Abstract
The pathway by which ubiquitin chains are generated on substrate via a cascade of enzymes consisting of an E1, E2 and E3 remains unclear. Multiple distinct models involving chain assembly on E2 or substrate have been proposed. However, the speed and complexity of the reaction have precluded direct experimental tests to distinguish between potential pathways. Here we introduce new theoretical and experimental methodologies to address both limitations. A quantitative framework based on product distribution predicts that the really interesting new gene (RING) E3s SCFCdc4 and SCFβ-TrCP work with the E2 Cdc34 to build polyubiquitin chains on substrates by sequential transfers of single ubiquitins. Measurements with millisecond time resolution directly demonstrate that substrate polyubiquitylation proceeds sequentially. Our results present an unprecedented glimpse into the mechanism of RING ubiquitin ligases and illuminate the quantitative parameters that underlie the rate and pattern of ubiquitin chain assembly.
Attachment of a polyubiquitin chain with at least four ubiquitins linked together through their lysine 48 residue (Lys48) targets proteins to the proteasome for degradation.1 A cascade of three enzymes carries out the synthesis of polyubiquitin chains: a ubiquitin activating enzyme (E1), a ubiquitin conjugating enzyme (E2), and a ubiquitin ligase (E3).2 RING (really interesting new gene) E3s catalyze the direct transfer of ubiquitin from an E2 to a lysine on a target protein.3 SCFCdc4 is the founding member of the largest family of E3s—the cullin-RING ubiquitin ligases (CRLs) that may comprise the majority of all human ubiquitin ligases.3 Thus, unraveling the mechanism of SCF will have broad functional ramifications for the preponderance of human E3s.
Different pathways for ubiquitin chain assembly by RING E3s have been envisioned based on indirect evidence. On the one hand, Cdc34-SCF ubiquitylates substrates bearing a single ubiquitin significantly faster than non-ubiquitylated substrates,4,5 suggesting that it processively builds polyubiquitin chains on substrates with an initial slow transfer of ubiquitin followed by rapid elongation into a Lys48-linked polyubiquitin chain. On the other hand, the E2 Ube2g2, a close relative of Cdc34, collaborates with the E3 gp78 to build a polyubiquitin chain on its active site cysteine that can be transferred en bloc to substrate.6,7 Various permutations of the en bloc mechanism have been entertained, in which the chain is built either from proximal to distal end or vice versa.8,9,10 Due to the rapid speed of ubiquitin chain synthesis, intermediates that would reveal the underlying pathway cannot be kinetically resolved. Thus, it has not been possible to establish definitively the pathway of chain assembly for any RING E3. Here we introduce new theoretical and experimental methodologies to address both limitations.
Quantitative analysis of product distribution
Processivity emerges from the relationships between reaction and dissociation rates for different product intermediates.11 To quantify the processivity of SCF, we established an assay capable of simultaneously monitoring the concentrations of substrate and its different ubiquitylated product intermediates. Our assay consisted of an engineered phosphopeptide substrate derived from human Cyclin E1 (CycE) and purified Saccharomyces cerevisiae Cdc34-SCFCdc4.4,5,12 CycE was selected because it is a defined, chemically homogeneous substrate that binds with high affinity to the substrate-binding pocket of SCFCdc4.12,13 Moreover, intact Cyclin E is a substrate of SCFCdc4 in vivo14 and the degron from CycE can support turnover in vivo of an engineered substrate, Sic1, from which the endogenous degrons have been eliminated.12 To examine the simplest system that recapitulated the processive behavior of Cdc34-SCFCdc4, we focused on single turnover reaction conditions containing an excess of SCFCdc4 over radiolabeled CycE. We initiated reactions by combining two pre-incubated mixtures: the ‘charged E2’ mixture containing ubiquitin, E1, ATP, and Cdc34 was pre-incubated for two minutes to ensure the formation of saturating concentrations of Cdc34~ubiquitin thioesters (E2~Ub), and the ‘substrate-ligase’ mixture containing SCF and radiolabeled substrate was pre-incubated to ensure the formation of an enzyme-substrate complex. To maximize resolution of ubiquitin conjugates, the reaction products were fractionated on long SDS-polyacrylamide gels. Consistent with previous assays performed with Sic1,5 conjugation of the Nedd8 homologue Rub1 to the Cdc53 subunit of budding yeast SCFCdc4 did not alter the ubiquitylation kinetics of CycE (Supplementary Fig. 1), and thus all subsequent SCFCdc4 assays were performed with unmodified E3.
