Abstract
In order to understand the role of plasma proteins in the rapid liver clearance of dextran-coated superparamagnetic iron oxide (SPIO) in vivo, we analyzed the full repertoire of SPIO-binding blood proteins using novel two-dimensional differential mass spectrometry approach. The identified proteins showed specificity for surface domains of the nanoparticles: mannan-binding lectins bound to the dextran coating, histidine-rich glycoprotein and kininogen bound to the iron oxide part, and the complement lectin and contact clotting factors were secondary binders. Nanoparticle clearance studies in knockout mice suggested that these proteins, as well as several previously identified opsonins, do not play a significant role in the SPIO clearance. However, both the dextran coat and the iron oxide core remained accessible to specific probes after incubation of SPIO in plasma, suggesting that the nanoparticle surface could be available for recognition by macrophages, regardless of protein coating. These data provide guidance to rational design of bioinert, long-circulating nanoparticles.
1 Introduction
There is an increasing interest in medical applications of nanomaterials. In this regard, thorough understanding of interactions of nanomaterials with the body milieu is mandatory. When nanomaterials are injected into the blood stream, extensive interactions with plasma proteins, cells, and other blood components take place (reviewed by Moghimi [1]). Liposomes are one example of nanocarriers where such interactions have been studied in detail. Phospholipids in the outer bilayer of liposomes attract some known opsonins such as immunoglobulins and complement [2, 3], and other plasma components such as lipoproteins [4]. These events have been shown to be important for clearance of liposomes by reticuloendothelial macrophages that reside in the liver and spleen.
Dextran-coated superparamagnetic iron oxide nanoparticles (SPIO) are widely used as magnetic resonance imaging contrast agents in the clinic (e.g., Ferridex™). These particles consist of two main chemical components: crystalline iron oxide core (magnetite) and low molecular weight dextran (~10 kDa). Some types of SPIO nanoparticles have been reported to exhibit prolonged circulation times, either due to their ultrasmall size (less than 20 nm) [5] or extensive surface crosslinking and PEGylation [6, 7]. Larger SPIO (50-150 nm: Ferridex, Micromod SPIO, Ferumoxides) with unmodified dextran coating are rapidly eliminated from circulation by the liver and spleen, and therefore these particles primarily enhance MR contrast in these organs [8]. It is important to better understand the mechanisms of this rapid clearance in order to design long-circulating (stealth) SPIO.
The mechanism whereby nanoparticles and liposomes accumulate in the liver and the spleen could be related to the nature of proteins that adsorb onto the surface of systemically administered nanoparticles [9]. It has been shown that dextran-iron oxide and dextran-poly(isobutylcyanoacrylate) nanoparticles are extensively coated in plasma with known opsonins such as complement, fibronectin and fibrinogen [10, 11]. However, the significance of these interactions in the nanoparticle clearance in vivo is not known. Some previous experiments suggested that dextran-iron oxide nanoparticles could be directly recognized through a yet-to-be-defined receptor mechanism, without plasma opsonin involvement [12]. The validity of this last claim in vivo is difficult to prove or disprove, in view of the constant presence of plasma proteins in the body.
In order to shed light on the role of plasma proteins in the SPIO clearance, we analyzed the spectrum of plasma proteins that bind to the nanoparticles and examined the role of these proteins as potential nanoparticle opsonins. In order to do that we developed a method for the proteomic analysis of the nanoparticle plasma coating without washing steps. Our analysis surprisingly showed the selectivity of plasma proteome towards SPIO surface dextran and exposed iron oxide. Using knockout mice, we show that these attached plasma proteins are unlikely to play a role in the in vivo clearance of SPIO. We further demonstrate that the plasma proteins do not mask completely the surface dextran and iron oxide of the nanoparticles, suggesting that the SPIO surface could be directly recognized by macrophages. This study provides insight to the mechanisms of nanoparticle uptake and gives an incentive to further understand the nanoparticle surface properties in order to design non-toxic stealth nanoparticles.
2 Materials and Methods
2.1 Plasma protein binding to nanoparticles
Superparamagnetic dextran iron oxide nanoparticles (SPIO) from various sources were used in this study. Amino-dextran SPIO of 50nm size were obtained from Micromod GmbH, Germany, and were labeled with fluorescein isothiocyanate (Sigma) to block the amino groups and to facilitate their detection with microscope. Alternatively, SPIO were prepared by the published method (magnetic nanoworms [7]) with the exception that no crosslinking or amination steps were performed. In both types of particles, the surface charge was similar (zeta potential −4.95 mV and −0.77 mV for nanoworms and FITC-Micromod-SPIO, respectively).
Mouse plasma was obtained from freshly drawn mouse blood by cardiac puncture using either citrate or heparin as anticoagulant, and was stored at −80°C before the experiments. Two hundred μg of SPIO were incubated with 300 μl of mouse plasma, containing 10 μl of Sigma tissue protease inhibitor cocktail, for 10 min under vortexing at room temperature. The unbound proteins were extensively washed away using MACS® Midi magnetic separation column (Miltenyi Biotec) or using 4 rounds of ultracentrifugation and resuspension in PBS, and the particles were boiled in 10% SDS for 60 min. Following this, iron oxide was pelleted using Beckman TLA-100 ultracentrifuge (70,000g for 10 min) and the supernatant with the eluted proteins precipitated in 5 volumes of ice cold acetone overnight at −20° C, washed and analyzed on SDS-PAGE or submitted as a whole for LC-MS/MS. Protein concentration was measured using Bio-Rad Protein assay. For two-dimensional mass spectrometry, nanoparticles were incubated with plasma as described above, with the exception that no washing was performed following the incubation. Particles were pelleted once to the bottom at 70,000×g for 7 min, the supernatant was removed as much as possible, the pellet volume was measured by pipette (usually <10 μl), and the pellet was incubated in 50 μl of 8M Urea/1M NaCl/1M imidazole at 80°C for 30 min. The particles were pelleted again and the supernatant was submitted as is for 2D mass spectrometry. The control was exactly the same volume of plasma, processed in exactly the same way.
