondrial pathway (intrinsic pathway), which is initiated by cleavage of Bid by caspase 8, is required for cellular apoptosis (reviewed in ref. 15).
Whilst the normal physiological role of TRAIL is not fully understood, the ligand's ability to trigger apoptosis in a variety of transformed cell lines suggests that it may be a physiological modulator of tumor cell apoptosis. Increasing evidence suggests that TRAIL may be an important player in immune surveillance against virally-infected cells and tumors,15–19 and in particular against haematological malignancies.20 Further studies have demonstrated that TRAIL also participates in the homeostasis of the lymphoid compartment of the immune system by inducing apoptosis in immune cells that have fulfilled their function.21–24 In line with a regulatory role in the homeostasis of the immune system, several studies have demonstrated that TRAIL may also function to attenuate autoimmune reactions.25–27
TRAIL-R1 and TRAIL-R2 are expressed at relatively high levels in tumor tissue relative to the levels observed in normal human tissues.28 This, combined with the ability of TRAIL to induce apoptosis in a wide variety of cancer cell lines whilst sparing normal cells, suggests that TRAIL may have therapeutic utility in the treatment of human cancer. Evidence in vitro and in vivo suggests that tumor cells are sensitive to treatment by TRAIL, and that effects are enhanced by concomitant treatment with chemotherapeutic agents.15,16 More recently, agonistic murine and rabbit monoclonal antibodies have been developed that selectively bind to either TRAIL-R1 or TRAIL-R2, and mimic the tumor killing properties of TRAIL.29–32 Agonistic monoclonal antibodies bring additional benefits over TRAIL as therapeutic reagents because of their prolonged half life in vivo and because their effects are not compromised by binding to TRAIL decoy receptors or potential escape mutants of TRAIL-R1 or TRAIL-R2.33 Consequently, recent efforts have focused on the development of fully human monoclonal antibodies to TRAIL-R1 and TRAIL-R2 that can be used to treat human cancer with minimal associated immunogenicity.34–36
We have applied phage display technology to isolate large panels of human scFv specific for TRAIL-R1 and TRAIL-R2, as well as a group of scFv that cross-react with both receptors. Using a novel high-throughput screen, we demonstrated that several TRAIL-R1 and TRAIL-R2 specific scFv possess potent anti-tumor properties, inducing cell death without the requirement of any secondary cross-linking agents. Moreover, we demonstrate tumor cell killing by purified monomeric Fab, indicating that avidity is not essential to induce apoptosis. Thus, agonistic scFv and Fab fragments, as well as whole IgG molecules, are able to induce apoptosis of human tumor cell lines through specific binding to TRAIL-R1 and TRAIL-R2.
These agonistic scFv have exquisite specificity for TRAIL-R1 or TRAIL-R2, they do not bind to any of the decoy receptors, and they also compete directly with TRAIL for binding to either TRAIL-R1 or TRAIL-R2. The high-throughput surrogate apoptosis assay we developed for the identification of scFv agonists also proved predictive of IgG agonism, since several scFv described have now been converted to IgG format, and have been confirmed to exhibit apoptotic activity both in vitro and in vivo.
Results
Isolation of scFv antibodies specific for TRAIL-receptors.
Purified recombinant TRAIL-R1 and TRAIL-R2 Fc-fusion proteins were used as target antigens for antibody isolation using a large, non-immunized, human scFv phage antibody library.37,38 TRAIL receptors were presented to the library either immobilised onto solid supports or in solution phase using biotinylated TRAIL-receptors and streptavidin coated magnetic beads (Dynal).39 Antibody isolation (‘selection’) strategies were designed to produce receptor specific scFv by performing selections on either TRAIL-R1 or TRAIL-R2 alone, or to produce cross-reactive scFv by performing alternate rounds of selection on both TRAIL-R1 and TRAIL-R2.
TRAIL-receptor specific scFvs from each selection strategy were identified by direct binding ELISA against TRAIL-R1, TRAIL-R2 and an irrelevant Fc-fusion protein. ScFvs that bound to one or both TRAIL-receptors with an ELISA signal at least 3-fold greater than an irrelevant control antibody were deemed to be specific for either TRAIL-R1 or TRAIL-R2, or cross-reactive with both receptors. In total, over 14,000 scFvs were analyzed by ELISA and 624 TRAIL-R1, 705 TRAIL-R2 and 1123 TRAIL-R1/-R2 cross-reactive scFvs were identified. The ELISA experiments demonstrated that selections on TRAIL-R1 and TRAIL-R2 alone yielded predominantly scFvs specific for each receptor. Very few (2%) scFvs isolated from selections on TRAIL-R1 cross-reacted with TRAIL-R2. Similarly, only 0.4% of scFvs isolated to TRAIL-R2 cross-reacted with TRAIL-R1. Thus the vast majority of TRAIL-R1 and TRAIL-R2 cross-reactive scFvs were derived from the hybrid selection approach where both TRAIL-R1 and TRAIL-R2 were utilized at alternate rounds of selection.
The nucleotide sequences of all 2452 TRAIL-receptor binding scFvs were determined and their respective variable heavy (VH) and variable light (VL) amino acid sequences translated. This analysis identified 637 scFvs to TRAIL-receptors that were all different by at least one amino acid; 234 scFvs to TRAIL-R1, 269 scFvs to TRAIL-R2 and 134 scFvs that were cross-reactive with both TRAIL-R1 and TRAIL-R2.
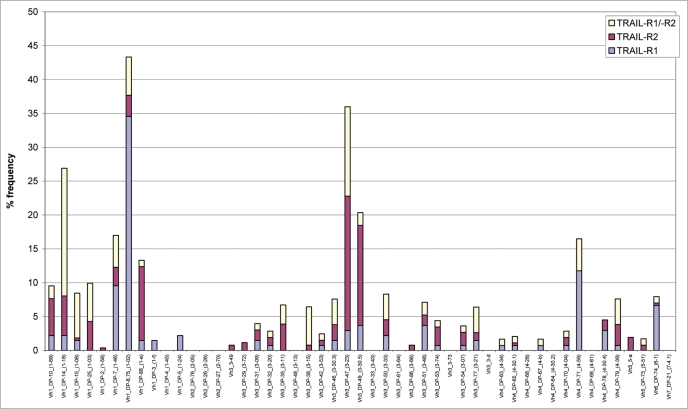
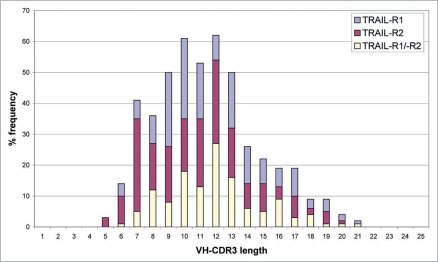
To illustrate the diversity of the scFvs produced to TRAIL-R1 and TRAIL-R2, each of the 637 different scFv sequences were aligned to human germline VH sequence databases using BLASTN (version 2.0.9). This analysis identified 27 different VH germlines utilized in the panel of 234 TRAIL-R1 scFv, 31 different VH germlines in the panel of 269 TRAIL-R2 scFv and 28 different VH germlines in the panel of 134 TRAIL-R1/-R2 cross-reactive antibodies (Fig. 1). ScFv diversity was also demonstrated by a study of VH-CDR3 sequence information. 158 (68%) of the 234 TRAIL-R1 specific scFvs, 192 (71%) of the 269 TRAIL-R2 specific scFvs and 130 (97%) of the 134 TRAIL-R1/-R2 cross-reactive scFvs all had unique VH-CDR3 sequences with lengths ranging from 5 to 21 amino acids (Fig. 2). This analysis demonstrated that the panel of scFvs isolated to TRAIL-R1, TRAIL-R2 or to both receptors were all highly diverse in terms of their VH germline gene usage and also their VH-CDR3 length.
