Abstract
Duplication of genomic segments provides a primary resource for the origin of evolutionary novelties. However, most previous studies have focused on duplications of complete protein-coding genes, whereas little is known about the significance of duplication segments that are entirely internal to genes. Our examination of six fully sequenced genomes reveals that internal duplications of gene segments occur at a high frequency (0.001–0.013 duplications/gene per million years), similar to that of complete gene duplications, such that 8–17% of the genes in a genome carry duplicated intronic and/or exonic regions. At least 7–30% of such genes have acquired novel introns, either because a prior intron in the same gene has been duplicated, or more commonly, because a spatial change has activated a latent splice site. These results strongly suggest a major evolutionary role for internal gene duplications in the origin of genomic novelties, particularly as a mechanism for intron gain.
Keywords: exons, genome evolution, intron evolution, splice site
Because gene duplication is considered to be a primary source of evolutionary novelties (1, 2), studies on the duplication process and its impact on genome architecture are critical for understanding basic evolutionary processes. Up to now, most studies have focused on duplications of complete protein-coding genes resulting from polyploidization or large segmental duplications (3–9). Those studies have revealed a high rate of gene duplication that is only slightly less than the mutation rate at silent sites, implying that on time scales of 100 million years (MY) or so, all genes within a typical eukaryotic genome will have duplicated at least once (2). Although most gene duplicates are eliminated from the population in just a few MY, a minority are maintained by processes of neofunctionalization and subfunctionization (2).
However, those studies do not reveal the full impact of duplication on genome evolution. Because there is a very strong negative relationship between the length of a duplication span and its frequency (10), far more duplication events involve gene segments than entire genes. We can thus be certain that prior quantitative studies based on complete gene duplication have underestimated the rate at which novel genes are created by duplicative processes, perhaps dramatically so. In this study, we examined another type of novel gene creation process, in which duplication events are internal to genes (designated internal gene duplication hereafter).
Although many individual genes with internally duplicated sequences have been observed, there have been very few genomewide studies, all of which focus only on duplicated exons (11, 12). Because of the lack of systematic study of internal gene duplication, the quantitative contribution of this process to genome evolution may have been overlooked. To fully appreciate the significance of internal gene duplication as an evolutionary force, we need to understand its dynamic patterns, gain and loss frequencies, and the effects of internal gene duplication on gene-structure evolution.
Compared with the gradual and minor functional divergence between complete duplicated genes, internal gene duplication can lead to the immediate acquisition of a novel function, on some occasions providing a substantial selective advantage (13). Also, many human genetic disorders are associated with internal gene duplications, such as breast cancer, Duchenne muscular dystrophy, and familial hypercholesterolemia (14–16). Thus, studies on the internal duplication process will facilitate a better understanding of not only the mechanisms of genome evolution but also the quantitative nature of such mutations as a major source of disease. Here, we present a study across six complete eukaryotic genomes of the dynamics of duplication events internal to genes and their impact on a structural aspect unique to eukaryotic genes, spliceosomal introns.
Although the sources of spliceosomal introns remain a mystery, several models have been proposed for their origin (17–19): (i) recruitment from an ancestral pool of group-II introns, with the current intron set representing the remnants of a large ancestral pool, (ii) insertion of preexisting spliced introns into genes through RNA and cDNA intermediates, (iii) insertion and decay of transposons, (iv) internal tandem duplication of coding sequences containing AGGT cryptic splice sites, (v) emergence from genomic regions that are subject to posttranscriptional surveillance pathways, and (vi) generation of novel splice sites by point mutations in coding regions. However, these hypotheses either have few examples to support them or have not been tested, and to date, the mechanisms of intron creation remain puzzling and controversial (20–22).
Among conserved orthologous genes across species, the rate of intron gain is thought to be low with intron loss rates being dominant in some lineages (23–26). Studies on intron dynamics in gene families suggest that the rates of intron gain/loss are slightly higher than in nonduplicated genes (27, 28). However, whether and how the process of duplication itself influences the intron creation process is unclear. In this study, we find that internal gene duplications often lead to the creation of new gene structures, in particular the origin of entirely new introns resulting from the creation or activation of latent splice sites after genome sequence rearrangement.
Results and Discussion
We characterized the rate of internal gene duplication in the genomes of six eukaryotes with high-quality annotated sequences, including mammals (Homo sapiens, Mus musculus), invertebrates (Drosophila melanogaster, Caenorhabditis elegans), and plants (Arabidopsis thaliana, Oryza sativa). By BLASTing the sequence of each annotated gene against itself (including both exon and intron sequences), we found that 8.3–16.6% of the genes in these species contain internally duplicated segments (see ratio 1 in Table 1 and Table S1). These ratios are slightly higher than those in a previous report that focused on genes with exon duplication (10.7% in H. sapiens, 7.1% in D. melanogaster, and 7.5% in C. elegans) (11).