For immunoblotting, the following antibodies were used: goat polyclonal anti-mouse kallikrein and goat polyclonal anti-mouse HPRG (R&D Systems), rat monoclonal anti-mouse MBL-A (Abcam) and rabbit polyclonal anti-mouse HMWK (generated by the laboratory of Dr. McCrae).
2.2 Protein identification and analysis using mass spectroscopy
The full details of sample preparation, the instrumental setup, data collection and analysis are provided in the Supplemental Methods section. Briefly, for protein identification using one-dimensional D LC/MS/MS, 5 μg of the protein were enzymatically cleaved by trypsin and the automated Nano LC LTQ MS/MS (Thermo Scientific, Waltham, MA) was performed as described by Salvesen et al [13]. For large-scale protein identification using two-dimensional proteomics, 400 μg mouse plasma proteins were digested with trypsin as described in Supplemental Methods, separated on SCX (strong cation exchange) column and each fraction (24 total) was subsequently analyzed on LTQ-Orbitrap.
The MS/MS spectra were analyzed by Sorcerer 2 (Sage-N Research Inc.) with SEQUEST (v.27, rev. 11) as the search program for peptide/protein identification. The relative abundance of each identified protein in different samples were analyzed by QTools, our in-house developed open source tool for automated differential peptide/protein spectral counting analysis (http://sr.burnham.org/sr/homepage/proteomics/links.html).
2.3 Proteomic data filtering
At present there is no ideal method to statistically analyze the differential proteomic data from few technical replicates. Our semi-arbitrary filtering approach was based on the recent publication by Nesvizhskii and coworkers [14]. The protein hits in each replicate after the spectral counting were filtered so that only enrichment ratios above 1.5 were included in the analysis. To increase the sensitivity of identification to >80% , the threshold of spectral counts for 1.5-2.0-fold enrichment was set to >20, and 2.0-4.0-fold enrichment was set to >10. For > 4-fold enrichment, the minimal threshold of spectral counts for each of the enriched protein was set to 5. Such filtering allowed to reduce the number of hits to a manageable list of about 50 proteins. Then, the enriched hits shared by both replicates were extracted to combine a common list of 24 proteins. The datasheets with the non-filtered and semi-filtered data are provided in the Supplement.
2.4 Probing the chemical domains on the nanoparticle surface
To quantify the binding of FITC-labeled anti-dextran antibody (Stem Cell Research), 50 μl of 1 μg/ml iron oxide was incubated with 10 μl of the antibody in presence of 50 μl PBS or heparinized mouse plasma. After 10 min incubation at RT, the particles were pelleted at 70,000×g and the percent of the Ab remaining in the supernatant was measured by fluorescence, using same concentration of the Ab in either PBS or 50% plasma, but without SPIO, as 100%. Because of the absence of information on the concentration of the antibody, the difference between the control and the sample supernatant fluorescence was used to calculate the relative binding.
In order to determine the degree of exposure of iron on the nanoparticle surface, 10 μg of histidine-rich D5 domain of high molecular weight kininogen (D5-GST fusion, from the laboratory of Dr. McCrae) was immobilized on 100 μl of glutathione agarose beads (Amersham). Ten micrograms of iron oxide were then preincubated with 50 μl PBS or mouse plasma for 10 min and then added to the beads. The slurry was incubated for another 10 min and washed 3 times with 1 ml PBS. The bound iron was measured using QuantiChrom Iron Assay (BioAssay Systems) as described in the instructions for the kit.
2.5 Mouse experiments
All the animal work was reviewed and approved by Burnham Institute’s Animal Research Committee or by institutional committees where the knockout experiments were performed. The list of the sources of knockout mouse strains is provided in the Supplemental Methods. Normal C57BL/6J mice were used as controls and were gender and age matched with the corresponding knockout mice.
The animals were anesthetized with Avertin, and nanoparticles (4 mg Fe/kg body weight) were injected into the tail vein in a total volume of up to 150 μl. Blood was collected from the periorbital vein by heparinized capillaries at different time points, and plasma was separated from cells by centrifugation at 5000 rpm for 2 min. Ten microliters of plasma were collected to measure iron levels by QuantiChrom Assay. When fluorescently labeled nanoparticles were used, plasma was diluted 100 times and FITC fluorescence was measured using LS-50B spectrofluorimeter (Perkin-Elmer) or FUJI FLA-5100 scanner. The data were fitted into mono-exponential decay curve using Prism 4 (GraphPad Software) to calculate half-lives in plasma. For histological analysis of the nanoparticle uptake, the animals were sacrificed 3 h post-injection by cervical dislodgement under anesthesia, and livers were dissected, fixed in formalin, cryosectioned and analyzed by fluorescent microscope.
2.6 In vitro experiments using isolated Kupffer cells
Kupffer cells were isolated from collagenase-perfused mouse liver by a differential centrifugation method [15]. Briefly, anesthetized mouse was perfused through the heart with 1% BSA/RPMI, followed by 7 ml of Sigma collagenase IV solution (0.5 mg/ml in 1% BSA/RPMI) through hepatic portal vein. The liver was then excised, minced and incubated in collagenase solution at 37°C for 15 minutes. The hepatocytes were depleted by centrifugations at 30×g for 5 minutes three times. The resulting non-parenchymal cells, which are about 70% Kupffer cells as verified by F4/80 immunostaining, was washed three times and used for further experiments.
To test the nanoparticle uptake by cells, 20 μg SPIO were incubated with 100 μl of 1% BSA/PBS, whole plasma, or SPIO opsonin-depleted plasma (by preincubation with 200 μg SPIO and pelleting the particles using ultracentrifuge). Then, 1×106 Kupffer cells were incubated at 37°C for 1h with SPIO. All the samples were supplemented with 4 mM Ca++ and, for inhibition of coagulation of plasma, 10 μM PPACK (Calbiochem) prior to mixing with SPIO. By the end of the incubation, the cells were washed in BSA/PBS 3 times, lysed with 10% SDS and iron oxide uptake was quantified as described above. Alternatively, after the incubation with nanoparticles the cells were seeded in slide chambers (NalgeNunc) to study the SPIO uptake by microscopy.