Figure 1.
Sequence diversity of 637 TRAIL-receptor binding scFv. VH germline gene segment utilization is shown for the panel of 234 TRAIL-R1 specific scFv (blue bars), 269 TRAIL-R2 specific scFv (red bars) and 134 TRAIL-R1/-R2 cross-reactive scFv (yellow bars). The frequency of VH germline utilization is expressed as a percentage of the total for each panel of scFv.
Identification of agonistic scFv to TRAIL-receptors using a surrogate assay of apoptosis.
To measure inhibition of tumor cell growth by TRAIL-receptor scFv in vitro, a surrogate apoptosis assay was developed that responds to the chemical reduction of growth medium resulting from cell proliferation. Agonistic scFv were identified in the assay by their ability to inhibit cell growth and prevent a colour change from blue to pink.
Apoptosis assays were developed for TRAIL-R1 using HeLa and ST486 cell lines and for TRAIL-R2 with HT1080 cells. In the first instance, the sensitivity of each cell line to TRAIL alone was measured, but minimal inhibition of cell proliferation was observed. Assay conditions were further optimised by adding TRAIL in combination with a sub-lethal dose of the protein synthesis inhibitor cycloheximide to potently induce apoptosis. The optimised conditions identified for each cell line were then applied to the testing of TRAIL-R1 and TRAIL-R2 scFv and Fab fragments for potential agonistic activity.
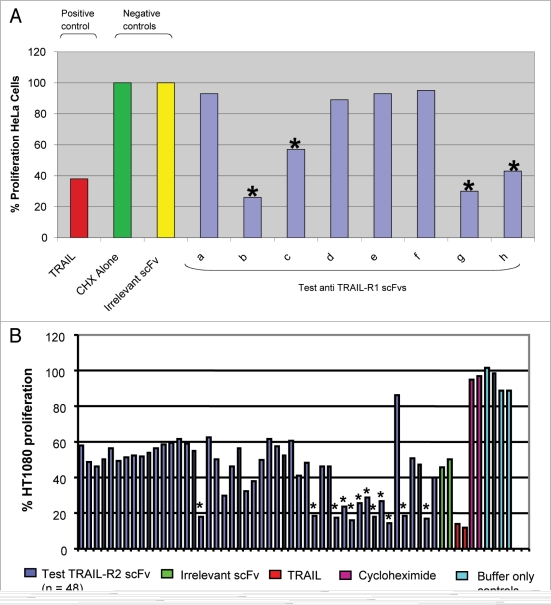
The panel of 234 TRAIL-R1 specific scFvs were tested for cytotoxic effects on TRAIL-R1 expressing HeLa cells. To identify scFv capable of inhibiting HeLa proliferation, all 234 TRAIL-R1 specific scFvs were purified by IMAC and tested in the surrogate apoptosis assay at a single concentration (600 nM) in combination with cycloheximide (Fig. 3A). In this preliminary screening format a total of 14 TRAIL-R1 specific scFv were identified as putative agonists that inhibited proliferation of HeLa cells.
Figure 3.
Identification of putative agonistic anti-TRAIL-R1 and anti-TRAIL-R2 scFv in the surrogate apoptosis assay. (A) Anti-proliferative effects of TRAIL-R1 specific scFv (at 600 nM, blue bars) in combination with cycloheximide (0.5 µg/ml) were measured in the surrogate apoptosis assay using HeLa cells. Putative scFv agonists were identified as those that significantly inhibited HeLa cell proliferation relative to the irrelevant control scFv (anti-BLyS, yellow bar). In the above example 4 scFvs were identified (marked *) as putative TRAIL-R1 agonists for further study. TRAIL (at 125 ng/ml, red bar) in combination with cycloheximide was included in the assay as a positive control. No effect was seen with cycloheximide treatment alone (green bar). (B) Anti-proliferative effects of TRAIL-R2 periplasmic scFv preparations investigated in a high-throughput Alamar blue assay using HT1080 cells in combination with cycloheximide. Putative scFv agonists were identified as those that significantly inhibited HT1080 cell proliferation relative to the irrelevant control scFv (anti-CEA6, green bar). In this example 48 anti-TRAIL-R2 scFv were tested and 12 inhibitors of HT1080 cell proliferation identified for further study (marked *). TRAIL (at 125 ng/ml) in combination with cycloheximide (red bar) was included in the assay as a positive control and cycloheximide was tested alone as a negative control (magenta bar).
To identify putative agonists of TRAIL-R2, the panel of 269 TRAIL-R2 specific scFvs were tested for cytotoxic effects on TRAIL-R2 expressing HT1080 cells. The surrogate apoptosis assay was further developed to enable direct screening of periplasmic scFv extracts in high throughput (96-well) format in combination with cycloheximide (Fig. 3B). Periplasmic scFv extracts are of lower purity than IMAC purified scFv, and consequently assay performance was slightly compromised with a 40% reduction in maximal cellular proliferation. However, a sufficient assay window remained to enable identification of agonistic scFv (Fig. 3B). In this preliminary screen, a total of 35 TRAIL-R2 specific scFv were identified as putative agonists that inhibited proliferation of HT1080 cells.
The panel of 134 TRAIL-R1/-R2 cross-reactive scFvs were tested for cytotoxic effects on both TRAIL-R1 expressing ST486 cells and TRAIL-R2 expressing HT1080 cells. The high-throughput TRAIL-R2 assay using HT1080 cells was again used to identify TRAIL-R1/-R2 cross-reactive scFv capable of inhibiting tumor cell proliferation through TRAIL-R2. In addition, a second high-throughput assay was developed to enable direct screening of TRAIL-R1/-R2 cross-reactive periplasmic scFv extracts for anti-proliferative properties against the TRAIL-R1 expressing cell line ST486. However, after analysis in both of these assays, none of the 134 TRAIL-R1/-R2 cross-reactive scFv were shown to inhibit proliferation of either ST486 or HT1080 cells (data not shown).
Characterization of anti-TRAIL-R1 agonistic scFvs.
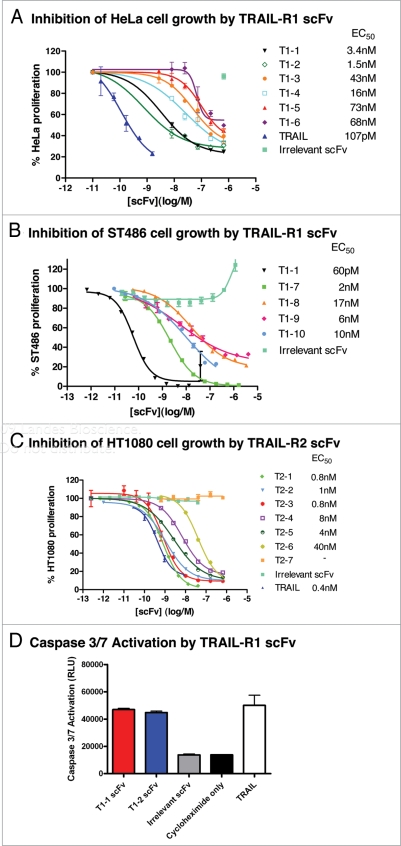
The 14 putative TRAIL-R1 agonistic scFv were characterized in the surrogate apoptosis assay to determine EC50 data. The agonistic activity of 10 scFvs was confirmed in the assay with the most potent scFvs having EC50s in the low nanomolar range (Fig. 4A). The two most potent scFv identified, T1-1 and T1-2, inhibited HeLa cell proliferation with EC50's of 3.4 nM and 1.5 nM respectively. A selection of TRAIL-R1 scFvs were also tested in the surrogate apoptosis assay for anti-tumor effects on a second TRAIL-R1 expressing cell line ST486. Again, the TRAIL-R1 scFvs potently inhibited tumor cell growth with the most potent scFv, T1-1, having an EC50 of 60 pM (Fig. 4B).