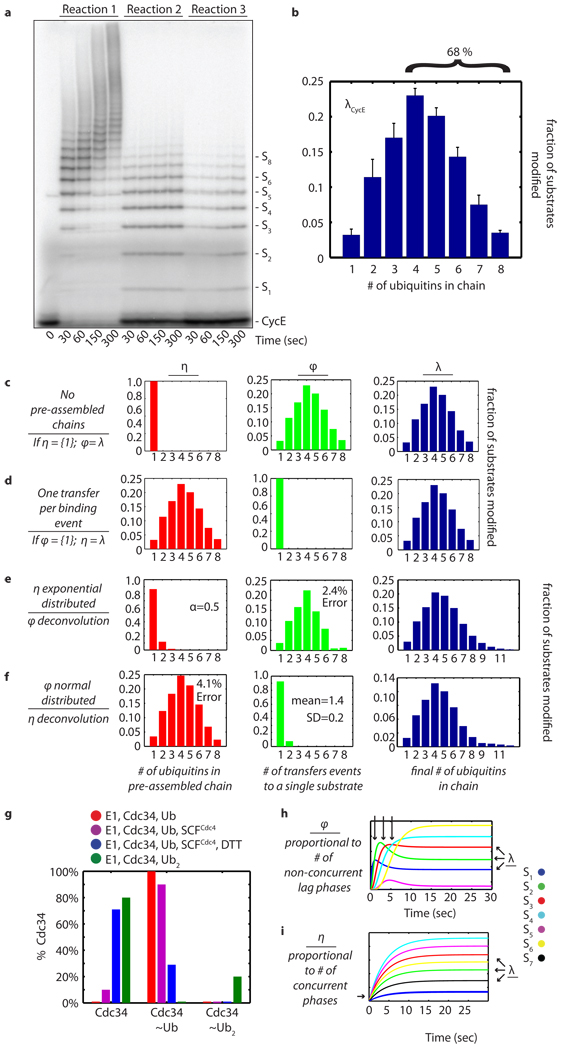
Under these reaction conditions, CycE was extensively polyubiquitylated by Cdc34-SCFCdc4 within 30 seconds (reaction 1, Fig. 1a), and products containing ≥6 ubiquitins were visible within 10 seconds (Supplementary Fig. 1). Thus, with the time resolution offered by manual mixing it was not apparent whether ubiquitin conjugates were formed by multiple sequential transfers of monoubiquitin or by en-bloc transfer of pre-formed chains. However, we reasoned that quantitative analysis of the length distribution of polyubiquitin chains attached to CycE during a single encounter with SCF might provide clues to the pathway of chain assembly. To determine the length distribution we carried out reactions with 1,000-fold excess chase of the unlabeled substrate peptide added to the ‘charged E2’ mixture (reaction 2, Fig. 1a). Under these conditions, radiolabeled substrate pre-bound to SCF was rapidly ubiquitylated, but upon dissociation further ubiquitylation occurred at a significantly reduced rate due to competition from the chase peptide. To evaluate the effectiveness of the chase, we carried out a parallel reaction in which the chase peptide was added to the ‘substrate-ligase’ mixture prior to initiation (reaction 3, Fig. 1a). The distribution of products in reaction 3 was subtracted from the distribution of products in reaction 2 at each time point (Supplementary Fig. 2) to yield the average distribution for substrate, λ (Fig. 1b). Three main points were highlighted by these experiments. First, it is evident from reaction 2 that the single encounter reaction was complete within 30 seconds. Second, 72% of CycE encounters with SCFCdc4 resulted in no ubiquitin modification (Fig. 1a and Supplementary Fig. 2). Third, of those substrates that were modified, 68% of CycE acquired a polyubiquitin chain with 4 or more ubiquitins (Fig. 1b).