3 Results
3.1 Proteomic analysis of opsonization of SPIO
To identify SPIO-binding proteins, we initially incubated the nanoparticles with mouse plasma, removed unbound proteins by extensive washing, and then eluted the bound proteins (Fig. 1A). Under these conditions, the nanoparticles absorbed as much as 1 mg protein per mg iron oxide. Gel analysis showed significant enrichment of the nanoparticles with plasma proteins (Fig. 1B). It should be noted that the protein binding profiles were identical for all types of SPIO nanoparticles, whether they were from commercial source or prepared in our laboratory.
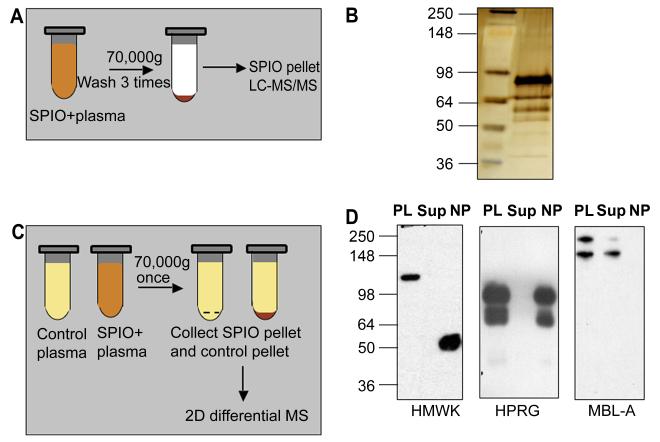
Figure 1. Protein binding and effect of plasma proteins on SPIO clearance and macrophage uptake.
A, Schematic representation of one-dimensional proteomic analysis of plasma proteins bound to SPIO. B, Silver stained SDS-PAGE. Proteins were eluted from SPIO using the above procedure and analyzed by LC-MS/MS (Table 1). Left lane, SeeBlue Plus2 standard size marker (Invitrogen); right lane, eluted proteins. C, Schematic representation of two-dimensional differential proteomics procedure used to identify plasma proteins that bind to SPIO. See Methods for complete description of the procedure. D, Depletion of some of the binding proteins from plasma after incubation with SPIO. The particles were incubated with plasma as described in Methods, pelleted using ultracentrifuge and the supernatant was analyzed by western blotting. Each gel included plasma proteins before incubation with SPIO (PL), supernatant after pelleting the particles (Sup) and the particles (NP) washed by additional 4 rounds of ultracentrifugation with PBS. HMWK undergoes cleavage on the particle surface (uncleaved size 120 kDa) to form activated kininogen (HKa). To detect MBL-A, the proteins were separated and detected under non-reducing conditions. Note that MBL-A was not detectable on SPIO, while it was depleted from plasma.
Mass spectrometric identification of these eluted proteins from one such experiment is shown in Table 1 (for complete proteomic data, see Supplementary Table 1). The most significant and reproducible hits were histidine-proline rich glycoprotein (HPRG), high molecular weight kininogen (HMWK), and plasma prekallikrein (KLK). The enrichment of these proteins on the particles was striking, considering their relatively low abundance in plasma.
Table 1. Significant plasma proteins eluted from SPIO and identified by 1D LC-MS.
Each protein entry shows the IPI (International Protein Index) number, link to the EMBL-EBI entry, and total peptide spectral counts. The particles were incubated with plasma, washed extensively with PBS and the tightly bound proteins were eluted as described in Methods. Additional proteomic data such as the peptides identified, and peptide and protein probabilities are provided in the Supplemental Table 1. The experiment was repeated 3 times.
| IPI # | Spectral count |
Protein identified | Function |
|---|---|---|---|
| IPI00177214 | 2 | IMMUNOGLOBULIN | Immunoglobulin |
| IPI00131695 | 10 | SERUM ALBUMIN. | Carrier protein |
| IPI00320239 | 14 | TETRANECTIN. | Lectin-like, sugar-binding |
| IPI00113057 | 24 | PLASMA KALLIKREIN. | Serine protease, intrinsic clotting activator |
| IPI00114958 | 113 | ISOFORM HMW OF KININOGEN-1. |
Surface-binding protein |
| IPI00322304 | 120 | HISTIDINE-PROLINE RICH GLYCOPROTEIN (HPRG). |
Adaptor protein |
Fibronectin, vitronectin and C3 complement have been shown to bind to SPIO [11]. However, C3 and fibronectin were not reproducibly present on plasma-treated SPIO in our mass spectrometry experiments. Also, immunoglobulin detection in the eluted proteins was not reproducible from experiment to experiment (not shown).
Minor variations in incubation and washing conditions could lead to carryover of non-bound proteins in the SPIO binding assay we used. Alternatively, rigorous washing could remove weakly interacting proteins, as has been observed with other types of nanoparticles [16]. In order to identify all the bound proteins we decided to employ a procedure that eliminates the bias of the washing steps (Fig. 1C, Methods). We incubated SPIO in plasma, quickly pelleted the particles in an ultracentrifuge, and analyzed the proteins enriched in the SPIO pellet. Because the proteins were present at their native concentration without washing steps to dilute them throughout the experiment, the weakly bound proteins presumably stayed bound to the particles. Because of the high complexity of the samples, we used a two-dimensional chromatographic separation in order to resolve the mixture [17]. Differential counting software was used to compare peptide intensities between the SPIO and control plasma samples. According to Table 2 and Supplementary Table 2, many more significant proteins were enriched on the nanoparticles, compared to the previous analysis. This process greatly enriched some of the proteins that were otherwise undetectable when the particles were extensively washed. There was a significant enrichment in the nanoparticle pellet of mannose-binding lectins (MBLs), MBL-associated serine proteases (MASPs), apolipoproteins, beta-2 glycoprotein and clotting factors FXI and FXII. The enrichment was more then 7-fold for the mannose-binding lectins A and C.
Table 2. Proteins enriched on SPIO as identified by 2D differential MS.