Figure 4.
Tumor cell killing by agonistic TRAIL-R1 and TRAIL-R2 scFvs. Example data demonstrating anti-TRAIL-R1 scFv induced tumor cell killing of (A) HeLa and (B) ST486 cells and anti-TRAIL-R2 scFv induced tumor cell killing of (C) HT1080 cells. Cell viability is presented as percent proliferation in the presence of increasing concentrations of different anti-TRAIL-R1 or TRAIL-R2 scFv, TRAIL, or irrelevant scFvs. The test TRAIL-R1 and TRAIL-R2 scFvs presented all inhibited proliferation of either HeLa, ST486 or HT1080 cells as shown (EC50 values given) with no inhibition observed by any of the irrelevant control scFvs. (D) Confirmation of the induction of apoptosis by TRAIL-R1 scFvs T1-1 and T1-2 in a caspase 3/7 activation assay, showing the effect of T1-1 (red) and T1-2 (blue) as compared to an irrelevant scFv (grey), TRAIL (white) or cycloheximide alone (black). All scFvs were assayed in triplicate at a single concentration of 100 nM.
To demonstrate that the effects of the anti-TRAIL-R1 scFvs on cell viability were due to apoptosis, T1-1 and T1-2 scFvs were tested in a caspase activation assay using HeLa cells. Both scFvs were able to induce a specific activation of caspase 3/7 that was not observed with either an irrelevant scFv or cycloheximide alone (Fig. 4D), demonstrating that the anti-TRAIL-R1 scFvs were inducing cell death through an apoptosis pathway.
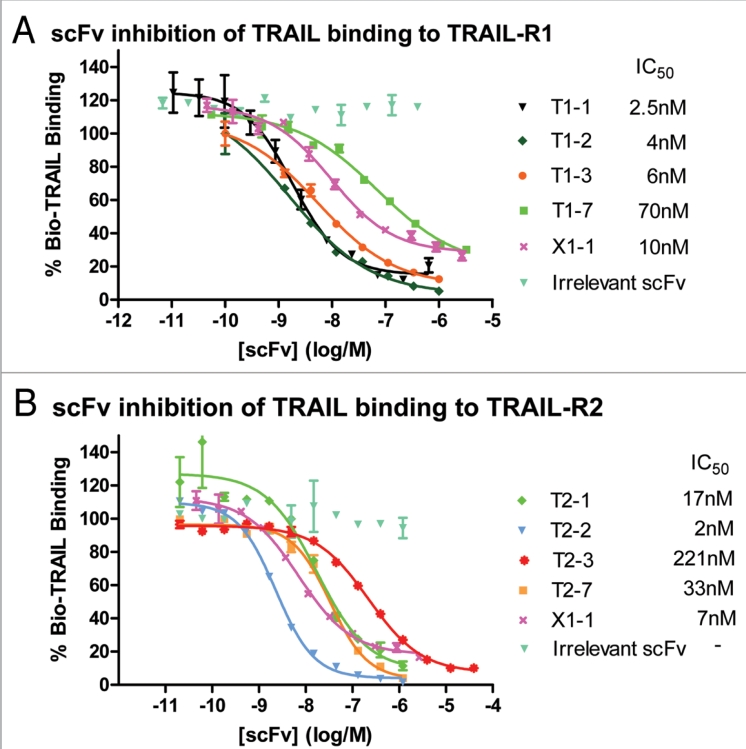
The ability of the panel of 10 TRAIL-R1 specific scFv to antagonise TRAIL binding to TRAIL-R1 was investigated using a biochemical ligand-binding assay. All of the agonistic scFv were shown to inhibit TRAIL binding to TRAIL-R1 (Fig. 5A). Again, T1-1 and T1-2 were the most potent anti-TRAIL-R1 scFv identified (IC50's of 2.5 nM and 4 nM, respectively) indicating that for TRAIL-R1 the most potent agonistic scFv were also the most potent antagonists of TRAIL binding.
Figure 5.
TRAIL-R1 and TRAIL-R2 scFv antagonise TRAIL binding to either TRAIL-R1 and/or TRAIL-R2. Example data demonstrating that (A) binding of TRAIL to TRAIL-R1 and (B) binding of TRAIL to TRAIL-R2 can be inhibited by increasing concentrations of different TRAIL-R1 scFv (T1-1, T1-2, T1-3 and T1-7), TRAIL-R2 scFv (T2-1, T2-2, T2-3 and T2-7) and one TRAIL-R1/-R2 cross-reactive scFv (X1-1) (IC50s shown).
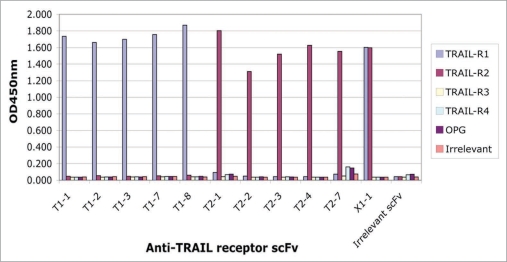
The 5 anti-TRAIL-R1 agonistic scFvs, T1-1, T1-2, T1-3, T1-7 and T1-8 were confirmed as specific for TRAIL-R1 by ELISA (Fig. 6), with no binding to other TRAIL receptor family members.
Figure 6.
Specificity of different scFv for TRAIL receptors. The specificity of anti-TRAIL receptor scFvs was verified by ELISA against a panel of related antigens: TRAIL-R1, TRAIL-R2, TRAIL-R3, TRAIL-R4 and OPG together with an unrelated “irrelevant” Fc-fusion protein. TRAIL-R1 scFv (T1-1, T1-2, T1-3, T1-7, T1-8) bound specifically to TRAIL-R1 (blue bars), TRAIL-R2 scFv (T2-1, T2-2, T2-3, T2-4, T2-7) bound specifically to TRAIL-R2 (red bars), and the TRAIL-R1/-R2 cross-reactive scFv (X1-1) bound to both TRAIL-R1 and TRAIL-R2. No binding was observed to any of the TRAIL decoy receptors (TRAIL-R3, TRAIL-R4, OPG) or to the irrelevant Fc-fusion protein control.
Characterization of TRAIL-R1 agonist T1-1 in Fab format.
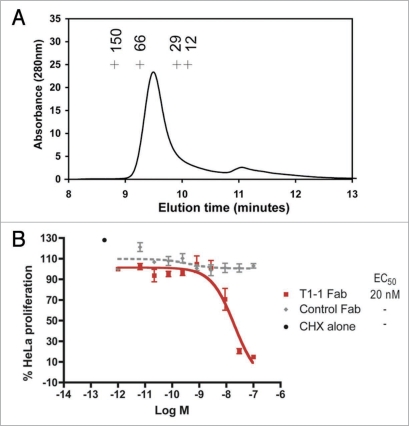
The anti-TRAIL-R1 scFvs preparations are likely to be monomeric, but may also contain small quantities of scFv dimeric or multimeric fractions. In order to test the effect of valency on the agonistic activity of TRAIL-R1 scFv, T1-1 was retested in the apoptosis assay in Fab format. To ensure that a purely monomeric Fab fragment was obtained, the T1-1 Fab was purified and subsequently re-assayed by size-exclusion chromatography. This demonstrated the presence of a monomeric peak at the expected molecular weight of around 50 kDa, with no evidence of any higher order species (Fig. 7A). In the surrogate apoptosis assay, this monovalent T1-1 Fab fragment was able to demonstrate nearly a 100% reduction of HeLa cell proliferation, with an EC50 of 20 nM (Fig. 7B).
Figure 7.