Figure 1. Final product distribution for SCFCdc4 and CycE.
a, In reaction 1, pre-incubated 32P-labeled CycE and SCFCdc4 were added to the charged E2 mix. In reactions 2 and 3, excess unlabeled CycE was pre-incubated with charged E2 mix and labeled CycE, respectively. b, The single encounter polyubiquitin chain length distribution, λCycE. Error bars: +/− SD, n=3. c, If η(1)=100%, then φ=λ. d, If φ(1)=100%, then η=λ. e, Deconvolution of λCycE and exponentially distributed η. f, Deconvolution of λCycE and normal distributed φ. g, Mass spectrometry of Cdc34 thioesterified for 2’ with indicated components. h, Simulated kinetics η(1)=100%. i, Simulated kinetics φ(1)=100%.
We next sought to develop a quantitative framework to address whether the experimentally determined product distribution λCycE (Fig. 1b) places constraints on the potential pathways of ubiquitin chain assembly. We considered three hypothetical situations. First, we imagined that only monoubiquitin was attached in each transfer event (Fig. 1c, ‘sequential’). Binning all of the transfer events per substrate gave the transfer distribution φ, which in this case would equal λ. Second, we imagined the other extreme in which only one transfer event occurs per substrate (Fig. 1d, ‘en-bloc’). In this case, λ would be equal to the distribution of pre-assembled polyubiquitin chains thioesterified to E2, which we named η. Third, we considered permutations that combined sequential and en bloc transfers. For example, there are eight possible ways of making substrate modified with four ubiquitins (Sn, where n=4), including two transfers of diubiquitin or transfer of monoubiquitin followed by transfer of triubiquitin, etc. From this analysis, a key point emerged: regardless of the type of distribution we started with, the family of η and φ distributions compatible with λCycE (see Supplementary Methods) was restricted to extreme cases where either η or φ was nearly equal to λCycE (Fig. 1e, f and Supplementary Fig. 3–7). Therefore, the vast majority of substrates either underwent one transfer per binding event or received a single ubiquitin per transfer event. Thus, accurately measuring product distribution constrained the number of possible pathways that could give rise to the reaction products we observed.
As a first test of whether ubiquitins were transferred all at once or sequentially, we measured the distribution of polyubiquitin chain lengths present on the active site of Cdc34 in the presence or absence of SCFCdc4 by intact mass spectrometry. Cdc34 subjected to our standard ‘charged E2’ pre-incubation was completely converted to thioesters carrying a single ubiquitin (Cdc34~Ub; Fig. 1g and Supplementary Fig. 10). In the presence of SCFCdc4, 89% of Cdc34 was detected as Cdc34~Ub and 11% was unmodified; no Cdc34 species with more than one ubiquitin attached was detected. A control experiment run with diubiquitin confirmed that our assay was able to detect diubiquitin chains thioesterified to the active site of Cdc34 (Cdc34~Ub2; Supplementary Fig. 11), but charging of Cdc34 with diubiquitin occurs with poor efficiency (~20%). Thus, our analysis of the product distribution λ coupled with measurement of the ubiquitin population thioesterified to Cdc34 under our reaction conditions (an estimate of η) strongly predicts that Cdc34–SCFCdc4 assembled ubiquitin chains on substrate primarily by sequential transfers of single ubiquitin molecules.
