Abstract
Semen is the main vector for HIV transmission worldwide. Recently, a peptide fragment (PAP248–286) has been isolated from seminal fluid that dramatically enhances HIV infectivity by up to four to five orders of magnitude. PAP248–286 appears to enhance HIV infection by forming amyloid fibers known as SEVI, which are believed to enhance the attachment of the virus by bridging interactions between virion and host-cell membranes. We have solved the atomic-level resolution structure of the SEVI precursor PAP248–286 using NMR spectroscopy in SDS micelles, which serve as a model membrane system. PAP248–286, which does not disrupt membranes like most amyloid proteins, binds superficially to the surface of the micelle, in contrast to other membrane-disruptive amyloid peptides that generally penetrate into the core of the membrane. The structure of PAP248–286 is unlike most amyloid peptides in that PAP248–286 is mostly disordered when bound to the surface of the micelle, as opposed to the α-helical structures typically found of most amyloid proteins. The highly disordered nature of the SEVI peptide may explain the unique ability of SEVI amyloid fibers to enhance HIV infection as partially disordered amyloid fibers will have a greater capture radius for the virus than compact amyloid fibers. Two regions of nascent structure (an α-helix from V262-H270 and a dynamic α/310 helix from S279-L283) match the prediction of highly amyloidogenic sequences and may serve as nuclei for aggregation and amyloid fibril formation. The structure presented here can be used for the rational design of mutagenesis studies on SEVI amyloid formation and viral infection enhancement.
Introduction
Despite the rapid progress of the AIDS pandemic, the HIV virus is a surprisingly weak pathogen in vitro with only <0.1% of virus particles succeeding in infecting a host cell.1, 2 Emerging evidence suggests that difficulties in virus attachment to the host cell in vitro, rather than intrinsic deficiencies in viral particles, are the source of this low infection rate. Since the half-life of free virions of HIV is very short in comparison to the probability of a virion randomly encountering the appropriate receptor, most virions will decay before initiating a successful infection cycle unless they can rapidly attach to the cell membrane surface where the search for the cell receptor proceeds by inherently more efficient two-dimensional diffusion.3, 4 Viral attachment to the cell surface entry of the virus into the host cell therefore appears to be the primary barrier to HIV infection in vitro. 5
The difference between in vitro and in vivo infection rates suggests this process is much more efficient in vivo than in vitro and that cofactors absent in vitro but present in vivo may be responsible for this difference. A clear candidate has emerged from a recent screening of a large library of peptides and low molecular weight proteins (<50 kDa) found in human semen. This screening identified SEVI (Semen-derived Enhancer of Viral Infection), a naturally occurring 39 amino acid fragment from Prostatic Acid Phosphatase (PAP) that enhanced the rate of HIV infection dramatically across a broad range of HIV phenotypes.6–8 Limiting dilution assays showed a truly remarkable enhancement by four to five orders of magnitude when HIV viral loads more closely resembling the actual conditions during sexual transmission were used, with only 1–3 virions necessary to establish a persistent infection in CEMx M7 dendritic cells in the presence of SEVI.6 Furthermore, SEVI’s ability to increase viral infectivity is not limited to the HIV virus, as it also exerts a similar effect on the XMRV retrovirus.9
Since the structures of SEVI are not known in any form, it is difficult to make predictions about the interactions of SEVI-with the cell membrane that facillitate HIV viral attachment. Some degree of aggregation of the peptide is necessary for activity as freshly prepared, monomeric solutions of SEVI are ineffective at promoting viral infectivity and the enhancement of HIV infectivity increases with time as SEVI is incubated in solution.6 It has been established that SEVI is an amyloidogenic peptide, and that amyloid fibers of the peptide are more effective than the monomeric peptide in promoting HIV cell binding and membrane fusion.6 This is in agreement with previous studies that have shown a more modest enhancement of infectivity of enveloped viruses by other amyloidogenic proteins, such as A and -synuclein.10 While some degree of oligomerization seems to be a requirement for SEVI-enhanced infectivity, amyloid formation does not seem to be an absolute requirement for the enhancement of HIV infection by PAP248–286 as it occurs well before the formation of amyloid fibers.6
These interactions are likely to be highly dependent on specific structural details as other amyloid proteins which share the same gross cross-beta sheet structure as SEVI show a much lower enhancement of infectivity. To understand the mechanism by which SEVI promotes the bridging of viral and host cell membranes, we have solved the high-resolution structure of the SEVI precursor PAP248–286 in a membrane mimicking envirionment (SDS micelles) using NMR spectroscopy. The structure of PAP248–286 possesses an unusual amount of disorder compared to other amyloid proteins, which may explain the much greater ability of SEVI to enhance HIV infection compared to amyloid fibers formed from other proteins.6 From the structure and bioinformatic analysis of the PAP248–286 sequence we have also identified a putative amyloidogenic region of the PAP248–286 sequence.
Materials and Methods
NMR Sample Preparation
SEVI refers to the amyloid form of peptides derived from fragments of Prostatic Acid Phosphatase (PAP). Of these peptides, the fragment PAP248–286 was chosen for this study, as amyloid fibers formed from PAP248–286 are the most effective at enhancing HIV infection. The PAP248–286 peptide was synthesized and purified to > 95% purity by Biomatik (Toronto, ON). Since the large size of the amyloid form of the peptide precludes its study by solution NMR, peptide aggregation must be prevented for the duration of the experiment to obtain a high-resolution structure of the peptide. The removal of preformed aggregates of the peptide that can act as nuclei for further peptide aggregation is essential to arrest this process, accordingly the peptide was dissolved first in a strongly disaggregating 1:1 solution of trifluoroacetic acid (TFA) and hexafluoroisopropanol (HFIP) at a concentration of 5 mg/ml to break up preformed amyloid fibers.11 The TFA/HFIP solution was evaporated by a stream of nitrogen gas and the resulting film redissolved in pure HFIP at a concentration of 2 mg/ml. HFIP was removed by lyophilization under high vacuum for 24 hours. Samples were prepared for NMR measurements by dissolving 3.4 mg of lyophilized peptide in 20 mM phosphate buffer at pH 7.3 containing 10% D2O, 120 mM NaCl, and 200 mM perdeuterated SDS (Cambridge Isotopes Laboratory) to a final concentration of 2.5 mM. The choice of SDS micelles, neutral pH, and the addition of 120 mM NaCl was based on previous experiments showing the electrostatic nature of PAP248–286 membrane binding and the ability of PAP248–286 to cause substantial vesicle aggregation, particularly at acidic pH.11 Samples prepared with zwitterionic DPC (dodecylphosphocholine) micelles aggregated rapidly and gave rise to a very broad and weak signal, while samples prepared without 120 mM NaCl displayed poorer resolution and showed visible aggregation within 24 hours.