Purification of monomeric Fab fragment T1-1 and inhibition of HeLa cell growth in the Alamar Blue® surrogate apoptosis assay. The relative molecular mass of the purified T1-1 Fab as determined by size exclusion chromatography is shown in (A). For calibration, the elution peaks for four standard proteins (alcohol dehydrogenase, Mw 150 kDa; bovine serum albumin, Mw 66 kDa; carbonic anhydrase, Mw 29 kDa and cytochrome C, Mw 12 kDa) are shown as crosses. A single peak is seen at the predicted Mw for a monomeric Fab fragment (∼50 kDa), with no evidence of higher Mw peaks. The effect of T1-1 Fab on HeLa cell viability is shown in (B). Cell survival, expressed as % HeLa cell proliferation, is shown following incubation with increasing concentrations of T1-1 Fab (filled squares, red) and an irrelevant Fab (filled diamonds, grey).
Characterization of anti-TRAIL-R2 agonistic scFvs.
The 35 putative TRAIL-R2 agonistic scFv identified in the high-throughput surrogate apoptosis assay were characterized more comprehensively in the assay and the activity of six agonistic TRAIL-R2 scFv confirmed (Fig. 4C). The most potent anti-TRAIL-R2 scFv identified, T2-1, T2-2 and T2-3, inhibited HT1080 tumor cell growth with EC50s of 0.8 nM, 1 nM and 0.8 nM respectively, comparing very favorably to homotrimeric TRAIL in the same assay (EC50 of 0.4 nM).
The ability of these six agonistic TRAIL-R2 scFv to antagonise TRAIL binding to TRAIL-R2 was also investigated using a biochemical ligand-binding assay. As with the TRAIL-R1 agonistic scFv, all of the agonistic anti-TRAIL-R2 scFv were also shown to inhibit TRAIL binding to TRAIL-R2 (Fig. 5B). However, in this instance the most potent agonists were not necessarily the most potent inhibitors of TRAIL binding to TRAIL-R2. The three most potent inhibitors of HT1080 proliferation, T2-1, T2-2 and T2-3 (EC50s of 0.8 nM, 1 nM and 0.8 nM), inhibited TRAIL binding in the TRAIL-R2 binding assay with contrasting IC50s of 17 nM, 2 nM and 221 nM, respectively (Fig. 5B). In addition, TRAIL-R2 specific scFv were also identified that blocked TRAIL binding to TRAIL-R2 but did not demonstrate any inhibition of HT1080 cell proliferation. For example, scFv T2-7 was a potent inhibitor of TRAIL binding to TRAIL-R2 (IC50 33 nM) (Fig. 5B), but no inhibition of HT1080 cell proliferation was observed with this scFv in the Alamar blue assay (Fig. 4C), which indicated that there was no functional agonism.
The specificity of the anti-TRAIL-R2 agonistic scFvs was confirmed by ELISA, with specific binding to TRAIL-R2 and no binding to either TRAIL-R1 or the other TRAIL-decoy receptors observed (Fig. 6).
Characterization of anti TRAIL-R1/-R2 cross reactive scFv.
Although none of the 134 TRAIL-R1/R2 cross-reactive scFv inhibited growth of either ST486 or HT1080 tumor cells in surrogate apoptosis assays, their potential to antagonise TRAIL binding to either TRAIL-R1 or TRAIL-R2 was investigated in the biochemical ligand-binding assays described above for TRAIL-R1 and TRAIL-R2. One scFv, X1-1, was identified that inhibited TRAIL binding to both TRAIL-R1 and TRAIL-R2 (Fig. 5) with IC50's of 10 nM and 7 nM respectively. The specificity of X1-1 was confirmed by ELISA, where specific binding to TRAIL-R1 and TRAIL-R2, but not the TRAIL-decoy receptors was observed (Fig. 6). The potential for X1-1 to inhibit tumor cell growth was again investigated using purified scFv in the ST486 and HT1080 surrogate apoptosis assays, but again X1-1 did not demonstrate any agonistic activity against either TRAIL-R1 or TRAIL-R2 (data not shown).
To better understand the sequence conservation between TRAIL-R1 and TRAIL-R2 and the likelihood of isolating a cross-reactive agonistic scFv to the ligand-binding interface of both receptors, conserved residues were plotted onto a surface representation of the solved TRAIL-R2 crystal structure.40 Two loops within the TRAIL receptor extracellular domains, the 50s and 90s loops, are known to bind TRAIL. The 50s loop has relatively low sequence conservation (44%) between TRAIL-R1 and TRAIL-R2, whereas conservation within the 90s loop is slightly higher (71%), and includes a stretch of four amino acids (“EMCR”) which are known to make direct contact with TRAIL.40 However, the conserved patches in the ligand-binding loops are relatively small and are surrounded by regions of non-conserved residues, which would likely make the isolation of a cross-reactive, ligand-blocking scFv extremely challenging. In contrast, the C-terminal regions of the receptor extracellular domains show 87% identity over a stretch of 30 amino acids, providing a large area of conserved surface residues on the face of the receptor. This region does not interact with TRAIL, and is a likely binding site for the panel of cross-reactive antibodies.
Discussion
Agonistic antibodies to TRAIL-R1 and TRAIL-R2 have been described that have potential therapeutic utility as agents for the treatment of human cancer.15,16 Several anti-TRAIL-receptor mAbs have been described with potent in vitro and in vivo anti-tumor properties, and these antibody-mediated effects have been accentuated by concomitant treatment with chemotherapeutic agents.29–32,35,36 We have demonstrated that monomeric antibody formats in the form of a scFv or Fab fragment, are also able to induce cell death through specific binding to either TRAIL-R1 or TRAIL-R2.
ScFv specific for TRAIL-R1 and TRAIL-R2 were isolated from large naïve libraries using phage display technology. Different antibody isolation strategies were employed to maximise the numbers and diversity of scFv produced to both TRAIL-R1 and TRAIL-R2. Selections were performed on recombinant TRAIL-receptor fusion proteins in solution or immobilized onto solid supports by passive adsorption or by covalent coupling. In total, 234 different scFv specific for TRAIL-R1, 269 different scFv specific for TRAIL-R2 and 134 scFv cross-reactive with both TRAIL-R1 and TRAIL-R2 were identified from these selection strategies. The scFv were highly diverse in sequence as demonstrated by their broad utilization of VH germline genes and also their VH-CDR3 length. Moreover, the sequences of the scFv were novel and did not contain any of the motifs reported in other anti-TRAIL receptor antibodies (data not shown).41,42 Taking sequence diversity as a surrogate measure of epitope binding diversity, these data suggest that scFv have been generated to multiple epitopes of both TRAIL-R1 and/or TRAIL-R2.
ScFv or Fab fragments isolated by phage display are typically characterized by antigen binding specificity and affinity, or by their ability to inhibit specific ligand-receptor interactions.43–45 In this study, we were seeking anti-TRAIL receptor scFv with potential agonistic activity, and therefore developed a novel apoptosis assay to screen scFv directly for function. The assay measured the ability of scFv to inhibit the proliferation of TRAIL-R1 or TRAIL-R2 expressing tumor cell lines (HeLa, ST486 or HT1080) as a surrogate measure of apoptosis, and was used successfully to identify putative agonistic scFv. All 637 anti-TRAIL receptor scFv were rapidly profiled in this assay, and a total of 10 TRAIL-R1 specific scFv and 6 TRAIL-R2 specific scFv shown to potently inhibit tumor cell growth in vitro. Comprehensive ELISA analysis of these agonistic scFv confirmed their specificity for either TRAIL-R1 or TRAIL-R2 and no binding was observed to any of the decoy receptors, TRAIL-R3, TRAIL-R4 or OPG. This is not unexpected given the low level of sequence identity (45–60%) shared between the extracellular domains of these receptors at the amino acid level.46,47
Cross-reactive TRAIL-receptor antibodies are relatively few, but include those reported to TRAIL-R3 and TRAIL-R4.48 We have described here the isolation of 134 cross-reactive scFv that recognise both TRAIL-R1 and TRAIL-R2; to our knowledge, this is the first report of a panel of antibodies with this specificity. Furthermore, the broad sequence diversity among these 134 scFvs, especially in the VHCDR3 loop, suggests that they bind to multiple cross-reactive epitopes on TRAIL-R1 and TRAIL-R2. Despite this, none of the cross-reactive scFv inhibited growth of tumor cells and only one, X1-1, was able to block binding of the ligand TRAIL, suggesting that the most common epitopes for cross-reactive TRAIL receptor scFv lay outside of the TRAIL binding site. A comparison of the TRAIL binding sites of TRAIL-R1 and TRAIL-R2 indeed demonstrated relatively low sequence homology (44 and 71% within the 50s and 90s loops respectively), especially when compared to distal regions such as the C-terminal juxtamembrane portion (87%). This latter region seems a much more likely candidate for the location of cross-reactive epitopes and, as it is distant from the TRAIL binding site, may explain why most cross-reactive scFv were unable to block TRAIL.