Millisecond kinetics of SCF
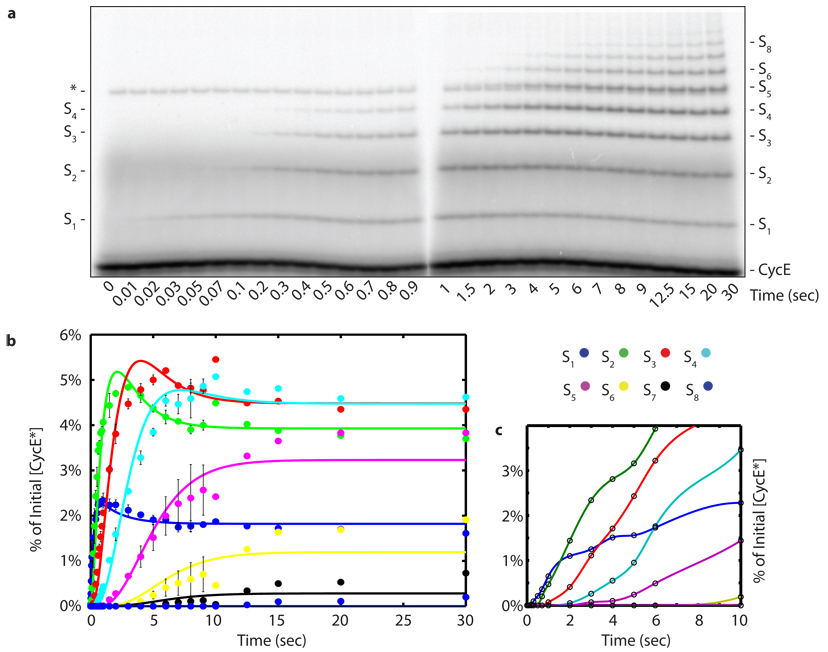
As a second, more definitive test of the hypothesis stated above, we sought to measure directly how the product distribution (Fig. 1b) developed as a function of time. During a single encounter between a RING ubiquitin ligase and substrate, each intermediate should either undergo a transfer event or dissociate. If monoubiquitin composed 100% of η as in Fig. 1c, the products of the reaction should appear sequentially in time starting with S1 and followed by S2, then S3, etc. Thus, the appearance of each sequential product should be delayed by a ‘lag’ phase (Fig. 1h). In contrast, if a single transfer composed 100% of φ as in Fig. 1d, then the pattern of ubiquitin chains attached to substrate at the earliest time-points should reveal the distribution of pre-assembled chains thioesterified to Cdc34 (Fig. 1i). Thus, products of increasing mass should accumulate sequentially if chain synthesis is sequential, but should accumulate contemporaneously if chains are transferred en bloc. Therefore, with sufficient time resolution a single encounter experiment would provide definitive data to distinguish between the alternative models. To achieve the necessary time resolution, we performed our single encounter reactions on a quench flow apparatus that allowed us to take measurements on a time scale ranging from 10 milliseconds to 30 seconds (Fig. 2a). To facilitate quantification of S2 and S5 in the CycE reaction the same reaction from Fig. 2a was fractionated on a gel with different resolving capabilities (Supplementary Fig. 12). Three major conclusions arose from these experiments. First, the product CycE–Ub (S1) was formed starting at the earliest time points (10–20 milliseconds) without a lag phase, indicating that E2~Ub binding to SCF was rapid. This is consistent with stopped-flow measurements carried out with SCFβ-TrCP and hCdc34.15 Second, each new ubiquitylated product appeared sequentially with non-concurrent lag phases (Fig. 2a, b and Supplementary Fig. 12). Third, the early reaction products S1–S3 ‘overshot’ their final levels indicating that these reaction intermediates serve as templates for the formation of subsequent products, supporting the model that polyubiquitin chains are built from multiple transfer events (Supplementary Fig. 16). Combined with the constraints on η and φ calculated above as well as our direct evaluation of the Cdc34~Ub pool (Fig. 1g), these data demonstrate that the underlying kinetic mechanism of our system was principally derived from sequential transfers of single ubiquitins.
Figure 2. Millisecond kinetics of a single encounter reaction reveal sequential processivity.
a, To achieve millisecond temporal resolution CycE reactions were performed on a quench flow apparatus and products were evaluated by SDS-PAGE and phosphorimaging. The reaction scheme matched reaction 2 of Fig. 1a. The asterisk marks a contaminant. Sn refers to CycE modified with n ubiquitins. b, Quantification shows successively longer lag phases for each additional ubiquitin added in the chain. The data was fit using closed form solutions refined by global regression analysis to a model with η=1. The error bars represent the range of values, n=2.
