Abstract
The tumor suppressor p53 is a member of the emerging class of proteins that have both folded and intrinsically disordered domains, which are a challenge to structural biology. Its N-terminal domain (NTD) is linked to a folded core domain, which has a disordered link to the folded tetramerization domain, which is followed by a disordered C-terminal domain. The quaternary structure of human p53 has been solved by a combination of NMR spectroscopy, electron microscopy, and small-angle X-ray scattering (SAXS), and the NTD ensemble structure has been solved by NMR and SAXS. The murine p53 is reported to have a different quaternary structure, with the N and C termini interacting. Here, we used single-molecule FRET (SM-FRET) and ensemble FRET to investigate the conformational dynamics of the NTD of p53 in isolation and in the context of tetrameric full-length p53 (flp53). Our results showed that the isolated NTD was extended in solution with a strong preference for residues 66–86 forming a polyproline II conformation. The NTD associated weakly with the DNA binding domain of p53, but not the C termini. We detected multiple conformations in flp53 that were likely to result from the interactions of NTD with the DNA binding domain of each monomeric p53. Overall, the SM-FRET results, in addition to corroborating the previous ensemble findings, enabled the identification of the existence of multiple conformations of p53, which are often averaged and neglected in conventional ensemble techniques. Our study exemplifies the usefulness of SM-FRET in exploring the dynamic landscape of multimeric proteins that contain regions of unstructured domains.
Keywords: natively disordered, domain–domain interaction, quaternary structure, FRET, time-resolved
The tumor suppressor p53 is a tetrameric, multidomain transcription factor that plays key roles in maintaining the integrity of the human genome and in DNA repair machinery (1, 2). p53 is a partly intrinsically disordered protein, containing two folded domains: the DNA-binding core domain (CD; residues 94–294) and the tetramerization domain (TetD; residues 323–360) (3, 4). The intrinsically disordered N-terminal domain (NTD; residues 1–94) and C-terminal domain (CTD; residues 360–393) (5, 6) mediate interactions with several proteins such as p300/CBP, MDM2, 14-3-3, and S100 family that in turn regulate the activity of p53. Moreover, the NTD and CTD are the target sites of numerous posttranslational modificiations that modulate the activity of p53.
High-resolution structures of the CD and the TetD have been solved by using X-ray crystallography and NMR spectroscopy (3, 4, 7, 8). But, the intrinsic instability and the presence of highly disordered regions in p53 have impeded the application of conventional structural studies on full-length p53 (flp53). A combination of NMR spectroscopy and small-angle X-ray scattering (SAXS) in solution with electron microscopy on immobilized samples was recently used to solve the quaternary structures of a mutationally stabilized human flp53 and its DNA complex (9). The ensemble structure of the NTD has further been analyzed in solution by residual dipolar couplings and SAXS (10). A radically different structure has been proposed from cryoelectron microscopy studies on murine p53 in which the N and C termini are suggested to interact (11).
We applied single-molecule FRET (SM-FRET) (12, 13) and time-resolved FRET (TR-FRET) (14) techniques to gain further information on the quaternary structure of p53, the disposition of its disordered regions, and domain–domain interactions. The unique advantage of SM-FRET over ensemble-averaged measurements is that it allows us to segregate the heterogeneity of a complex ensemble, which is especially important in assigning structure–function relationship to proteins that are conformationally mobile, such as the denatured state of proteins (15–18). Owing to its unique advantages, SM-FRET has been widely applied to monitor structural changes (19–22), dynamics (23), and molecular interactions of biomolecules (24). However, there are only a few studies to date on large multidomain proteins and protein complexes using SM-FRET (25–27). This paucity is possibly because of the difficulty of selective and specific labeling, the distance limitations of FRET measurements, and the inherent complexity (such as stability) of the systems. Recent advancements with larger Förster critical distance donor/acceptor pairs and the development of more sophisticated alternating excitation techniques have allowed the extension of distance limitations (28). Although large dyes are used in these systems, TR-FRET allows the use of small fluorescence probes to study protein dynamics and conformational distribution (29). Based on the SM-FRET and TR-FRET experiments, we discuss our results in the context of the inherent conformational heterogeneity present in a single tetrameric p53 molecule while highlighting in the consistencies observed between ensemble and single-molecule measurements.
Results
Establishing the FRET System.
As there are no Cys residues in the NTD of p53, we introduced Cys residues as labeling anchors for FRET studies at different positions so that distances between different residues could be measured: p53N(1C91C) (M1C W91C); p53N(10C56C) (V10C E56C); and p53N(56C91C) (E56C W91C). The two Cys of each mutant were labeled with Alexa Fluor 488 (AF488; FRET donor) and Alexa Fluor 647 (AF647; FRET acceptor) randomly, i.e., the position of the donor and the acceptor may exchange. Random labeling has been adopted throughout the study for all of the samples under investigation. To check the effect of random labeling, we used the B-domain of protein A (BDPA) as a control, where we can introduce labels either selectively or randomly (21). The similarity of the histograms for the selectively and randomly labeled BDPA showed that this exchange did not noticeably affect the SM-FRET efficiency histogram.