Each protein entry shows the IPI number, link to the EMBL-EBI entry, total peptide spectral counts and enrichment ratio. Only significantly enriched hits (enrichment ratio >1.5 or unique) are shown. Full proteomic data such as the peptides identified, peptide and protein probabilities and unfiltered data are provided in the Supplement. The experiment was done in duplicate.
| IPI# | Identity | control rep 1 | enrich rep 1 | control rep 2 | enrich rep 2 | enrichment rep 1 |
enrichment rep 2 |
|---|---|---|---|---|---|---|---|
| IPI00350772 | APOLIPOPROTEIN B PRECURSOR. | 36 | 55 | 35.0 | 88 | 1.5 | 2.5 |
| IPI00330843 | COAGULATION FACTOR XII. | 26 | 42 | 27.0 | 46 | 1.6 | 1.7 |
| IPI00114958 | ISOFORM HMW OF KININOGEN-1. | 26 | 45 | 39.0 | 87 | 1.7 | 2.2 |
| IPI00129755 | ALPHA-1-ANTITRYPSIN 1-2. | 16 | 29 | 16.0 | 49 | 1.8 | 3.1 |
| IPI00113057 | PLASMA KALLIKREIN. | 20 | 51 | 16.0 | 61 | 2.6 | 3.8 |
| IPI00135560 | PHOSPHOLIPID TRANSFER PROTEIN. | 3 | 8 | 3.0 | 11 | 2.7 | 3.7 |
| IPI00283862 | PROTEASOME SUBUNIT ALPHA TYPE-1. | 4 | 8 | 0.0 | 5 | 2.0 | “unique” |
| IPI00136556 | TRANSCOBALAMIN-2. | 4 | 8 | 4.0 | 11 | 2.0 | 2.8 |
| IPI00322463 | BETA-2-GLYCOPROTEIN 1. | 29 | 59 | 54 | 80 | 2.0 | 1.5 |
| IPI00322304 | HISTIDINE-RICH GLYCOPROTEIN HRG. | 48 | 245 | 39 | 247 | 5.1 | 6.3 |
| IPI00113457 | MANNOSE-BINDING PROTEIN C. | 23 | 155 | 20 | 141 | 6.7 | 7.1 |
| IPI00467068 | ISOFORM 1 OF MANNAN-BINDING LECTIN SERINE PROTEASE 2. |
6 | 41 | 11 | 51 | 6.8 | 4.6 |
| IPI00652394 | ISOFORM D OF PROTEOGLYCAN 4. | 0 | 14 | 1 | 19 | “unique” | 19.0 |
| IPI00475209 | MANNAN-BINDING LECTIN SERINE PEPTIDASE 1, ISOFORM CRA_B. |
0 | 13 | 5 | 13 | “unique” | 2.6 |
| IPI00348094 | TUBULIN, BETA 1. | 0 | 11 | 0 | 8 | “unique” | “unique” |
| IPI00118413 | THROMBOSPONDIN 1. | 0 | 10 | 0 | 15 | “unique” | “unique” |
| IPI00405227 | VINCULIN. | 0 | 9 | 0 | 14 | “unique” | “unique” |
| IPI00169543 | ZYMOGEN GRANULE MEMBRANE PROTEIN 16. | 0 | 8 | 0 | 9 | “unique” | “unique” |
| IPI00131398 | MANNOSE-BINDING PROTEIN A. | 0 | 8 | 0 | 7 | “unique” | “unique” |
| IPI00109061 | TUBULIN BETA-2B CHAIN. | 0 | 5 | 0 | 9 | “unique” | “unique” |
| IPI00330594 | C-TYPE LECTIN DOMAIN FAMILY 4 MEMBER F. | 0 | 5 | 0 | 8 | “unique” | “unique” |
| IPI00465786 | TALIN-1. | 0 | 5 | 0 | 6 | “unique” | “unique” |
| IPI00119019 | COAGULATION FACTOR XI. | 0 | 5 | 0 | 5 | “unique” | “unique” |
There was also some enrichment (1.2-1.5-fold) with downstream complement factors such as C1q, C2, C5, clotting factors FIX, FX, and immunoglobulins (Supplemental Table 2) but there was not enough statistical power in two replicates in order to consider these proteins significantly enriched.
Hemoglobin and hemoglobin-binding hemopexin (proteoglycan-4) were also found to be enriched on the SPIO (Supplemental Table 2), probably as a result of mildly hemolytic plasma, but the significance of this enrichment in vivo is not clear. Some cellular proteins such as vinculin, tubulin-1 and talin-1were also enriched, possibly as a consequence of contamination of plasma with cell debris, but it is not clear to what extent this phenomenon happens in the circulation.
In order to confirm that some of the enriched proteins bind to SPIO, we performed western blotting analysis of plasma supernatant after pelleting the nanoparticles with ultracentrifuge. According to Fig. 1D, HPRG and HMWK are depleted from plasma but are also recoverable from the washed nanoparticles. The dimeric form of MBL-A (but not the monomeric one, which presumably has lower affinity) was also depleted from plasma after incubation with SPIO. However, when SPIO pellet was washed extensively and the bound proteins were analyzed, MBL-A was no longer detectable. These experiments underscore the problem of weakly bound plasma proteins that could be missed during washing steps as suggested [16].
3.2 Interaction between SPIO surface domains and plasma proteins
The identified proteins showed remarkable specificity towards the surface domains of SPIO. Thus, HPRG and HMWK possess extensive histidine-rich sequences [18]. These proteins are known to bind to metal ions and negatively-charged surfaces through histidine-rich domains [18, 19]. In general, his-tagged proteins are routinely isolated using bivalent nickel chelates [20]. Prekallikrein usually circulates in plasma in equimolar complex with HMWK, and binds to foreign surfaces through kininogen [18]. To test whether these histidine-rich sequences have high affinity for SPIO, we employed a peptide composed of 6 histidine residues. The His6 peptide showed strong binding to SPIO with 400 bound peptide molecules per mg iron (Fig 2A). Of two control peptides, CREKA showed no significant binding, while a positively charged 34-amino acid peptide F3 that contains no histidine bound weakly. None of the peptides bound to neutral dextran. These data suggest that histidine-rich sequences of plasma HMWK and HPRG are responsible for D-SPIO binding, likely through surface-exposed iron oxide. Loose dextran coating has been previously shown to incompletely mask the iron oxide core of Ferumoxides [21].