NMR Data Collection and Processing
All NMR spectra of PAP248–286 embedded in SDS micelles were recorded at 37°C using a 600 MHz Bruker Avance NMR spectrometer equipped with a TXI cryoprobe. Backbone and side-chain assignment of peptide was achieved using 2D 1H-1H TOCSY (Total Correlation Spectroscopy) recorded at a 80 ms mixing time and 2D 1H-1H NOESY (Nuclear Overhauser Enhancement Spectroscopy) recorded at 100 and 300 ms mixing times. Both spectra were recorded using 16 scans and 512 points in the indirect dimension, with a recycle delay of 1.5 s. Complex data points were acquired for quadrature detection in both frequency dimensions for the 2D experiments. All spectra were zero-filled in both dimensions to yield matrices of 2048 × 2048 points. Proton chemical shifts were referenced to the water proton signal at 4.7 ppm. All 2D spectra were processed using NMRPIPE and TopSpin software and analyzed using SPARKY.12, 13. Resonance assignments were carried out using a standard approach reported elsewhere.14
Structure Calculations
Structure calculations were performed using the X-PLOR-NIH program. A total of 286 NOEs, derived from the NOESY acquired at 300 ms mixing time, were categorized into three distance categories based on the cross-peak volumes obtained from SPARKY analysis: strong (1.8–2.9 Å), medium (1.8–4.5 Å), and weak (1.8–6.0 Å). These restraints were further used in the structure calculations: 145 restraints were intra-residue and 141 were inter-residue NOEs (Table 1). The torsion angle restraints were obtained from the TALOS program using the Hα chemical shift values.15 An extended structure of PAP248–286 was used as a starting point for the hybrid molecular dynamics simulated annealing (SA) protocol at a temperature of 4000 K for the generation of an initial 100 structures.16, 17 Explicit hydrogen bond restraints were not used in the calculations. Subsequently, these structures were refined using a further SA step and energy minimization. The final refinement was carried out using the refine_gentle.inp protocol, which gradually introduces the van der Waals radii. The few ambiguous angles found in the loop region were assigned an additional 60° of conformational freedom compared to their predicted values. Out of the 100 structures generated, an ensemble of 8 conformers with low RMSD was selected for further analysis. The covalent geometry of the conformers generated was analyzed using PROCHECK_NMR.18
Table 1.
Statistical information for the PAP248–286 structural ensemble
| Distance Constraints | |
| Total | 286 |
| Intra-residual | 145 |
| Inter-residual | 141 |
| Sequential (i – j = 1) | 106 |
| Medium (i – j = 2, 3, 4) | 35 |
| Structural Statistics | |
| NOE violations (Å) | 0.0318 ± 0.0045 |
| Dihedral angle, restraint | 0.7816 ± 0.1550 |
| violations (°) | 0.0024 ± 0.0003 |
| RMSD for bond deviations (Å) | 0.4037 ± 0.0160 |
| RMSD for angle deviations (°) | |
| RMSD of all backbone atoms (Å) | 0.15 ± 0.05 |
| Val 262 – His 270 | 0.74 ± 0.43 |
| Pro 278 – Leu 283 | |
| RMSD of all heavy atoms (Å) | 0.57 ± 0.12 |
| Val 262 – His 270 | 1.60 ± 0.60 |
| Pro 278 – Leu 283 | |
| Ramachandran Plot | |
| Residues in most favored region (%) | 65.8 |
| Residues in additionally allowed region (%) | 29.8 |
| Residues in generously allowed region (%) | 3.7 |
| Residues in disallowed region (%) | 0.7 |
Results
The structure of PAP248–286 in SDS micelles was determined using solution NMR through 2D 1H-1H TOCSY and NOESY data collected as mentioned in the Materials and Methods section. Sample preparation was the key step in obtaining high-resolution spectra of PAP248–286. In an effort to optimize spectral resolution and sensitivity, several 1D 1H chemical shift spectra were acquired over a temperature range of 25–45°C (spectra not shown) and the best compromise between sensitivity and resolution was found to be near 37°C.
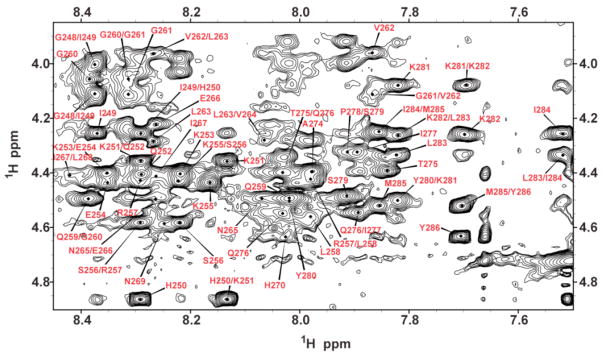
The 2D 1H-1H NOESY spectra displays numerous, relatively well-resolved cross-peaks, with a significant degree of spectral dispersion in the amide region indicating the presence of at least a partial degree of secondary structure throughout the peptide (Fig. 1). This is an indication of secondary structure formation in the peptide, as a completely disordered structure would have both poor chemical shift dispersion due to the similarity of chemical shift values in the random coil state and poor NOE cross-peak intensity due to the high mobility of the structure. In the V262-H270 and Y280-I284 regions, inter-residue dαHNH(i, i + 3), dαHNH(i, i + 4), and dαHβH(i, i + 3) NOE connectivities along with negative chemical shift indices provide unequivocal evidence for helical structure (Figs. 2 and 3). Similarly, the CD spectra of PAP248–286 in SDS micelles and trifluroethanol solutions exhibit the double minima at 208 and 222 nm characteristic of partial α-helical content (Fig. S1 in the Supplementary Materials section). The remainder of the peptide has chemical shift index values consistent with disordered conformations, although the NOE crosspeak intensity pattern, which is more sensitive to transient structure formation, shows strong sequential dNHNH(i,i+1) peaks indicative of transient α-helical or β-sheet formation.
Figure 1.
The finger print region of a 2D 1H-1H NOESY spectrum of SDS micelles containing PAP248–86 showing NOE connectivities among Hα nuclei.
Figure 2.
NOE intensity plot for the amino acid residues of PAP248–286 showing the NOE connectivity among residues. Thicker lines correspond to stronger NOE intensities.