Table 1.
Number of internally duplicated genes and the new introns associated with internal gene duplications
| Section | Category | A. thaliana | O. sativa | C. elegans | D. melanogaster | M. musculus | H. sapiens |
|---|---|---|---|---|---|---|---|
| I | Total protein-coding genes | 26,514 | 42,802 | 17,830 | 9,221 | 23,739 | 21,481 |
| Internally duplicated protein-coding genes | 2,868 | 6,952 | 1,478 | 978 | 2,703 | 3,571 | |
| Ratio 1 | 10.8% | 16.2% | 8.3% | 10.6% | 11.4% | 16.6% | |
| II | Internally duplicated genes with orthologs in two closely related species (Ratio 2) | 299 (10.4%) | 1,163 (16.7%) | 90 (6.1%) | 155 (15.8%) | 601 (22.2%) | 930 (26.0%) |
| Internally duplicated genes with orthologs and new introns (number of new introns)* | 46 (112) | 90(204) | 27 (55) | 24 (48) | 39 (46) | 68 (105) | |
| Ratio 3 | 15.4% | 7.7% | 30.0% | 15.5% | 6.5% | 7.3% | |
| Ratio 4 | 9.4% | 3.6% | 6.7% | 10.3% | 5.0% | 4.5% |
The total number of protein-coding genes and genes with internal duplications are summarized in section I. The numbers of internally duplicated genes with new introns, and the ratios of genes with intron gain to all internally duplicated genes, are summarized in section II. Ratio 1 = internally duplicated protein-coding genes/total protein-coding genes. Ratio 2 = internally duplicated genes with orthologs in two closely related species/internally duplicated protein-coding genes. Ratio 3 = internally duplicated genes with orthologs and new introns/internally duplicated genes with orthologs in two closely related species. Ratio 4 = cDNA-supported internally duplicated genes with orthologs and new introns (Table 3)/internally duplicated genes with orthologs in two closely related species.
*The numbers of new introns identified from internally duplicated genes with orthologs are listed in parentheses. Some genes have multiple new introns.
Internal Duplication Commonly Reflects a Steady-State Birth and Death Process.
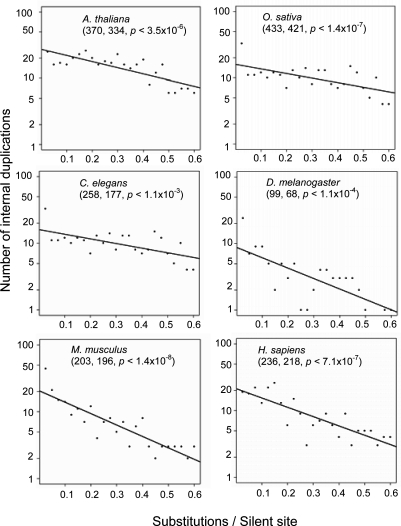
To better understand the power of internal gene duplication as an evolutionary force, we studied the demographic features of this process. First, we examined the age distribution of internal duplications in each species. To date the age of internal duplications, we estimated the number of substitutions between duplicated regions at sites generally assumed to undergo no selection (S), i.e., synonymous sites in coding regions, and all sites in introns, except for the 5 nucleotides at the two intronic ends, which are likely to be under functional constraint for efficient intron splicing. When the numbers of internal duplications are plotted against their age on a scale of S, internal duplications display an approximately negative exponential age distribution for all six genomes (Fig. 1), consistent with a steady-state birth and death process (2, 7), implying that internal gene duplications have been originating and disappearing in these genomes at approximately constant rates for many millions of years.
Fig. 1.
Age distributions of internal gene duplications in six eukaryotic genomes based on internal gene duplications for which S < 0.6. Numbers of internal duplications are plotted against their ages (substitutions per silent site). The bin size of the horizontal axis is 0.025 substitutions per silent site. The number of internal duplication events and the number of internally duplicated genes with the S < 0.6 value are listed as the first two numbers in parentheses in each plot. Least-squares regression provides the linear relationship between the natural logarithm of the numbers of internal duplications and their ages, revealing a negative exponential age distribution for the internal duplications. All regressions are significant at the 1% level, and each P value is listed in parentheses in each plot. The fitted parameters are shown in Table 2.