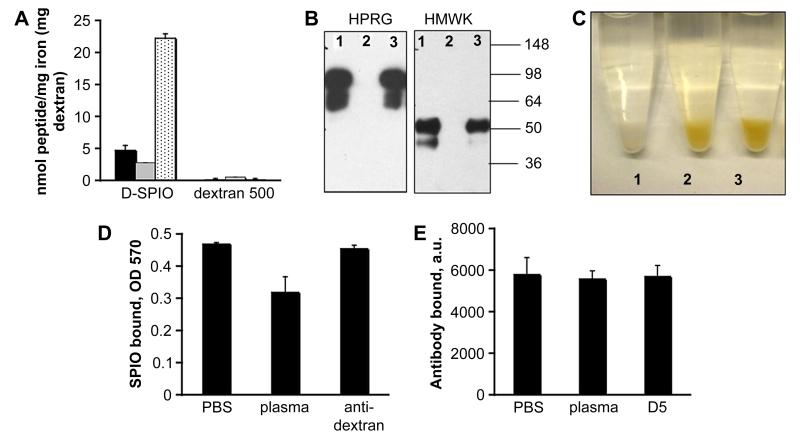
Figure 2. Probing accessibility of dextran and iron oxide on SPIO.
A, Binding of various peptide sequences to SPIO. A highly basic 34-amino acid peptide, F3 [35] (black bars), CREKA [34] (grey bars), and His-6 (dotted bars) were used. The peptides were FITC-labeled to allow quantification. SPIO were incubated with the peptides for 10 min, pelleted in an ultracentrifuge, and the amount of unbound and bound peptide was determined. As control, peptides were incubated with dextran 500 kDa (Sigma), and the amount of bound peptide was determined after separation of dextran by filter column. B, The nanoparticles were incubated with mouse plasma only (lane 1), in the presence of histidine-rich domain 5 (D5) of HMWK (lane 2) or with BSA (lane 3) for 10 min. Particle-bound proteins were eluted and separated by SDS gel electrophoresis as described in Methods. C, Binding of SPIO to D5 immobilized on glutathione-agarose. See Methods section for description of the procedure. Tube labels: 1, plain glutathione-agarose + SPIO particles (control); 2, D5-agarose + SPIO, 3, D5-agarose + SPIO +plasma. D, Binding of particles to D5-agarose was quantified by the iron assay (see Methods). SPIO were preincubated with either PBS, plasma or anti-dextran antibody before addition to the beads. Note that plasma only partially reduces the binding to D5-agarose, and that anti-dextran antibody does not interfere with the binding. Average of 3 experiments is shown. E, Binding of FITC-labeled anti-dextran antibody to SPIO. SPIO were incubated with either PBS, plasma or D5 fragment and subsequently with anti-dextran antibody. The binding of the antibody was quantified as described in Methods. The D5 treatment and plasma treatment had no effect on the antibody binding.
To test whether the binding of HPRG and HMWK to SPIO could be inhibited by histidine-rich peptide, we used a 30 kDa D5 domain of mouse kininogen-1 fused to GST. Precoating SPIO with this protein completely abolished binding of HMWK and HPRG from plasma, while GST or BSA did not have any effect on the protein binding (Fig. 2B).
In order to test the effect of plasma proteins on the accessibility of iron oxide core, we immobilized D5 protein through the GST fusion domain onto glutathione-agarose beads, and the beads were incubated with SPIO in the presence of 10-fold excess of plasma or in PBS. According to Fig. 2C-D, the binding of SPIO to the D5-beads was only minimally reduced in the presence of plasma, suggesting that plasma proteins do not coat completely the surface of nanoparticles. Dextran did not bind to D5-agarose (not shown), confirming that the binding of dextran-iron oxide nanoparticles to histidine-rich motifs took place through the exposed iron oxide core.
Next, we probed the accessibility of dextran chains in plasma using an anti-dextran IgG. Incubating the particles in plasma or in PBS did not significantly affect the binding of the antibody to SPIO (Fig. 2E), indicating that dextran-binding plasma proteins, similar to the iron oxide-binding proteins, do not shield the surface of the nanoparticles.
Importantly, the dextran coat and exposed iron oxide core domains showed no cross-reactivity: D5 protein did not affect the binding of anti-dextran IgG to SPIO, and vice versa (Fig. 2D-E).
3.3 SPIO-binding proteins and plasma opsonins in nanoparticle clearance
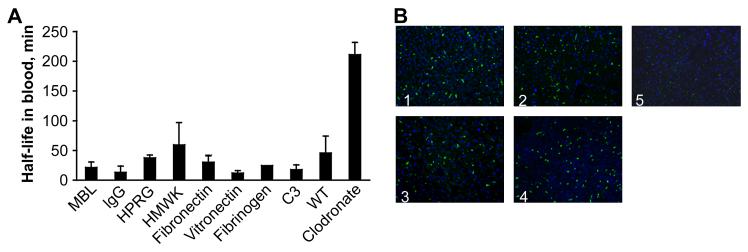
We tested some of the SPIO-binding proteins revealed by the proteomics analyses and some additional known opsonins for their effect on nanoparticle clearance and liver uptake by using knockout mice for each protein. The data shown in Figure 3A, indicate that SPIO clearance is normal in mice lacking C3, immunoglobulin, MBL-A, MBL-C, HPRG, HMWK and Fetuin-A. Kallikrein does not bind to SPIO in the HMWK knockout mice (our unpublished observation), so its role could be also excluded. Similarly, MASPs circulate in complex with MLBs [22] and are not supposed to be recruited to the particle surface in the MBL-deficient mice. In addition, mice null for other known opsonins such as fibronectin, fibrin, and vitronectin were similar to wild-type controls in SPIO clearance (Fig. 3A). The liver is the main organ responsible for nanoparticle uptake from the blood, and in accordance with earlier results, selective depletion of Kupffer macrophages in the liver using clodronate liposomes [23] produced a 5 to 10 fold increase in SPIO circulation time (Fig. 3A). The liver accumulation of the nanoparticles was similar between knockout mice and controls, but was only minimal in clodronate liposome-treated mice (Fig. 3B).