The Förster critical distance (R0) for AF488/AF647 is ≈52 Å, which allows distances between 42 and 66 Å, corresponding to FRET efficiency of 0.8 and 0.2, respectively, to be accurately measured. For much larger or much smaller distances, the measurement becomes insensitive and may introduce large errors. To measure larger distances, we used another donor/acceptor pair, Alexa Fluor 546/Alexa Fluor 647 (AF546/AF647), which has a Förster critical distance of 63 Å. The Förster critical distances, we measured, are different from those reported by Invitrogen (56 and 74 Å for AF488/AF647 and AF546/AF647, respectively), probably because of the change in the fluorescence quantum yield and spectra upon labeling.
flp53 has 10 Cys residues, three of which are buried and involved in Zn2+ coordination (Cys-176, Cys-238, and Cys-242). Some of the other seven Cys residues are buried or partly exposed (Cys-124, Cys-135, Cys-141, and Cys-275), and the exposed residues include Cys-182, Cys-229, and Cys-277 (30). To avoid mislabeling, we mutated all of the exposed and partly exposed Cys to Ala, i.e., C124A, C182A, C275A, and C277A. We used two mutant forms of p53 harboring labeling positions in the NTD and the DNA binding domain (56C229C and 56C292C). For the mutant p53 (56C292C), we mutated Cys-229 to Ala. These mutations were introduced in the tetrameric flp53 (residues1–393) and a monomeric construct lacking the oligomerization and C-terminal regulatory domain (p53NCD; residues 1–292), for labeling and subsequent single-molecule experiments. FRET pairs AF488/AF647 and AF546/AF647 were used for all mutants. The protein and DNA binding control experiments showed that the mutant p53 and wild-type p53 had similar DNA and protein binding constants, suggesting that the dyes had little effect on the protein functionality and structure (see SI Text for details).
Distance Between Residues in the NTD of p53 Using SM-FRET.
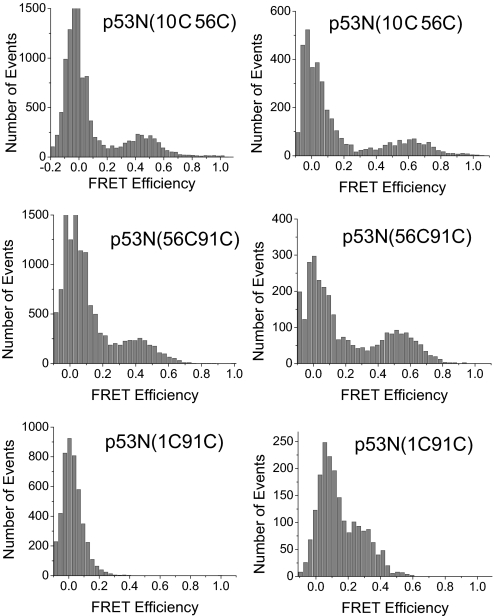
p53N(10C56C) and p53N(56C91C) with AF488 as FRET donor and AF647 as acceptor gave a single peak (apart from the zero peak) in the SM-FRET efficiency histogram at FRET efficiency of 0.45 and 0.41, respectively (Fig. 1). The single peak pattern of the SM-FRET efficiency histogram suggested that there was only one ensemble of conformations that could be observed in the NTD of p53. No peak was observed between 0.2 and 1 in the SM-FRET efficiency histogram of p53N(1C91C), suggesting the distance between residues 1 and 91 was too large for the AF488/AF647 pair as any peak with FRET efficiency <0.2 is not resolvable with our setup (because of the strong zero peak). Using AF546 as a FRET donor, the histogram peak shifted from low to high FRET efficiency. The peak for p53N(10C56C) and p53N(56C91C) shifted to 0.61 and 0.53, respectively (Table 1). A peak at 0.25 for p53N(1C91C) was also observed, because of the larger Förster critical distance for the AF546/AF647 pair. The apparent average distance between dyes was calculated with the FRET efficiency at the peak using the equation r = (1/Epeak − 1)1/6R0. The apparent average distances obtained from AF546/AF647 system were greater than those obtained from the AF488/AF647 system (Table 1), which reflects the broad distance distribution of the flexible NTD. Previous work (31) suggests that a systematic positive deviation would be observed in a loose flexible system when a steady-state FRET technique is applied and the Förster critical distance is longer than the distance to be measured, i.e., R0 > r, whereas a negative deviation would be expected when R0 < r. The systematic deviation is minimized when the FRET efficiency is close to 0.5. Because the FRET efficiency for p53N(10C56C) and p53N(56C91C) was slightly <0.5 when AF488 was used as donor and slightly >0.5 when AF546 acted as donor, the actual distance should be in between the values obtained with these two donor/acceptor pairs, i.e., 54–58 Å for p53N(10C56C) and 56–62 Å for p53N(56C91C). For p53N(1C91C), the FRET efficiency was only 0.25; a negative deviation is, therefore, expected, i.e., the actual distance may be >76 Å. Assuming that the NTD is an ideal chain, the radius of gyration (Rg) can be calculated from the FRET end-to-end distance with the equation Rg = r/, where r is the end-to-end distance. The calculated Rg value of 31.0 Å is similar to the value obtained from SAXS experiments (27 Å). The slightly greater value from FRET may result from the linker between the dye and the protein but may also be an indication of the nonideal chain behavior of the NTD, caused by the high helix propensity of the proline-rich region (10).
Fig. 1.