Current understanding of TRAIL-R activation argues that it is an oligomerization event mediated by homotrimeric TRAIL40 or by bivalent agonistic IgG31,35 that is the key event in receptor activation, and therefore it is perhaps surprising that a monomeric scFv fragment can reproduce these effects. Examples of scFv agonists are rare, but include those described to the CD40 receptor49 and the MuSK tyrosine kinase receptor.50 The anti-MuSK receptor scFv were also isolated by phage display using the same antibody libraries described here together with selection on recombinant MuSK-Fc fusion protein. It was postulated that the dimeric nature of the antigen enabled selection of scFv binding to the interface across neighbouring MuSK receptors thus promoting receptor dimerization. Similarly, our TRAIL-receptor selection strategies also utilized dimeric TRAIL-receptor Fc-fusion proteins and therefore the anti-tumor activity we observe could be due to a similar mechanism of action. This could therefore provide a generic strategy for the selection of antibodies to multimeric cell surface receptors.
For some anti-TRAIL-receptor antibodies, the anti-tumor properties are reliant upon or are enhanced by the addition of cross-linking agents to promote receptor oligomerisation.31,51 The scFv identified in our study, however, did not require the addition of a secondary cross-linking agent for activity, and when cross-linking was investigated for one scFv (T1-1) no further gains in tumoricidal activity were observed (data not shown). Another possibility is that the scFv fragments may have spontaneously formed into dimers or higher order multimers,52,53 thus enhancing their potential cross-linking activity and potential for receptor activation. To further investigate this we purified TRAIL-R1 specific scFv T1-1 by FPLC chromatography and demonstrated that both a monomeric scFv (data not shown) and, importantly, a monomeric Fab fragment could inhibit proliferation of HeLa cells independently of any higher order species. To our knowledge, this is the first example of a pure, monomeric Fab fragment acting as an agonist.
Recent studies have suggested that a domain in TNF receptors mediates ligand-independent receptor assembly and that TRAIL-R1 and TRAIL-R2 may be preformed complexes that do not require TRAIL for cross-linking.13,54,55 Regardless of whether or not TRAIL binds to preformed receptor complexes, it is interesting to compare the binding of the agonistic scFv described here to that of TRAIL. Indeed, all 16 of the agonistic scFv in our study were also able to antagonise the binding of TRAIL to either TRAIL-R1 or TRAIL-R2 suggesting that they bind to epitopes overlapping with the TRAIL binding site. Therefore, our scFv and Fab fragments may function by binding to residues within the TRAIL binding site and exerting their anti-tumor effects by mimicking the apoptosis-inducing mechanism of TRAIL. Indeed, for two TRAIL-R1 specific scFv described here, we were able to demonstrate receptor activation via the caspase pathway, indicating that the growth inhibitory effect was due to the induction of apoptosis rather than any other type of cell death.
The scFv described here have been reformatted as IgG and shown to potently inhibit tumor cell growth thus demonstrating that the screen developed to identify scFv agonists was also predictive of agonism as whole IgG. In vitro studies confirmed that the mode of action was via agonism of TRAIL-R1 and TRAIL-R2, with subsequent activation of cellular caspases, Bid and cleavage of PARP leading to apoptosis and cell death.34,36 As IgG, these TRAIL receptor antibodies showed improved potencies relative to their scFv counterparts and also demonstrated anti-tumor properties in the absence of either chemotherapeutic or cross-linking agents. However, it was noticeable that the effects of these IgGs' were significantly enhanced by simultaneous treatment with either chemotherapy or irradiation.34,36,56 Moreover, significant anti-tumor activity in multiple in vivo xenograft models of cancer has been demonstrated,34,36 and subsequently two antibodies from this group are now being studied extensively in clinical trials for the treatment of human cancers.
Materials and Methods
Reagents and cell lines.
Recombinant human TRAIL-R1, TRAIL-R2, TRAIL-R3 and TRAIL-R4 Fc fusion proteins were produced by HGSI (Rockville, USA). OPG Fc fusion protein and an irrelevant Fc fusion protein were supplied by R&D. Biotinylated TRAIL-R1 and TRAIL-R2 were prepared using NHS-biotin coupling chemistry (Pierce) at a molar ratio of 5:1 biotin to protein.
Tumor cell lines HeLa (cervix epitheloid carcinoma), ST486 (Burkitt's lymphoma) and HT1080 (fibrosarcoma) were obtained from ATCC. HeLa cells express both TRAIL-R1 and -R2, ST486 express TRAIL-R1 only and HT1080 express TRAIL-R2 only (data not shown). Cycloheximide was obtained from Sigma (Cat. No. R750107) and Alamar Blue™ from Serotec (Cat. No. BUF 012A). Recombinant TRAIL was purchased from Peprotech (Cat. 310-04).
Antibody isolation.
Phage display technology was used to isolate scFv to TRAIL-R1, TRAIL-R2 and scFv capable of cross-reacting with both receptors. A large non-immunised human scFv (single chain variable fragment) phage display library, now expanded from 1010 to 1011 binding members (Lloyd et al. in press), was used for antibody isolation (‘selections’), as described previously.37 Briefly, human TRAIL-R1 or TRAIL-R2 Fc-fusion proteins, at 10 µg/ml in PBS, were immobilised directly onto MaxiSorp plates (Nunc) or covalently coupled to Protein Immobilizer plates (Nunc), overnight at 4°C. Three to four rounds of panning selection were performed for each TRAIL receptor using the human scFv phage antibody library. Soluble selections were carried out as previously described39 using biotinylated TRAIL-R1 or biotinylated TRAIL-R2 at a concentration of 100 nM. For both the immobilisation and soluble selection strategies, a deselection step was performed using a 10-fold molar excess of an irrelevant Fc-fusion protein to minimize the possibility of isolating scFv to the Fc domain of the fusion protein.
Phage ELISA.
TRAIL-R1, TRAIL-R2, TRAIL-R3, TRAIL-R4, OPG and an irrelevant Fc-fusion protein, all at 1 µg/ml in PBS, were immobilised onto 96 well Maxisorp plates (Nunc) overnight at 4°C. Anti-TRAIL receptor scFv were prepared as phage supernatants and screened by phage ELISA as described previously.43,57
DNA sequencing.
DNA sequencing was performed at the DNA sequencing facility at Human Genome Sciences, Inc. Full length VH and VL sequences of all the TRAIL-receptor specific scFv identified in ELISA were determined as described previously.43
Preparation of bacterial periplasmic extracts.