From the age distributions presented in Fig. 1, we estimated the birth and death rates of internally duplicated genes with a previously published evolutionary demographic method (7), confining the analyses to recent internal duplications (S < 0.6) for which the saturation of substitutions per site is not a substantial problem (Table 2). We find that internal duplication processes occur at rates comparable to duplications of complete genes (29), i.e., 10−3 duplications per gene over a time span equivalent to 1% divergence at neutral sites (B in Table 2). Using the estimated molecular clock for silent sites in different species [2.5 substitutions/site per billion years (BY) for mammals; 15.6 substitutions/site per BY for invertebrates; 9.9 substitutions/site per BY for plants (29), (the plant substitution rate is corrected according to ref. 30]; the rates of internal duplications range from 0.001 to 0.013 events/gene per MY (Table 2). Thus, over the course of 77–1,000 MY (mean at 541 MY), essentially all genes in an average eukaryotic lineage will have experienced at least one internal duplication event. The birth-rate estimates define the rates of duplication origin per gene copy, not the rate of fixation at the population level, because the youngest duplicates in our analyses are almost certainly not fixed at the population level, given their levels of divergence.
Table 2.
Estimated death rates, half-lives, and birth rates, based on internal gene duplications for which S < 0.6
| Species | Internal gene duplication |
Complete gene duplication |
||||||
|---|---|---|---|---|---|---|---|---|
| Death rates |
Half-lives |
Birth rates |
B | D | ||||
| D (SE) | d (SE) | S0.5 (SE) | Years, 106 (SE) | B (SE) | B*(SE) | |||
| A. thaliana | 0.577 (0.059) | 86.0 (14.0) | 0.0081 (0.0013) | 0.661 (0.108) | 0.0011 (0.0001) | 0.002 (0.00013) | 0.0032 | 0.033 |
| O. sativa | 0.716 (0.047) | 125.9 (16.6) | 0.0055 (0.0007) | 0.451 (0.059) | 0.0016 (0.0006) | 0.002 (0.00007) | - | - |
| C. elegans | 0.474 (0.089) | 64.3 (17.0) | 0.0108 (0.0028) | 0.346 (0.091) | 0.0021 (0.0002) | 0.006 (0.00049) | 0.0028 | 0.229 |
| D. melanogaster | 0.764 (0.072) | 144.4 (30.3) | 0.0048 (0.0010) | 0.154 (0.032) | 0.0041 (0.0005) | 0.013 (0.00150) | 0.0011 | 0.136 |
| M. musculus | 0.793 (0.037) | 157.3 (18.0) | 0.0044 (0.0005) | 0.882 (0.101) | 0.0023 (0.0002) | 0.001 (0.00008) | 0.0030 | 0.134 |
| H. sapiens | 0.725 (0.052) | 129.0 (18.9) | 0.0054 (0.0008) | 1.074 (0.157) | 0.0012 (0.0001) | 0.001 (0.00004) | 0.0049 | 0.081 |
For internally duplicated genes, D (death rate of internal duplication per gene) and B (birth rate of internal duplication per gene) are estimated on a time scale of divergence at silent sites of 1% (S = 0.01). d is the instantaneous loss rate of internal duplications. B* is the birth rate of internal duplications per gene per MY. Standard errors are shown in parentheses. The data for complete gene duplications are from ref. 29.
After a birth event, the survivorship of an internally duplicated gene determines the extent of its potential future impact on genome evolution. We found that the death rates of internally duplicated genes are 2- to 17-fold higher than for complete gene duplications (Table 2), suggesting that alleles with internal duplications are on average more deleterious than completely duplicated genes. Although complete gene duplications may cause dosage defects, internal duplications frequently alter gene-coding sequences (e.g., introducing frame shifts and premature stop codons), presumably to a large enough extent to be purged from the population by selection. However, internally duplicated genes can acquire mutations at birth during the duplication process (e.g., indels accompanied by sequence rearrangement) that may occasionally provide raw genetic materials for adaptive evolution, whereas completely duplicated genes must await the gradual arrival of new mutations that may contribute to preservation by subfunctionalization or neofunctionlization.
Internal Duplication Creates New Introns.
A central question with respect to long-term evolution concerns the probability of establishment of novel structural variants resulting from internal duplication events. Although likely predominantly deleterious, the insertion of internally duplicated fragments into a gene has the potential to occasionally create novel gene structures. To determine whether internal gene duplications play a role in the creation of novel introns, for each of the six genomes, we focused on internally duplicated genes with internal-duplication-free orthologs (identified as “orthologs” hereafter) in two other species (one closely related and one further out-group; see Table S2), so that the ancestral gene structures before internal duplication could be safely inferred. The fraction of internally duplicated genes that fulfill this criterion is a function of availability of reasonable close out-group species, but 6–26% of the internally duplicated protein-coding genes that we identified met this criterion (ratio 2 in Table 1). For this subset of genes, based on parsimony, introns unique to the internally duplicated genes are considered to be new introns. To avoid inferring intron gain incorrectly by only comparing orthologs from two relatively closely related species, we also searched for orthologs of internally duplicated genes, if available, in all other species used in this study, which have long evolutionary distances to the studied genome. We confirmed that such internal duplications are lineage specific, suggesting that the unique introns resulted from intron gain in internally duplicated genes, rather than from intron loss in the other two closely related species. Because we are interested in identifying those introns created by internal duplication, only those new introns flanking or within the duplicated regions are reported here. An average 13.7% of internally duplicated genes appear to be associated with the creation of new introns (ratio 3 in Table 1; see Dataset S1).