Figure 3. Summary of circulation half-lives in mice.
A, Knockout mice and matched wild type controls were injected with FITC-labeled or unlabeled SPIO, and particle half-lives were determined as described in Methods. Mice injected with clodronate liposomes served as a control for SPIO circulation time in mice with impaired liver uptake (right bar). The values are averages from 2-3 mice per group. B, Liver histology of some of the knockout mice sacrificed 3h post-injection of SPIO showing that there is no difference in accumulation of FITC-labeled SPIO in Kupffer macrophages (green dots). Panel labels: 1, HMWK-deficient; 2, wild type; 3, complement C3-deficient; 4, MBL-A/C-deficient; 5, wild type clodronate-treated. Objective: ×20.
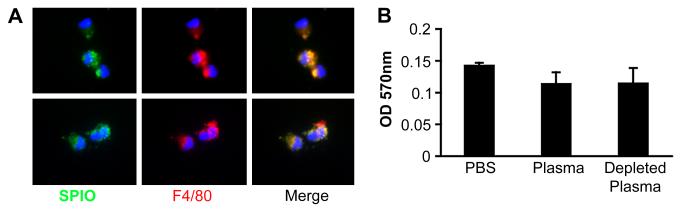
In vitro experiments using isolated Kupffer cells showed that the binding and uptake of SPIO particles by Kupffer cells is unchanged or slightly inhibited in presence of whole mouse plasma, or plasma that was depleted of SPIO-binding proteins (Figs. 4A-B). These combined data provide no support for an involvement of plasma opsonins in SPIO removal from the blood, and suggest that plasma protein coating does not prevent interaction with macrophage receptors.
Figure 4. Cellular uptake of SPIO in isolated Kupffer cells.
Non-labeled SPIO were preincubated with plasma or PBS and added to a suspension of isolated Kupffer cells in PBS/BSA. A, The effect of plasma on nanoparticle uptake as determined by microscopy. The particles were detected with a FITC-labeled anti-dextran antibody (green). Macrophages were stained with F4/80 antibody (red). The cells were examined by fluorescent microscopy. Objective, ×60. Upper panel, PBS-treated particles: lower panel, plasma-treated particles. B, Quantification of SPIO uptake by mouse Kupffer cells in the presence or absence of mouse plasma. As an additional control, mouse plasma depleted for SPIO-binding proteins was used (right bar). Average of 2 experiments are shown.
4 Discussion
Knowledge of the interactions of nanomaterials with host proteins is instrumental to understanding the biological and medical effects of these materials. The most valid study of nanoparticle opsonization would be to recover the injected nanoparticles from circulation and analyze the repertoire of bound proteins, but it was not technically feasible. Using modern mass spectrometry tools, we demonstrate in vitro that SPIO particles selectively bind certain plasma proteins onto their surface.
We demonstrated the existence of three distinct sets of proteins that bind to SPIO (Fig. 5). A set of iron oxide-binding proteins include strongly binding proteins HPRG and kininogen-1. These proteins appear to bind to exposed parts of the negatively charged iron oxide core in SPIO, directly through histidine-rich domains [18]. The binding of these proteins was clearly selective, as indicated by the resistance of the interaction to stringent washing of the particles and the high degree of enrichment relative to more abundant plasma proteins, such as albumin. In addition, β-2 glycoprotein and apolipoprotein B are known to bind to negatively-charged surfaces [24, 25]. As such, it is likely that they also are attracted to iron oxide core, but additional experiments would be necessary to confirm that.
Figure 5.
Schematic representation of the assembly of SPIO-binding proteins on the NP surface.
Another set of proteins binds to dextran component of SPIO. These proteins were mostly enriched in the pellet separated from plasma without washing of the particles. The enrichment of some of the SPIO-bound minor plasma proteins was as high as 7-fold. Interestingly, some of those enriched proteins were the mannose-binding lectins A and C. MBLs are known to bind strongly to mannose polysaccharides on the bacterial surface [26], but may also interact with the D-glucose units of the dextran coating, albeit less avidly. Immunoglobulins were also found, although their enrichment was not as significant (Supplemental Table 2). Sugar-binding antibodies in plasma have been reported before [27].
The last set of proteins appears to become associated with SPIO trough attachment to primary binders. Thus, kallikrein binds to foreign surfaces through HMWK, and does not bind to SPIO in HMWK-deficient plasma (our unpublished observation). Some of the weakly-binding proteins are also likely to be recruited through HMWK, such as coagulation factors XI and XII. In a similar fashion, MBL-associated serine proteases (MASP-1 and MASP-2) circulate in complexes with MBLs in plasma [26], suggesting that their binding to SPIO is indirect.
Interestingly, transferrin and albumin, the most abundant plasma protein, showed no significant enrichment on the SPIO, even under these less stringent binding conditions. These results reveal a previously unrecognized subtlety in protein binding to nanoparticles; proteins significant to the biological effects of nanomaterials could easily be missed under certain experimental conditions, producing incomplete “opsonome maps”.
We also provide data indicating that these bound plasma proteins are unlikely to promote SPIO clearance by liver macrophages. Depending on the surface properties, both stimulatory and inhibitory effects on nanoparticle uptake by serum have been previously reported [11, 12, 28, 29]. Because serum has undergone blood clotting, it may produce a somewhat different nanoparticle coating than plasma, which we have studied. However, in view of the apparent lack of any striking effect by plasma in vitro, and our results with knockout mice deficient in various candidate opsonins, we suggest that plasma protein opsonization is not responsible for the uptake of SPIO by the liver and spleen.