A 96 well plate containing bacterial glycerol stocks was replicated into a 2 ml (deepwell) 96 well plate containing 500 µl of 2TY, 100 µg/ml ampicillin (Becton Dickinson, Oxford, UK), 0.1% glucose (VWR BDH, Lutterworth, UK). The culture was incubated at 37°C for approximately 5 hr at 250 rpm and then scFv expression induced by the addition of 100 µl of 0.2 mM IPTG (VWR BDH, Lutterworth, UK) in 2TY. Incubation was continued overnight at 30°C with agitation at 250 rpm. The bacteria were pelleted by centrifugation at 2,500 rpm for 10 minutes at 4°C. Post-centrifugation, the supernatant was removed and the bacterial pellet resuspended in 300 µl of TES pH 7.4 (50 mM Tris-HCl, 0.5 mM EDTA, 0.5 M Sucrose). The microtiter plates were left on ice for 30 minutes. The scFv enriched bacterial periplasmic extract was then prepared by clarification of the suspension using centrifugation at 4,000 rpm for 10 minutes at 4°C.
Purification of scFv and Fab molecules.
scFv were prepared from Escherichia coli periplasmic extracts and then purified by immobilised metal affinity chromatography (IMAC) as described previously.58 For expression of Fab molecules in E. coli, the VH and VL regions were cloned from the phage display vector pCan-tab6 into a Fab expression vector pFab, which expresses the heavy and light chains of the Fab under the control of the Lac promoter. Fabs were expressed and purified with the same methods used for scFvs except that an additional size exclusion chromatography step was included to ensure the purification of purely monomeric Fab fragments, as described previously.59 The relative molecular mass of the purified Fab was assessed by size-exclusion gel chromatography on a Superose 12 HR 10/30 column (Pharmacia) in PBS, pH 7.4, calibrated with standard proteins (alcohol dehydrogenase, Mw 150 kDa; bovine serum albumin, Mw 66 kDa; carbonic anhydrase, Mw 29 kDa and cytochrome C, Mw 12 kDa). The flow-rate was 0.5 ml/min and the absorbance of the effluent stream was monitored at 280 nm.
Tumor cell proliferation assay.
Tumor cell lines were seeded in culture medium onto 96 well tissue culture plates the day prior to the assay (HeLa, 3 × 104/well or HT1080, 1 × 105 cells/well) and grown overnight at 37°C and 5% CO2. ST486 cells were plated at 5 × 104/well on the same day as the assay. TRAIL-receptor scFv/Fabs were analyzed in one of two formats: (1) as scFv prepared directly from periplasmic extracts or (2) as purified scFv or Fab fragments. ScFv were added to the tumor cells in combination with a sub-lethal dose of the sensitising agent, cycloheximide (500 ng/ml) and the cells incubated for 16–18 hours at 37°C, 5% CO2. Fab fragments were added to the tumor cells in combination with 33 µg/ml cycloheximide. Irrelevant scFv or Fab fragments served as negative controls and recombinant TRAIL (125 ng/ml) as a positive control. After incubation of scFv or Fab with the tumor cell lines, Alamar Blue™ was aseptically added in an amount equal to 10% of the culture volume. The plates were returned to the incubator for an additional 4 hrs at 37°C and viability assessed by measuring fluorescence on a Wallac 1420 workstation at 560 nm excitation and 590 nm emission. The EC50 for the binding of the scFv or Fab fragment to TRAIL-R1 or TRAIL-R2 was determined and compared with that of TRAIL.
TRAIL inhibition assay.
The ability of individual TRAIL-receptor scFvs to inhibit the binding of biotinylated-TRAIL to immobilised TRAIL-R1 or TRAIL-R2 was assessed in a biochemical receptor inhibition assay. TRAIL-R1 or TRAIL-R2 Fc fusion proteins were coated onto Nunc 96-well Maxisorp plates (Nunc) at 25 ng TRAIL receptor/well. IMAC-purified scFv (from 30 µg/ml to 0.01 µg/ml) were added to each well in the presence of 120 ng/ml biotinylated TRAIL. Binding of biotinylated TRAIL was then detected via streptavidin-DELFIA® technology (Wallac) and read on a Wallac 1420 workstation at 340 nm excitation and 615 nm emission. The IC50 value for the competition of scFv for TRAIL binding to TRAIL-R was determined using Graphpad Prism® software.
Caspase activation assay.
HeLa cells were seeded in culture medium onto 96 well tissue culture plates the day prior to the assay at a density of 3 × 104/well and grown overnight at 37°C and 5% CO2. The scFv were added to the tumor cells in combination with a sub-lethal dose of the sensitising agent, cycloheximide (33 µg/ml) and the cells incubated for 6 hours at 37°C, 5% CO2. Irrelevant scFv or cycloheximide alone served as negative controls. The assay was performed according to the manufacturer's instructions (Caspase-Glo® 3/7 Assay from Promega). Following incubation of HeLa cells with scFv, 100 µl Caspase-Glo® reagent was added and the plate incubated for 1 hour at room temperature before reading on a luminometer.
Structural alignment of TRAIL-R1 and TRAIL-R2.
Structural illustrations using the X-ray co-ordinates of TRAIL-R2 from PDB entry 1d0g40 were performed using the molecular graphics package PYMOL.60
Figure 2.
VH-CDR3 lengths of scFv isolated to TRAIL-R1 (blue bars), TRAIL-R2 (red bars) or cross-reactive with both receptors (yellow bars). VH-CDR3 sequence lengths ranged from 5 to 21 amino acids with a mean length of 11.5 amino acids.
Acknowledgements
We thank the contributions of members of the DNA sequencing facility at Human Genome Sciences, Inc.
Abbreviations
- TRAIL
TNF-related apoptosis-inducing ligand
- TRAIL-R1
TRAIL receptor 1
- TRAIL-R2
TRAIL receptor 2
- TRAIL-R3
TRAIL receptor 3
- TRAIL-R4
TRAIL receptor 4
- OPG
osteoprotegerin
- scFv
single chain Fv fragments
- TNF
tumor necrosis factor
- DISC
death inducing signalling complex
- VH
variable heavy
- VL
variable light
- CDR
complementarity determining region
- IMAC
immobilised metal affinity chromatography
- PBS
phosphate buffered saline
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/10057
Supplementary Material
References
- 1.Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem. 1996;271:12687–12690. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- 2.Ashkenazi A, Pai RC, Fong S, Leung S, Lawrence DA, Marsters SA, et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 4.Pan G, O'Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, et al. The receptor for the cytotoxic ligand TRAIL. Science. 1997;276:111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 5.Walczak H, Degli-Esposti MA, Johnson RS, Smolak PJ, Waugh JY, Boiani N, et al. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J. 1997;16:5386–5397. doi: 10.1093/emboj/16.17.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhary PM, Eby M, Jasmin A, Bookwalter A, Murray J, Hood L. Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NFkappaB pathway. Immunity. 1997;7:821–830. doi: 10.1016/s1074-7613(00)80400-8. [DOI] [PubMed] [Google Scholar]
- 7.Pan G, Ni J, Wei YF, Yu G, Gentz R, Dixit VM. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science. 1997;277:815–818. doi: 10.1126/science.277.5327.815. [DOI] [PubMed] [Google Scholar]