Single-molecule FRET histogram for isolated NTD (p53NTD) with dyes labeled at various positions as shown. (Left) AF488/AF647 FRET pair was used. (Right) AF546/AF647 FRET pair was used. Data were acquired at 20 °C in phosphate buffer (pH 7) with 20 mM phosphate plus 150 mM NaCl.
Table 1.
Distances obtained from SM-FRET measurements
| Observable | p53N10C56C | p53N56C91C | p53N1C91C | p53NCD56C229C | p53NCD56292C |
|---|---|---|---|---|---|
| E488647 | 0.45 | 0.40 | NA | 0.63 | 0.60 |
| E546647 | 0.61 | 0.53 | 0.25 | 0.77 | 0.70 |
| R488647, Å | 54 | 56 | NA | 48 | 49 |
| R546647,Å | 58 | 62 | 76 | 52 | 55 |
E488–647, E546–647, R488–647, and R547–647 are the FRET efficiency and corresponding distance obtained with AF488 and AF546 as donor, respectively. The error in distance is estimated to be 10%, which is the upper limit of the error after considering all the errors in the calculation of Förster distance, the peak position of the histogram, and the γ value.
Distance Between Residues in the NTD of p53 Using TR-FRET.
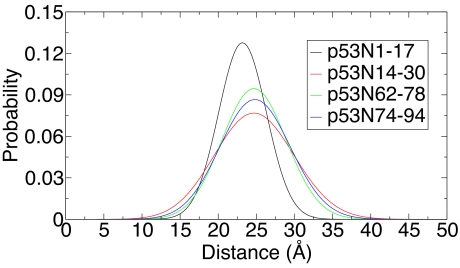
To get average distances and the distance distribution between residues in the NTD of p53, we synthesized a series of short peptides: residues 1–17 (p53N1–17), residues 14–30 (p53N14–30), residues 62–78 (p53N62–78), and residues 74–92 (p53N74–92) (see SI Text for sequences). In the peptides p53N1–17, p53N62–78, and p53N74–92, naphthylalanine (Nal-Ala) at the C terminus was used as FRET donor and 5-(((acetylamino)ethyl)amino) naphthalene-1-sulfate (EDANS) at the N terminus acted as FRET acceptor. This pair has a Förster critical distance of 22 Å. Peptide p53N14–30 has Trp at position 23, which has similar excitation and emission spectra to those of Nal-Ala. To avoid the influence of the fluorescence from Trp on FRET measurement, we used another donor–acceptor pair of EDANS/Dabsyl (4-dimethylaminoazobenzene-4′-sulfonyl), which has a Förster critical distance of 30 Å. Time-resolved fluorescence spectra of the FRET donor were acquired in the presence and absence of FRET, which were then fitted numerically to a differential equation using a skewed Gaussian as radial distribution function (see SI Text). The equilibrium distance distributions are shown in Fig. 2(compare Fig. S1 for the fit of raw data) and all of the fitted parameters are listed in Table 2. No obvious systematic deviations were found in the residuals (see SI Text) and a more complex model (two skewed Gaussian radial distribution function) did not improve the χ2 significantly, whereas the random coil model (Gaussian model) also provided larger χ2 values. The same data were also fitted with another program (32) with and without acceptor signal to check for any fitting errors. We could not improve the determination of the parameters by global fitting with both the donor and the acceptor decay data. The fitting without using the acceptor signal gave similar results as shown in Table 2 (see Table S1). All of the peptides gave broad end-to-end distance distributions, which suggested all of the peptides truncated from NTD do not have a rigid conformation, supporting the notion that NTD is intrinsically disordered (5, 10). The distance distribution of the p53N1–17 peptide was narrower than the other peptides and peaked at a smaller value. The distance distributions of p53N14–30, p53N62–78, and p53N74–92 were very similar. The similarity between the three peptides may be caused by their propensity to form helical secondary structure (10, 33). The high helix propensity for p53N74–92 was consistent with the circular dichroism spectrum, which showed clear characteristics of polyproline II (PPII) structure, i.e., a positive peak at 227 nm (see Fig. S2). The circular dichroism spectra for the other peptides did not show any obvious evidence for secondary structure, which, however, does not exclude the possibility for the existence of residual secondary structures. Although indication for PPII structure was observed (10), the end-to-end distance of p53N62–78 and p53N74–92 was significantly shorter than expected from ideal PPII structure [similar to results from SM-FRET data of p53N(56C91C)]. The existence of cis-proline (34, 35) and flexible residues (36) may significantly decrease the end-to-end distance observed by FRET. A compacted conformation of PPII-forming peptides is also observed with other techniques (37). All four peptides gave very small intramolecular diffusion coefficient values, which are much smaller than the reported values for very flexible peptides (38, 39) but similar to the values observed for other protein's internal chain segments (40). It is reasonable to observe a very small diffusion coefficient caused by the presence of Pro residues in p53N1–17 (four Pro), p53N62–78 (six Pro), and p53N74–92 (eight Pro), the α-helix formation propensity of p53N14–30, and the strong sequence dependence of peptide flexibility (36, 41).
Fig. 2.
Distance distribution for synthetic peptides derived from the NTD of p53. The distributions were obtained by fitting the time-resolved fluorescence of the donor in the presence of FRET with assumption of single skewed Gaussian distribution. Data were acquired at 20 °C in phosphate buffer (pH 7) with 20 mM phosphate plus 150 mM NaCl.