Novel Splice Sites Can Be Activated from Latent Sites After Internal Gene Duplication.
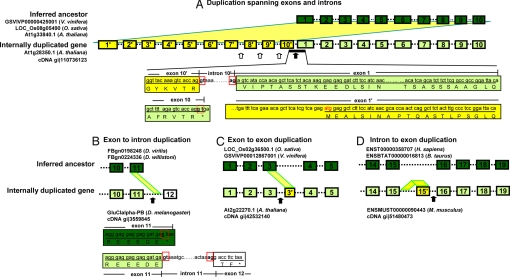
Novel introns in internally duplicated genes can emerge in two ways (Fig. 2). First, an existing intron in the ancestral gene can be duplicated together with flanking exons or exon fragments, i.e., as simple copies of an existing intron without the creation of novel splice sites. Less than half of the new introns are of this type (Table 3). The majority of new introns are of a second type, introns flanked by at least one new splice site (Table 3). Therefore, through internal gene duplication, new introns arise not only by simple duplication of preexisting introns, but more frequently by the creation of novel splice sites. This pattern is validated by cDNA confirmations of many new introns (Table 3). In some cases, the new splice sites can still be identified as originating from previous latent splice sites. Some cDNA confirmed examples are shown in Fig. 2. In Fig. 2 A and B, the ancestral gene had a nucleotide G before the stop codon TAA, and after the internal duplication the latent GT dinucleotide at the stop-codon junction was evoked. This activated upstream splice site now pairs with a downstream AG in the duplicated sequence, with the sequence in between being recognized as a novel intron.
Fig. 2.
Examples of cDNA confirmed introns created by internal gene duplication. Genes with internal duplications and their inferred ancestral genes are shown. (A) Duplication spanning exons and introns. (B) Exon to intron duplication. (C) Exon to exon duplication. (D) Intron to exon duplication. The ancestral genes were inferred from orthologous genes in two out-group species (the IDs of orthologous genes are listed by the referred ancestors) and an internal-duplication-free paralogous gene if available (such as in A). For the purpose of illustration in these examples, the duplicated copy in internally duplicated genes that best aligns with the inferred ancestor gene is considered as the original sequence, and the unaligned duplicated copy is considered derived. Exons and introns are shown with filled boxes and dotted lines, respectively. Exons that are found in both the inferred ancestor (dark green) and the internally duplicated gene (light green) have the same labels. The newly duplicated exons (yellow) are named after the original exon that they were duplicated from, with a prime suffix (e.g., exon 1′ to 10′ in A and exon 3′ in C). Exon 12 in B, filled with white color, is a new exon, but not derived from duplicated sequence directly. Exon 15′ in D is duplicated from sequences located in intron 15. Because of the fast evolution in intron sequences, the intron 15 sequences in internal-duplication-free orthologs (ENST00000358707 in H. sapiens and ENSBTAT00000016813 in B. taurus) do not share significant similarity with intron 15 in the internally duplicated gene (ENSMUST00000090443), nor with the new exon 15′, but the intron and exon in the duplicated gene still show similarity. Duplicated regions are projected as yellow shadows from ancestral genes to derived copies (in A–C), or from an intron of the same gene (in D). Examples of introns created by new splice sites are marked by solid black arrows. Hollow arrows point to examples of new introns created by duplications of preexisting introns. All of these new introns are confirmed by cDNA evidence (GenBank accession numbers are shown). In some cases, the source of the new splice sites can still be identified. The exon–intron boundary sequences around the new splice sites are displayed. In A, all of the exon and intron sequences in the ancestral gene were duplicated. The joining between exon 10′ and exon 1 yielded a new intron sequence: assuming exon 10 represents the last exon in the ancestral gene, the 5′ splice site GT (in a red box) in exon 10′ is activated from a previous latent splice site (G before the stop codon TAA, in a dotted red box) in the ancestral sequence of exon 10. In B, the 5′ splice site GT (a solid red box) is activated from a previous latent splice site (dotted red box) in the exon 11 in the inferred ancestral sequences.
Table 3.