- 8.Screaton GR, Mongkolsapaya J, Xu XN, Cowper AE, McMichael AJ, Bell JI. TRICK2, a new alternatively spliced receptor that transduces the cytotoxic signal from TRAIL. Curr Biol. 1997;7:693–696. doi: 10.1016/s0960-9822(06)00297-1. [DOI] [PubMed] [Google Scholar]
- 9.Degli Esposti MA, Smolak PJ, Walczak H, Waugh J, Huang CP, DuBose RF, et al. Cloning and characterization of TRAIL-R3, a novel member of the emerging TRAIL receptor family. J Exp Med. 1997;186:1165–1170. doi: 10.1084/jem.186.7.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsters SA, Sheridan JP, Pitti RM, Huang A, Skubatch M, Baldwin D, et al. A novel receptor for Apo2L/TRAIL contains a truncated death domain. Curr Biol. 1997;7:1003–1006. doi: 10.1016/s0960-9822(06)00422-2. [DOI] [PubMed] [Google Scholar]
- 11.Emery JG, McDonnell P, Burke MB, Deen KC, Lyn S, Silverman C, et al. Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J Biol Chem. 1998;273:14363–14367. doi: 10.1074/jbc.273.23.14363. [DOI] [PubMed] [Google Scholar]
- 12.Sheridan JP, Marsters SA, Pitti RM, Gurney A, Skubatch M, Baldwin D, et al. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277:818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 13.Clancy L, Mruk K, Archer K, Woelfel M, Mongkolsapaya J, Screaton G, et al. Preligand assembly domain-mediated ligand-independent association between TRAIL receptor 4 (TR4) and TR2 regulates TRAIL-induced apoptosis. Proc Natl Acad Sci USA. 2005;102:18099–18104. doi: 10.1073/pnas.0507329102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprick MR, Weigand MA, Rieser E, Rauch CT, Juo P, Blenis J, et al. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity. 2000;12:599–609. doi: 10.1016/s1074-7613(00)80211-3. [DOI] [PubMed] [Google Scholar]
- 15.Kelley SK, Ashkenazi A. Targeting death receptors in cancer with Apo2L/TRAIL. Curr Opin Pharmacol. 2004;4:333–339. doi: 10.1016/j.coph.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Yagita H, Takeda K, Hayakawa Y, Smyth MJ, Okumura K. TRAIL and its receptors as targets for cancer therapy. Cancer Sci. 2004;95:777–783. doi: 10.1111/j.1349-7006.2004.tb02181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeda K, Smyth MJ, Cretney E, Hayakawa Y, Kayagaki N, Yagita H, et al. Critical role for tumor necrosis-factor realted apoptosis-inducing ligand in immune surveillance against tumor development. J Exp Med. 2002;195:161–169. doi: 10.1084/jem.20011171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cretney E, Takeda K, Yagita H, Glaccum M, Peschon JJ, Smyth MJ. Increased susceptibility to tumor initiation and metastasis in TNF-related apoptosis-inducing ligand-deficient mice. J Immunol. 2002;168:1356–1361. doi: 10.4049/jimmunol.168.3.1356. [DOI] [PubMed] [Google Scholar]
- 19.Sedger LM, Glaccum MB, Schuh JCL, Kanaly ST, Williamson E, Kayagaki N, et al. Characterization of the in vivo function of TNFalpha-related apoptosis-inducing ligand, TRAIL/Apo2L, using TRAIL/Apo2L gene-deficient mice. Eur J Immunol. 2002;32:2246–2254. doi: 10.1002/1521-4141(200208)32:8<2246::AID-IMMU2246>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 20.Zerafa N, Westwood JA, Cretney E, Mitchell S, Waring P, Iezzi M, et al. Cutting edge: TRAIL deficiency accelerates hematological malignancies. J Immunol. 2005;175:5586–5590. doi: 10.4049/jimmunol.175.9.5586. [DOI] [PubMed] [Google Scholar]
- 21.Ursini-Siegel J, Zhang W, Altmeyer A, Hatada EN, Do RK, Yagita H, et al. TRAIL/Apo-2 ligand induces primary plasma cell apoptosis. J Immunol. 2002;169:5505–5513. doi: 10.4049/jimmunol.169.10.5505. [DOI] [PubMed] [Google Scholar]
- 22.Kayagaki N, Yamaguchi N, Abe M, Hirose S, Shirai T, Okumura K, et al. Suppression of antibody production by TNF-realted apoptosis-inducing ligand (TRAIL) Cell Immunol. 2002;219:82–91. doi: 10.1016/s0008-8749(02)00602-0. [DOI] [PubMed] [Google Scholar]
- 23.Hayakawa Y, Screpanti V, Yagita H, Grandien A, Ljunggren HG, Smyth MJ, et al. NK cell TRAIL eliminates immature dendritic cells in vivo and limits dendritic cell vaccination efficiency. J Immunol. 2004;172:123–129. doi: 10.4049/jimmunol.172.1.123. [DOI] [PubMed] [Google Scholar]
- 24.Lünemann JD, Waiczies S, Ehrlich S, Wendling U, Seeger B, Kamradt T, et al. Death ligand TRAIL induces no apoptosis but inhibits activation of human (auto) antigen-specific T cells. J Immunol. 2002;168:4881–4888. doi: 10.4049/jimmunol.168.10.4881. [DOI] [PubMed] [Google Scholar]
- 25.Lamhamedi-Cherradi S-E, Zheng S-J, Maguschak KA, Peschon JJ, Chen YH. Defective thymocyte apoptosis and accelerated autoimmune diseases in TRAIL-/- mice. Nat Immunol. 2003;4:255–260. doi: 10.1038/ni894. [DOI] [PubMed] [Google Scholar]
- 26.Song K, Chen Y, Goke R, Wilmen A, Seidel C, Goke A, et al. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is an inhibitor of autoimmune inflammation and cell cycle progression. J Exp Med. 2000;191:1095–1104. doi: 10.1084/jem.191.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilliard B, Wilmen A, Seidel C, Liu TS, Goke R, Chen Y. Roles of TNF-related apoptosis-inducing ligand in experimental autoimmune encephalomyelitis. J Immunol. 2001;166:1314–1319. doi: 10.4049/jimmunol.166.2.1314. [DOI] [PubMed] [Google Scholar]
- 28.Daniels RA, Turley H, Kimberley FC, Liu XS, Mongkolsapaya J, Ch'En P, et al. Expression of TRAIL and TRAIL receptors in normal and malignant tissues. Cell Res. 2005;15:430–438. doi: 10.1038/sj.cr.7290311. [DOI] [PubMed] [Google Scholar]
- 29.Ichikawa K, Liu W, Zhao L, Wang Z, Liu D, Ohtsuka T, et al. Tumoricidal activity of a novel anti-human DR5 monoclonal antibody without hepatocyte cytotoxicity. Nat Med. 2001;7:954–960. doi: 10.1038/91000. [DOI] [PubMed] [Google Scholar]
- 30.Griffith TS, Rauch CT, Smolak PJ, Waugh JY, Boiani N, Lynch DH, et al. Functional analysis of TRAIL receptors using monoclonal antibodies. J Immunol. 1999;162:2597–2605. [PubMed] [Google Scholar]
- 31.Chuntharapai A, Dodge K, Grimmer K, Schroeder K, Marsters SA, Koeppen H, et al. Isotype-dependent inhibition of tumor growth in vivo by monoclonal antibodies to death receptor 4. J Immunol. 2001;166:4891–4898. doi: 10.4049/jimmunol.166.8.4891. [DOI] [PubMed] [Google Scholar]
- 32.Guo Y, Chen C, Zheng Y, Zhang J, Tao X, Liu S, et al. A novel anti-human DR5 monoclonal antibody with tumoricidal activity induces caspase-dependent and caspase-independent cell death. J Biol Chem. 2005;280:41940–41952. doi: 10.1074/jbc.M503621200. [DOI] [PubMed] [Google Scholar]
- 33.Bin L, Thorburn J, Thomas LR, Clark PE, Humphreys R. Tumor-derived mutations in the TRAIL receptor DR5 inhibit TRAIL signaling through the DR4 receptor by competing for ligand binding. J Biol Chem. 2007;282:28189–28194. doi: 10.1074/jbc.M704210200. [DOI] [PubMed] [Google Scholar]