To ensure that our conclusions were not an artifact of the reaction design, we changed the order of addition in our reactions. SCFCdc4 was pre-incubated with the ‘charged E2’ mixture for 2 minutes (in which case 89% of Cdc34 is present in thioesterified form; Fig 1g) and reactions were initiated by combining with radiolabeled CycE. Products appeared following non-concurrent lag phases of increasing duration (Fig 2c), analogous to that observed when the reaction was initiated by addition of Cdc34~Ub to CycE prebound to SCFCdc4 (Fig 2a). Thus, regardless of whether CycE first encountered Cdc34~Ub–SCF or Cdc34~Ub encountered CycE–SCF, single ubiquitins were transferred to substrate in a sequential manner. Interestingly, reactions initiated by addition of CycE were delayed compared with those initiated by addition of Cdc34~Ub, indicating that Cdc34~Ub productively associates with SCFCdc4 faster than does CycE.
SCFβ-TrCP is sequentially processive
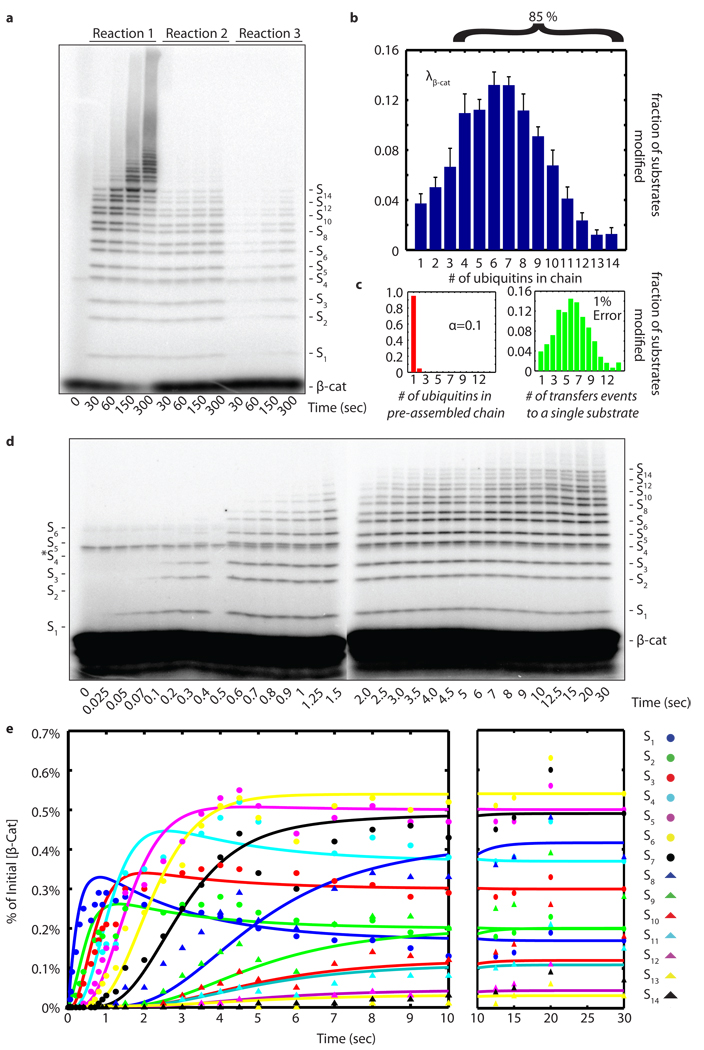
We next sought to test whether the sequential processive chain assembly we observed for SCFCdc4 is unique or illuminates a general principle of SCF ubiquitin ligase mechanism. To address this issue, we evaluated ubiquitylation of a phosphopeptide derived from β-Catenin (β-Cat) by its cognate E2–E3 complex, hCdc34 and human SCFβ-TrCP. Nedd8 conjugated E3 (N8-SCFβ-TrCP) was used for these experiments, because prior work demonstrated a potent stimulation of β-Cat ubiquitylation upon Nedd8 conjugation.4 As was seen with CycE–SCFCdc4, β-Cat was rapidly modified by N8-SCFβ-TrCP and it was not possible to resolve intermediates in chain assembly by manual mixing4 (Fig. 3a). Quantification of product distribution λβ-Cat revealed that 6% of β-Cat molecules were modified in a single encounter with N8-SCFβ-TrCP, of which 85% received ≥4 ubiquitins (Fig. 3b and Supplementary Fig. 2). Distribution analysis of λβ-Cat (Fig. 3c) and kinetic resolution of β-Cat ubiquitylation by quench-flow (Fig. 3d, e) revealed sequential appearance of intermediates analogous to those observed with CycE ubiquitylation by SCFCdc4.