Table 2.
Parameters of the distance distribution, diffusion coefficient, and χ2 obtained from the fit of TR-FRET data
| Parameter | p53N1–17 | p53N14–30 | p53N62–78 | p53N74–92 |
|---|---|---|---|---|
| Mean, Å | 22.3−0.5+0.3 | 22.3−0.8+0.0 | 23.2−0.4+0.3 | 23.0−0.5+0.3 |
| Standard deviation, Å | 3.2−1.4+0.8 | 5.4−0.0+0.4 | 4.4−1.4+0.2 | 4.8−1.1+0.3 |
| Diffusion coefficient, Å2/ns | 0.0−0.0+0.1 | 0.0−0.0+0.2 | 0.4−0.3+0.0 | 0.6−0.4+0.0 |
| χ2 | 1.24 | 1.28 | 1.48 | 1.24 |
The error on the parameters is defined by the projection of the region in parameter space where χ2 < 1.05 χ2min.
SM-FRET Efficiency Histogram of the Monomeric Subunit of p53.
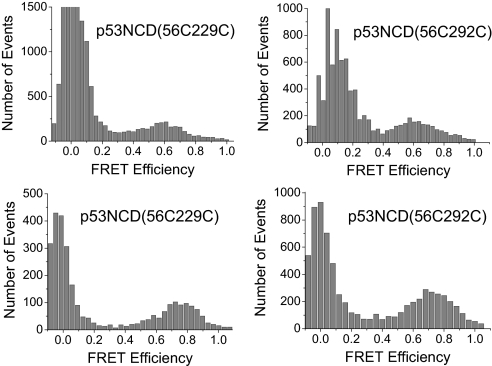
The NTD plus CD of p53 (p53NCD; residues 1–292) does not contain the TetD and the unstructured C terminus and is a paradigm for studying the behavior of monomeric p53. The SM-FRET efficiency histogram of p53NCD gave a single broad peak (apart from the zero peak) (Fig. 3) for both the FRET pair mutants studied. When AF488/AF647 were used, its peak centred at a FRET efficiency of 0.63 for p53NCD(56C229C) and 0.60 for p53NCD(56C292C), corresponding to apparent average distances of 48 ± 2 and 49 ± 2 Å, respectively. Larger apparent average distances were obtained when AF546/AF647 was used for similar reasons discussed above (Table 1). Because the deviation can be minimized and neglected when the FRET efficiency is ≈0.5, apparent average distance values corresponding to FRET efficiency close to 0.5 are, therefore, regarded to closer to the actual mean distances. We therefore took 48 ± 2 and 49 ± 2 Å as the apparent average distances between fluorophores in p53NCD(56C229C) and p53NCD(56C292C), respectively.
Fig. 3.
Single-molecule FRET histogram for the NTD plus DNA binding CD (p53NCD). (Upper) AF488/AF647 FRET pair was used. (Lower) AF546/AF647 FRET pair was used. Data were acquired at 20 °C in phosphate buffer (pH 7) with 20 mM phosphate plus 150 mM NaCl.
It is very interesting to observe smaller apparent average distances between the dyes in p53NCD (≈48–49 Å) than that in the isolated p53N(56C91C) (56–62 Å). This was a clear indication of an interaction between the NTD and CD, because the interdye distance in isolated NTD is expected to be smaller than that in p53NCD without this interaction. The NTD is extended and highly flexible in solution and possibly exists in a dynamic equilibrium between the free state and the bound state where NTD associates with CD. The single peak appearing in the SM-FRET efficiency histogram could arise from either (i) multiple conformational ensembles that exchange faster than 1 ms−1 (hence not resolvable on SM time scale) or (ii) two (or more) ensembles with slow exchange rate but either one has much greater distances and so is not observable in SM-FRET experiments or the ensembles have similar FRET efficiency and, therefore, give a broad peak. To distinguish the possibilities clearly, further investigation will be required, as, for example, using alternating excitation SM-FRET, which can detect longer distances (28).
Quaternary Structure of flp53 and Domain–Domain Interactions.