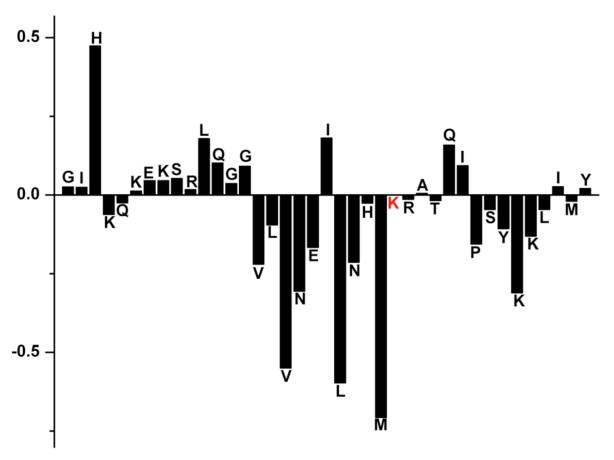
Figure 3.
Alpha proton chemical shift index (CSI) for PAP248–286 showing the disordered N-terminal end (CSI near zero) and the central and C-terminal helical regions (CSI <−0.1). The Ha chemical shift of K272 was not detected (red). The CSI was calculated by subtracting the Ha chemical shifts values measured for the peptide from the random coil shifts values for the respective amino acid reported in the literature.
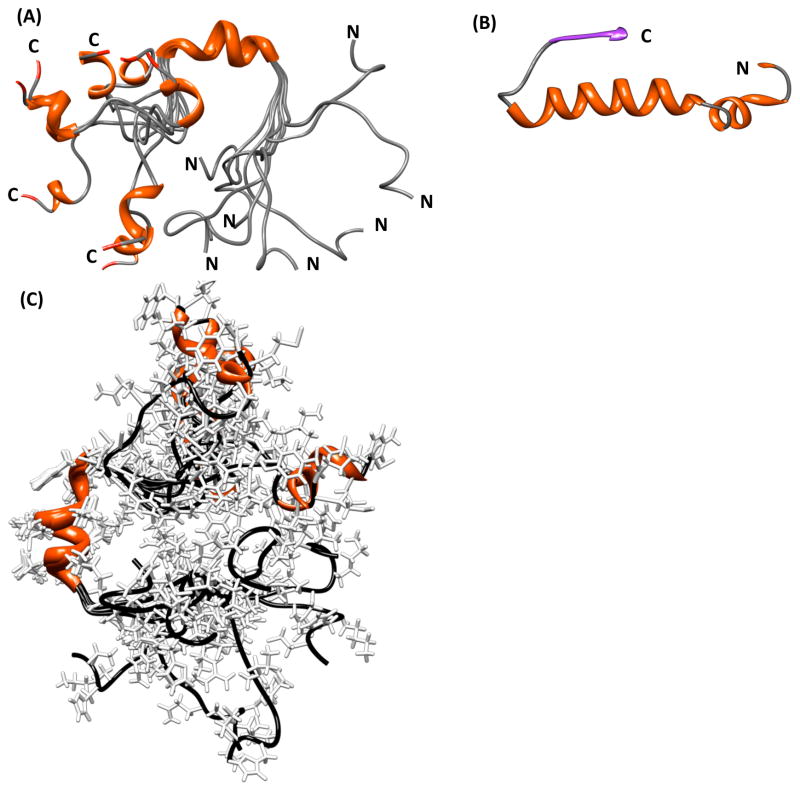
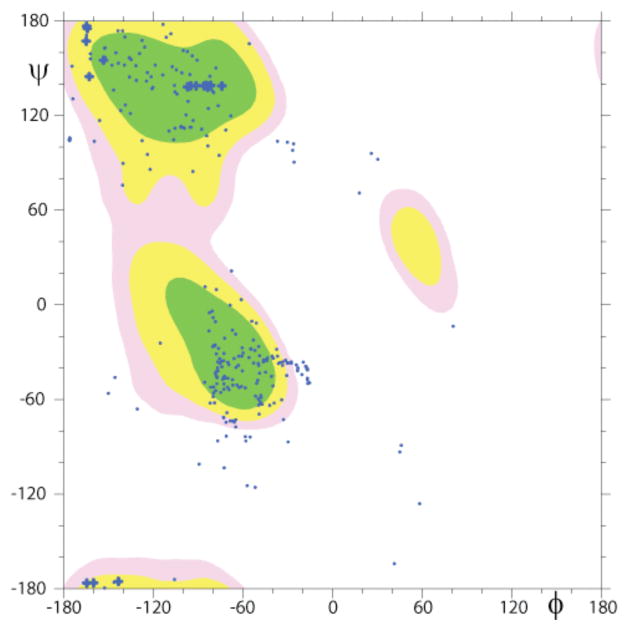
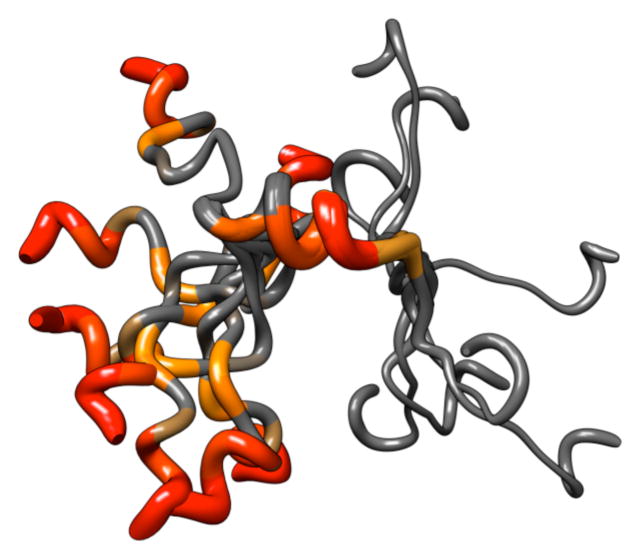
To create the structure of PAP248–286, all NOEs were converted into distances and modeled using the classical simulated annealing protocol built in XPLOR-NIH. Out of the 100 refined PAP248–286 structures, 26 did not have NOE violations of > 0.5 Å, dihedral angle restraint violations of >5°, have bonds that deviate from ideality by a RMS difference of > 0.01 Å, or have bond angles that deviate from ideality by a RMS difference of > 2°. Out of the structures that passed the acceptance criteria, an ensemble of 8 conformers were further selected for the final analysis as a representation of the transient structure formed by the disordered PAP248–286 on the micelle. Statistical information on the peptide structure along with a summary of the backbone and side chain NOEs used for secondary structure assignment is provided in Table 1 and Fig. 4. The secondary structure representation of PAP248–286 from these 8 conformers is shown in Fig. 5A. The structure of the PAP248–286 peptide can be separated into three distinct regions: a highly flexible N-terminal region (G248 - G261), a rigid α-helical central region (V262-H270), and a flexible C-terminal region (M271 - Y286) containing a short 310 or α-helix (S279-L283). The RMSD for the C- and N-terminal ends is very large, reflecting a high degree of disorder in these regions. The central region from V262-H270 is more ordered and a superposition of the 8 low-energy conformers gives an RMSD of 0.15 ± 0.05 Å for the backbone atoms from V262-H270, and an RMSD of 0.57 ± 0.12 Å for all heavy atoms. Analysis of the Ramachandran plot, for the final 8 conformers of the PAP248–286 shows that 65.8% of the residues fall in the most favored region while the rest of the residues fall in the additionally allowed (29.8 %), generously allowed (3.7%), and disallowed (0.7%) regions of the plot as shown in Fig. 6.