Figure 3. Human Cdc34-SCFβ-TrCP is sequentially processive.
a, same as Fig. 1a, except that human Cdc34 and Nedd8-conjugated SCFβ-TrCP were assayed with 32P-labeled β-Cat substrate. b, Product distribution (λβ-Cat) was quantified as in Fig. 1b. Error bars: +/− SD, n=3. c, The poisson distribution of φ using λβ-Cat that deviated the most from φ(1)=100% within our set error bounds with α=0.2. d, β-Cat reactions with the scheme of reaction 2 (Fig 1a) performed on a quench flow apparatus. e, Quantification shows successively lengthening lag phases for each additional ubiquitin added in the chain. The data was fit as in Fig. 2b.
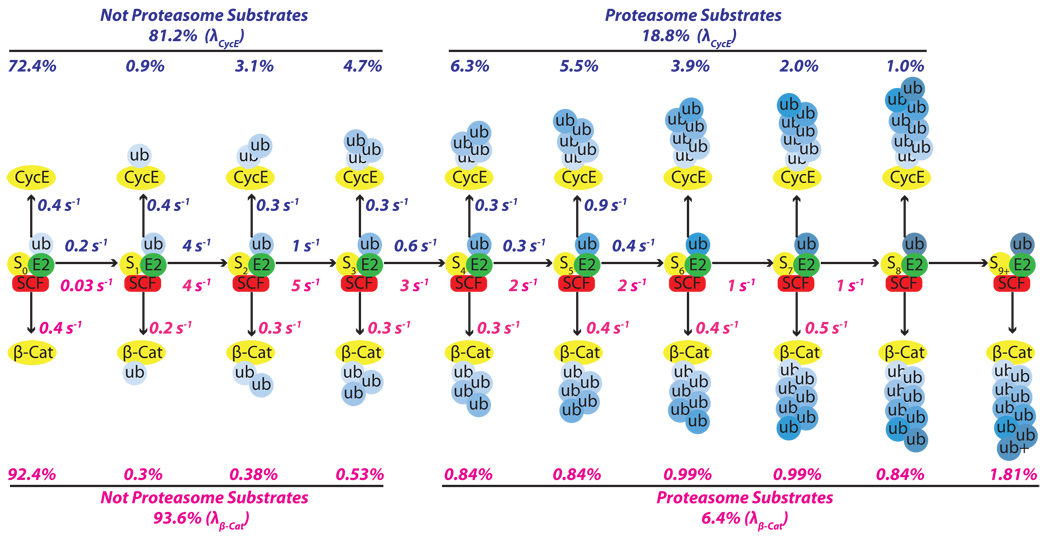
Although the general behavior of SCFCdc4 and N8-SCFβ-TrCP were similar, the enzymes differed in the extent to which they converted bound substrate to product and elongated ubiquitin chains. Using a kinetic model in which monoubiquitin composed 100% of η, we were able to employ methods borrowed from the study of nucleic acid polymerases16 to extrapolate estimates for the individual reaction and dissociation rate constants from our single encounter quench-flow experiments (Fig. 4, Supplementary Fig. 13–15).
Figure 4. Kinetic basis for Cdc34-SCF processivity.
The millisecond kinetics of a single encounter reaction were fit to a sequential model revealing estimates for individual transfer and dissociation rates for each intermediate in the generation of polyubiquitylated CycE (blue numbers) and β-Cat (red numbers) products. The percentages listed above or below each product were the percentages from the final product distributions (λ) shown in Fig. 1b and Fig. 3b.