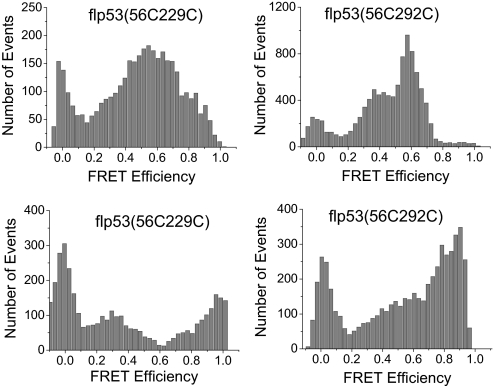
To explore the quaternary structure of flp53 and potential domain-domain interactions, we prepared several mutants: flp53(56C229C), flp53(56C292C), flp53(2C229C), and flp53(2C394C), all of which were based on the quadruple Cys → Ala mutant construct. To avoid FRET between different monomers within the same tetramer, the concentration of labeled protein and the unlabeled protein was controlled to be ≈100 pM and ≈1 μM, respectively, so that the chance of observing tetramers with two labeled subunits is statistically insignificant. SM-FRET experiments for flp53(2C229C) suggested that the N terminus of the NTD is distant from the CD because no peak with FRET efficiency between 0.2 and 1 was observed when AF488/AF647 was used. Similarly, no peak was observed between 0.2 and 1 in the SM-FRET efficiency histogram of flp53(2C394C), when AF488/AF647 or AF546/AF647 were used, which suggested that the distance between the two ends of the flp53 was at least >80 Å. This experiment excluded the possibility of interactions between the N and C termini within the same monomer. We then designed another experiment to probe any possible interaction between the N and C termini of different monomers in the same tetramer. The concentration of the labeled protein, 100 pM, is too low to form tetramers in the single-molecule experiments because the dissociation constant of tetrameric p53 to dimers is ≈100 nM (42). We designed bulk measurements to maintain labeled p53 at tetrameric levels. In the new flp53 mutant, we introduced the tetracysteine motif (CCPGCC) to the C terminus of p53, through which ReAsH (a fluorescent dye that selectively labels CCPGCC; Invitrogen) was labeled to the protein with close to 100% labeling efficiency (43). AF488 was then introduced to Cys-2 as FRET donor to establish a FRET system with ReAsH, which gives a Förster critical distance of 59 Å (calculated from fluorescence quantum yield and spectral overlap). A FRET efficiency of 0.04−0.02+0.08 was obtained from the steady-state fluorescence intensity measurements, corresponding to an apparent average distance of 100 ± 20 Å. Because there are four acceptors in a tetramer with a FRET donor, this experiment suggested that the distance between the N terminus and any of the C termini in the same tetramer was >100 ± 20 Å and hence excluded the possibility of N and C termini interactions between different subunit. Although Okorokov et al. (11) propose that the N and C termini of murine p53 interact within a tetramer, our FRET study clearly showed no indication of such interactions within the human protein. The FRET result is consistent with the previous SAXS model of p53 (9) but is different from the cryoelectron microscopy results (11).
The SM-FRET histogram of both flp53(56C229C) and flp53(56C292C) showed multiple peaks and broad distribution (Fig. 4), implying multiple conformations for the flp53 that interconvert in >1 ms. The multiple peaks cannot be assigned to FRET between different monomers within the same tetramer because it was experimentally excluded by controlling the concentration of the labeled and unlabeled protein (as mentioned above). The labeling sites for p53NCD and corresponding flp53 are exactly the same, but the histograms are obviously different, suggesting the organization of the domains is different in the presence and absence of the TetD and CTD. p53NCD gave apparently only one peak between 0.2 and 1 in the SM-FRET efficiency histogram. This peak, although still present in the SM-FRET efficiency histogram of flp53, was flanked with new peaks appearing at higher and lower FRET efficiency. The comparison of the histograms of p53NCD and flp53 (Figs. 3 and 4) strongly suggested that tetramerization influenced the relative position of the NTD and the CD significantly. If the NTD interacts with the CD, it is more likely that it not only binds to the CD from the same monomer but also to the CDs from the other monomers within the same tetramer because the NTD is long and flexible enough to make contacts with CD's of other subunits (Fig. 5). TetD plays an important role in bringing the CDs close in space for a proximal interaction with NTDs. In the simplest scenario, if the NTD binds to all of the CDs in the same tetramer but differentially, then four different conformations with different distances would be expected (Fig. 5), which would be reflected by the appearance of multiple peaks in the SM-FRET efficiency histogram. Because the peak observed in the SM-FRET efficiency histogram for the p53NCD corresponds to the NTD–CD interacting conformer, the distance between position 56 and 229 or 292 in flp53 may even be shorter when the NTD interacts with the neighboring CDs.
Fig. 4.
Single-molecule FRET histogram for flp53. (Upper) AF488/AF647 FRET pair was used. (Lower) AF546/AF647 FRET pair was used. Data were acquired at 20 °C in phosphate buffer (pH 7) with 20 mM phosphate plus 150 mM NaCl.
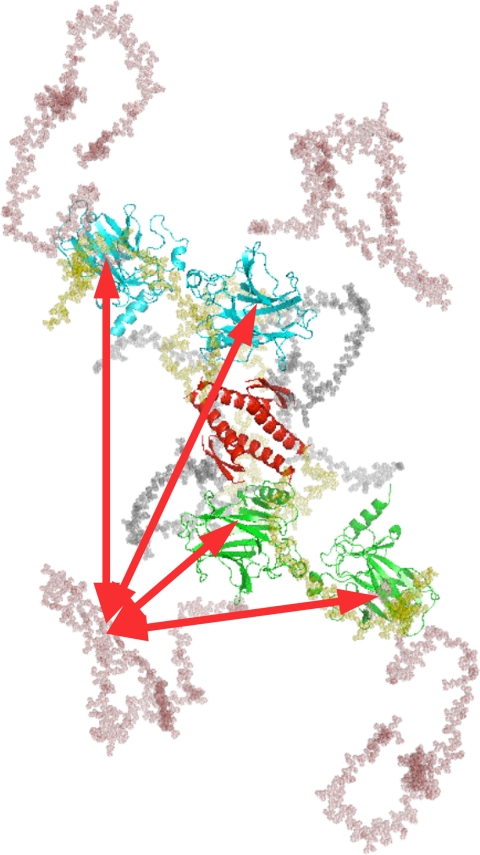
Fig. 5.