We favor the alternative hypothesis that there is a direct interaction between SPIO and cellular receptors. Indeed, our experiments showed the lack of any masking effect of plasma on SPIO, suggesting that the nanoparticle surface is still accessible for receptor recognition in vivo. Pattern recognition receptors, such as Toll-like receptors, are known to bind bacterial and fungal polysaccharides [30]. Scavenger receptors have been suggested to play a role in macrophage uptake of several types of nanoparticles, including iron oxide and polystyrene particles, and adenovirus [12, 29, 31]. Multiple receptors with overlapping specificities could be also involved in the recognition of a single particle type. Receptor identification will be an important future task, as it would allow rational design of inhibitors for the macrophage uptake process.
The proteins that bind to the surface of nanoparticles could play a role in nanoparticle-induced toxicity. MBLs and MASPs bound to the surface are known to activate the lectin-complement pathway [26], while immunoglobulins can trigger the classical complement pathway. The complement activation was previously reported for other types of nanoparticles coated with dextran [32], but the underlying mechanism of such activation has not been explored.
In addition to complement activation, the undesirable effects of nanomaterials can include thrombosis and inflammation [33]. The binding of kallikrein, kininogen, coagulation Factors XI and XII onto SPIO surface could potentially initiate the activation of the intrinsic pathway of blood clotting. We have previously described clotting in tumor vessels upon systemic administration of SPIO that were targeted to tumors with a clot-binding peptide [34]. However, the intravascular clotting was restricted to tumor vessels, although a major portion of the particles were taken up by the liver and spleen, indicating that inherent clot-promoting activity, if any, of the particles was not sufficient to initiate the clotting.
5 Conclusions
In this study, we demonstrate that dextran-coated iron oxide nanoparticles specifically interact with metal-binding and sugar-binding plasma proteins. Despite coating with plasma proteins, nanoparticles are cleared by resident liver and spleen macrophages through opsonin-independent mechanism. This suggests that either dextran or exposed iron oxide could be directly recognized by macrophages. The absorbed plasma proteins might be responsible for toxic effects of nanoparticles. The results of this study explain the role of plasma proteins in the macrophage recognition of short-circulating SPIO and perhaps should not be extrapolated to other types of SPIO such as crosslinked or PEGylated nanoparticles. These data are important in designing SPIO with stealth-like properties and modifying the thrombogenic and complement-activating potential of nanomaterials.
Supplementary Material
6 Acknowledgements
We thank Drs. Robert Rickert (Burnham Institute), and Cora Schäfer, Silke Enssen and Willi Jahnen-Dechent (Aachen University Clinic) for knockout mice and help with testing them. We also thank Drs. Khatereh Motamedchaboki and Lawrence Brill of Burnham Proteomics Facility for help with 2D proteomics experiments and analysis of the data. This work was supported by National Cancer Institute Grant CA119335 and, in part, by CA124427.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
8 References
- 1.Moghimi SM, Hunter AC, Murray JC. Long-circulating and target-specific nanoparticles: theory to practice. Pharmacol Rev. 2001;53(2):283–318. [PubMed] [Google Scholar]
- 2.Moghimi SM, Szebeni J. Stealth liposomes and long circulating nanoparticles: critical issues in pharmacokinetics, opsonization and protein-binding properties. Prog Lipid Res. 2003;42(6):463–78. doi: 10.1016/s0163-7827(03)00033-x. [DOI] [PubMed] [Google Scholar]
- 3.Chonn A, Semple SC, Cullis PR. Association of blood proteins with large unilamellar liposomes in vivo. Relation to circulation lifetimes. J Biol Chem. 1992;267(26):18759–65. [PubMed] [Google Scholar]
- 4.Yan X, Kuipers F, Havekes LM, Havinga R, Dontje B, Poelstra K, et al. The role of apolipoprotein E in the elimination of liposomes from blood by hepatocytes in the mouse. Biochem Biophys Res Commun. 2005;328(1):57–62. doi: 10.1016/j.bbrc.2004.12.137. [DOI] [PubMed] [Google Scholar]
- 5.Weissleder R, Bogdanov A, Jr., Neuwelt EA, Papisov M. Long-circulating iron oxides for MR imaging. Advanced Drug Delivery Reviews. 1995;16:321–334. [Google Scholar]
- 6.Park JH, von Maltzahn G, Zhang L, Derfus AM, Simberg D, Harris TJ, et al. Systematic Surface Engineering of Magnetic Nanoworms for In vivo Tumor Targeting. Small. 2009 doi: 10.1002/smll.200801789. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JH, von Maltzahn G, Zhang L, Schwartz MP, Ruoslahti E, Bhatia S, et al. Magnetic Iron Oxide Nanoworms for Tumor Targeting and Imaging. Adv Mater. 2008;20:1630–1635. doi: 10.1002/adma.200800004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulte JW, Kraitchman DL. Iron oxide MR contrast agents for molecular and cellular imaging. NMR Biomed. 2004;17(7):484–99. doi: 10.1002/nbm.924. [DOI] [PubMed] [Google Scholar]
- 9.Kamps JA, Scherphof GL. Receptor versus non-receptor mediated clearance of liposomes. Adv Drug Deliv Rev. 1998;32(12):81–97. doi: 10.1016/s0169-409x(97)00133-6. [DOI] [PubMed] [Google Scholar]
- 10.Bertholon I, Ponchel G, Labarre D, Couvreur P, Vauthier C. Bioadhesive properties of poly(alkylcyanoacrylate) nanoparticles coated with polysaccharide. J Nanosci Nanotechnol. 2006;6(910):3102–9. doi: 10.1166/jnn.2006.418. [DOI] [PubMed] [Google Scholar]
- 11.Moore A, Weissleder R, Bogdanov A., Jr. Uptake of dextran-coated monocrystalline iron oxides in tumor cells and macrophages. J Magn Reson Imaging. 1997;7(6):1140–5. doi: 10.1002/jmri.1880070629. [DOI] [PubMed] [Google Scholar]