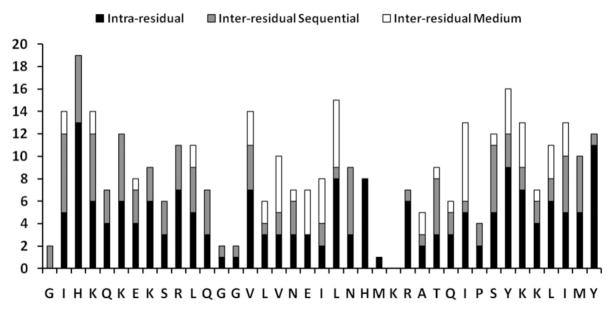
Figure 4.
Histogram of the number of NOEs detected versus the residue number showing the number of intra-residue, sequential (i – j = 1), and medium range (i – j = 2, 3, 4) NOEs detected. Long-range (i – j>4) NOEs were not observed.
Figure 5.
(A) Secondary structure representation of an overlaid ensemble of NMR-derived conformers for PAP248–286 showing the helical region from V262-H270, the highly flexible N- and C-termini, and the short helix from S279-L283. (B) Secondary structure representation of PAP248–286 extracted from the crystal structure of human PAP. In the crystal structure, residues L283-Y286 are in a β-sheet conformation, while residues K251-I277 are in a helical conformation with a distortion at G261. (C) All atom representation of the PAP248–286 conformational ensemble.
Figure 6.
Ramachandran plot showing the Phi and Psi angles for the ensemble of conformers of the PAP248–286 peptide.
Positioning of the PAP248–286 in the micelle
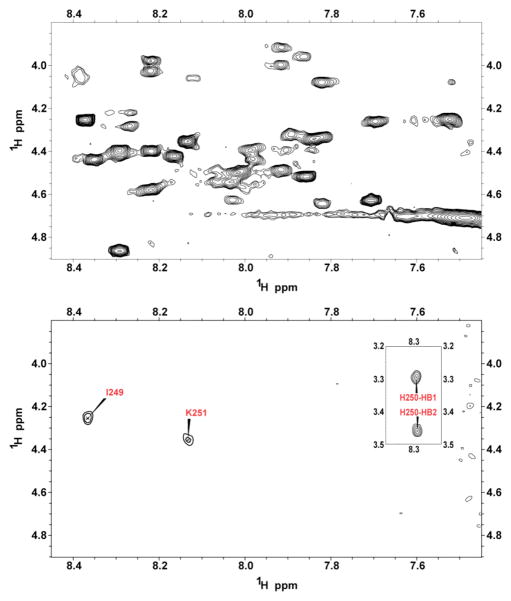
The paramagnetic quencher Mn2+ was used to determine the membrane orientation of the PAP248–286. The signal intensities of the nuclei that are in close proximity to the Mn2+ ion are decreased due to the increase in the relaxation rate of the nuclei. Since manganese ions cannot penetrate into the hydrophobic interior of the micelle, changes in signal intensity reflect the exposure of the amino acid residues of the peptide to the solvent. To identify the exposure of specific residues to solvent more precisely, 2D 1H-1H TOCSY spectra were used to monitor the changes in the chemical shift of the α-proton peaks after the addition of 0.8 mM MnCl2. It is interesting to note that all of the peaks were completely quenched by a moderate (0.8 mM) concentration of Mn2+, except the α-protons of Ile 249 and Lys 251 and the β-protons of His 250 as shown in Fig. 7. The near complete quenching of the residues of PAP248–286 is in contrast to other amyloidogenic peptides which are bound to the surface of detergent micelles but show only a partial reduction in the peak intensity after the addition of as much as 1.2 mM MnCl2, and suggests the depth of penetration of PAP248–286 is significantly less 19, 20 Partial rather than complete quenching of the residues near the N-terminus of the peptide suggests this cluster of residues is either buried further into the micelle than the remainder of the peptide or the mobility of these residues is altered so that the rotational reorientation time of these residues is shorter than the electron spin relaxation time.21–23
Figure 7.
Finger print region of a 2D 1H-1H TOCSY spectrum of SDS micelles containing PAP248–286 after the addition of 0.8 mM MnCl2. The complete quenching of almost all peaks indicates that the peptide is well exposed to the solvent and does not penetrate deeply into the micelle.