Functional implications of our model
The model shown in Fig. 4 reveals the kinetic basis of processive polyubiquitin chain synthesis by budding yeast Cdc34-SCFCdc4 and human Cdc34-SCFβ-TrCP and accounts for the differences in their behavior. Most encounters of substrate and SCF are unproductive because koff is faster than kUb1. This is particularly exaggerated for β-Cat owing to its low value for kUb1. Once a single ubiquitin is attached, the majority of substrates are committed to polyubiquitylation due to the drastic increase in kUb2 relative to a nearly constant koff. This gives rise to the high percentage of modified substrates with four or more ubiquitins in their chain (68% for CycE and 85% for β-Cat). The overall chain length is limited by the progressive decrease in transfer rates (kUbn) as the chain becomes longer matched against the relatively constant rate at which product intermediates dissociate. This reduction in transfer rate most likely arises because the distal end of the flexible chain samples a progressively larger volume as it increases in length17. The longer chains on β-Cat are a result of a less dramatic decline in kUbn after the second ubiquitin is attached. We do not understand the basis for this difference. Meanwhile, the constant rate of dissociation for both CycE and β-Cat implies that ubiquitin chains of increasing length do not change the intrinsic affinity of these substrates for SCF.
Casual inspection of our model suggests that modest changes in the ratio kUb1/koff for the first step would substantially alter the fraction of substrate that acquires a chain of ≥4 ubiquitins in a single encounter with SCF. This in turn provides a simple basis for SCF to modulate a substrate’s degradation half-life (i.e. the larger koff is or smaller kUb1 is, the lower the probability that a substrate is modified in a single encounter with SCF, which would translate to a longer half-life). Comparison of CycE and β-Cat, which have distinct kUb1/koff ratios, underscores how the efficiency and pattern of substrate ubiquitylation can be tuned by these parameters. Despite these differences, it is remarkable how similar the reaction parameters are for two different enzymes from organisms separated by over 1 billion years of evolution. In both cases koff was ~0.4 sec−1 and the fastest rate of ubiquitin chain elongation was 4–5 sec−1. This suggests that true substrates are tuned to dissociate within a few seconds and that a transfer rate of 5 sec−1 may be imposed by a conserved rate-limiting step. It will be of great interest to determine what molecular event enforces this speed limit.
We conclude that polyubiquitin chains are built on SCF substrates by sequential transfers of single ubiquitins. We establish a mechanistic framework that can be applied to other CRLs and RING ubiquitin ligases to obtain individual rate constants for substrate dissociation and ubiquitin transfer at each step in the process of chain assembly. Our model indicates that the processivity, efficiency, and pattern of ubiquitylation is governed by the sharp discontinuity in rates between the first transfer and subsequent transfers, contrasted with the shared dissociation rate among substrate and product intermediates.
Methods Summary
Proteins
CycE and β-Cat phosphopeptide were purchased from New England Peptide. Ubiquitin and K48 diubiquitin were purchased from Boston Biochem. Uba1 and SCFCdc4 were prepared and purified as described.5 Full length yeast Cdc34 was purified as described.18 His7-Rub1 was purified from E. coli inclusion bodies4 and human E1, UbcH3B (hCdc34), and Nedd8-SCFβ-TrCP were prepared and purified as described.4 Yeast Ubc12 and Ula1–Uba3 were purified as described.19 Rub1, Ubc12, Ula1–Uba3, and ATP were incubated with immobilized SCFCdc4 to make Rub1-conjugated SCFCdc4. PKA was purchased from New England Biolabs.