- 12.Raynal I, Prigent P, Peyramaure S, Najid A, Rebuzzi C, Corot C. Macrophage endocytosis of superparamagnetic iron oxide nanoparticles: mechanisms and comparison of ferumoxides and ferumoxtran-10. Invest Radiol. 2004;39(1):56–63. doi: 10.1097/01.rli.0000101027.57021.28. [DOI] [PubMed] [Google Scholar]
- 13.Timmer JC, Enoksson M, Wildfang E, Zhu W, Igarashi Y, Denault JB, et al. Profiling constitutive proteolytic events in vivo. Biochem J. 2007;407(1):41–8. doi: 10.1042/BJ20070775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi H, Fermin D, Nesvizhskii AI. Significance Analysis of Spectral Count Data in Label-free Shotgun Proteomics. Mol Cell Proteomics. 2008;7(12):2373–85. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lepay DA, Nathan CF, Steinman RM, Murray HW, Cohn ZA. Murine Kupffer cells. Mononuclear phagocytes deficient in the generation of reactive oxygen intermediates. J Exp Med. 1985;161(5):1079–96. doi: 10.1084/jem.161.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, et al. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc Natl Acad Sci U S A. 2007;104(7):2050–5. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vollmer M, Nagele E, Horth P. Differential proteome analysis: two-dimensional nano-LC/MS of E. coli proteome grown on different carbon sources. J Biomol Tech. 2003;14(2):128–35. [PMC free article] [PubMed] [Google Scholar]
- 18.Colman RW, Schmaier AH. Contact system: a vascular biology modulator with anticoagulant, profibrinolytic, antiadhesive, and proinflammatory attributes. Blood. 1997;90(10):3819–43. [PubMed] [Google Scholar]
- 19.Jones AL, Hulett MD, Parish CR. Histidine-rich glycoprotein: A novel adaptor protein in plasma that modulates the immune, vascular and coagulation systems. Immunol Cell Biol. 2005;83(2):106–18. doi: 10.1111/j.1440-1711.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 20.Bornhorst JA, Falke JJ. Purification of proteins using polyhistidine affinity tags. Methods Enzymol. 2000;326:245–54. doi: 10.1016/s0076-6879(00)26058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung CW. Surface properties of superparamagnetic iron oxide MR contrast agents: ferumoxides, ferumoxtran, ferumoxsil. Magn Reson Imaging. 1995;13(5):675–91. doi: 10.1016/0730-725x(95)00023-a. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi M, Iwaki D, Kanno K, Ishida Y, Xiong J, Matsushita M, et al. Mannose-binding lectin (MBL)-associated serine protease (MASP)-1 contributes to activation of the lectin complement pathway. J Immunol. 2008;180(9):6132–8. doi: 10.4049/jimmunol.180.9.6132. [DOI] [PubMed] [Google Scholar]
- 23.Van Rooijen N, Sanders A. Liposome mediated depletion of macrophages: mechanism of action, preparation of liposomes and applications. J Immunol Methods. 1994;174(12):83–93. doi: 10.1016/0022-1759(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 24.Schousboe I. beta 2-Glycoprotein I: a plasma inhibitor of the contact activation of the intrinsic blood coagulation pathway. Blood. 1985;66(5):1086–91. [PubMed] [Google Scholar]
- 25.Maher VM, Kitano Y, Neuwirth C, Gallagher JJ, Thompson GR, Myant NB. Effective reduction of plasma LDL levels by LDL apheresis in familial defective apolipoprotein B-100. Atherosclerosis. 1992;95(23):231–4. doi: 10.1016/0021-9150(92)90026-d. [DOI] [PubMed] [Google Scholar]
- 26.Fujita T, Matsushita M, Endo Y. The lectin-complement pathway--its role in innate immunity and evolution. Immunol Rev. 2004;198:185–202. doi: 10.1111/j.0105-2896.2004.0123.x. [DOI] [PubMed] [Google Scholar]
- 27.Chacko BK, Appukuttan PS. Dextran-binding human plasma antibody recognizes bacterial and yeast antigens and is inhibited by glucose concentrations reached in diabetic sera. Mol Immunol. 2003;39(15):933–9. doi: 10.1016/s0161-5890(03)00028-2. [DOI] [PubMed] [Google Scholar]
- 28.Furumoto K, Nagayama S, Ogawara K, Takakura Y, Hashida M, Higaki K, et al. Hepatic uptake of negatively charged particles in rats: possible involvement of serum proteins in recognition by scavenger receptor. J Control Release. 2004;97(1):133–41. doi: 10.1016/j.jconrel.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Nagayama S, Ogawara K, Minato K, Fukuoka Y, Takakura Y, Hashida M, et al. Fetuin mediates hepatic uptake of negatively charged nanoparticles via scavenger receptor. Int J Pharm. 2007;329(12):192–8. doi: 10.1016/j.ijpharm.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 30.Rappleye CA, Goldman WE. Fungal stealth technology. Trends Immunol. 2008;29(1):18–24. doi: 10.1016/j.it.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Kanno S, Furuyama A, Hirano S. A murine scavenger receptor MARCO recognizes polystyrene nanoparticles. Toxicol Sci. 2007;97(2):398–406. doi: 10.1093/toxsci/kfm050. [DOI] [PubMed] [Google Scholar]
- 32.Lemarchand C, Gref R, Passirani C, Garcion E, Petri B, Muller R, et al. Influence of polysaccharide coating on the interactions of nanoparticles with biological systems. Biomaterials. 2006;27(1):108–18. doi: 10.1016/j.biomaterials.2005.04.041. [DOI] [PubMed] [Google Scholar]
- 33.Dobrovolskaia MA, McNeil SE. Immunological properties of engineered nanomaterials. Nat Nanotechnol. 2007;2(8):469–78. doi: 10.1038/nnano.2007.223. [DOI] [PubMed] [Google Scholar]
- 34.Simberg D, Duza T, Park JH, Essler M, Pilch J, Zhang L, et al. Biomimetic amplification of nanoparticle homing to tumors. Proc Natl Acad Sci U S A. 2007;104(3):932–6. doi: 10.1073/pnas.0610298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porkka K, Laakkonen P, Hoffman JA, Bernasconi M, Ruoslahti E. A fragment of the HMGN2 protein homes to the nuclei of tumor cells and tumor endothelial cells in vivo. Proc Natl Acad Sci U S A. 2002;99(11):7444–9. doi: 10.1073/pnas.062189599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.