Discussion
Like many amyloidogenic short peptides and some amyloidogenic proteins, monomeric PAP248–286 is predominantly unstructured in solution. However, PAP248–286 is most active in the form of large aggregates with the characteristic β-sheet conformation of amyloid proteins (SEVI fibers). The structure of the final SEVI product is an obvious target for therapeutic intervention to block the action of SEVI, but the structures of intermediates along the aggregation pathway of PAP248–286 may ultimately be just as fruitful as therapeutic targets. The significance of the intermediate state is due to the energetics of the aggregation process; amyloid fibers are thermodynamically very stable but are kinetically difficult to form due to the low probability that multiple aggregation prone segments will align in the correct orientation required for the precise self-assembly of the very ordered amyloid fiber. For this reason, aggregation of completely disordered proteins usually gives rise to amorphous aggregates rather than structured oligomers,24–26 a fact that has been exploited by small molecule inhibitors that stabilize the disordered state of the monomer and divert aggregation from potentially toxic protofibrillar species to non-toxic amorphous aggregates.8, 27 Preorganization of the monomeric state can overcome some of the difficulties associated with self-assembly by restricting the degrees of freedom of motion in the native state and aligning aggregation-prone sites in the molecule in the proper orientation for intermolecular association.28, 29
Taken these considerations into account, it can be seen that knowledge of the conformational preferences of the monomeric state of an amyloidogenic peptide can give substantial insight into the pathways of aggregation and eventual structure of the amyloid fiber.19, 20, 30–32 However, the transient structure present in intrinsically disordered proteins is difficult to detect experimentally. Amyloidogenic proteins, particularly unstructured ones, are notoriously difficult to crystallize. The structural investigation of largely unstructured proteins by solution NMR is difficult due to poor chemical shift dispersion and the structural heterogeneity of the ensemble which gives rise to substantial line-broadening due to conformational exchange among members of the ensemble on the submillisecond time-scale.33 The most structured regions of the peptide, which are the regions of most interest for the reasons mentioned above, are usually the victim of the most severe line-broadening. Another serious complication is the aggregation of the peptide, which severely aggravates the line-broadening due to conformational exchange and severely limits the observation time available before the aggregated protein is too large for solution NMR experiments. These limitations can be partially overcome by using trifluoroethanol or hexafluoroisopropanol solvent systems which stabilize helical states of the peptide and reduce or eliminate aggregation at high concentrations.34–37 The structure promoting solvents are used to stabilize transient structures in the natural conformational ensemble, which is overall disordered.38 However, the use of these solvent systems can be questioned as they can give rise to artificial levels of structure that are not present in the biological system.39–42
Many of these limitations can be overcome by the use of detergent micelles to stabilize the monomeric protein, which offer a more biologically relevant environment than organic solvents as amyloid proteins are often found in intimate association with cellular membranes and many of their biological effects derive from this association. 43–49 SEVI in particular is thought to act by binding to the cell membrane surface and neutralizing the natural repulsion that exists between the membranes of the HIV virion and the target cell.6, 7 Unlike mixtures of organic solvents and water, detergent micelles are heterogeneous like real cell membranes, having a hydrophobic core similar to the phospholipid bilayer and a polar interface between the micelle and solution. Detergents also have the additional advantage of trapping the aggregation-prone protein in a monomeric state due to the small size of the detergent micelle and the large electrostatic repulsion that exists between each micelle. This property is essential for limiting the rapid loss of signal due to aggregation of the peptide.
PAP248–286 is Largely Disordered on the Membrane Surface, Unlike Many Amyloid Peptides
PAP248–286 binds to the surface of the SDS micelle in a largely disordered manner. The complete quenching of the signal of almost every residue at a relatively low concentration of Mn+2 suggests the binding of PAP248–286 is almost entirely electrostatic, as previously inferred based by differential scanning calorimetry and the loss of PAP248–286 infection promoting activity in mutants lacking cationic residues.7, 11 The superficial binding of PAP248–286 to the surface may explain the apparent lack of toxicity of PAP248–286 as the ability of amyloid peptides to disrupt membranes is frequently correlated with the penetration of the peptide into the hydrophobic part of the bilayer membrane.6, 19, 20, 50, 51
Most unstructured amyloidogenic peptides readily adopt an α-helical conformation when bound to membranes due to the amphipathic and hydrophobic nature of most amyloidogenic sequences. A common motif among amyloid peptides bound to detergent micelles is the helix-turn-helix motif, in which an amphipathic helix is separated from a more polar helix by a short flexible linker region. Most high-resolution structures of amyloidogenic peptides bound to detergent micelles show some variation of this motif, although a considerable degree of conformational flexibility is present in most of the peptides as the location of the linker region and the length of the helical regions of the peptide can vary substantially depending on the experimental conditions.52–55 The structure of PAP248–286 is consistent with this general trend, however the helical content is lower than other amyloid peptides and the structure is dominated by long flexible regions joining short helices rather than by the well-defined helices separated by short linker regions that are typical of other amyloid peptides.
While the α-helical regions of the membrane-bound monomer of amyloid peptides are not found in the final amyloid product, regions which are disordered in the membrane-bound state frequently remain so throughout the aggregation pathway. For example, the N-terminus of Aβ1–42 and the C-terminus of α-synuclein is disordered in both SDS micelles and in the structure of the amyloid fiber.56–61 However, the disordered regions of these peptides are composed of negatively charged residues and not poised to interact favorably with the negatively charged cell membrane. On the other hand, the disordered N-terminus of PAP248–286 has a high percentage of positively charged lysine and arginine residues which have a high affinity for surface of cell membranes. If the conformational flexibility and superficial electrostatic membrane binding found in monomeric PAP248–286 is also present in the structure of the membrane-bound SEVI amyloid-fiber, these structural characteristics may help explain the unique proficiency of SEVI among amyloid proteins to enhance HIV infection. Flexible, highly charged cationic polymers like polylysine share SEVI’s ability to enhance retroviral infection rates, and share many common characteristics with the structure and membrane binding mode proposed here.11, 62, 63 Like PAP248–286, these polymers bind exclusively electrostatically to the membrane surface without penetration into the headgroup region, undergo helix-to-β-sheet transitions, and retain a considerable amount of flexibility when bound to the surface.64–66 Disordered regions frequently have higher interaction rates than structured regions in proteins because the interaction volume (radius of capture) is larger for the disordered state than a compact, folded one.67, 68 This is especially true if, like the association of proteins with lipids, binding is based on multiple weak, non-specific interactions rather than a single specific interaction site. 69 Although a considerable degree of conformational reorganization is required to form the SEVI amyloid fiber, an unusual degree of disorder in the SEVI amyloid fiber may facilitate the membrane bridging interactions implicated in both the enhancement of HIV infection by SEVI and polylysine.68 In addition, disordered regions in the amyloid fiber may facilitate the endocytosis of SEVI fibers. Many endocyotic proteins have extensive disordered regions which appear to allow the capture of endocyotic nucleation factors across a large cytosolic volume.69 A high degree of disorder in the SEVI amyloid fiber would likely enhance the rate of endocyotic internalization relative to other amyloid fibers, which internalize the HIV virion if it is bound to the SEVI fiber at the time.70 The structure of SEVI amyloid fibers remains unsolved and only limited mutagenesis work has been done.6, 7 Further structural work on the SEVI amyloid fiber is likely to yield informative insights into the mechanism of HIV enhancement by SEVI and serve as a test of this hypothesis.