Ubiquitylation assay
CycE (200 nM) or β-Cat (2 µM) was incubated with γ-[32P]-ATP (132 nM) and PKA for 45 minutes at 30°C to make radiolabeled CycE or β-Cat. Yeast ubiquitylation reactions contained ATP (2mM), ubiquitin (60 µM), Uba1 (0.8 µM), Cdc34 (10 µM), SCFCdc4 (150 nM), and radiolabeled CycE (10 nM). Human ubiquitylation reactions contained ATP (2mM), ubiquitin (60 µM), E1 (1 µM), Cdc34 (10 µM), SCFβ-TrCP (500 nM), and radiolabeled β-Cat (100 nM). As indicated, single encounter reactions contained an unlabeled CycE chase (10 µM) or β-Cat chase (100 µM). Millisecond reactions were performed on a quench flow apparatus (Kintek RQF-3 Rapid Quench Flow). Reactions contained a buffer previously described20 at 23°C. Reactions were quenched with SDS-PAGE buffer with βME and run on 20 cm 5–20% Tricine gels (CycE) or Glycine gels (β-Cat) that were quantified with a phosphor screen (Molecular Devices). Thioester formation assays contained Cdc34 (10 µM), Uba1 (1 µM), ATP (2mM), ubiquitin or K48 diubiquitin (15 µM), and SCFCdc4 (100 nM) as indicated. After 2 minutes, reactions were stopped with excess 5% acetic acid and analyzed on an Agilent LC-MSD.
Analysis
Deconvolutions and regression were performed in Matlab. Global fitting was performed with KinTek Global Kinetic Explorer. Mass spec data was processed using the Chemstation software package.
Supplementary Material
Acknowledgements
We thank J. Vielmetter of the Caltech Protein Expression Facility for providing SCFCdc4, β-TrCP-Skp1, and human E1. S. Hess, R.L.J. Graham, and the Proteome Exploration Laboratory for providing assistance with mass spec of CycE and Cdc34 thioester. We thank S. Schwarz for gifts of reagents. We thank D. Sprinzak and all the members of the Deshaies and Shan lab for support and helpful discussions. N.W.P. was supported by the Gordon Ross Fellowship, and an NIH Training Grant. R.J.D. is an Investigator of the HHMI. This work was supported in part by NIH GM065997.
Footnotes
Author Contributions
N.W.P. performed all computational modeling and experiments except G.K. performed the mass spec experiments in Fig. 1g. N.W.P., R.J.D., and S.O.S. conceived the experiments. N.W.P. and R.J.D. wrote the manuscript with editorial input from the other authors.
References
- 1.Thrower JS, Hoffman L, et al. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dye BT, Schulman B. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annual review of biophysics and biomolecular structure. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 3.Petroski M, Deshaies R. Function and regulation of cullin–RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 4.Saha A, Deshaies R. Multimodal Activation of the Ubiquitin Ligase SCF by Nedd8 Conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petroski M, Deshaies R. Mechanism of Lysine 48-Linked Ubiquitin-Chain Synthesis by the Cullin-RING Ubiquitin-Ligase Complex SCF-Cdc34. Cell. 2005;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 6.Ravid T, Hochstrasser M. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat Cell Biol. 2007;9:422–427. doi: 10.1038/ncb1558. [DOI] [PubMed] [Google Scholar]
- 7.Li W, Tu D, et al. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- 8.Hochstrasser M. Lingering Mysteries of Ubiquitin-Chain Assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 9.Li W, Tu D, et al. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci USA. 2009;106:3722–3727. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 11.Fersht A. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. New York: W. H. Freeman and Company; 1999. [Google Scholar]
- 12.Nash P, Tang X, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 13.Orlicky S, Tang X, et al. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 14.Strohmaier H, Spruck CH, et al. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 15.Kleiger G, Saha A, Deshaies R. Rapid E2–E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009 doi: 10.1016/j.cell.2009.10.030. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kati WM, Johnson KA, et al. Mechanism and fidelity of HIV reverse transcriptase. J Biol Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 17.Petroski M, Kleiger G, et al. Evaluation of a Diffusion-Driven Mechanism for Substrate Ubiquitination by the SCF-Cdc34 Ubiquitin Ligase Complex. Molecular Cell. 2006;24:523–534. doi: 10.1016/j.molcel.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Feldman RM, Correll CC, et al. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 19.Kamura T, Conrad M, et al. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes & Development. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petroski M, Deshaies R. In Vitro Reconstitution of SCF Substrate Ubiquitination with Purified Proteins. Methods in Enzymology. 2005;398:143–158. doi: 10.1016/S0076-6879(05)98013-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.