Schematic SAXS model of flp53 (9), where the DNA binding CDs are in green or cyan, the TetD is in red, linkers arein gray, the NTD is in salmon, and C terminus is in yellow. The red arrows indicate possible interactions between the NTD and CD in the p53 tetramer.
Extensive control experiments using fluorescence correlation spectroscopy (FCS) (Fig. S3) and time-resolved fluorescence anisotropy (Fig. S4) were carried out (see SI Text for details). These experiments supported the conclusion that the multiple peaks observed in the single-molecule FRET histogram were evidence for the existence of subpopulations of conformers but not a result of protein aggregates or restricted fluorophores, as observed in previous work with protein–chaperone interactions (44).
Discussion
The SM-FRET studies are consistent with the previous structural work on human flp53, have confirmed that the N and C termini are very far apart, have revealed structural details about the NTD, and have detected weak interactions that were not visible to bulk measurements.
Expanded NTD.
It has been proposed that the proline-rich region in the NTD forms PPII structure (10). The mean end-to-end distance obtained for the synthetic peptide derived from the proline-rich region (residues 74–92) was much shorter than the expected distance for a PPII structure with the same number of residues, probably because of the presence of cis-Pro or flexible residues in this region. For both residual dipolar couplings and SAXS experiments, a population of <20% of cis-Pro would be at the limit of detection. Conversely, SM-FRET and TR-FRET yield detailed distributions of distances and subpopulations, which enables detection of minor conformers. Moreover, the r−6 dependence of FRET efficiency allows, when proper pairs of dyes are used, to emphasize the contribution of minor conformations. The SM-FRET experiments showed that the apparent end-to-end distance of the NTD was large, suggesting an extended NTD in isolation. The N terminus of NTD was found to be distant from the CD, which is in agreement with the results obtained from the SAXS experiments (9). However, an obvious indication for the existence of interactions between the NTD and the CD was obtained from the observation of shorter distance between residues 56 and 229 (or 292) in the p53NCD than between residues 56 and 91 in isolated NTD. The experiments further suggested that the NTD interacted with CD with a correlation time >1 ms. This interaction seemed not to significantly reduce the radius of gyration of the whole molecule, which means that the NTD maintains loose and flexible characteristics. It is known that NTD can act as ssDNA mimic and the isolated NTD interacts weakly with isolated CD (45).
Multiple Conformations of flp53.
The observation of multiple peaks in the SM-FRET efficiency histogram provided direct evidence for the existence of multiple conformations of flp53, which did not exchange on time scale <1 ms. Whereas only a single but broad peak was observed in the SM-FRET efficiency histogram for p53NCD, we obtained complex histograms for flp53, indicative of effects of forming the tetramer. NMR experiments suggest that the structure of the CD does not change upon tetramerization (46). TetD holds p53 as a loosely packed dimer of dimers, making the NTD of each monomer accessible to the CD of the other monomers in a tetrameric subunit, resulting in multiple conformations (Fig. 5). The FRET data reported here showed the existence of weak interactions between the NTD and the CDs within the same tetramer, which are not directly detectable in NMR experiments (46).
The FRET results were, in general, consistent with the proposed structure of flp53 (9). The interaction between the NTD and the CD observed in the SM-FRET experiments was not detected in the SAXS experiments, which localized only the quaternary structures of the folded domains, and measured the overall radius of gyration, which is insensitive to the disordered N termini. Because both the SAXS and FRET experiments suggested that the NTD has a very extended conformation even in the presence of CD, the interaction between NTD and CD is probably weak. However, such interactions may play a functional role in mediating interactions with other proteins or response elements.
Materials and Methods
Protein Expression, Purification, and Labeling.
Human p53 NTD (residues 1–91), p53 NTD plus CD (residues 1–292) and flp53 (residues 1–393) were expressed in Escherichia coli and purified as described (46, 47). In p53 NTD, two cysteine residues were introduced at either positions 1 and 91, positions 10 and 56, or positions 56 and 91. All of the mutants of NTD plus CD and flp53 were based on a stabilized mutant with quadruple mutation (48). Exposed Cys (Cys-182, Cys-275, and Cys-227) and the partly buried one (Cys-124) were mutated to Ala. An additional Cys was then introduced into appropriate exposed positions so that fluorophores could be attached to p53 through Cys residues. The Cys residues were labeled with a FRET donor/acceptor pair, either AF488 and AF647 or AF546 and AF647. The proteins were labeled as described (21). After the reaction being quenched with 1 mM β-mercaptoethanol, the labeled protein was separated from the free dyes on a G-25 desalting column. No evidence of multiple labeling of the purified protein was observed in mass spectra. The labeling of flp53 with ReAsH was carried out in the presence of DTT as described (43). Excess ReAsH was removed on a G-25 desalting column.
Single-Molecule FRET Measurements.
Details of single-molecule experiments have been described (21). In brief, the FRET pair (AF488/AF647 or AF546/AF647)-labeled proteins were excited with either the 488 line (for AF488) or 514 line (for AF546) from an argon ion laser (model 35LAP321–230; Melles Griot). Fluorescence from the donor and acceptor was separately detected with two photon-counting modules (SPCM-AQR14; PerkinElmer). All of the single-molecule experiments were carried out at 20 °C in phosphate buffer (pH 7) with 20 mM phosphate plus 150 mM NaCl (both highest grade from Sigma/Aldrich). To prevent the labeled protein from sticking to the surface of the cover-glass chamber, we added 1 μM unlabeled protein to the solution.