The origin and cDNA confirmation of new introns derived from internal gene duplication
| Section | Category | A. thaliana | O. sativa | C. elegans | D. melanogaster | M. musculus | H. sapiens | |
|---|---|---|---|---|---|---|---|---|
| I | Annotated new introns | Duplicated splice sites | 48 (12) | 31 (13) | 19 (12) | 9 (2) | 2 (1) | 22 (7) |
| Novel splice sites | 64 (41) | 173 (87) | 36 (25) | 39 (23) | 44 (38) | 83 (64) | ||
| Total | 112 (46) | 204 (90) | 55 (27) | 48 (24) | 46 (39) | 105 (68) | ||
| Confirmed new introns (with cDNA) | Duplicated splice sites | 20 (8) | 15 (6) | 4 (3) | 8 (4) | 2 (1) | 22 (7) | |
| Novel splice sites | 27 (23) | 54 (39) | 5 (4) | 23 (14) | 32 (29) | 44 (38) | ||
| Total | 47 (28) | 69 (42) | 9 (6) | 31 (16) | 34 (30) | 66 (42) | ||
| Ratio 1 (confirmed introns/annotated introns) | 42.0% | 33.8% | 16.4% | 64.6% | 73.9% | 62.9% | ||
| II | New introns with alternative splicing | Duplicated splice sites | 9 (7) | 4 (3) | 1 (1) | 2 (1) | 0 (0) | 5 (3) |
| New splice sites | 11 (8) | 15 (15) | 0 (0) | 9 (9) | 21 (18) | 32 (28) | ||
| Total | 20 (15) | 19 (18) | 1 (1) | 11 (10) | 21 (18) | 37 (30) | ||
| Ratio 2 (alternative spliced new introns/all confirmed new introns) | 42.6% | 27.5% | 11.1% | 35.5% | 61.8% | 56.1% | ||
Annotated new introns and new introns with cDNA confirmation are further categorized in section I based on the origins of their splice sites. The alternative splicing patterns of novel introns are analyzed in section II. The numbers of genes where the new introns reside are listed in parentheses.
These observations suggest that internal gene duplications harbor substantial potential for the spontaneous production of new introns via alterations in the spatial configurations of latent splice sites. Although the activation of latent splice sites is a necessary condition for intron creation by this mechanism, this alone is unlikely to be sufficient, in that other cis-elements, such as a polypyrimidine track, branch-point sequence, exonic splicing enhancers or silencers, and intronic splicing enhancers or silencers may also be required/involved in intron creation (31). Unfortunately, the generally diffuse nature of the sequence signatures of such elements makes it very difficult to map their locations (and hence trace their origins) in new introns. However, as with the splice sites themselves, small latent cis-elements are likely widespread in genomes, enhancing the likelihood that spatial changes through internal gene duplication will occasionally activate them as splicing signals.
Although intron gain events have been previously reported in different species (24, 27, 28, 32), in very few cases has the sequence origin of introns and/or the evolution of their splice sites been uncovered. A very few spliceosomal introns are known to derive from mobile elements in plant, fly, and human (33–37). In C. elegans, a few de novo introns are thought to have originated from internal exonic sequences, with their splice sites created by point mutations (19). In human, de novo introns have been reported in isolated cases to result from fortuitous splice-site creation (38, 39). Rogers (40) proposed that duplicating a segment containing the AGGT may generate 5′ and 3′ splice sites, and some introns have been found in human with perfect matching sequences at 5′ and 3′ exon-intron boundaries (20), in support of this model. However, it is still under debate as to whether such introns are authentic or their sequences are actually absent in cDNA (but present in mRNA) resulted from template switching during the reverse transcription in the preparation of cDNA/EST libraries (see also refs. 21 and 41). In our study, the underlying mechanism of novel intron birth through internal gene duplication is distinct from the Rogers model, because the splice sites of novel introns generally do not locate in direct duplicated repeats.
Novel Introns Are Supported by cDNA Evidence.
Of the annotated introns that we have identified as novel, an average 48.9% were confirmed by cDNA sequences (ratio 1 in Table 3; see Dataset S1 for details), validating their active recognition by the spliceosomal machinery. The support level is variable across the six genomes (ranging from 16.4% in C. elegans to 73.9% in M. musculus), but this may largely be a consequence of the variable comprehensiveness of the cDNA data among species and should not be interpreted to imply distinct difference among species. However, the cDNA support does provide a lower bound for the ratio of novel introns that can be effectively recognized. For these novel introns in internally duplicated genes, the frequency of alternative splicing is comparable with that in other genes (42) (ratio 2 in Table 3), which is a further indicator that the treatment of these novel introns by the splicesome is not unusual.
Because the biological significance of internal gene duplication on genome evolution might be questioned if internally duplicated genes have no functions, we also searched for the evidence of functionality of these internally duplicated genes with new introns. Although experimental evidence is likely to be the only completely accurate way to explore gene function, such information is still very limited in eukaryotes. Here, we infer the functional significance of these internally duplicated genes through their nonsynonymous to synonymous substitution rates (N/S). After constructing phylogenetic trees of internally duplicated genes with orthologous genes in two other close-related species (Table S2), we performed likelihood-ratio tests to compare two models: N/S of the internally duplicated genes are fixed to 1 (null hypothesis), and N/S of the internally duplicated genes are estimated to be <1 (alternative model) (43). Seventy percent of all internally duplicated genes with new introns have N/S significantlly <1, indicating that they are under strong purifying selection [false discovery rate = 0.05 (44)]. The remaining 30% of internally duplicated genes containing new introns either are evolving a neutral fashion or lack enough power to reject the null hypothesis model. Therefore, the majority of the internally duplicated genes with new introns are suggested to be under purifying selection.