- 34.Pukac L, Kanakaraj P, Humphreys R, Alderson R, Bloom M, Sung C, et al. HGS-ETR1, a fully human TRAIL-receptor 1 monoclonal antibody, induces cell death in multiple tumor types in vitro and in vivo. Br J Cancer. 2005;92:1430–1441. doi: 10.1038/sj.bjc.6602487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motoki K, Mori E, Matsumoto A, Thomas M, Tomura T, Humphreys R, et al. Enhanced apoptosis and tumor regression induced by a direct agonist antibody to tumor necrosis factor-related apoptosis-inducing ligand receptor 2. Clin Cancer Res. 2005;11:3126–3135. doi: 10.1158/1078-0432.CCR-04-1867. [DOI] [PubMed] [Google Scholar]
- 36.Marini P, Denzinger S, Schiller D, Kauder S, Welz S, Humphreys R, et al. Combined treatment of colorectal tumors with agonistic TRAIL receptor antibodies HGS-ETR1 and HGS-ETR2 and radiotherapy: enhanced effects in vitro and dose-dependent growth delay in vivo. Oncogene. 2006;25:5145–5154. doi: 10.1038/sj.onc.1209516. [DOI] [PubMed] [Google Scholar]
- 37.Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, et al. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol. 1996;14:309–314. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 38.Lloyd C, Lowe D, Edwards B, Welsh F, Dilks T, Hardman C, et al. Modelling the human immune response: performance of a 1011 human antibody repertoire against a broad panel of therapeutically relevant antigens. Protein Eng Des Sel. 2009;22:159–168. doi: 10.1093/protein/gzn058. [DOI] [PubMed] [Google Scholar]
- 39.Hawkins RE, Russell SJ, Winter G. Selection of phage antibodies by binding affinity. Minicking affinity maturation. J Mol Biol. 1992;226:889–896. doi: 10.1016/0022-2836(92)90639-2. [DOI] [PubMed] [Google Scholar]
- 40.Hymowitz SG, Christinger HW, Fuh G, Ultsch M, O'Connell M, Kelley RF, et al. Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with death receptor 5. Mol Cell. 1999;4:563–571. doi: 10.1016/s1097-2765(00)80207-5. [DOI] [PubMed] [Google Scholar]
- 41.Fellouse FA, Li B, Compaan DM, Peden AA, Hymowitz SG, Sidhu SS. Molecular Recognition by a Binary Code. J Mol Biol. 2005;348:1153–1162. doi: 10.1016/j.jmb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 42.Li B, Russell SJ, Compaan DM, Totpal K, Marsters SA, Ashkenazi A, et al. Activation of the Proapoptotic Death Receptor DR5 by Oligomeric Peptide and Antibody Agonists. J Mol Biol. 2006;361:522–536. doi: 10.1016/j.jmb.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 43.Edwards BM, Barash SC, Main SH, Choi GH, Minter R, Ullrich S, et al. The reamarkable flexibility of the human antibody repertoire; isolation of over one thousand different antibodies to a single protein, BLyS. J Mol Biol. 2003;334:103–118. doi: 10.1016/j.jmb.2003.09.054. [DOI] [PubMed] [Google Scholar]
- 44.Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, et al. Isolation of high affinity human antibodie directly from large synthetic repertoires. EMBO J. 1994;13:3245–3260. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheets MD, Amersdorfer P, Finnern R, Sargent P, Lindquist E, Schier R, et al. Efficient construction of a large nonimmune phage antibody library: the production of high-affinity human single-chain antibodies to protein antigens. Proc Natl Acad Sci USA. 1998;95:6157–6162. doi: 10.1073/pnas.95.11.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashkenazi A. DR4, DR5, DcR1, DcR2. In: Oppenheim JJ, Feldmann M, editors. Cytokine Reference. Academic Press; 2000. pp. 1733–1742. [Google Scholar]
- 47.Degli-Esposti MA. To die or not to die—the quest for TRAIL receptors. J Leukoc Biol. 1999;65:535–542. doi: 10.1002/jlb.65.5.535. [DOI] [PubMed] [Google Scholar]
- 48.Ch'en PF, Xu XG, Liu XS, Liu Y, Song CJ, Screaton GR, et al. Characterisation of monoclonal antibodies to the TNF and TNF receptor families. Cell Immunol. 2005;236:78–85. doi: 10.1016/j.cellimm.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 49.Ledbetter JA, Francisco JA, Siegall CB, Gilliland LK, Hollenbaugh D, Aruffo A, et al. Agonistic activity of a CD40-specific single-chain Fv constructed from the variable regions of mAb G28-5. Crit Rev Immunol. 1997;17:427–435. [PubMed] [Google Scholar]
- 50.Xie MH, Yuan J, Adams C, Gurney A. Direct demonstration of MuSK involvement in acetylcholine receptor clustering through identification of agonist scFv. Nat Biotechnol. 1997;15:768–771. doi: 10.1038/nbt0897-768. [DOI] [PubMed] [Google Scholar]
- 51.Mori E, Thomas M, Motoki K, Nakazawa K, Tahara T, Tomizuka K, et al. Human normal hepatocytes are susceptible to apoptosis signal mediated by both TRAIL-R1 and TRAIL-R2. Cell Death Differ. 2004;11:203–207. doi: 10.1038/sj.cdd.4401331. [DOI] [PubMed] [Google Scholar]
- 52.Kortt AA, Malby RL, Caldwell JB, Gruen LC, Ivancic N, Lawrence MC, et al. Recombinant anti-sialidase single-chain variable fragment antibody. Characterization, formation of dimer and higher-molecular-mass multimers and the solution of the crystal structure of the single-chain variable fragment/sialidase complex. Eu J Biochem. 1994;221:151–157. doi: 10.1111/j.1432-1033.1994.tb18724.x. [DOI] [PubMed] [Google Scholar]
- 53.Schier R, Bye J, Apell G, McCall A, Adams GP, Malmqvist M, et al. Isolation of high-affinity monomeric human anti-c-erbB-2 single chain Fv using affinity-driven selection. J Mol Biol. 2006;255:28–43. doi: 10.1006/jmbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 54.Chan FK, Chun HJ, Zheng L, Siegel RM, Bui KL, Lenardo MJ. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science. 2000;288:2351–2354. doi: 10.1126/science.288.5475.2351. [DOI] [PubMed] [Google Scholar]
- 55.Wassenaar TA, Quax WJ, Mark AE. The conformation of the extracellular binding domain of death receptor 5 in the presence and absence of the activating ligand TRAIL: A molecular dynamics study. Proteins. 2007;70:333–343. doi: 10.1002/prot.21541. [DOI] [PubMed] [Google Scholar]
- 56.Georgakis GV, Li Y, Humphreys R, Andreeff M, O'Brien S, Younes M, et al. Activity of selective fully human agonistic antibodies to the TRAIL death receptors TRAIL-R1 and TRAIL-R2 in primary and cultured lymphoma cells: induction of apoptosis and enhancement of doxorubicin- and bortezomib-induced cell death. Br J Haematol. 2005;130:501–510. doi: 10.1111/j.1365-2141.2005.05656.x. [DOI] [PubMed] [Google Scholar]
- 57.Lou J, Marzari R, Verzillo V, Ferrero F, Shenq M, Yang C, et al. Antibodies in haystacks: how selection strategy influences the outcome of selection from molecular diversity libraries. J Immunol Methods. 2001;253:233–242. doi: 10.1016/s0022-1759(01)00385-4. [DOI] [PubMed] [Google Scholar]
- 58.Osbourn JK, McCafferty J, Derbyshire EJ, Waibel R, Chester KA, Boxer G, et al. Isolation of a panel of human anti-CEA single chain Fv from a large phage display library. Tumor Target. 1999;4:150–157. [Google Scholar]
- 59.Casey JL, Keep PA, Chester KA, Robson L, Hawkins RE, Begent RH. Purification of bacterially expressed single chain Fv antibodies for clinical applications using metal chelate chromatography. J Immunol Methods. 1995;179:105–116. doi: 10.1016/0022-1759(94)00278-5. [DOI] [PubMed] [Google Scholar]
- 60.DeLano WL. MACPYMOL. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.