TR-FRET Measurements.
TR-FRET experiments were carried out with a single photon counting setup (LifeSpec-ps; Edingburgh Instruments) with a light-emitting diode featuring ≈500-ps pulse width at 282 nm. The peptides were measured in phosphate buffer (20 mM phosphate plus 100 mM NaCl, pH 7) at 50 μM. Details on data analysis are available in SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. Mark Wells for assistance in peptide design and synthesis, Dr. Grace W. Yu (MRC Centre for Protein Engineering) for MDM2, and Dr. Daniel P. Teufel (MRC Centre for Protein Engineering) for Taz2. S.R. is supported by a fellowship from the Cambridge Commonwealth Trust. L.Y. is supported by a Biotechnology and Biological Sciences Research Council David Phillips Research Fellowship. E.L. is supported by the Israel Science Foundation grant (to E.H., grant No. 1102/06).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909644106/DCSupplemental.
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Hainaut P, Wiman K. 25 Years of p53 Research. New York: Springer; 2005. [Google Scholar]
- 3.Clore GM, et al. Refined solution structure of the oligomerization domain of the tumour suppressor p53. Nat Struct Biol. 1995;2:321–333. doi: 10.1038/nsb0495-321. [DOI] [PubMed] [Google Scholar]
- 4.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: Understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 5.Dawson R, et al. The N-terminal domain of p53 is natively unfolded. J Mol Biol. 2003;332:1131–1141. doi: 10.1016/j.jmb.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Müller-Tiemann BF, Halazonetis TD, Elting JJ. Identification of an additional negative regulatory region for p53 sequence-specific DNA binding. Proc Natl Acad Sci USA. 1998;95:6079–6084. doi: 10.1073/pnas.95.11.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joerger AC, Hwee CA, Veprintsev DB, Blair CM, Fersht AR. Structures of p53 cancer mutants and mechanism of rescue by second-site suppressor mutations. J Biol Chem. 2005;280:16030–16037. doi: 10.1074/jbc.M500179200. [DOI] [PubMed] [Google Scholar]
- 8.Lee W, et al. Solution structure of the tetrameric minimum transforming domain of p53. Nat Struct Biol. 1994;1:877–890. doi: 10.1038/nsb1294-877. [DOI] [PubMed] [Google Scholar]
- 9.Tidow H, et al. Quaternary structures of tumor suppressor p53 and a specific p53-DNA complex. Proc Natl Acad Sci USA. 2007;104:12324–12329. doi: 10.1073/pnas.0705069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wells M, et al. Structure of tumour suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc Natl Acad Sci USA. 2008;105:5762–5767. doi: 10.1073/pnas.0801353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okorokov AL, et al. The structure of p53 tumor suppressor protein reveals the basis for its functional plasticity. EMBO J. 2006;25:5191–5200. doi: 10.1038/sj.emboj.7601382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss S. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nat Struct Biol. 2000;7:724–729. doi: 10.1038/78941. [DOI] [PubMed] [Google Scholar]
- 13.Schuler B, Eaton WA. Protein folding studied by single-molecule FRET. Curr Opin Struc Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beechem JM, Haas E. Simultaneous determination of intramolecular distance distributions and conformational dynamics by global analysis of energy transfer measurements. Biophys J. 1989;55:1225–1236. doi: 10.1016/S0006-3495(89)82918-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman E, Haran G. Coil-globule transition in the denatured state of a small protein. Proc Natl Acad Sci USA. 2006;103:11539–11543. doi: 10.1073/pnas.0601395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laurence TA, Kong X, Jäger M, Weiss S. Probing structural heterogeneities and fluctuations of nucleic acids and denatured proteins. Proc Natl Acad Sci USA. 2005;102:17348–17353. doi: 10.1073/pnas.0508584102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc Natl Acad Sci USA. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merchant KA, Best RB, Louis JM, Gopich IV, Eaton WA. Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy any molecular simulations. Proc Natl Acad Sci USA. 2007;104:1528–1533. doi: 10.1073/pnas.0607097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deniz AA, et al. Single-molecule protein folding: Diffusion fluorescence resonance energy transfer studies of the denaturation of chymotrypsin inhibitor 2. Proc Natl Acad Sci USA. 2000;97:5179–5184. doi: 10.1073/pnas.090104997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuler B, Lipman EA, Eaton WA. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 21.Huang F, Sato S, Sharpe TD, Ying LM, Fersht AR. Distinguishing between cooperative and unimodal downhill protein folding. Proc Natl Acad Sci USA. 2007;104:123–127. doi: 10.1073/pnas.0609717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrecka J, et al. Single-molecule tracking of mRNA exiting from RNA polymerase II. Proc Natl Acad Sci USA. 2008;105:135–140. doi: 10.1073/pnas.0703815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henzler-Wildman KA, et al. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 24.Sharma S, et al. Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell. 2008;133:142–153. doi: 10.1016/j.cell.2008.01.048. [DOI] [PubMed] [Google Scholar]