Conclusions
Our conclusion that internal duplication is a creative force for the origin of novel splice sites has significant implications for the estimation of rates of intron gains. In previous studies, only a few intron gains have been identified in the six species that we have studied (23–26), yielding estimated rates of intron gain of <0.04 per gene per BY (23). Although such low rates appear to be at odds with our observations, previous studies have been restricted to a small subset of highly conserved orthologs, and only introns with conserved flanking exon sequences were studied, which would specifically exclude genes that have experienced dynamic sequence rearrangement. Given that such genes compose ≈8–17% of the total pool of protein-coding genes in multicellular species, our results suggest that many preceding analyses may not reflect the complete picture of intron origin.
Our study of internal gene duplication reveals a remarkable and previously underappreciated contributor to genome evolution. Internal duplication occurs at a steady, high rate, exhibiting patterns of evolutionary demography similar to that previously found for complete-gene duplicates. The death rates of internally duplicated genes are relatively high, but their half-lives are sufficiently long to occasionally contribute significant resources to longer-term evolutionary processes. More importantly, internal duplications lead directly to the creation of new gene architectural features, such as spliceosomal introns, a hallmark of eukaryotic gene structure. As the likely mechanism for creating new introns in the cases we have uncovered is the evocation of cryptic splice sites after sequence rearrangements, our results shed light on one of the greatest mysteries in genome evolution: the mechanisms of intron creation.
Materials and Methods
Details about data resources, calculation of number of substitutions per silent site in internally duplicated genes, birth and death rates of internal duplications, identification cDNA evidence for new introns, and purifying selection analysis of internally duplicated genes can be found in SI Text.
Identification of Internally Duplicated Genes.
First, repetitive elements were masked with RepeatMasker (www.repeatmasker.org). Then, every exon and intron sequence was compared with all of the exon and introns sequences within the same gene, using TBLASTX and BLASTN (45). A gene was identified as containing an internal duplication if at least one of the following conditions was satisfied: (i) an exon sequence had similarity to that of a different exon sequence, (ii) an intron sequence had similarity to an exon sequence, and/or (iii) two different regions of the same exon have similarity to each other. Significantly similar sequences were determined by TBLASTX or BLASTN alignment with E-value <10−5. However, if both similar sequences were identified within intron regions (i.e., similar sequences are in different introns or in different positions of same intron), such cases were not considered as internal duplications in this analysis because of the possibility that repeats found in introns only are novel repetitive elements, which were not masked by RepeatMasker. Observing such unidentified repeat elements in exon regions is assumed to be much less likely because selection pressure would preserve gene integrity. However, to provide a general idea of the prevalence of three classes of duplications (exon–exon, exon–intron, and intron–intron), we summarize the statistics for all six genomes in Table S3.
The estimated duplicated length from the blast results ranges from 11 to 17,103 bp with medians ≈100 bp (95 bp in A. thaliana, 74 bp in O. sativa, 107 bp in C. elegans, 128 bp in D. melanogaster, 119 bp in M. musculus, and 110 bp in H. sapiens). If a single internal gene duplication event spans multiple exons and introns, the full length of duplication is the sum of duplicated segments in all exons and intron. Therefore, the duplication length calculated above from each individual exon and intron duplication only provide a lower bound estimation of the internal gene duplication length span.
After producing the dataset of internally duplicated genes, we applied the following filters to the dataset: (i) if there were alternative transcripts containing internal duplications from a gene locus, only one such transcript was randomly chosen for its age calculation to avoid inflated counting; and (ii) we restricted our subsequent analysis to protein-coding genes, because it was not clear how to define the nucleotide positions under neutral evolution in noncoding genes. Table S1 shows the number of genes after each filtering process and the final dataset.
Identification of Intron Gains in Internally Duplicated Genes.
To infer intron gains in the internally duplicated genes, we needed to infer the structure of the ancestral gene before the internal duplication event. To this end, we used internal-duplication-free orthologs from one closely related species, and an additional out-group species (all reference species are listed in Table S2). Informative orthologs were required to meet the following criteria: (i) absence of internal duplication and (ii) high similarity in overall sequence instead of just a domain alignment. The alignment coverage was required to be ≥50% of the total length of the internally duplicated gene and ≥80% of the length of the homologous gene from reference genomes, after merging all of the individual aligned regions identified by BLASTP (E ≤ 10−5).