Nascent Helical Structure in A274-I284 and V262-H270 May Nucleate Amyloid Formation
Two regions of nascent helical structure may be nucleation sites for amyloid formation. The first region consists of a stable, regular α-helix extending from V262 to H270, the second region consists of two dynamic or transient α- or 310 helices extending from A274-Q276 and Y280-I284 separated by a kink or distortion at residues I277-S279. The propensity for helical conformations in these regions may be essential for fiber formation, as a helix-to-β-strand transition is a common intermediate step in the amyloidogenesis of many amyloid proteins.30, 71 The importance of helical intermediates in amyloid formation can be seen by the fact that the induction of helical structure through either the addition of a helix-promoting solvent or by membrane-binding can dramatically enhance the rate of amyloidogenesis.71 Mutational analyses of amyloid proteins with helical intermediates have shown that the helical intermediate should be metastable to efficiently promote β-strand formation. The stability of the helices in these regions may be critical for determining the ultimate rate of amyloidogenesis; if the helices are excessively stabilized by mutations in the sequence or by helix stabilizing drugs, the peptide can be kinetically trapped as a helical intermediate - conversely a very unstable helix is insufficiently populated to impact the kinetics.72, 73
The limited amount of mutagenesis data currently available on SEVI suggests the nascent, dynamic helix at the C-terminus is essential for amyloidogenesis. Analyses of truncated versions of the PAP248–286 peptide have shown that deletions at the C-terminal end of the peptide in the region of the dynamic helix significantly reduce both amyloid formation and HIV promotion. 6 On the other hand, deletions at the N-terminal end, which is disordered in the NMR structure, have little effect. The metastable 310 helices that are present in some of the structures at the C-terminus are particularly suited for promoting intermolecular interactions and β-strand formation.30 Due to the more extended conformation of the 310 helix compared to an α-helix, the grooves of the 310 helix are larger and more easily accommodates the side-chains of another peptide. Furthermore, the β-turns that comprise the 310 helix are less stable than the α-helix and can be easily destabilized to form β-strands, 30 as shown by the crystal structure of PAP in which L283-Y286 is part of a β-strand on the surface of the protein.74
The Proposed Amyloid Nucleation Sites are Consistent with Predictions of Amyloidogenic Regions
The increasing number of known amyloidogenic proteins has made possible the use of bioinformatic tools to predict amyloidogenic regions within proteins. The AGGRESCAN algorithm uses the inherent aggregation propensities of individual amino acids determined from mutational data on the Aβ peptide to find “hot spots”, short (~5 residue) stretches of amino acid that serve as nucleating centers for amyloid formation.75, 76 A color-mapped representation of the aggregation propensity from AGGRESCAN on the PAP248–286 structure is provided in Fig. 8. As shown in Fig. 8, the predictions of AGGRESCAN substantially match the helical regions of the NMR structure. AGGRESCAN predicts two aggregation “hot spots” in PAP248–286: G260-E266, which is approximately within the V262-H270 helical region, and K281-Y286 in the area of both the dynamic helix at the C-terminal end and the L283-Y286 sequence identified by activity studies on truncated peptides. The PASTA algorithm uses pair-wise energy functions to determine the probability that a given stretch of residues can be in a parallel or anti-parallel beta-strand.77, 78 Similar to the prediction of AGGRESCAN, PASTA predicts that residues 262–270 are prone to form parallel beta-sheets (energy score of -6 for residues 262–270 in a parallel alignment, stretches of residues in the beta-sheet conformation in known fiber structures generally have a score of > 4).
Figure 8.
Prediction of the amyloidogenic propensity of the PAP248–286 sequence by AGGRESCAN. Red, orange, and grey colors indicate regions of high, moderate, and low amyloidogenic propensity respectively.
Since amyloid formation is almost always deleterious to a protein’s normal function and general cellular health, there is substantial evolutionary pressure against amyloid formation. One method of preventing amyloid formation is to flank particularly amyloidogenic sequences with “gatekeeper residues”, evolutionarily conserved sequences which are either enriched in residues that are either strong β-sheet breakers (glycine or proline residues) or highly charged sequences which enhance the electrostatic repulsion between proteins and therefore minimize aggregation.79 The ordered 262–270 region of PAP248–286 fits this pattern, being terminated on the N-terminal side by a double glycine repeat (GG) and flanked by a highly charged sequence (QKEK) nearby. Similarly, the C-terminal side of this sequence is flanked by a proline residue (P31) and a nearby lysine repeat (KK), which would also serve to limit the aggregation of the Y280-Y286 amyloidogenic region. This pattern of gatekeeper residues is conserved in all PAP sequences determined so far, suggesting that one of the functions of these residues is to limit the propensity of PAP to aggregate. The observation that PAP248–286 with all the cationic amino acids mutated to alanine has a higher tendency to aggregate gives some support to this hypothesis.7
Conclusion
We have solved the first high-resolution structure of the SEVI precursor peptide PAP248–286 as a first step towards understanding the structural factors that drive PAP248–286 to form the amyloid fibers that are identified as the form responsible for increasing HIV infectivity. PAP248–286 has an unusual amount of structural disorder when bound to the membrane compared to other amyloid proteins which may facilitate the binding of virus particles by maximizing the radius of capture for incoming virus particles. The disordered N-terminal end also has a high percentage of positively charged residues which can interact with multiple lipids on the virus surface. Furthermore, the combination of structural disorder and high positive charge at the N-terminus of PAP248–286 may be linked to the high affinity of the SEVI amyloid fiber for the HIV virus and the remarkable enhancement of HIV infectivity by the SEVI amyloid fiber if this disorder persists in the amyloid state as predicted. Although the peptide is mostly disordered, two ordered regions of the peptide are consistent with bioinformatic predictions of high amyloidogenic propensity. The regions of nascent structure in the peptide bear further investigation, especially by mutational analysis to identify residues that are particularly responsible for controlling SEVI formation.
Supplementary Material
Acknowledgments
This study was supported by funds from the National Institutes of Health (DK078885, GM084018 and RR023597 to A.R.).