- 25.Diez M, et al. Proton-powered subunit rotation in single membrane-bound F 0F1-ATP synthase. Nat Struct Mol Biol. 2004;11:135–141. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]
- 26.Mori T, Vale RD, Tomishige M. How kinesin waits between steps. Nature. 2007;450:750–754. doi: 10.1038/nature06346. [DOI] [PubMed] [Google Scholar]
- 27.Majumdar DS, et al. Single-molecule FRET reveals sugar-induced conformational dynamics in LacY. Proc Natl Acad Sci USA. 2007;104:12640–12645. doi: 10.1073/pnas.0700969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapanidis AN, et al. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci USA. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haas E. The study of protein folding and dynamics by determination of intramolecular distance distributions and their fluctuations using ensemble and single-molecule FRET measurements. ChemPhysChem. 2005;6:858–870. doi: 10.1002/cphc.200400617. [DOI] [PubMed] [Google Scholar]
- 30.Joerger AC, Allen MD, Fersht AR. Crystal structure of a superstable mutant of human p53 core domain: Insights into the mechanism of rescuing oncogenic mutations. J Biol Chem. 2004;279:1291–1296. doi: 10.1074/jbc.M309732200. [DOI] [PubMed] [Google Scholar]
- 31.Huang F, Settanni G, Fersht AR. Fluorescence resonance energy transfer analysis of the folding pathway of Engrailed homeodomain. Protein Eng Des Sel. 2008;21:131–146. doi: 10.1093/protein/gzm069. [DOI] [PubMed] [Google Scholar]
- 32.Haran G, Haas E, Szpikowska BK, Mas MT. Domain motions in phosphoglycerate kinase: Determination of interdomain distance distributions by site-specific labeling and time-resolved fluorescence energy transfer. Proc Natl Acad Sci USA. 1992;89:11764–11768. doi: 10.1073/pnas.89.24.11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 34.Best RB, et al. Effect of flexibility and cis residues in single-molecule FRET studies of polyproline. Proc Natl Acad Sci USA. 2007;104:18964–18969. doi: 10.1073/pnas.0709567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doose S, Neuweiler H, Barsch H, Sauer M. Probing polyproline structure and dynamics by photoinduced electron transfer provides evidence for deviations from a regular polyproline type II helix. Proc Natl Acad Sci USA. 2007;104:17400–17405. doi: 10.1073/pnas.0705605104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang F, Nau WM. A conformational flexibility scale for amino acids in peptides. Angew Chem Int Ed. 2003;42:2269–2272. doi: 10.1002/anie.200250684. [DOI] [PubMed] [Google Scholar]
- 37.Zagrovic B, et al. Unusual compactness of a polyproline type II structure. Proc Natl Acad Sci USA. 2005;102:11698–11703. doi: 10.1073/pnas.0409693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Möglich A, Joder K, Kiefhaber T. End-to-end distance distributions and intrachain diffusion constants in unfolded polypeptide chains indicate intramolecular hydrogen bond formation. Proc Natl Acad Sci USA. 2006;103:12394–12399. doi: 10.1073/pnas.0604748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haas E, Katchalski-Katzir E, Steinberg IZ. Brownian motion of the ends of oligopeptide chains in solution as estimated by energy transfer between the chain ends. Biopolymers. 1978;17:11–31. [Google Scholar]
- 40.Navon A, Ittah V, Landsman P, Scheraga HA, Haas E. Distributions of intramolecular distances in the reduced and denatured states of bovine pancreatic ribonuclease A. Folding initiation structures in the C-terminal portions of the reduced protein. Biochemistry. 2001;40:105–118. doi: 10.1021/bi001946o. [DOI] [PubMed] [Google Scholar]
- 41.Huang F, Hudgins RR, Nau WM. Primary and secondary structure dependence of peptide flexibility assessed by fluorescence-based measurement of end-to-end collision rates. J Am Chem Soc. 2004;126:16665–16675. doi: 10.1021/ja0466053. [DOI] [PubMed] [Google Scholar]
- 42.Sakamoto H, Lewis MS, Kodama H, Appella E, Sakaguchi K. Specific sequences from the carboxyl terminus of human p53 gene product form anti-parallel tetramers in solution. Proc Natl Acad Sci USA. 1994;91:8974–8978. doi: 10.1073/pnas.91.19.8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams SR, et al. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: Synthesis and biological applications. J Am Chem Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 44.Hillger F, et al. Probing protein-chaperone interactions with single-molecule fluorescence spectroscopy. Angew Chem Int Ed. 2008;47:6184–6188. doi: 10.1002/anie.200800298. [DOI] [PubMed] [Google Scholar]
- 45.Rajagopalan S, Andreeva A, Teufel DP, Freund SM, Fersht AR. Interaction between the transactivation domain of p53 and PC4 exemplifies acidic activation domains as single-stranded DNA mimics. J Biol Chem. 2009;284:21728–21737. doi: 10.1074/jbc.M109.006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veprintsev DB, et al. Core domain interactions in full-length p53 in solution. Proc Natl Acad Sci USA. 2006;103:2115–2119. doi: 10.1073/pnas.0511130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu GW, et al. The central region of HDM2 provides a second binding site for p53. Proc Natl Acad Sci USA. 2006;103:1227–1232. doi: 10.1073/pnas.0510343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikolova PV, Henckel J, Lane DP, Fersht AR. Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability. Proc Natl Acad Sci USA. 1998;95:14675–14680. doi: 10.1073/pnas.95.25.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.