We have tested different cutoff criteria of coverage length for defining orthologous genes, and our result remains similar. For example, when more stringent criteria, 80% coverage for both the internally duplicated gene and the homologous gene, were applied, only a minor portion (6%) of internally duplicated genes was eliminated from current dataset.
The intron positions of internally duplicated genes were compared with the putative ancestral states derived from orthologous genes. Based on the principle of parsimony, the unique introns identified in the internally duplicated genes were considered as new introns, following the method described in ref. 28 [we used T-Coffee global alignment tool (46) instead of ClustalW (47) for aligning orthologous protein sequences]. After screening with automated scripts, manual checking was performed to verify the results. During this process, we excluded some internally duplicated genes whose orthologs identified did not share similar exon/intron structures. If only the closely related out-group ortholog shared most intron positions with the internally duplicated gene and the further out-group ortholog had a much reduced shared profile of intron position, the S value of the internal duplicated gene was used to confirm that the internal duplication occurred subsequent to the two speciation events.
There are two potential categories of intron gain in an internally duplicated gene: (i) duplication of an existing intron, flanked by the original splice sites; and (ii) creation of an entirely novel intron, flanked by novel splice sites (5′, 3′ or both). Because intron sequences evolve rapidly, we examined the flanking exon sequences to differentiate the above two categories. Specifically, for each new intron in our list, we joined its flanking exon sequences (50 amino acid residues from each side), and BLASTed against the remaining sequences of the same gene. If exon sequences flanking the novel intron were also matched to the flanking region of a different intron in the same gene, we considered that the new intron was simply duplicated from an existing one. Otherwise, the new intron was classified as novel.
Supplementary Material
Acknowledgments.
We thank Dr. Francesco Catania, Dr. J. Ignacio Lucas-Lledó, and Dr. Thomas Doak for helpful discussions and Dr. Yves Van de Peer and Dr. Devin Scannell for valuable comments. This work was supported by National Institutes of Health Postdoctoral Fellowship F32GM083550 (to X.G.) and National Science Foundation Grant EF-0827411 (to M.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911093106/DCSupplemental.
References
- 1.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 2.Lynch M. The Origins of Genome Architecture. Sunderland, MA: Sinauer; 2007. [Google Scholar]
- 3.Bowers JE, Chapman BA, Rong J, Paterson AH. Unraveling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 4.Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- 5.Wolfe KH. Yesterday's polyploids and the mystery of diploidization. Nat Rev Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Z, et al. A genomewide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- 7.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 8.Gu X, Wang Y, Gu J. Age distribution of human gene families shows significant roles of both large- and small-scale duplications in vertebrate evolution. Nat Genet. 2002;31:205–209. doi: 10.1038/ng902. [DOI] [PubMed] [Google Scholar]
- 9.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katju V, Lynch M. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics. 2003;165:1793–1803. doi: 10.1093/genetics/165.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letunic I, Copley RR, Bork P. Common exon duplication in animals and its role in alternative splicing. Hum Mol Genet. 2002;11:1561–1567. doi: 10.1093/hmg/11.13.1561. [DOI] [PubMed] [Google Scholar]
- 12.Kondrashov FA, Koonin EV. Origin of alternative splicing by tandem exon duplication. Hum Mol Genet. 2001;10:2661–2669. doi: 10.1093/hmg/10.23.2661. [DOI] [PubMed] [Google Scholar]
- 13.Patty L. Protein Evolution. Oxford: Blackwell; 1999. [Google Scholar]
- 14.Hogervorst FB, et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 2003;63:1449–1453. [PubMed] [Google Scholar]
- 15.Weiss C, et al. Tandem duplication of DMD exon 18 associated with epilepsy, macroglossia, and endocrinologic abnormalities. Muscle Nerve. 2007;35:396–401. doi: 10.1002/mus.20705. [DOI] [PubMed] [Google Scholar]
- 16.Hu X, Worton RG. Partial gene duplication as a cause of human disease. Hum Mutat. 1992;1:3–12. doi: 10.1002/humu.1380010103. [DOI] [PubMed] [Google Scholar]