Footnotes
SUPPORTING INFORMATION AVAILABLE: CD spectra of PAP248–286 solution in the presence of SDS or TFE and complete reference 6. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dimitrov DS, Willey RL, Sato H, Chang LJ, Blumenthal R, Martin MA. J Virol. 1993;67:2182–2190. doi: 10.1128/jvi.67.4.2182-2190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rusert P, Fischer M, Joos B, Leemann C, Kuster H, Flepp M, Bonhoeffer S, Gunthard HF, Trkola A. Virology. 2004;326:113–129. doi: 10.1016/j.virol.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 3.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 4.Berg HC, Purcell EM. Biophys J. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckert DM, Kim PS. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 6.Munch J, et al. Cell. 2007;131:1059–1071. doi: 10.1016/j.cell.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 7.Roan NR, Munch J, Arhel N, Mothes W, Neidleman J, Kobayashi A, Smith-McCune K, Kirchhoff F, Greene WC. J Virol. 2009;83:73–80. doi: 10.1128/JVI.01366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hauber I, Hohenberg H, Holstermann B, Hunstein W, Hauber J. Proc Natl Acad Sci U S A. 2009;106:9033–9038. doi: 10.1073/pnas.0811827106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong SH, Klein EA, Das Gupta J, Hanke K, Weight CJ, Nguyen C, Gaughan C, Kim KA, Bannert N, Kirchhoff F, Munch J, Silverman RH. J Virol. 2009;83:6995–7003. doi: 10.1128/JVI.00268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wojtowicz WM, Farzan M, Joyal JL, Carter K, Babcock GJ, Israel DI, Sodroski J, Mirzabekov T. J Biol Chem. 2002;277:35019–35024. doi: 10.1074/jbc.M203518200. [DOI] [PubMed] [Google Scholar]
- 11.Brender JR, Hartman K, Gottler LM, Cavitt ME, Youngstrom DW, Ramamoorthy A. Biophys J. doi: 10.1016/j.bpj.2009.08.034. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 13.Goddard TD, Kneller DG. SPARKY. Vol. 3. University of California; San Francisco: 1999. [Google Scholar]
- 14.Wuthrich K. NMR of Proteins and Nucleic Acids. John Wiley and Sons; New York: 1986. [Google Scholar]
- 15.Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 16.Nilges M, Gronenborn AM, Brunger AT, Clore GM. Protein Eng. 1988;2:27–38. doi: 10.1093/protein/2.1.27. [DOI] [PubMed] [Google Scholar]
- 17.Stein EG, Rice LM, Brunger AT. J Magn Reson. 1997;124:154–164. doi: 10.1006/jmre.1996.1027. [DOI] [PubMed] [Google Scholar]
- 18.Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 19.Nanga RP, Brender JR, Xu J, Veglia G, Ramamoorthy A. Biochemistry. 2008;47:12689–97. doi: 10.1021/bi8014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nanga RP, Brender JR, Xu J, Hartman K, Subramanian V, Ramamoorthy A. J Am Chem Soc. 2009;131:8252–8261. doi: 10.1021/ja9010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuzi S, Hasegawa J, Kawaminami R, Naito A, Saito H. Biophys J. 2001;81:425–434. doi: 10.1016/S0006-3495(01)75710-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bloembergen N. J Chem Phys. 1957;27:572–573. [Google Scholar]
- 23.Solomon I. Phys Rev. 1955;99:559–565. [Google Scholar]
- 24.Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y. Nat Struct Mol Biol. 2007;14:1157–1164. doi: 10.1038/nsmb1345. [DOI] [PubMed] [Google Scholar]
- 25.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL. Proc Natl Acad Sci U S A. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreira ST, Vieira MNN, De Felice FG. IUBMB Life. 2007;59:332–345. doi: 10.1080/15216540701283882. [DOI] [PubMed] [Google Scholar]
- 27.Ehrnhoefer DE, Bieschke J, Boeddrich A, Herbst M, Masino L, Lurz R, Engemann S, Pastore A, Wanker EE. Nat Struct Mol Biol. 2008;15:558–566. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- 28.Knight JD, Hebda JA, Miranker AD. Biochemistry. 2006;45:9496–9508. doi: 10.1021/bi060579z. [DOI] [PubMed] [Google Scholar]
- 29.Hall D, Hirota N, Dobson CM. J Mol Biol. 2005;351:195–205. doi: 10.1016/j.jmb.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 30.Harrison RS, Sharpe PC, Singh Y, Fairlie DP. Rev Physiol Biochem Pharmacol. 2007;159:1–77. doi: 10.1007/112_2007_0701. [DOI] [PubMed] [Google Scholar]
- 31.Bertoncini CW, Rasia RM, Lamberto GR, Binolfi A, Zweckstetter M, Griesinger C, Fernandez CO. J Mol Biol. 2007;372:708–722. doi: 10.1016/j.jmb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Schweitzer-Stenner R, Measey T, Hagarman A, Eker F, Griebenow K. Biochemistry. 2006;45:2810–2819. doi: 10.1021/bi052282r. [DOI] [PubMed] [Google Scholar]
- 33.Hsu STD, Bertoncini CW, Dobson CM. J Am Chem Soc. 2009;131:7222–7223. doi: 10.1021/ja902307q. [DOI] [PubMed] [Google Scholar]
- 34.Tomaselli S, Esposito V, Vangone P, van Nuland NAJ, Bonvin A, Guerrini R, Tancredi T, Temussi PA, Picone D. ChemBioChem. 2006;7:257–267. doi: 10.1002/cbic.200500223. [DOI] [PubMed] [Google Scholar]
- 35.Valerio M, Porcelli F, Zbilut JP, Giuliani A, Manetti C, Conti F. ChemMedChem. 2008;3:833–843. doi: 10.1002/cmdc.200700324. [DOI] [PubMed] [Google Scholar]
- 36.Crescenzi O, Tomaselli S, Guerrini R, Salvadori S, D’Ursi AM, Temussi PA, Picone D. Eur J Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- 37.Cort JR, Liu Z, Lee GM, Huggins KN, Janes S, Prickett K, Andersen NH. Protein Eng Des Sel. 2009;22:497–513. doi: 10.1093/protein/gzp029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jao SC, Ma K, Talafous J, Orlando R, Zagorski MG. Amyloid. 1997;4:240–252. [Google Scholar]
- 39.Buck M. Q Rev Biophys. 1998;31:297–355. doi: 10.1017/s003358359800345x. [DOI] [PubMed] [Google Scholar]
- 40.Hong DP, Hoshino M, Kuboi R, Goto Y. J Am Chem Soc. 1999;121:8427–8433. [Google Scholar]
- 41.Gast K, Siemer A, Zirwer D, Damaschun G. Eur Biophys J Biophys Lett. 2001;30:273–283. doi: 10.1007/s002490100148. [DOI] [PubMed] [Google Scholar]
- 42.Kumar S, Modig K, Halle B. Biochemistry. 2003;42:13708–13716. doi: 10.1021/bi035330l. [DOI] [PubMed] [Google Scholar]
- 43.Hebda JA, Miranker AD. Ann Rev Biophys. 2009;38:125–152. doi: 10.1146/annurev.biophys.050708.133622. [DOI] [PubMed] [Google Scholar]
- 44.Brender JR, Durr UHN, Heyl D, Budarapu MB, Ramamoorthy A. Biochim Biophys Acta. 2007;1768:2026–2029. doi: 10.1016/j.bbamem.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engel MF. Chem Phys Lipids. 2009;160:1–10. doi: 10.1016/j.chemphyslip.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 46.Bystrom R, Aisenbrey C, Borowik T, Bokvist M, Lindstrom F, Sani MA, Olofsson A, Grobner G. Cell Biochem Biophys. 2008;52:175–189. doi: 10.1007/s12013-008-9033-4. [DOI] [PubMed] [Google Scholar]
- 47.Aisenbrey C, Borowik T, Bystrom R, Bokvist M, Lindstrom F, Misiak H, Sani MA, Grobner G. Eur Biophys J. 2008;37:247–255. doi: 10.1007/s00249-007-0237-0. [DOI] [PubMed] [Google Scholar]
- 48.Beyer K. Cell Biochem Biophys. 2007;47:285–299. doi: 10.1007/s12013-007-0014-9. [DOI] [PubMed] [Google Scholar]
- 49.Lashuel HA, Lansbury PT. Q Rev Biophys. 2006;39:167–201. doi: 10.1017/S0033583506004422. [DOI] [PubMed] [Google Scholar]
- 50.Smith PE, Brender JR, Ramamoorthy A. J Am Chem Soc. 2009;131:4470–4478. doi: 10.1021/ja809002a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brender JR, Hartman K, Reid KR, Kennedy RT, Ramamoorthy A. Biochemistry. 2008;47:12680–12688. doi: 10.1021/bi801427c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coles M, Bicknell W, Watson AA, Fairlie DP, Craik DJ. Biochemistry. 1998;37:11064–11077. doi: 10.1021/bi972979f. [DOI] [PubMed] [Google Scholar]
- 53.Patil SM, Xu S, Sheftic SR, Alexandrescu AT. J Biol Chem. 2009;284:11982–1991. doi: 10.1074/jbc.M809085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ulmer TS, Bax A, Cole NB, Nussbaum RL. J Biol Chem. 2005;280:9595–603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 55.Motta A, Andreotti G, Amodeo P, Strazzullo G, Morelli MAC. Proteins. 1998;32:314–323. [PubMed] [Google Scholar]
- 56.Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Proc Natl Acad Sci U S A. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vilar M, Chou HT, Luhrs T, Maji SK, Riek-Loher D, Verel R, Manning G, Stahlberg H, Riek R. Proc Natl Acad Sci U S A. 2008;105:8637–8642. doi: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Del Mar C, Greenbaum EA, Mayne L, Englander SW, Woods VL. Proc Natl Acad Sci U S A. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shao HY, Jao SC, Ma K, Zagorski MG. J Mol Biol. 1999;285:755–773. doi: 10.1006/jmbi.1998.2348. [DOI] [PubMed] [Google Scholar]
- 60.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Doeli H, Schubert D, Riek R. Proc Natl Acad Sci U S A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. Proc Natl Acad Sci U S A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davis HE, Rosinski M, Morgan JR, Yarmush ML. Biophys J. 2004;86:1234–1242. doi: 10.1016/S0006-3495(04)74197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Landazuri N, Le Doux JM. J Gene Med. 2004;6:1304–1319. doi: 10.1002/jgm.618. [DOI] [PubMed] [Google Scholar]
- 64.Tribet C, Vial F. Soft Matter. 2008;4:68–81. doi: 10.1039/b708431p. [DOI] [PubMed] [Google Scholar]
- 65.Zloh M, Ramaswamy C, Sakthivel T, Wilderspin A, Florence AT. Magn Reson Chem. 2005;43:47–52. doi: 10.1002/mrc.1508. [DOI] [PubMed] [Google Scholar]
- 66.Lee JH, Gustin JP, Chen TH, Payne GF, Raghavan SR. Langmuir. 2005;21:26–33. doi: 10.1021/la048194+. [DOI] [PubMed] [Google Scholar]
- 67.Shoemaker BA, Portman JJ, Wolynes PG. Proc Natl Acad Sci U S A. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tompa P. FEBS J. 2009;276:5406–5415. doi: 10.1111/j.1742-4658.2009.07250.x. [DOI] [PubMed] [Google Scholar]
- 69.Dafforn TR, Smith CJ. EMBO Rep. 2004;5:1046–1052. doi: 10.1038/sj.embor.7400276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. Cell. 2009;137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abedini A, Raleigh DP. Phys Biol. 2009;6:15005. doi: 10.1088/1478-3975/6/1/015005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brender JR, Lee EL, Cavitt MA, Gafni A, Steel DG, Ramamoorthy A. J Am Chem Soc. 2008;130:6424–6429. doi: 10.1021/ja710484d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nerelius C, Sandegren A, Sargsyan H, Raunak R, Leijonmarck H, Chatterjee U, Fisahn A, Imarisio S, Lomas DA, Crowther DC, Stromberg R, Johansson J. Proc Natl Acad Sci U S A. 2009;106:9191–9196. doi: 10.1073/pnas.0810364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jakob CG, Lewinski K, Kuciel R, Ostrowski W, Lebioda L. Prostate. 2000;42:211–218. doi: 10.1002/(sici)1097-0045(20000215)42:3<211::aid-pros7>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 75.de Groot NS, Pallares I, Aviles FX, Vendrell J, Ventura S. BMC Struct Biol. 2005;5:18–32. doi: 10.1186/1472-6807-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Conchillo-Sole O, de Groot NS, Aviles FX, Vendrell J, Daura X, Ventura S. BMC Bioinformatics. 2007;8:65–81. doi: 10.1186/1471-2105-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trovato A, Seno F, Tosatto SCE. Prot Eng Des Sel. 2007;20:521–523. doi: 10.1093/protein/gzm042. [DOI] [PubMed] [Google Scholar]
- 78.Trovato A, Chiti F, Maritan A, Seno F. PLoS Comput Biol. 2006;2:1608–1618. doi: 10.1371/journal.pcbi.0020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reumers J, Maurer-Stroh S, Schymkowitz J, Rousseau F. Hum Mutat. 2009;30:431–437. doi: 10.1002/humu.20905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.