- 17.Roy SW, Gilbert W. The evolution of spliceosomal introns: Patterns, puzzles, and progress. Nat Rev Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 18.Catania F, Lynch M. Where do introns come from? PLoS Biol. 2008;6:e283. doi: 10.1371/journal.pbio.0060283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irimia M, et al. Origin of introns by “intronization” of exonic sequences. Trends Genet. 2008;24:378–381. doi: 10.1016/j.tig.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Zhuo D, Madden R, Elela SA, Chabot B. Modern origin of numerous alternatively spliced human introns from tandem arrays. Proc Natl Acad Sci USA. 2007;104:882–886. doi: 10.1073/pnas.0604777104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy SW, Irimia M. When good transcripts go bad: Artifactual RT-PCR “splicing” and genome analysis. BioEssays. 2008;30:601–605. doi: 10.1002/bies.20749. [DOI] [PubMed] [Google Scholar]
- 22.Cocquet J, Chong A, Zhang G, Veitia RA. Reverse transcriptase template switching and false alternative transcripts. Genomics. 2006;88:127–131. doi: 10.1016/j.ygeno.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 23.Roy SW, Penny D. Patterns of intron loss and gain in plants: Intron loss-dominated evolution and genomewide comparison of O. sativa and A. thaliana. Mol Biol Evol. 2007;24:171–181. doi: 10.1093/molbev/msl159. [DOI] [PubMed] [Google Scholar]
- 24.Coulombe-Huntington J, Majewski J. Intron loss and gain in Drosophila. Mol Biol Evol. 2007;24:2842–2850. doi: 10.1093/molbev/msm235. [DOI] [PubMed] [Google Scholar]
- 25.Coulombe-Huntington J, Majewski J. Characterization of intron loss events in mammals. Genome Res. 2007;17:23–32. doi: 10.1101/gr.5703406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy SW, Penny D. Smoke without fire: Most reported cases of intron gain in nematodes instead reflect intron losses. Mol Biol Evol. 2006;23:2259–2262. doi: 10.1093/molbev/msl098. [DOI] [PubMed] [Google Scholar]
- 27.Castillo-Davis CI, Bedford TB, Hartl DL. Accelerated rates of intron gain/loss and protein evolution in duplicate genes in human and mouse malaria parasites. Mol Biol Evol. 2004;21:1422–1427. doi: 10.1093/molbev/msh143. [DOI] [PubMed] [Google Scholar]
- 28.Lin H, Zhu W, Silva JC, Gu X, Buell CR. Intron gain and loss in segmentally duplicated genes in rice. Genome Biol. 2006;7:R41. doi: 10.1186/gb-2006-7-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch M, Conery JS. The evolutionary demography of duplicate genes. J Struct Funct Genomics. 2003;3:35–44. [PubMed] [Google Scholar]
- 30.Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proc Natl Acad Sci USA. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Burge CB. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coghlan A, Wolfe KH. Origins of recently gained introns in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2004;101:11362–11367. doi: 10.1073/pnas.0308192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwamoto M, Nagashima H, Nagamine T, Higo H, Higo K. A tourist element in the 5′-flanking region of the catalase gene CatA reveals evolutionary relationships among Oryza species with various genome types. Mol Gen Genet. 1999;262:493–500. doi: 10.1007/s004380051110. [DOI] [PubMed] [Google Scholar]
- 34.Giroux MJ, et al. De novo synthesis of an intron by the maize transposable element dissociation. Proc Natl Acad Sci USA. 1994;91:12150–12154. doi: 10.1073/pnas.91.25.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wessler SR. The splicing of maize transposable elements from pre-mRNA: A minireview. Gene. 1989;82:127–133. doi: 10.1016/0378-1119(89)90037-1. [DOI] [PubMed] [Google Scholar]
- 36.Purugganan M, Wessler S. The splicing of transposable elements and its role in intron evolution. Genetica. 1992;86:295–303. doi: 10.1007/BF00133728. [DOI] [PubMed] [Google Scholar]
- 37.Sela N, et al. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol. 2007;8:R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Courseaux A, Nahon JL. Birth of two chimeric genes in the Hominidae lineage. Science. 2001;291:1293–1297. doi: 10.1126/science.1057284. [DOI] [PubMed] [Google Scholar]
- 39.Fablet M, Bueno M, Potrzebowski L, Kaessmann H. Evolutionary origin and functions of retrogene introns. Mol Biol Evol. 2009;26:2147–2156. doi: 10.1093/molbev/msp125. [DOI] [PubMed] [Google Scholar]
- 40.Rogers JH. How were introns inserted into nuclear genes? Trends Genet. 1989;5:213–216. doi: 10.1016/0168-9525(89)90084-x. [DOI] [PubMed] [Google Scholar]
- 41.Chabot B, Elela SA, Zhuo D. Comment on “When good transcripts go bad: artifactual RT-PCR ‘splicing’ and genome analysis.”. BioEssays. 2008;30:1256. doi: 10.1002/bies.20844. author reply 1257–1258. [DOI] [PubMed] [Google Scholar]
- 42.Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35:125–131. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z, Nielsen R, Hasegawa M. Models of amino acid substitution and applications to mitochondrial protein evolution. Mol Biol Evol. 1998;15:1600–1611. doi: 10.1093/oxfordjournals.molbev.a025888. [DOI] [PubMed] [Google Scholar]
- 44.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 46.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 47.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.